Abstract
All land plants contain at least one class II diterpene cyclase (DTC), which utilize an acid-base catalytic mechanism, for the requisite production of ent-copalyl diphosphate (ent-CPP) in gibberellin A (GA) phytohormone biosynthesis. These ent-CPP synthases (CPSs) are hypothesized to be derived from ancient bacterial origins and, in turn, to have given rise to the frequently observed additional DTCs utilized in more specialized plant metabolism. However, such gene duplication and neo-functionalization has occurred repeatedly, reducing the utility of phylogenetic analyses. Support for evolutionary scenarios can be found in more specific conservation of key enzymatic features. While DTCs generally utilize a DxDD motif as the catalytic acid, the identity of the catalytic base seems to vary depending, at least in part, on product outcome. The CPS from Arabidopsis thaliana has been found to utilize a histidine-asparagine dyad to ligate a water molecule that serves as the catalytic base, with alanine substitution leading to production of 8β-hydroxy-ent-CPP. Here this dyad and effect of Ala substitution is shown to be specifically conserved in plant CPSs involved in GA biosynthesis, providing insight into plant DTC evolution and assisting functional assignment. Even more strikingly, while GA biosynthesis arose independently in plant-associated bacteria and fungi, the catalytic base dyad also is specifically found in the relevant bacterial, but not fungal, CPSs. This suggests functional conservation of CPSs from bacteria to plants, presumably reflecting an early role for derived diterpenoids in both plant development and plant-microbe interactions, eventually leading to GA, and a speculative evolutionary scenario is presented.
Introduction
The embryophytes (land plants) are often prolific producers of labdane-related diterpenoids, whose biosynthesis is defined by the activity of class II diterpene cyclases, DTCs [1]. The basis for the observed diversity of labdane-related diterpenoids presumably stems from the fact that all land plants contain at least one DTC. Specifically, for phytohormone biosynthesis, which requires bicyclization of the general diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP, 1) to ent-labdadienyl/copalyl diphosphate (ent-CPP, 2) by such an enzyme [2]. At least in vascular plants (tracheophytes) the relevant phytohormone is the well-known gibberellin A, GA [3], while ent-kaurenoic acid, an early intermediate in GA biosynthesis, serves a similar role in the earlier diverging bryophytes [4]. The requisite presence of an ent-CPP synthase (CPS) has been hypothesized to serve as a genetic reservoir that gave rise to the observed diversity of land-plant derived labdane-related diterpenoids. In particular, via gene duplication and neo-functionalization most embryophytes contain several paralogous DTCs, many of which no longer produce ent-CPP [2]. However, phylogenetic analysis does not cleanly distinguish between plant DTCs involved in GA biosynthesis relative to those dedicated to more specialized metabolism (Figure 1). While this complexity presumably arises from repeated such evolutionary derivation, it also limits the utility of phylogenetic comparison for functional assignment. In addition, it has been further hypothesized that the plant DTCs are derived from an ancient bacterial origin [5, 6], but the specifics of this acquisition are completely opaque.
Figure 1:

Representative phylogenetic tree for plant DTC family. Green lines indicate branches containing CPSs involved in GA biosynthesis, with the role of those indicated by green text having been verified by genetic approaches (i.e., knock-out lines). Red lines indicate branches where product outcomes differs (i.e., is no longer ent-CPP, 2), with those DTCs indicated by red text having been verified to be involved in more specialized metabolism by genetic approaches, despite also making ent-CPP. Also indicated are the major divisions in the plant kingdom mentioned in the text, as well as asterisks (*) indicating loss of class I (most likely KS) activity from bifunctional ancestors – i.e., to generate monofunctional DTCs. Scale bar refers to expected substitutions per site.
The reactions catalyzed by DTCs are highly exothermic, enzymatic containment of which requires significant structural support [7]. Maintenance of this structural integrity provides additional selective pressure that seems to have overwhelmed the usual phylogenetic signature for derivation of alternative enzymatic activity. Nevertheless, more specific conservation of key enzymatic features for particular functions might provide insight into the evolution of these enzymes. In particular, DTCs utilize a general acid-base mechanism. Initial protonation of GGPP leads to bicyclization that forms the eponymous labda-13E-en-8-yl+ diphosphate. This carbocation intermediate can be produced in four distinct stereo-configurations and undergo subsequent rearrangement, as well as be subjected to the addition of water, prior to terminating deprotonation [1]. DTCs contain a characteristic aspartate-rich DxDD motif, which has been shown to cooperatively serve as the catalytic acid required for olefin protonation [8](although preceding epoxidation relieves this requirement [9]). By contrast, the residues that make up the catalytic base required to quench the final carbocationic intermediate seem to vary, based at least in part on specific product outcome. For example, it has been suggested that all ent-CPP producing DTCs from plants utilize a water molecule that is tightly ligated, specifically including hydrogen-bonds to the side-chains of a conserved histidine-asparagine dyad, enabling it to serve as a general base (Figure 2). This arrangement was first suggested by determination of high-resolution crystal structures for the CPS from Arabidopsis thaliana, AtCPS [10, 11]. Further support was provided by the finding that alanine substitution for either (or both) residues in AtCPS led to addition of water and appearance of a hydroxyl group in place of the usually generated olefin – i.e., production of 8β-hydroxy-ent-CPP (3) rather than ent-CPP [12]. Other DTCs that mediate different product outcome have been similarly shown to utilize alternative residues as their catalytic bases [13–16].
Figure 2:
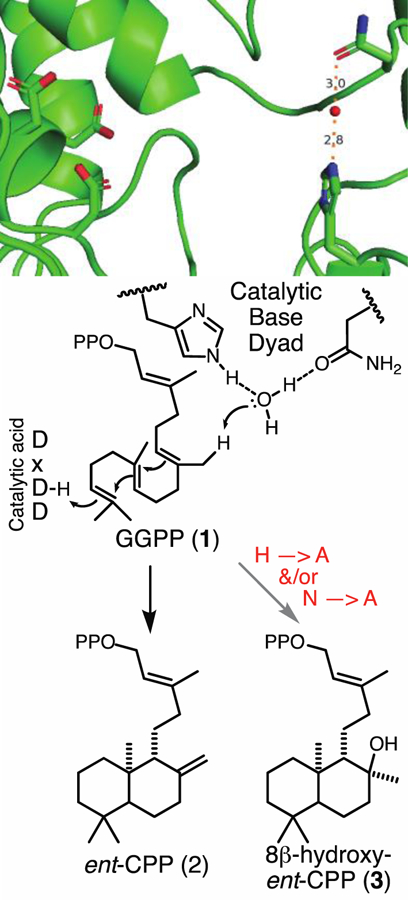
Active site of AtCPS showing arrangement of the highly conserved DxDD catalytic acid motif and His-Asn dyad coordinated water that acts as the catalytic base. Schematic below depicts the use of these acid-base groups for (bi)cyclization of GGPP (1) to ent-CPP (2) or, upon substitution of Ala for the His and/or Asn of the catalytic base dyad, 8β-hydroxy-ent-CPP (3).
The ancient derivation of plant DTCs from bacteria was first suggested on the basis of sequence similarity, including conservation of the DxDD motif [5, 17]. This hypothesis was later further supported by the observation of structural homology between plant and bacterial DTCs. Specifically, resemblance between the recently reported crystal structure for an ent-CPP (2) producing CPS from the bacterium Streptomyces platensis, SpCPS [18], and those previously determined for plant DTCs [10, 19]. However, while SpCPS contains the characteristic DxDD motif, its catalytic base remains unknown. Indeed, there is a distinct lack of enzymatic structure-function relationship investigations with bacterial DTCs. Moreover, the proposed homology is complicated by the fact that DTCs from both plants and fungi exhibit an extended sequence reflecting ancient fusion of these with subsequently acting class I diterpene synthases. At least in plants this appears to have been fusion of a CPS with an ent-kaurene synthase, KS [5, 17]. While the resulting bifunctional CPS/KS seems to have been retained in some bryophytes [20, 21], and bifunctional diterpene synthases involved in more specialized (resin acid) metabolism are still found in gymnosperms [22] – e.g., the abietadiene synthase from Abies grandis, AgAS [23] – in the expansive tracheophyte lineage this CPS/KS seems to have undergone early gene duplication and sub-functionalization. Specifically, loss of CPS activity in one copy and KS activity in the other, but with retention of the original sequence/domains (α, associated with class I activity, and the βγ didomains associated with class II DTC activity) in both [17]. These monofunctional CPSs and KSs then gave rise to the DTCs and extensive terpene synthase (TPS) families found in tracheophytes [24], albeit with early loss of the N-terminal γ domain and subsequent radiation leading to most of the other class I TPS sub-families, which generally act on shorter substrates – i.e., in 15-carbon sesquiterpenoid or 10-carbon monoterpenoid biosynthesis [17]. Altogether, this complex evolutionary history further complicates the question of the more specific ancient origins of plant DTCs.
Notably, in addition to endogenous production by vascular plants, GA also is produced by certain plant-associated microbes, both fungi and bacteria [3]. The identity of the relevant enzymes, particularly the oxygenases acting after initial production of the common ent-kaurene metabolite, demonstrates that GA biosynthetic pathways arose independently in each of these biological kingdoms [25]. For example, while the later steps are catalyzed by 2-oxoglutarate dependent dioxygenases in plants, they are catalyzed by cytochromes P450 monooxygenases in microbes, with these clearly separated into the class I (soluble) and class II (membrane anchored) families associated with bacteria and eukaryotes (fungi), respectively. However, the existence of some (albeit distant) sequence homology between the relevant domains comprising the CPS (βγ didomains) and KS (α domain) from these three biological kingdoms was noted and suggested to imply a common origin [5], but the specifics of such conservation has remained uncertain.
Here biochemical characterization of the catalytic base from selected examples across an extensive phylogenetic range of CPSs is reported. The results indicate conservation of the His-Asn dyad (and immediately surrounding residues for each) not only throughout the CPSs from embryophyte phytohormone metabolism, but also those from bacterial, although not fungal, GA biosynthesis. Also demonstrated here is the use of alternative residues to form the catalytic base in other ent-CPP producing CPSs, which indicates that the His-Asn catalytic base dyad is more specifically conserved for GA production in both plants and plant-associated bacteria. This conservation pattern not only provides signature motifs for plant CPSs involved in phytohormone metabolism, but also hints at an early and retained role for labdane-related diterpenoids in both plant development and plant-microbe interactions, and a speculative scenario for ancient acquisition of CPSs from bacteria during the transition from unicellular algae to multicellular organisms in plant evolution is presented.
Experimental
All reagents were purchased from Fisher Scientific unless noted otherwise. Alignments, and the phylogenetic tree (Figure 1), were generated with CLC Main Workbench (v8.1.3; Qiagen Aarhus A/S), with the tree visualized for presentation with FigTree (v1.4.3). Given the extensive phylogenetic range covered here these were generated using amino acid sequences. For the plant DTC family, the shown representative tree (Figure 1) and alignment (Figure 3) were generated with the following DTCs, given with presented abbreviation (GenBank accession), species name and relevant reference(s) for characterization: AgAS (Q38710), Abies grandis [23]; AtCPS (NP_192187), Arabidopsis thaliana [26]; GbLS (Q947C4), Gingko biloba [27]; JsCPS/KS (BAJ39816), Jungermannia subulata [21]; MpCPS (APP91795), Marchantia polymorpha [28]; OsCPS1 (XP_015624005), Oryza sativa [29]; OsCPS2 (AY602991), O. sativa [30]; OsCPS4 (Q6E7D7), O. sativa [31]; PgCPS (ADB55707), Picea glauca [32]; PpCPS/KS (XP_024380398), Physcomitrella patens [20]; PsCPS (O04408), Pisum sativum [33]; TaCPS1 (BAH56558), TaCPS2 (BAH56559) & TaCPS3 (BAH56560), Triticum aestivum [34]; TaCPS4 (BAP01383), T. aestivum [35]; DsCPS1 (ABV57835), Salvia miltiorhizza, also known as Dan-shen [36]; DsCPS2 (AHJ59322), DsCPS4 (AKN91186) & DsCPS5 (AHJ59324), Dan-shen [37]; SmCPS/KSL (AEK75338), Selaginella moellendorffii [38]; SmCPS (JX413782), S. moellendorffi [39]; ZmCPS1 (AAA73960), Zea mays [40]; ZmCPS2 (AAT70083), Z. mays [41]. For the microbial CPSs the shown alignment (Figure 7) was generated with: BjCPS (NP_768789), Bradyrhizobium japonicum [5]; EtCPS (WP_020322919), Erwinia tracheiphila [42]; MlCPS (NP_106893), Mesorhizobium etli; SfCPS (NP_443949), Sinorhizobium fredii; ReCPS (NP_659791), Rhizobium etli [43]; SpCPS (ACO31276), Streptomyces platensis [44]; GfCPS/KS (Y15013), Gibberella fujikuroi [45].
The recombinant plant CPSs used in this study were expressed as pseudo-mature constructs: AtCPS-Δ84, OsCPS1-Δ123, OsCPS2-Δ75, and PpCPS/KS-Δ121. Truncations for OsCPS1, OsCPS2, and PpCPS/KS were made to match the previously described optimal truncation site for AtCPS [46]. Similarly, the recombinant AtKS used in this study also was expressed as the previously reported pseudo-mature construct [47]. All cloning was carried out using the Invitrogen Gateway system. Generation of the pENTR/SD/D-TOPO based constructs has been described for AtCPS [46], OsCPS1 & 2 [30], PpCPS/KS and GfCPS/KS [14], ReCPS [43] and AtKS [48]. Similarly, synthetic genes for EtCPS and SpCPS, codon optimized for expression in E. coli were obtained (Invitrogen), and cloned by directional topoisomerization into pENTR/SD/D-TOPO.
The AtCPS mutants used in this study have been previously described [12]. The KS knock-out mutants used in this study were designed based on those previously reported for PpCPS/KS [20], and a related fungal CPS/KS in the case of GfCPS/KS [49], with the loss of KS activity in both verified here by the resulting lack of ent-kaurene production. All mutants were generated by whole-plasmid PCR amplification with overlapping mutagenic primers of the relevant pENTR clone and verified by complete gene sequencing prior to transfer by directional recombination into the expression vector pDEST14. To enable co-expression, AtKS was similarly transferred from pENTR into a previously described pACYCDuet1-DEST vector [50].
While the studies described here were initiated with use of a previously described modular metabolic engineering system [51], it was discovered that this led to further derivatization of dephosphorylated DTC products. In particular, relative to previous work [12], a change in amount of the presumed epimer of 8β-hydroxy-ent-CPP produced by the AtCPS mutants was traced to the use of different sources for the antibiotic chloramphenicol (Chl) utilized to select for the GGPP synthase carrying pGG-DEST vector. Indeed, employment of an alternative system to engineer the production of GGPP in E. coli without the use of Chl led the complete disappearance of this presumed epimer (c.f., Figure 4 here and Figure 1 from [12]), while addition of the corresponding empty vector pACYCDuet1 (and use of Chl) was sufficient for its reappearance (data not shown). This effect also appears to give rise to previously observed derivatives of ent-copalol (the dephosphorylated derivative of ent-CPP) and other DTC products. Given that these derivatives are only observed in the absence of any subsequently acting class I diterpene synthases (e.g., KS), the relevant reaction is inefficient and does not interfere with use of the original system with such downstream enzymes. Nevertheless, it was necessary to employ an alternative system for analysis of DTC activity. Here a system was developed utilizing the GGPP synthase and isopentenyl diphosphate isomerase from the GA biosynthetic operon found in Erwinia tracheiphila where the encoding genes are adjacent to each other [42]. The corresponding fragment of the operon was amplified by PCR, cloned by directional topoisomerization into pENTR/SD/D-TOPO, with verification by complete sequencing prior to transfer into the previously described expression vector pCDFDuet1-DEST [50], via an LR reaction. This provided efficient flux of GGPP to the DTCs expressed from the compatible pDEST14 expression vector, as well as the subsequently acting AtKS expressed from the additionally compatible pACYCDuet1-DEST.
The relevant plasmids were transformed into the BL21-Star strain of E. coli and the resulting recombinant strains were used for functional analysis via metabolic engineering. Each was inoculated into 45 mL of liquid TB media (10 g casein, 10 g NaCl, 5 g yeast extract, in 1 L H2O, and pH adjusted to 7.0) with the appropriate antibiotics and shaken at 37 °C until an OD600 of 0.4 – 0.6 was reached. At this time, phosphate buffer (pH 7.5) was added to 100mM and MgCl2 to 0.5mM (final concentrations) and the cultures were transferred to a 16°C shaker. These cultures were let shake at 16 °C for half an hour or until the OD600 reached 0.6 and were induced with 1 mM IPTG. After 3 days fermentation at 16 °C, enzymatic products were extracted by addition of equal volume of hexanes and gentle swirling. The organic solvent was separated out and then dried under N2, with the residue resuspended in fresh hexanes and analyzed by GC-MS. Analysis of products by GC-MS was carried out as previously described [52]. Briefly, using a 3900 GC with Saturn 2100T ion trap MS (Varian), equipped with a HP-5MS column (Agilent, 0.25 μm, 0.25 ID, 30 m) with a He flow rate of 1.2 mL/min, and the following oven temperature program: 50 °C for 3 min, 15 °C/min to 300 °C, with 3 min. hold. MS data was collected from m/z of 90 to 500, starting at 12 min. until the end of the run. Samples (1 μL) were introduced by splitless injection at 250 °C.
Results
Rather than an early split between CPSs involved in GA biosynthesis and DTCs that are instead dedicated to more specialized metabolism, phylogenetic analysis of the plant DTC family reveals a complex evolutionary pattern (Figure 1). For example, there are two major clades within the monocot Poaceae (cereal) plant family. One is clearly devoted to more specialized metabolism, indicative of an early duplication and neo-functionalization event. Nevertheless, the other contains both CPSs from GA biosynthesis and additional paralogs that have been shown to be involved in more specialized metabolism, indicating that such duplication and neo-functionalization has occurred repeatedly within this family [53]. Similar complexity also exists in the dicots – e.g., in the Lamiaceae family [54]. Indeed, characterization of TPSs from the liverwort Marchantia polymorpha suggests that the ancient gene duplication and sub-functionalization of an ancestral CPS/KS to form separate CPS and KS in the tracheophyte lineage may have independently occurred in the bryophyte lineage as well [28]. While this complexity seems to reflect repeated derivation of DTCs for more specialized metabolism from the CPSs involved in GA biosynthesis, it confounds bioinformatic assignment of catalytic and/or physiological function.
To investigate the hypothesis that more specific conservation of key enzymatic features for GA phytohormone biosynthesis might provide insight into DTC evolution, a more detailed investigation of the relevant CPSs was undertaken here. In particular, although it has been previously suggested that all ent-CPP (2) producing CPSs from plants utilize the His-Asn dyad ligated water molecule as their catalytic base [12], more extensive sequence alignment demonstrated that this is not the case. For example, in the DTC phylogenetic clade containing those from cereal plants devoted to more specialized metabolism, both rice (Oryza sativa) and wheat (Triticum aestivum) encode DTCs known to produce 2, OsCPS2 [30, 55] and TaCPS1 [35], respectively, neither of which contain this dyad. Specifically, although the His is present, both have a cysteine in place of the Asn. By contrast, the His-Asn dyad is present in OsCPS1, which is required for rice GA biosynthesis [29], while OsCPS2 has been shown to be involved in more specialized metabolism instead [56]. This suggests that the dyad might be more specifically retained in those DTCs involved in such phytohormone metabolism. Indeed, the dyad appears to be conserved not only in such CPSs from tracheophytes but also those from the bryophytes as well, including in bifunctional CPS/KSs (Figure 3).
Figure 3:
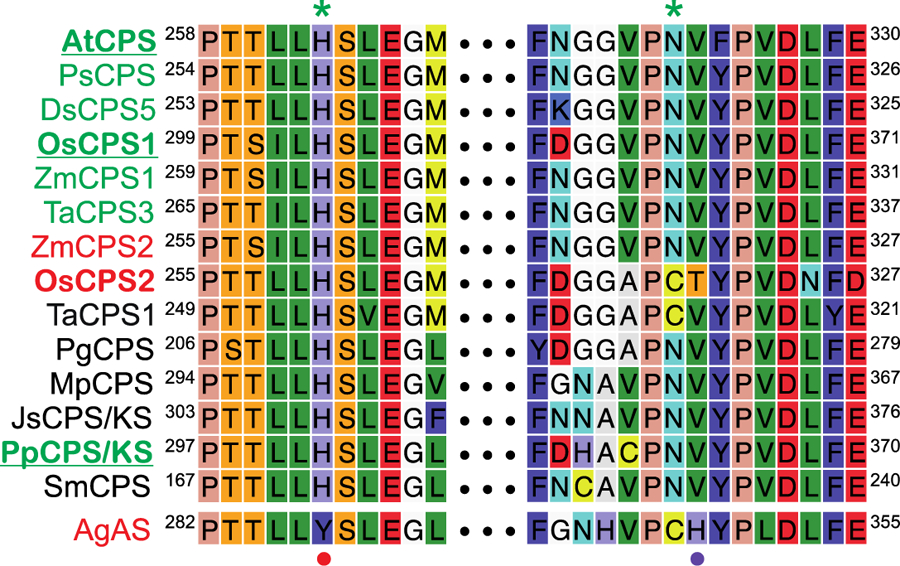
Alignment of selected plant DTCs focused on the catalytic base dyad (with numbering for the first and last residue in each case). Those whose names are in green are known to be involved in GA biosynthesis, while those named in red are known to function in more specialized metabolism, those in bold are mentioned in the text and those underlined further investigated here. The residues comprising the catalytic base dyad in the CPSs from GA biosynthesis are indicated by green asterisks (*) above the alignment, those making up the alternative dyad in AgAS, which produces enantiomeric (normal) CPP (differentiating it from all the others shown here, which produce ent-CPP), are indicated by red (•) and purple (•) dots below the alignment, with the purple dot also indicating the Thr that seems to pair with the ancestral His to make up an alternative catalytic base dyad in OsCPS2.
Given the production of 8β-hydroxy-ent-CPP (3) by Ala substitution for either the His or Asn from the catalytic base dyad in AtCPS [12], it was reasoned that similar effects would provide support for their functional conservation in other CPSs. Accordingly, the equivalent mutants were made in selected CPSs spanning the phylogenetic range of plants. Specifically, as AtCPS is from a dicot, OsCPS1 was chosen as an exemplar for monocots and the bifunctional CPS/KS from the moss Physcomitrella patens (PpCPS/KS) to represent the bifunctional enzymes from the earlier diverging bryophytes. In the case of PpCPS/KS these mutants were constructed in the presence of an additional mutation (D635A) that knocks out the KS activity (PpCPS/KS), enabling observation of the product from just the DTC activity [20]. The resulting mutants were all assayed in a modular metabolic engineering system, with heterologous expression in Escherichia coli also engineered to produce GGPP, leading to observation of the resulting DTC products as the dephosphorylated derivatives produced by endogenous phosphatases, which can be readily extracted from these cultures with organic solvents and analyzed by GC-MS. Note that, although it was previously reported that the AtCPS mutants produced an epimeric mixture of 8-hydroxy-ent-CPP, more careful analysis revealed that only the 8β epimer (3) was actually produced. Similarly, the OsCPS1 and PpCPS/KS mutants all were found to primarily produce 3 (Figure 4). These results support functional conservation of these residues as a catalytic base in the CPSs involved in hormonal metabolism. Thus, this His-Asn dyad appears to be ancestral in at least the plant DTC family.
Figure 4:
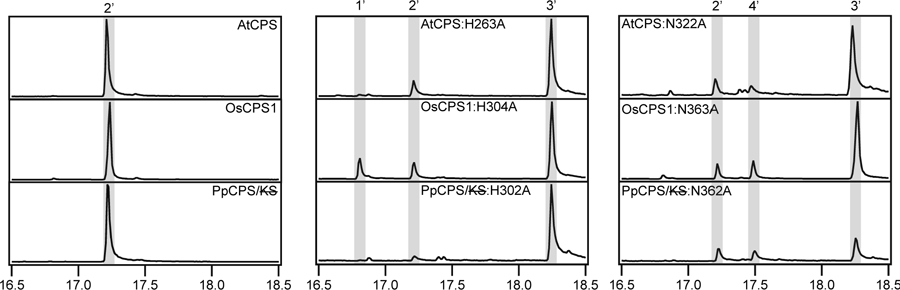
GC-MS chromatograms of dephosphorylated class II products from wild-type or mutant CPSs and CPS/KSs from plants (1’, dephosphorylated derivative of GGPP, geranylgeraniol; 2’, dephosphorylated derivative of ent-CPP, ent-copalol, 3’, dephosphorylated derivative of 8β-hydroxy-ent-CPP, 8β-hydroxy-ent-copalol; 4’, dephosphorylated derivative of ent-kolavenyl diphosphate, ent-kolavenol).
Intriguingly, in each of these cases Ala substitution for the Asn also led to production of small amounts of ent-kolavenyl diphosphate (ent-KPP, 4), resulting from a series of 1,2-shifts, alternating between hydride and methyl groups, before terminating deprotonation (presumably by removal of the originally added proton by the catalytic DxDD motif as previously demonstrated for production of 4 [57]). This requires further conservation in positioning of the initially formed intermediate, ent-labda-13E-en-8-yl+ diphosphate, such that it cannot be deprotonated at an alternative position by the remaining His of the catalytic base dyad, as well as the absence of any active site feature (residue or water) capable of deprotonating the intervening carbocation intermediates (Figure 5).
Figure 5:
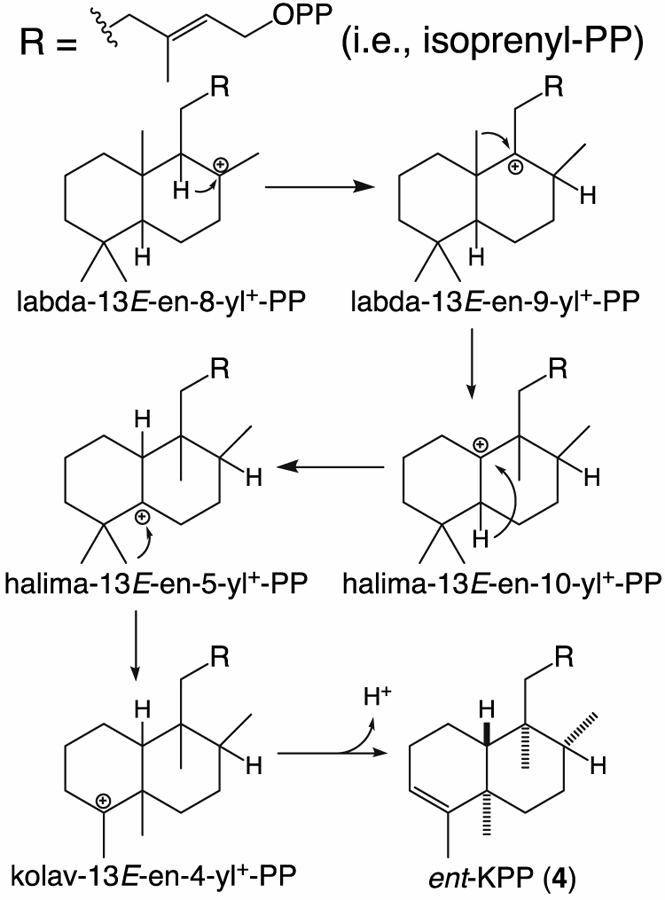
Scheme depicting carbocation intermediates required for rearrangement of labda-13E-8-yl+ diphosphate (PP) to yield kolavenyl-PP (KPP) – e.g., the ent-KPP (4) observed here.
As noted above, OsCPS2 produces ent-CPP, but does not contain the Asn of the ancestral catalytic base dyad, with a Cys at this position instead. In contrast to Ala substitution for the retained His, which leads to primary production of 8β-hydroxy-ent-CPP (3), Ala substitution for this Cys does not significantly alter product outcome (Figure 6), suggesting use of an alternative residue in the OsCPS2 catalytic base dyad. Notably, it has been previously shown that AgAS utilizes an alternative catalytic base dyad in its DTC active site [14]. While consistent with the distinct production of the normal stereoisomer of CPP by AgAS, this also demonstrates the flexibility of DTCs in forming their catalytic base. The residues that make up the catalytic base dyad in AgAS have some similarities to the ancestral Asn-His pair. For example, AgAS also utilizes a His [13], but rather than occupying the ancestral/first (His) position, this His immediately follows the ancestral/second (Asn) position, and is directly hydrogen-bound to the Tyr that is found in place of the ancestral His – i.e., is in the first position (Figure 3). Such flexible interaction is consistent with the location of these residues on loops in DTC protein structures. Given the presence of a threonine immediately following the Cys in OsCPS2, this was hypothesized to pair with the ancestral His. Indeed, Ala substitution for this Thr led to primary production of 3 (Figure 6). Coupled with the lack of such effect on product outcome upon Ala substitution for the Cys that corresponds to the ancestral Asn, these results indicate that OsCPS2 utilizes an alternative His-Thr dyad as its catalytic base.
Figure 6:
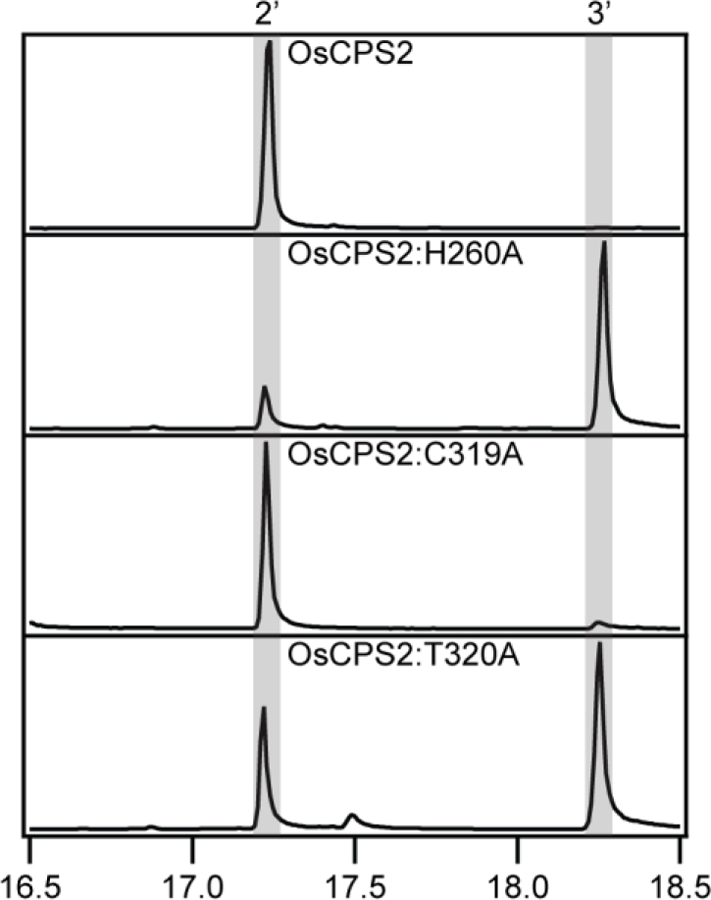
GC-MS chromatograms of dephosphorylated class II products from wild-type or mutant OsCPS2 (2’, ent-copalol; 3’, 8β-hydroxy-ent-copalol).
Beyond plants, it has been previously noted that the His from the catalytic base dyad is conserved in not only PpCPS/KS but also the bifunctional CPS/KS involved in GA biosynthesis by the fungus Gibberella fujikuroi, GfCPS/KS [14]. However, the Asn does not appear to be present in GfCPS/KS, nor in the bacterial ent-CPP producing SpCPS. By contrast, both residues appear to be present and well-conserved in the CPSs involved in GA biosynthesis by bacteria (Figure 7). Particularly given the otherwise low sequence identity between the plant and bacterial CPSs, this hints at the possibility that the catalytic base dyad might be conserved between them.
Figure 7:

Alignment of selected microbial ent-CPP producing DTCs focused on the catalytic base dyad (numbering for first and last residues indicated in each case). Only SpCPS is clearly not involved in GA biosynthesis, as indicated by the red text. Those whose names are underlined were investigated here. The residues comprising the ancestral catalytic base dyad in the bacterial CPSs from GA biosynthesis are indicated by green asterisks (*) above the alignment, while the functionally conserved histidine in GfCPS/KS and SpCPS is indicated by a red dot (•) below the alignment, and the alternative paired residue indicated by purple dots (•) below the relevant residue.
The hypothesis that the His-Asn catalytic base dyad is functionally conserved in these bacterial CPSs from GA biosynthesis was tested by substituting Ala for these residues in the relevant CPS from Erwinia tracheiphila (EtCPS), which falls within the same Enterobacteriaceae family as E. coli, and is readily expressed therein [42]. Ala substitution for the His led to predominant production of 8β-hydroxy-ent-CPP (3), while that for the Asn led to less, but still substantial such addition of water – i.e., production of 3 (Figure 8). Although this latter mutation exerts a smaller effect on product outcome, these results nevertheless suggest that the His-Asn dyad may be functionally equivalent in the bacterial and plant CPSs involved in GA biosynthesis.
Figure 8:
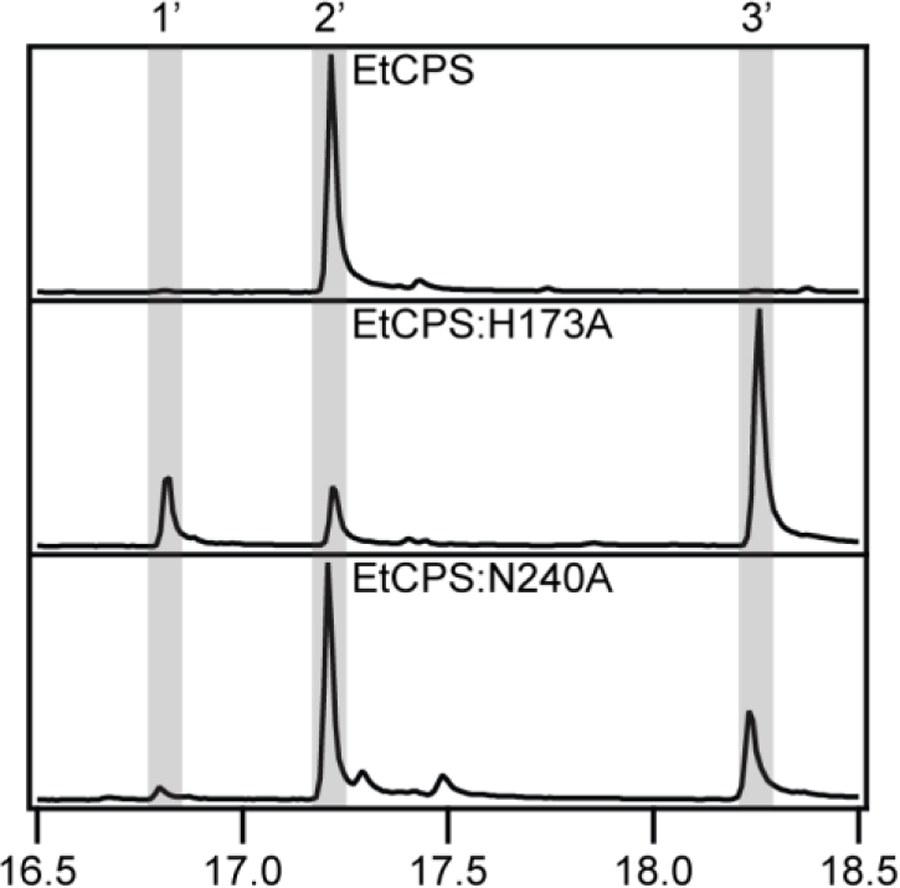
GC-MS chromatograms of dephosphorylated class II products from wild-type or mutant EtCPS (1’, geranylgeraniol; 2’, ent-copalol; 3’, 8β-hydroxy-ent-copalol).
While the catalytic base dyad is otherwise well-conserved, the Asn has been substituted by serine in a few of these bacterial CPSs from the Rhizobium genus, which form a monophyletic clade. Notably, while that from Rhizobium etli (ReCPS) has been previously reported to produce at least some ent-CPP (2) [43], re-examination of its product profile here demonstrated that it actually primarily produces 8β-hydroxy-ent-CPP (3). As previously described, due to poor heterologous expression of ReCPS its activity was assessed by coupling to the KS from A. thaliana (AtKS). While this leads to the previously reported observation of ent-kaurene (5) [43], the resulting cultures were found to also yield even larger amounts of ent-13-epi-manoyl oxide (6) (Figure 9), which AtKS has previously been shown to produce from 3 [14]. Previous phylogenetic analysis has demonstrated that ReCPS exhibits an appreciably longer branch length relative to the other characterized bacterial CPSs from the Rhizobiales order [43], suggesting that this CPS may be subjected to less selective pressure, perhaps reflecting its reduced ability to act in GA biosynthesis. Indeed, Asn substitution for this Ser is not sufficient to restore ReCPS to production of only 2 (Figure 9), consistent with further drift away from this original activity. Nevertheless, the overall sequence homology and conserved genomic context (i.e., in the relevant operon) of ReCPS and the related Ser containing Rhizobium CPSs clearly indicates that these had at least an ancestral role in GA biosynthesis.
Figure 9:
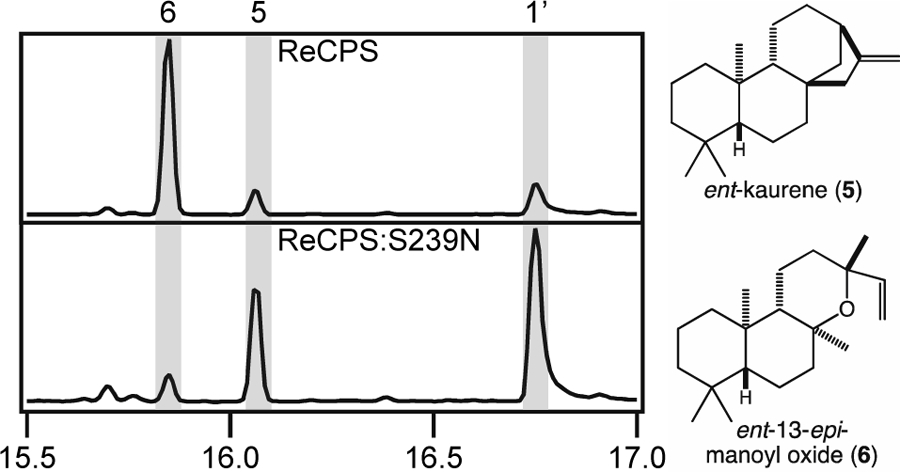
GC-MS chromatograms of products from co-expression of wild-type or mutant ReCPS with AtKS (1’, geranylgeraniol; 5, ent-kaurene; 6, ent-12-epi-manoyl oxide, which are depicted here).
As noted above, the fungal GfCPS/KS does not contain the Asn of the catalytic base dyad found in the plant and bacterial CPSs from GA biosynthesis, with a glycine at this position instead (Figure 7). This Gly is preceded by a Ser and followed by a Thr. The conserved His retains functional importance, as Ala substitution has already been suggested to lead to production of 8β-hydroxy-ent-CPP (3), albeit this was only indirectly inferred from production of the further cyclized ent-12-epi-manoyl oxide (6) by the KS active site [14]. Given the use of such an adjacent residue to form the alternative catalytic base dyad identified here in OsCPS2, it was hypothesized that the Ser or Thr flanking the Gly found at the second (Asn) position might interact with the conserved His. Use of such an alternative catalytic base dyad also was investigated by Ala substitution here. Just as with PpCPS/KS, to eliminate the KS activity these substitutions were made in the presence of an additional mutation (D668A), which knocks out the KS activity (GfCPS/KS). Notably, while Ala substitution for the Ser does not change product outcome, Ala substitution for the His led to primary production of 3, and Ala substitution for the Thr also led to substantial such addition of water (Figure 10). Moreover, in all the fungal CPS/KSs identified to-date [58], both the His and Thr (but not the Ser) are completely conserved. Hence, it appears that the fungal CPS/KSs utilize a His-Thr catalytic base dyad that differs from the His-Asn dyad found in the plant and bacterial CPSs involved in GA biosynthesis not only in composition but also positioning (i.e., of the second (Thr) residue), which then resembles that found in OsCPS2, albeit these almost certainly independently arose via convergent evolution.
Figure 10:
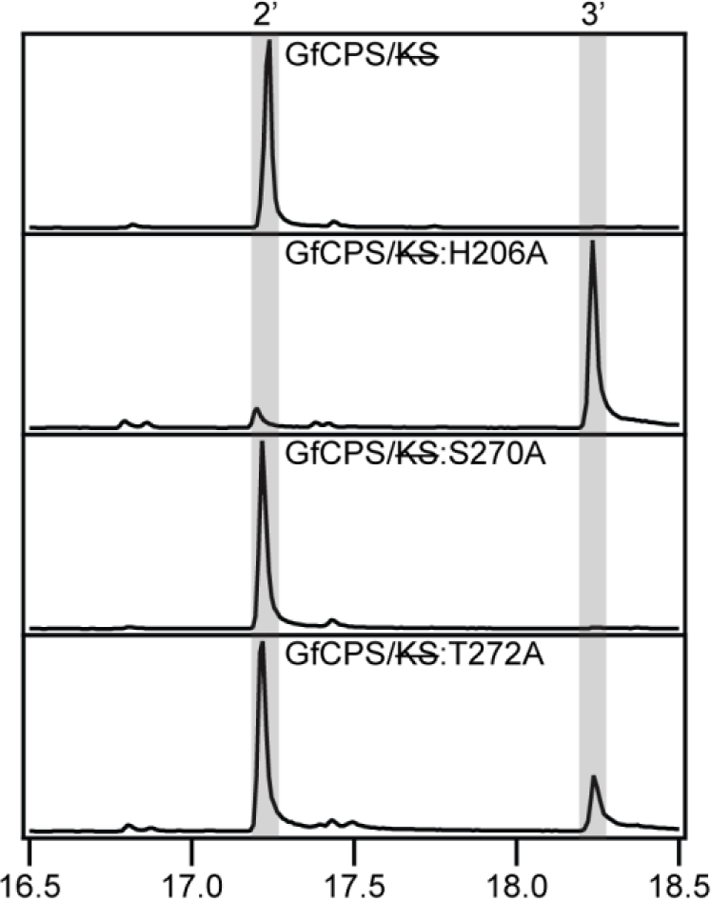
GC-MS chromatograms of dephosphorylated class II products from wild-type or mutant GfCPS/KS class I knockout (2’, ent-copalol; 3’, 8β-hydroxy-ent-copalol).
While the more generally conserved His is retained in the phylogenetically distant, but functionally analogous (i.e., 2 producing) bacterial SpCPS (Figure 7), this appears to utilize an even more divergent catalytic base dyad. In particular, inspection of the SpCPS crystal structure indicates that rearrangement of two loops may have led to formation of an alternative catalytic base dyad, with an Asp from a distinct C-terminal loop directly hydrogen-bonded to the retained His (Figure 11A). The direct interaction of these residues is analogous to the direct interaction between the Tyr and His that make up the catalytic base dyad in AgAS [14]. Accordingly, this His-Asp pair was hypothesized to form the catalytic base dyad in SpCPS, which also was investigated by Ala substitution, along with the Ser found at the second (Asn) position of the His-Asn dyad found in the plant and bacterial CPSs from GA biosynthesis. While Ala substitution for this Ser did not alter product outcome, Ala substitution for either the His or Asp led to primary production of 8β-hydroxy-ent-CPP (3) rather than 2 (Figure 11B). Accordingly, SpCPS uses an even more divergent His-Asp catalytic dyad to achieve the same production of ent-CPP (2) as the other CPSs investigated here.
Figure 11:
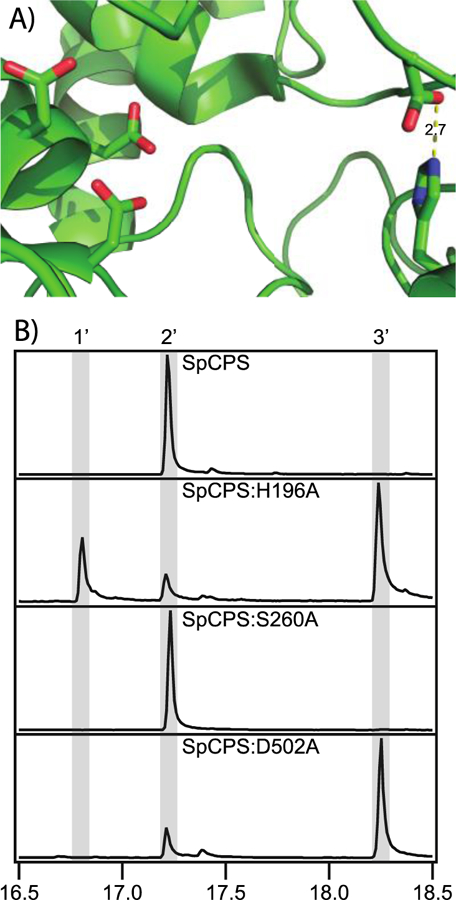
A) Active site of SpCPS showing arrangement of the highly conserved DxDD catalytic acid motif and alternative His-Asp dyad shown here to directly act as the catalytic base. B) GC-MS chromatograms of dephosphorylated class II products from wild-type or mutant SpCPS, as indicated (1’, geranylgeraniol; 2’, ent-copalol; 3’, 8β-hydroxy-ent-copalol).
Discussion
The results reported here not only strongly support the hypothesized repeated derivation of DTCs for more specialized metabolism from the CPSs involved in embryophyte phytohormone metabolism (e.g., GA), but also provide a means to assist bioinformatic resolution of these. In particular, given the conservation of the His-Asn catalytic base dyad in the CPSs from such phytohormone biosynthesis (Figure 3), as evidenced by the effect of Ala substitution for these residues on product outcome (Figure 4). However, although the presence of this ancestral His-Asn dyad seems to imply production of the relevant ent-CPP (2), such conservation does not necessarily indicate a primary role in GA phytohormone biosynthesis. For example, genetic analyses indicates that ZmCPS2 (also known as An2), which produces 2 [41], and contains the His-Asn dyad (Figure 3), is primarily involved in more specialized labdane-related diterpenoid metabolism rather than GA biosynthesis [59].
Nevertheless, loss of the His-Asn dyad does not rule out production of ent-CPP (2). For example, OsCPS2, which also produces 2 [30], has lost the ancestral Asn and seems to utilize an alternative His-Thr catalytic base dyad instead (Figure 6). Intriguingly, TaCPS1, which similarly produces 2 [35], and also has lost the ancestral Asn, seems to have independently evolved an alternative catalytic base, as the Thr from OsCPS2 is not present (Figure 3). This suggests independent re-evolution of such product outcome. In particular, OsCPS2 and TaCPS1 fall within the clade of monocot DTCs solely involved in more specialized metabolism (Figure 1). This clade almost certainly underwent loss of the ancestral His-Asn dyad during the relevant early neo-functionalization event, presumably reflecting divergence of product outcome, consistent with the recently reported loss of the ancestral dyad and diversity of products exhibited by the characterized DTCs [60, 61]. By contrast, ZmCPS2 appears to have arisen from more recent duplication of an ancestral CPS involved in GA biosynthesis as it falls within the same clade as the monocot CPSs known to be involved in such metabolism, and still produces 2, presumably due to retention of the ancestral His-Asn dyad. Notably, the only two members of this clade that no longer produce 2 have alterations to the His-Asn dyad, specifically substitution of Gly for the Asn, presumably leading to the observed production of 8β-hydroxy-ent-CPP (3)[60].
Thus, although it is possible to re-evolve production of 2, initial loss of the ancestral His-Asn dyad seems to generally lead to alteration of product outcome if not outright inactivation. Given that the derived phytohormone is required for normal plant growth and development, this provides strong selective pressure for retention of these residues, consistent with the observed conservation of the His-Asn catalytic base dyad in the plant CPSs involved in such biosynthesis. This further highlights the utility of this dyad as a requisite motif for bioinformatic assignment of such physiological function (although not absolute proof). At the least, retention of this dyad provides confident assignment of catalyzed product outcome (i.e., 2).
Strikingly, the His-Asn catalytic base dyad is not only present in these plant CPSs but also can be found in the CPSs from bacterial GA biosynthesis (Figure 7), with mutation of these residues to Ala exerting analogous functional consequences as well (Figure 8). Similar to the situation in plants, this His-Asn dyad is not required for the production of 2 by bacterial CPSs, as SpCPS does not contain the Asn from the dyad (Figure 7). Indeed, SpCPS seems to utilize a particularly divergent catalytic base dyad (Figure 11). Thus, while it is possible that the bacterial CPSs involved in their production of GA acquired the His-Asn dyad in a parallel evolutionary process, its presence hints at the possibility of long-standing conservation of this key enzymatic feature. Further consistent with this evolutionary scenario is the observed broader conservation of the immediately surrounding residues (Figures 3 & 7), including the preceding Leu and following Ser for the His (i.e., a LHS motif), and the flanking proline and Val, respectively, for the Asn (i.e., a PNV motif).
The presence of the His-Asn dyad in the bacterial CPSs contrasts with the loss of at least the Asn and flanking residues in the fungal bifunctional CPS/KS involved in GA biosynthesis in that biological kingdom (Figure 7). Instead, these seem to utilize an alternative His-Thr catalytic base dyad (Figure 10). Altogether, the results reported here highlight the potential for long-standing conservation of this His-Asn catalytic base dyad from bacteria to plants, suggesting that the CPSs involved in GA biosynthesis from these two distinct biological kingdoms may share a common evolutionary origin, and have been subject to similarly stringent selective pressure for the production of 2.
Such conservation encourages speculation about the original acquisition of CPS by plants from bacteria. Intriguingly, a labdane-related diterpenoid has been implicated as an aggregation signal for sea lettuce (Ulva), but appears to be made by a bacterial epiphyte (Zobellia uliginosa) rather than the alga itself [62]. This hints at an ancient role for labdane-related diterpenoids in plant evolution, potentially during the transition from unicellular algae to multicellular organisms, perhaps similarly dependent on such plant-microbe interactions. Assuming a key role for ent-CPP (2) in such biosynthesis, such a proto-plant could have then acquired the associated CPS from the relevant bacteria, perhaps in the transition to living on land. This early land-plant DTC would have then been retained for its role in developmental processes. In addition, the CPS also could have been retained in plant-associated bacteria for its selective advantage in interactions with these multicellular hosts, consistent with the conservation pattern observed here.
Regardless of such speculation, as discussed above the results reported here provide insight into the evolution of the plant DTC family, strongly supporting the hypothesized repeated derivation of additional DTCs for more specialized metabolism from the CPS involved in biosynthesis of embryophyte phytohormones such as GA. Moreover, the results should be useful in future bioinformatic-directed functional assignment and investigations. In particular, conservation of the His-Asn dyad (as LHS and PNV motifs) seems to characterize the CPSs involved in phytohormone metabolism, while alterations to these residues implies a role for the DTC in more specialized metabolism instead. Accordingly, plant CPSs with the ancestral His-Asn can be presumed to produce 2, with tentative assignment of a role in embryophyte phytohormone biosynthesis, while the absence of these residues highlights those DTCs that can be expected to exhibit alternative product outcome, providing an enriched source to mine for novel catalytic activity.
Acknowledgments
Funding
This work was supported by a grant from the NIH (GM076324) to R.J.P.
Footnotes
Conflicts of interest
R.J.P. is a member of the scientific advisory board for Manus Bio, Inc.
References
- 1.Peters RJ (2010) Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep 27, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zi J, Mafu S and Peters RJ (2014) To Gibberellins and Beyond! Surveying the Evolution of (Di)Terpenoid Metabolism. Annu. Rev. Plant Biol 65, 259–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedden P and Sponsel V (2015) A Century of Gibberellin Research. J. Plant Growth Regul 34, 740–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki S, Nakajima M and Kawaide H (2015) Hormonal diterpenoids derived from ent-kaurenoic acid are involved in the blue-light avoidance response of Physcomitrella patens. Plant Signal. Behav 10, e989046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K and Peters RJ (2009) Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583, 475–480 [DOI] [PubMed] [Google Scholar]
- 6.Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Mudock MP, Mead ME, Prisic S, Wang K, Lin K-Y, Chang T-K, Peters RJ and Oldfield E (2010) Diterpene cyclases and the nature of the isoprene fold. Proteins 78, 2417–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wendt KU, Schulz GE, Corey EJ and Liu DR (2000) Enzyme mechanisms for polycyclic triterpene formation. Angew. Chem. Int. Ed 39, 2812–2833 [PubMed] [Google Scholar]
- 8.Prisic S, Xu J, Coates RM and Peters RJ (2007) Probing the role of the DXDD motif in class II diterpene cyclases. Chembiochem. 8, 869–874 [DOI] [PubMed] [Google Scholar]
- 9.Dong LB, Rudolf JD, Deng MR, Yan X and Shen B (2018) Discovery of the Tiancilactone Antibiotics by Genome Mining of Atypical Bacterial Type II Diterpene Synthases. Chembiochem. 19, 1727–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köksal M, Hu H, Coates RM, Peters RJ and Christianson DW (2011) Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol 7, 431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köksal M, Potter K, Peters RJ and Christianson DW (2014) 1.55A-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. Biochim. Biophys. Acta 1840, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter K, Criswell J and Peters RJ (2014) Novel product chemistry from mechanistic analysis of ent-copalyl diphosphate synthases from plant hormone biosynthesis. Angew. Chem. Int. Ed 53, 7198–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Criswell J, Potter K, Shephard F, Beale MB and Peters RJ (2012) A single residue change leads to a hydroxylated product from the class II diterpene cyclization catalyzed by abietadiene synthase. Org. Lett 14, 5828–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mafu S, Potter KC, Hillwig ML, Schulte S, Criswell J and Peters RJ (2015) Efficient heterocyclisation by (di)terpene synthases. Chem. Commun. (Camb) 51, 13485–13487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potter KC, Jia M, Hong YJ, Tantillo DJ and Peters RJ (2016) Product rearrangement from altering a single residue in the rice syn-copalyl diphosphate synthase. Org. Lett 18, 1060–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte S, Potter K, Lemke C and Peters RJ (2018) Catalytic bases and stereo-control in Lamiaceae class II diterpene cyclases. Biochemistry. 57, 3473–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Honzatko RB and Peters RJ (2012) Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat. Prod. Rep. 29, 1153–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolf JD, Dong LB, Cao H, Hatzos-Skintges C, Osipiuk J, Endres M, Chang CY, Ma M, Babnigg G, Joachimiak A, Phillips GN Jr. and Shen B (2016) Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase. J. Am. Chem. Soc 138, 10905–10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB and Peters RJ (2012) Insights into diterpene cyclization from the structure of the bifunctional abietadiene synthase. J. Biol. Chem 287, 6840–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A and Nozaki H (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett 580, 6175–6181 [DOI] [PubMed] [Google Scholar]
- 21.Kawaide H, Hayashi K, Kawanabe R, Sakigi Y, Matsuo A, Natsume M and Nozaki H (2011) Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J 278, 123–133 [DOI] [PubMed] [Google Scholar]
- 22.Hall DE, Zerbe P, Jancsik S, Quesada AL, Dullat H, Madilao LL, Yuen M and Bohlmann J (2013) Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol 161, 600–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stofer Vogel B, Wildung MR, Vogel G and Croteau R (1996) Abietadiene synthase from grand fir (Abies grandis). J. Biol. Chem 271, 23262–23268 [DOI] [PubMed] [Google Scholar]
- 24.Chen F, Tholl D, Bohlmann J and Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66, 212–229 [DOI] [PubMed] [Google Scholar]
- 25.Nett RS, Montanares M, Marcassa A, Lu X, Nagel R, Charles TC, Hedden P, Rojas MC and Peters RJ (2017) Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol 13, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun T-P and Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schepmann HG, Pang J and Matsuda SP (2001) Cloning and characterization of Ginkgo biloba levopimaradiene synthase, which catalyzes the first committed step in ginkolide biosynthesis. Arch. Biochem. Biophys 392, 263–269 [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Kempinski C, Zhuang X, Norris A, Mafu S, Zi J, Bell SA, Nybo SE, Kinison SE, Jiang Z, Goklany S, Linscott KB, Chen X, Jia Q, Brown SD, Bowman JL, Babbitt PC, Peters RJ, Chen F and Chappell J (2016) Molecular Diversity of Terpene Synthases in the Liverwort Marchantia polymorpha. Plant Cell. 28, 2632–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M and Matsuoka M (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134, 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prisic S, Xu M, Wilderman PR and Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136, 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Hillwig ML, Prisic S, Coates RM and Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39, 309–318 [DOI] [PubMed] [Google Scholar]
- 32.Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S and Bohlmann J (2010) Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol 152, 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ait-Ali T, Swain SM, Reid JB, Sun T and Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11, 443–454 [DOI] [PubMed] [Google Scholar]
- 34.Toyomasu T, Kagahara T, Hirose Y, Usui M, Abe S, Okada K, Koga J, Mitsuhashi W and Yamane H (2009) Cloning and characterization of cDNAs encoding ent-copalyl diphosphate synthases in wheat: insight into the evolution of rice phytoalexin biosynthetic genes. Biosci. Biotechnol. Biochem 73, 772–775 [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Zhou K, Toyomasu T, Sugawara C, Oku M, Abe S, Usui M, Mitsuhashi W, Chono M, Chandler PM and Peters RJ (2012) Functional characterization of wheat copalyl diphosphate synthases elucidates the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry. 84, 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B and Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett 11, 5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, Snyder JH, Song J, Chen S, Huang L, Peters RJ and Qi X (2015) Functional divergence of diterpene synthases in the medicinal plant Salvia miltiorrhiza. Plant Physiol 169, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mafu S, Hillwig ML and Peters RJ (2011) A novel labda-7,13e-dien-15-ol-producing bifunctional diterpene synthase from Selaginella moellendorffii. Chembiochem 12, 1984–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Kollner TG, Yin Y, Jiang Y, Chen H, Xu Y, Gershenzon J, Pichersky E and Chen F (2012) Nonseed plant Selaginella moellendorffi has both seed plant and microbial types of terpene synthases. Proc. Natl. Acad. Sci. U.S.A 109, 14711–14715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB and Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell. 7, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T and Peters RJ (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol. Biol 59, 881–894 [DOI] [PubMed] [Google Scholar]
- 42.Nagel R and Peters RJ (2017) Investigating the Phylogenetic Range of Gibberellin Biosynthesis in Bacteria. Mol. Plant Microbe Interact 30, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hershey DM, Lu X, Zi J and Peters RJ (2014) Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J. Bact 196, 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR and Shen B (2011) Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc. Natl. Acad. Sci. U.S.A 108, 13498–13503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tudzynski B, Kawaide H and Kamiya Y (1998) Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene. Curr. Gent 34, 234–240 [DOI] [PubMed] [Google Scholar]
- 46.Prisic S and Peters RJ (2007) Synergistic substrate inhibition of ent-copalyl diphosphate synthase: A potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol 144, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi S, Sun T, Kawaide H and Kamiya Y (1998) The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol 116, 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu M, Wilderman PR and Peters RJ (2007) Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. U.S.A 104, 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawaide H, Sassa T and Kamiya Y (2000) Functional analysis of the two interacting cyclase domains in ent-kaurene synthase from the fungus Phaeosphaeria sp. L487 and a comparison with cyclases from higher plants. J. Biol. Chem 275, 2276–2280 [DOI] [PubMed] [Google Scholar]
- 50.Morrone D, Lowry L, Determan MK, Hershey DM, Xu M and Peters RJ (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol 85, 1893–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cyr A, Wilderman PR, Determan M and Peters RJ (2007) A Modular Approach for Facile Biosynthesis of Labdane-Related Diterpenes. J. Am. Chem. Soc 129, 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrone D, Chen X, Coates RM and Peters RJ (2010) Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J 431, 337–344 [DOI] [PubMed] [Google Scholar]
- 53.Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K and Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79, 659–678 [DOI] [PubMed] [Google Scholar]
- 54.Johnson SR, Bhat WW, Bibik J, Turmo A, Hamberger B, Evolutionary Mint Genomics C and Hamberger B (2019) A database-driven approach identifies additional diterpene synthase activities in the mint family (Lamiaceae). J. Biol. Chem 294, 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T and Toyomasu T (2004) Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J 39, 886–893 [DOI] [PubMed] [Google Scholar]
- 56.Lu X, Zhang J, Brown B, Li R, Rodriguez-Romero J, Berasategui A, Liu B, Xu M, Luo D, Pan Z, Baerson SR, Gershenzon J, Li Z, Sesma A, Yang B and Peters RJ (2018) Inferring Roles in Defense from Metabolic Allocation of Rice Diterpenoids. Plant Cell. 30, 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Potter KC, Zi J, Hong YJ, Schulte S, Malchow B, Tantillo DJ and Peters RJ (2016) Blocking Deprotonation with Retention of Aromaticity in a Plant ent-Copalyl Diphosphate Synthase Leads to Product Rearrangement. Angew. Chem. Int. Ed 55, 634–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bomke C and Tudzynski B (2009) Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry. 70, 1876–1893 [DOI] [PubMed] [Google Scholar]
- 59.Vaughan MM, Christensen S, Schmelz EA, Huffaker A, McAuslane HJ, Alborn HT, Romero M, Allen LH and Teal PE (2015) Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ 38, 2195–2207 [DOI] [PubMed] [Google Scholar]
- 60.Pelot KA, Chen R, Hagelthorn DM, Young CA, Addison JB, Muchlinski A, Tholl D and Zerbe P (2018) Functional Diversity of Diterpene Synthases in the Biofuel Crop Switchgrass. Plant Physiol 178, 54–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy KM, Ma LT, Ding Y, Schmelz EA and Zerbe P (2018) Functional Characterization of Two Class II Diterpene Synthases Indicates Additional Specialized Diterpenoid Pathways in Maize (Zea mays). Front. Plant Sci 9, 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuo Y, Imagawa H, Nishizawa M and Shizuri Y (2005) Isolation of an Algal Morphogenesis Inducer from a Marine Bacterium. Science. 307, 1598. [DOI] [PubMed] [Google Scholar]


