Abstract
Public health interventions to control the current epidemic of carbapenem-resistant Klebsiella pneumoniae are reliant upon a comprehensive understanding of their emergence and spread over a wide range of geographical scales. We analysed the genome sequences and epidemiological data of >1700 K. pneumoniae, isolated from patients in 244 hospitals in 32 countries, during the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE). We demonstrate that carbapenemase acquisition is the main cause of carbapenem resistance and has occurred across diverse phylogenetic backgrounds. However, 477/682 (69.9%) of carbapenemase-positive isolates are concentrated in four clonal lineages, sequence types (ST) 11, 15, 101, 258/512, and their derivatives. Combined analysis of the genetic and geographic distances between isolates with different beta-lactam resistance determinants suggests that the propensity of K. pneumoniae to spread in hospital environments correlates with the degree of resistance and that carbapenemase-positive isolates have the highest transmissibility. Indeed, we found that over half of hospitals contributing carbapenemase-positive isolates likely experienced within-hospital transmission, and inter-hospital spread is far more frequent within, rather than between, countries. Finally, we propose a value of 21 for the number of single nucleotide polymorphisms (SNPs) that optimises discrimination of hospital clusters, and detail the international spread of the successful epidemic lineage, ST258/512.
Keywords: Klebsiella pneumoniae, carbapenem resistance, carbapenemase-producing, Enterobacteriaceae, European survey, genomics
Introduction
Klebsiella pneumoniae, a major cause of both hospital- and community-acquired infections, is listed by the World Health Organisation (WHO) as one of the critical priority antibiotic-resistant bacterial pathogens for which new antibiotics are urgently needed [1]. Indeed, a recent study showed that carbapenem-resistant K. pneumoniae represents the fastest growing antibiotic resistance threat in Europe, in terms of human morbidity and mortality [2]. It is therefore critical to identify priority areas upon which to intensify public health intervention strategies.
Rapid expansion of carbapenem resistance in K. pneumoniae has been attributed to the acquisition of carbapenemase enzymes which hydrolyse carbapenems, a last-line class of antibiotics, and other beta-lactam antibiotics to varying degrees. An important feature that has facilitated widespread dissemination of carbapenemase genes is that they are associated with mobile elements that can spread horizontally within, and between, bacterial species [3, 4]. In addition to its importance as a major pathogen itself, K. pneumoniae has been identified as a crucial entry point of antibiotic resistance genes into the Enterobacterales (Enterobacteriaceae) family [5–7].
Emergence of carbapenem resistance seems to occur in clinical [8], urban (9) and agricultural settings (10) and is a worldwide phenomenon [11–13]. However, biased and fragmented surveillance, combined with the lack of standardisation used to characterise isolates, has limited the ability to discern the primary reservoirs and transmission dynamics of this global epidemic. The European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE) was the first study to systematically determine the incidence and epidemiology of carbapenem-non-susceptible K. pneumoniae at a continental scale, enrolling 455 hospitals across Europe and neighbouring countries [14]. Between November 2013 and May 2014, hospital laboratories were asked to submit the first ten consecutive carbapenem-non-susceptible clinical isolates of either K. pneumoniae or E. coli, together with one carbapenem-susceptible same-species clinical isolate (per non-susceptible isolate) to serve as a comparator. Isolates were obtained from clinical specimens submitted for diagnostic purpose, while samples obtained for screening purposes were excluded.
To elucidate the European-wide population structure and determine the epidemiology of carbapenem-non-susceptible K. pneumoniae with maximum resolution, we analysed the genomes of 1717 K. pneumoniae isolates submitted during the survey. This unprecedented sample provides an unbiased, continental and contemporaneous population snapshot of representative clinical isolates from hospitalised patients, and links lineage abundance and spatial expansion to variable genomic resistance determinants.
Results
Population structure and carbapenem resistance determinants
Genome sequences were obtained for 1717 clinical isolates that originated from 244 hospitals in 32 countries (Fig. 1, Supplementary Fig. 1 & Supplementary Tables 1, 2, 3, 4). 944/1717 (55.0%) isolates were submitted by hospital laboratories as carbapenem-non-susceptible, while 773/1717 (45.0%) were submitted as carbapenem-susceptible.
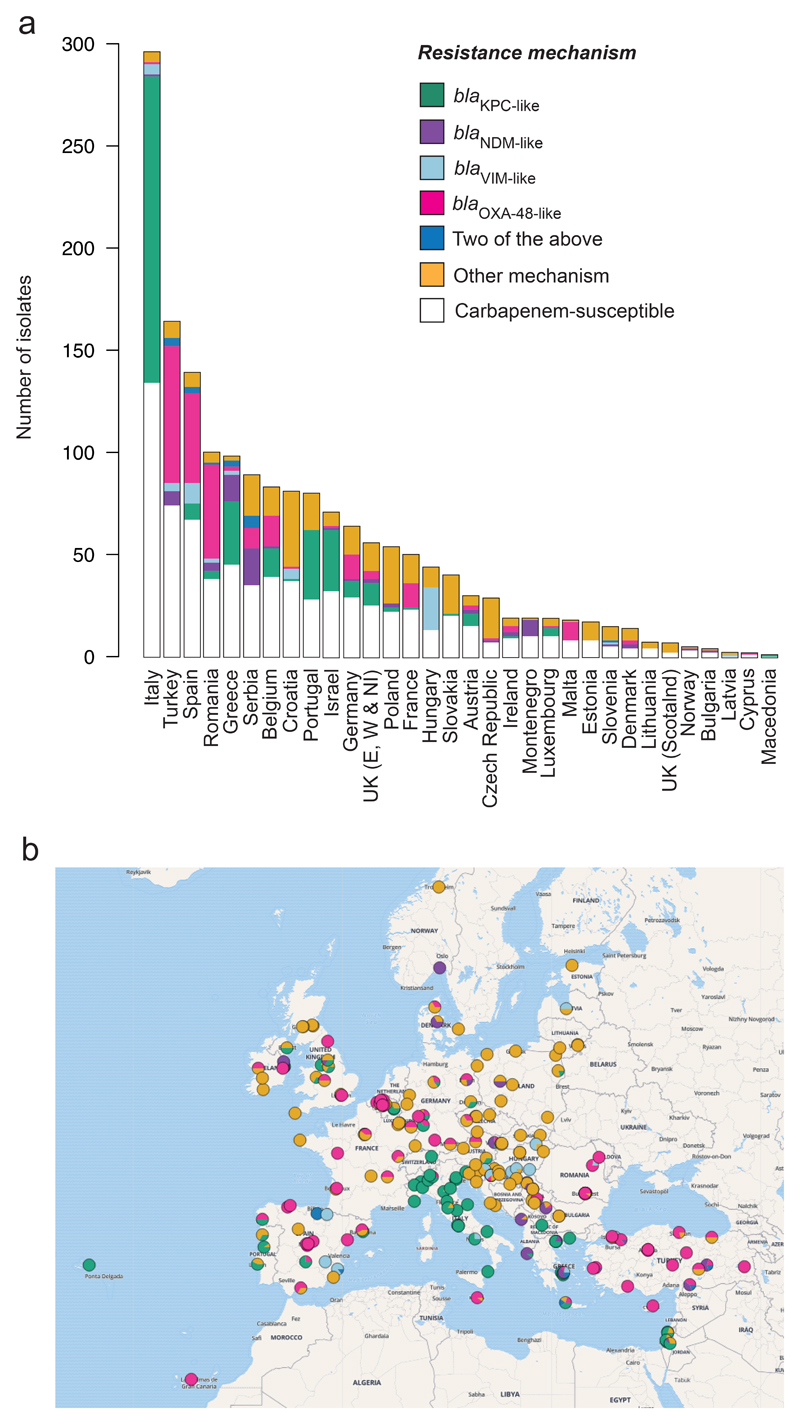
Figure 1. Geographical distribution of carbapenem resistance mechanisms amongst isolates submitted during the EuSCAPE survey.
a, Total number of isolates submitted by participating countries that were analysed in this study. Isolates are partitioned into those that possess one or more of four major carbapenemase genes (blaKPC-like, blaOXA-48-like, blaNDM-like and blaVIM-like) regardless of whether they were submitted as carbapenem-non-susceptible or -susceptible, those which lack any of the four genes that were submitted as carbapenem-non-susceptible (“Other mechanism”), and those submitted as carbapenem-susceptible. b, Pie charts representing the proportion of isolates submitted as carbapenem-non-susceptible from participating hospitals with different resistance mechanisms as in a.
Core genome phylogenetic analysis revealed a partitioning into four species, supporting recent taxonomic classifications [15, 16] (Fig. 2a). 1649/1717 (96.0%) belong to K. pneumoniae sensu stricto, 48/1717 (2.8%) to K. variicola, 17/1717 (1.0%) to K. quasipneumoniae, and 3/1717 (0.2%) to a newly-described species, K. quasivariicola [16]. 939/944 (99.5%) isolates submitted as carbapenem-non-susceptible belong to K. pneumoniae sensu stricto, 3/944 (0.3%) to K. variicola and 2/944 (0.2%) to K. quasipneumoniae.
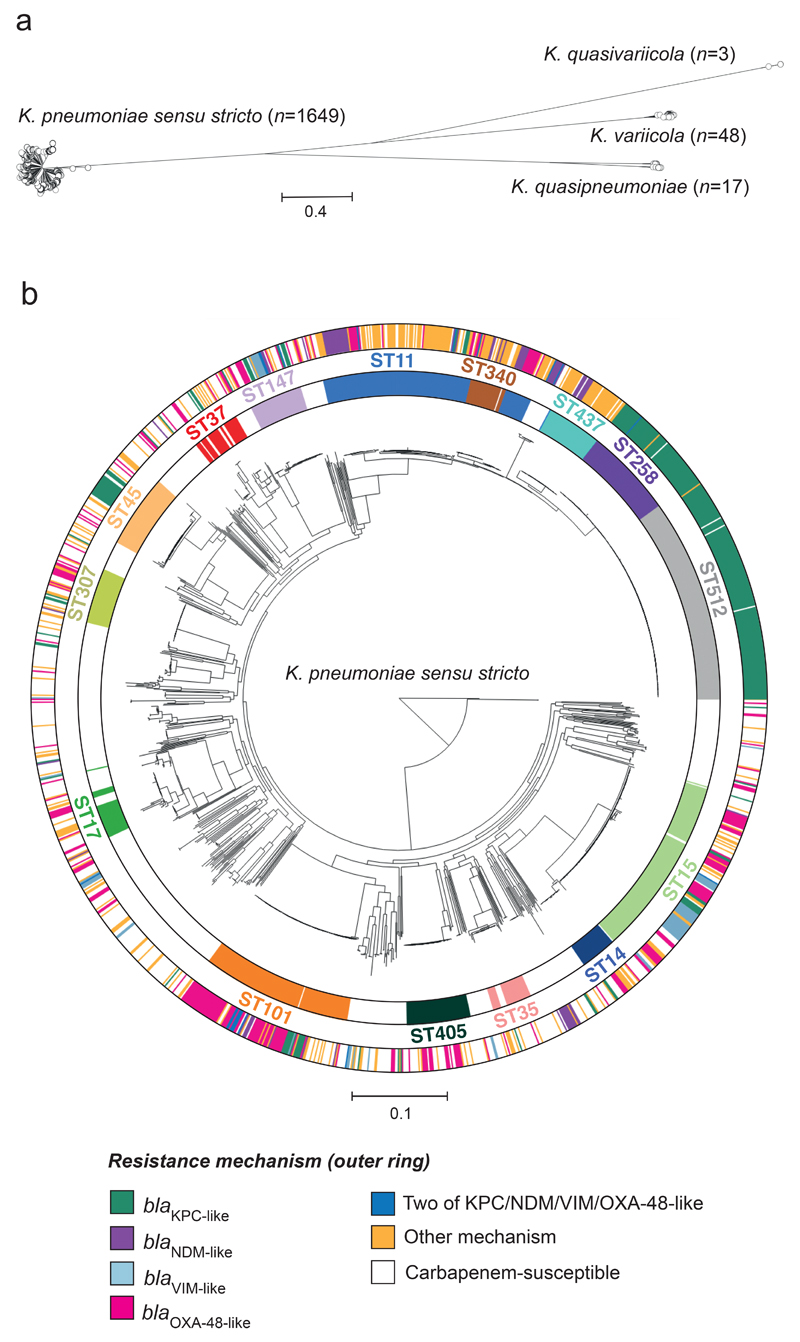
Figure 2. Carbapenemase-positive isolates are concentrated in major clonal lineages of K. pneumoniae.
a, Phylogenetic tree of 1717 EuSCAPE isolates analysed in this study, constructed using 2539 core genes, demonstrates division into four species. b, Phylogenetic tree of 1649 isolates belonging to K. pneumoniae sensu stricto with STs containing more than 20 isolates shown in the inner ring and labelled. The outer ring colours isolates by resistance mechanism as in Figure 1a. The scale in both trees represents the number of SNPs per variable site. Similar visualisations are available at: https://microreact.org/project/EuSCAPE_Kp/bc81fffe (a) and https://microreact.org/project/EuSCAPE_Kp/2585de34 (b)
We defined phylogenetic lineages in K. pneumoniae sensu stricto by their multi-locus sequence type (ST) [17]. 254 different STs were identified amongst isolates of this species, of which 94 (37.0%) contain isolates submitted as carbapenem-non-susceptible. 15/254 (5.9%) STs contain >20 isolates (Fig. 2b). Two of the most numerous STs, 258 (n=73) and 512 (n=163), which are single-locus variants (SLV), can be considered as a single lineage since there is no significant evolutionary separation between them (Fig. 2b).
We searched the genomes for beta-lactam resistance determinants which could be relevant to carbapenem resistance, and divided the entire collection into five beta-lactam resistome groups. Group (i) contained one or more of any carbapenemase gene published in the literature prior to April 2018 (see Methods) regardless of other mechanisms, group (ii) contained any extended spectrum beta-lactamase (ESBL) gene and/or an AmpC gene, in combination with porin defects, group (iii) contained any ESBL gene and/or an AmpC gene without any obvious porin defects, group (iv) showed absence of any ESBL or AmpC gene but presence of porin defects and, in group (v), all of the above determinants were absent.
Six hundred and eighty four isolates carried one or more carbapenemase genes and were assigned to group (i). Of these, all but two belonged to K. pneumoniae sensu stricto, which contained blaKPC-like (n=311), blaOXA-48-like (n=248), blaNDM-like (n=79), blaVIM-like (n=56), and blaIMP-like (n=3) genes in 28, 44, 13, 13, and 1 ST, respectively (Table 1 & Supplementary Table 4). A blaOXA-48-like gene was also found in one K. quasipneumoniae genome and a blaKPC-like gene in one K. variicola genome. Eighteen K. pneumoniae sensu stricto isolates carried two carbapenemase genes, the most commonly observed combination being that of blaNDM-like and blaOXA-48-like genes, which was found in 10 isolates. Furthermore, in silico detection of the blaKPC-like, blaOXA-48-like, blaNDM-like, and blaVIM-like carbapenemase genes showed 98.3-99.0% concordance with PCR results obtained previously by the national expert laboratories (NELs) from individual countries (Supplementary Table 5). Of all 684 group (i) isolates, 657 (96.1%) were submitted as carbapenem-non-susceptible, including 655/682 (96.0%) of those belonging to K. pneumoniae sensu stricto (Table 1). Upon central retesting with reference broth microdilution, we found that group (i) isolates possessed the highest phenotypic resistance to meropenem with a median MIC value of 32 (Fig. 3a & Supplementary Fig. 2).
Table 1.
Characteristics of all submitted K. pneumoniae sensu stricto isolates with different beta-lactam resistance determinants. NA – not applicable. CI = confidence intervals.
| Isolate subset | No. submitted isolates (% of total) | No. isolates submitted as carbapenem-non-susceptible (%) | No. of STs (and STs with >10% isolates)* | Simpson’s diversity of STs (and 95% CI)* | No. of countries (and countries with >10% isolates) | Carbapenemase gene variants (and no. of isolates) |
|---|---|---|---|---|---|---|
| blaKPC-like | 311 (18.9%) | 304 (97.7%) | 28 (ST258, n=69; ST512, n=157) | 0.68 (0.63-0.73) | 17 (Italy, n=150; Portugal, n=34; Greece, n=34) | KPC-2 (n=78), KPC-3 (n=232), KPC-12 (n=1) |
| blaOXA-48-like | 248 (15.0%) | 237 (95.6%) | 44 (ST15, n=38; ST101, n=67) | 0.88 (0.85-0.91) | 20 (Romania, n=47; Spain, n=46; Turkey, n=70) | OXA-48 (n=240), OXA-204 (n=1), OXA-162 (n=2), OXA-181 (n=1), OXA-232 (n=4) |
| blaNDM-like | 79 (4.8%) | 76 (96.2%) | 13 (ST11, n=25; ST101, n=12; ST274, n=10; ST395, n=8) | 0.84 (0.78-0.89) | 19 (Serbia, n=24; Greece, n=13; Turkey, n=10; Montenegro, n=8) | NDM-1 (n=79) |
| blaVIM-like | 56 (3.4%) | 53 (94.6%) | 13 (ST15, n=27; ST147, n=7) | 0.73 (0.61-0.85) | 9 (Hungary, n=21; Spain, n=12) | VIM-1 (n=33), VIM-4 (n=23) |
| blaIMP-like | 3 (0.2%) | 3 (100%) | 1 (ST15, n=3) | 0 (0-0) | 1 (Turkey, n=3) | IMP-1 (n=3) |
| Any carbapenemase gene (group i)** | 682 (41.4%) | 655 (96.0%) | 69 (ST15, n=78; ST101, n=84; ST258, n=69; ST512, n=157 | 0.89 (0.88-0.91) | 29 (Italy, n=157; Turkey, n=85) | See above |
| ESBL and/or AmpC gene + porin defects, but no carbapenemase (group ii) | 150 (9.1%) | 114 (76.0%) | 35 (ST11, n=30; ST15, n=18) | 0.92 (0.90-0.95) | 25 (Slovakia, n=19) | NA |
| ESBL and/or AmpC gene but no carbapenemase/porin defects (group iii) | 400 (24.3%) | 123 (30.8%) | 90 (ST11, n=52; ST15, n=44) | 0.95 (0.94-0.96) | 27 (Italy, n=43; Serbia, n=43) | NA |
| Porin defects but no carbapenemase/ESBL/AmpC (group iv) | 37 (2.2%) | 24 (64.9%) | 21 (ST17, n=5; ST437, n=4; ST512, n=4) | 0.96 (0.93-0.98) | 18 (Croatia, n=4; Italy, n=4; Portugal, n=4; United Kingdom, n=5) | NA |
|
No carbapenemase/ESBL/ AmpC/ porin defects (group v) |
380 (23.0%) | 23 (6.1%) | 161 | 0.98 (0.98-0.99) | 27 (Italy, n=71; Spain, n=41; Turkey, n=49) | NA |
Ambiguous STs (i.e. those with one or more uncertain alleles) were excluded for these calculations.
These include 3 isolates that were found to carry blaOXA-23 or blaOXA-58. However, since these genes are usually found only in Acinetobacter baumannii and they were found at low level in our samples, we suspect that they are contaminants.
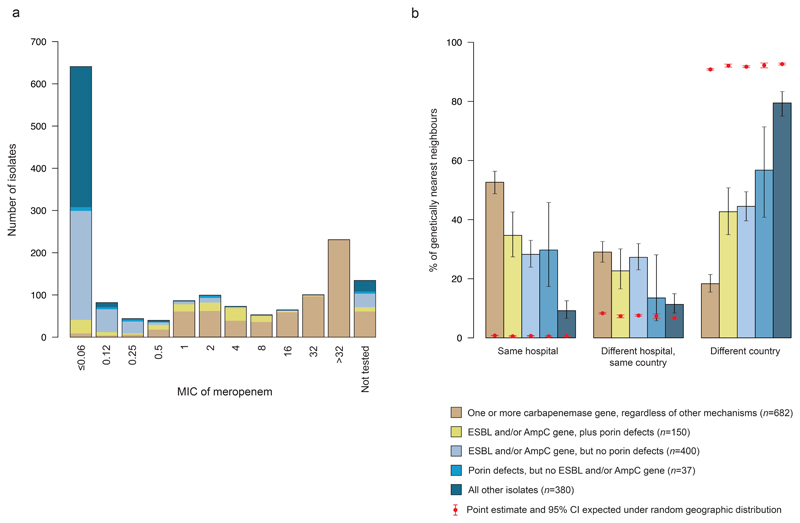
Figure 3. Carbapenemase-positive isolates show strong geographical clustering.
a, Barplots show the distribution of minimum inhibitory concentration (MIC) values amongst 1518 re-tested K. pneumoniae sensu stricto isolates with different beta-lactam resistance determinants. b, Barplots show the proportion of genetically nearest neighbours (gNNs) for K. pneumoniae sensu stricto isolates with different beta-lactam resistance determinants that are from different geographic contexts (the same hospital, a different hospital in the same country, or a hospital in a different country). The black error bars represent 95% confidence intervals (CI) of these proportions. The proportions of gNNs in the absence of geographic structure were calculated from 100 repeat analyses in which the hospital codes were randomly permutated, and the mean and 95% CI of these results are shown in red.
We found 150 isolates belonging to group (ii) that lacked a carbapenemase but harboured an extended spectrum beta-lactamase (ESBL) gene and/or an AmpC gene, in combination with porin defects (see Methods; Table 1 & Supplementary Table 4). These resistance determinants can also account for a carbapenem-non-susceptible phenotype. All belong to K. pneumoniae sensu stricto and 114/150 (76.0%) were submitted as carbapenem non-susceptible, a lower proportion than that observed for group (i) isolates. Central re-testing of these isolates also demonstrated lower phenotypic resistance to meropenem than group (i) isolates with a median MIC value of 1 (Fig. 3a & Supplementary Fig. 2).
A further 402, 40 and 441 isolates were assigned to groups (iii), (iv) and (v), respectively, including 400, 37 and 380 isolates from K. pneumoniae sensu stricto. 30.6%, 65.0% and 5.4% of isolates from these three groups were submitted as carbapenem-non-susceptible, respectively (or 30.8%, 64.9%, and 6.1% from K. pneumoniae sensu stricto only) (Table 1). However, based on conventional classification, isolates from these three groups were expected to be susceptible. Indeed, upon central re-testing, the three groups are associated with lower MIC values to meropenem than groups (i) and (ii) (Fig. 3a & Supplementary Fig. 2), with median MIC values of ≤0.06, 0.25 and ≤0.06. Thus, discrepancies between the classification of participating laboratories and gene contents may reflect heterogeneity in the antibiotic susceptibility testing, subsequent loss of resistance elements post-submission, or incorrect classification.
Limited genetic diversity among carbapenemase-positive isolates
Carbapenemase-positive isolates are concentrated in major clonal lineages of K. pneumoniae sensu stricto (Table 1 and Fig. 2b). In particular, 477/682 (69.9%) of all carbapenemase-positive isolates in this species belong to four lineages comprising STs 11, 15, 101 and ST258/512, as well as their derivatives (i.e. other closely-related STs that have evolved from these major STs). Among these STs (without their derivatives), the majority of isolates were carbapenemase-positive, except for ST11 (ST258/512 – 97.5%, ST101 – 69.9%, ST15 – 55.9%, ST11 – 37.9%), whilst the proportion of carbapenemase-positive isolates amongst the total K. pneumoniae sensu stricto sample was 39.8%. The major lineages are also characterised by low levels of core genome diversity, reflected by an average nucleotide identity (ANI) of 99.9-100% between same-ST pairs of STs 15, 101, and 258/512. This indicates a recent and common evolutionary descent. Furthermore, ST258 and ST512 harbour less diversity overall than each of the other three STs individually, supporting their combined grouping. However, ST11, also a SLV of ST258, consists of several sub-lineages that are distinct from ST258, and exhibits the highest diversity of the major STs with an ANI of 99.7-100% between pairs. For context, the ANI ranges from 94.1-95.4% between genomes of the four different species, and from 98.9-100% between genomes of K. pneumoniae sensu stricto. Despite an overall limited genetic diversity, STs 11, 15, 101, 258/512 are widely distributed across Europe, having been submitted from 22, 19, 15, and 15 countries, respectively.
Hence the five carbapenemase genes identified among isolates in K. pneumoniae sensu stricto are - for the majority - confined to recently emerging and vastly successful clonal backgrounds. This can also be seen by the limited population diversity, whereby diversity indices (ID) for carbapenemase-positive isolates were significantly lower than for the other beta-lactam resistome groups (Table 1). The only exception is blaOXA-48-like-containing isolates where the ID was slightly higher and confidence intervals overlapped with group (ii) isolates (ESBL/AmpC-positive isolates with porin defects).
Geographic spread
To explore the differential ecological success of K. pneumoniae sensu stricto with different beta-lactam resistance determinants, we measured the genetic relatedness of isolates belonging to the five previously defined beta-lactam resistome groups with respect to three geographic contexts: (a) from the same hospital, (b) from the same country but different hospitals, and (c) from different countries. We found that for 359 of the 682 (52.6%) carbapenemase-positive (group i) isolates, the genetically nearest neighbour (gNN) in the collection originated from patients treated in the same hospital (Fig. 3b). When stratifying by carbapenemase, we observed that the gNN was from the same hospital for 159/311 (51.1%) isolates carrying blaKPC-like genes, 125/248 (50.4%) carrying blaOXA-48-like genes, 43/79 (54.4%) carrying blaNDM-like genes and 42/56 (75.0%) carrying blaVIM-like genes. Yet isolates from other beta-lactam resistome groups (ii-v) had a lower proportion (34.7%, 28.3%, 29.7%, 9.2%) of gNNs obtained from same hospital patients with a significant downward trend that coincided with the decreasing ability to express carbapenem resistance (p-value<0.01, chi-squared test for trend) (Fig. 3b). ANI among gNNs was very high (>99.95-100%), irrespective of the beta-lactam resistome group. We tested the observed proportions against the null hypothesis of a random geographic distribution by permutation of the hospital codes for all isolates and obtained expected ranges by 100 repeat analyses. We could thus infer that, in the absence of geographic structure, the proportions of gNNs from the same hospital were expected to be 67.4x, 56.9x, 41.6x, 58.2x and 14.4x lower for all the resistome groups (i-v) considered. Assuming that gNNs with a high ANI share a recent common ancestor, our findings suggest that carbapenemase-positive strains have the highest transmissibility and epidemic potential and that the propensity of K. pneumoniae sensu stricto to spread in hospitals correlates with the degree of resistance. Isolates with an ESBL/AmpC gene and/or porin defects occupy an intermediate position between carbapenemase-positive isolates and those for which none of these beta-lactam resistance determinants were detected. We also observed a similar but less pronounced distribution for gNNs originating from different hospitals as long as they were from the same country. Notably, the lower bound of the confidence interval of gNNs in the group of isolates with none of the beta-lactam resistance determinants (group v) approaches that of the random distribution in this geographic context (Fig. 3b).
Thresholds for discriminating likely transmission events
Of the 171 hospitals that contributed carbapenemase-positive isolates, 96 (56.1%) had at least two isolates with a nearest neighbour relationship as described above, suggestive of within-hospital transmission. Given this high frequency of nosocomial spread of carbapenemase-positive K. pneumoniae sensu stricto, the ability to identify likely transmission events within hospitals directly from SNP distances would present a significant advance with respect to both immediate infection control priorities and retrospective outbreak investigations. Due to the widespread sampling that allows comparison across increasing geographic scales, the EuSCAPE collection provides an opportunity to investigate levels of diversity in relation to potential transmission.
We focused on the largest epidemic clone, ST258/512, within the EuSCAPE collection and first analysed how pairwise SNP differences vary for the same geographic contexts used previously. For each of these three contexts, we calculated the pairwise minimum SNP differences between isolates (Fig. 4a-c). The minimum SNP differences showed a central tendency and a shift of the mode to the right from 0 in the same hospital context (a), to 27 in the different hospital context (b) and 45 in the different country context (c). These results lend support to the notion that, amongst carbapenemase-positive isolates, geographic and evolutionary distance correspond, and also provide an indication that pairwise SNP differences between isolates from a given hospital provide meaningful information about transmissions within an institution. Under the assumption that isolates submitted from different hospitals/countries are less likely than those from the same hospital to be closely linked in a transmission chain, we would expect an optimum SNP threshold for defining an institutional transmission to be lower than the number of SNPs typically seen between isolates from different hospitals/countries.
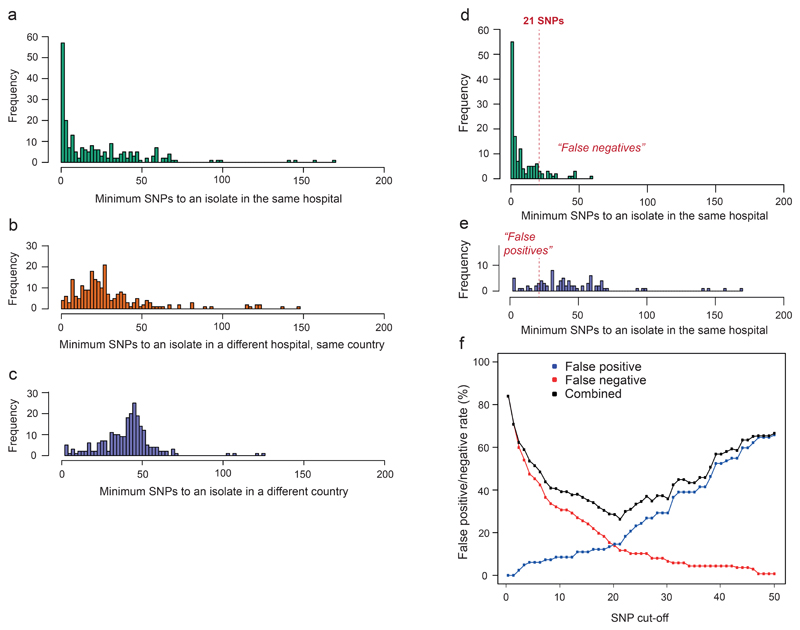
Figure 4. Determination of a SNP cut-off to aid outbreak investigations of ST258/512.
a-c, Number of core chromosomal SNPs between each of 236 ST258/512 isolates submitted during EuSCAPE and the most closely related isolate submitted by the same hospital (a), a different hospital in the same country (b), and a hospital in a different country (c). d-e, Number of SNPs between each ST258/512 isolate and the most closely related isolate submitted by the same hospital, when isolates are predicted to have been involved in intra-hospital (d) or inter-hospital (e) transmission. The dotted lines are drawn at the number of SNPs whereby the combined false positive and negative rates for correctly identifying intra-hospital transmission, calculated using the predictions as the reference, are at a minimum. f, False positive and negatives rates when different SNP cut-offs are used.
We next used a statistical method [18] to distinguish between intra- and inter-hospital transmission, through the comparison of SNP profiles of a given query isolate against a reference database. We used all ST258/512 genomes from the EuSCAPE collection as the database, representing isolates from 59 hospitals, and removed one isolate at a time to be the query. Using a naïve Bayesian classifier, the method determines the likelihood of any given query isolate originating from each of the hospitals represented in the reference database based on the presence of SNPs called against a standard reference genome. Intra-hospital transmission events are assigned in those cases where the most likely source hospital of a query isolate coincides with the hospital from which the isolate was actually recovered. In contrast, if a given query isolate is found to be most similar to those from a hospital which is different to the one from which it was sampled, then it is assigned as corresponding to an inter-hospital transmission, although it is not possible to assign directionality.
Using these predictions as a reference, we were able to determine an optimal value of 21 SNPs to discriminate hospital clusters, which minimises the number of false positives (pairs of isolates erroneously assigned to intra-hospital transmission) and false negatives (cases of missed intra-hospital transmission) (Fig. 4d-f). At the same time, this method provides a measure of uncertainty at different thresholds, which equates to a false positive rate of 14.6% and a false negative rate of 11.7% at 21 SNPs.
International spread of the epidemic ST258/512 clone
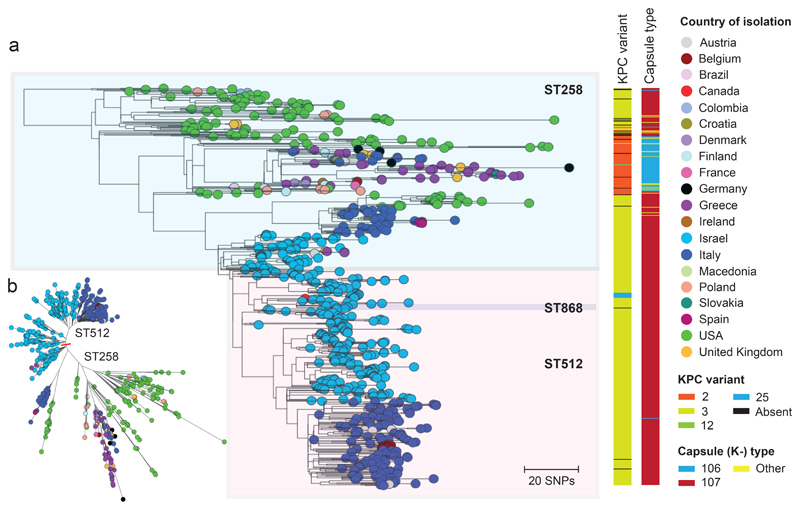
Finally, to determine the ancestral relationships between the dispersed populations of the ST258/512 clone, we analysed 236 ST258/512 genomes from the EuSCAPE collection together with 415 publicly available draft genomes of STs 258, 512, and another derivative, ST868 (Supplementary Table 6), which were isolated in 20 countries across Europe, the Middle East, and North and South America. After mapping sequence reads to the ST258 reference genome (accession number CP006923) we identified 68 recombined regions larger than 1kb. These included a 57.1kb region encompassing the capsular locus that introduced 563 SNPs, thereby accounting for the capsular (K-type) switch from K107 to K106, which coincides with the previously described evolutionary transition from “clade I” to “clade II” [19] (Supplementary Fig. 3). Another two recombination events over the capsular region affecting single isolates could also be traced to switches in K-type.
We then removed the recombined regions and used the resulting alignment to reconstruct a maximum likelihood tree of all 651 isolates (Fig. 5). ST258 isolates from the USA are basal in the phylogenetic tree, supporting the conventional view that this lineage emerged in the USA. All but three isolates (n=31) from Greece, sampled between 2007 and 2014, fall into a single clade (bootstrap support, 100%) suggesting one major introduction into Greece from the USA, and subsequent nationwide spread. Many isolates from other European countries (e.g. United Kingdom, Germany) also cluster amongst these Greek isolates, representing spread of this lineage out of Greece, likely via human travel. All 268 isolates from Israel, sampled between 2007 and 2014, also cluster together in a single clade (bootstrap support, 97%) suggesting one successful introduction followed by within-country dissemination. The most basal ST512 isolates are also from Israel, implying emergence of ST512 in Israel. The tree structure demonstrates a single successful introduction of ST512 from Israel to Italy (bootstrap support, 97%), although separate introductions of ST258 to Italy are also evident. The potential of ST258/512 to spread out of endemic countries and spawn outbreaks elsewhere is also exemplified by an ST258 cluster in Spain (4 patients in 1 hospital) and ST512 clusters in Belgium (7 patients in 2 nearby hospitals) and Austria (3 patients in 1 hospital). All three clusters likely originated from Italy based on phylogenetic inference.
Figure 5. International spread of the epidemic ST258/512 clone.
a, Phylogenetic tree of 651 isolates of ST258 and 512 (and a derivative of ST512, ST868), comprising 236 isolates submitted during the EuSCAPE survey and 415 isolates with publicly available sequence data. The tree was rooted using an ST11 isolate that was later removed. The isolate tips are coloured by the country of isolation, and metadata columns show the blaKPC-like variant and capsule (K-) type. b, An unrooted version of the same tree as in A. Similar visualisations are available at: https://microreact.org/project/EuSCAPE_ST258/bbafcc1c (a) and https://microreact.org/project/EuSCAPE_ST258/dd960284 (b).
Discussion
The emergence of carbapenem resistance is a major setback in the ability to effectively treat Gram-negative bacterial infections [20]. In K. pneumoniae, carbapenemases are the main contributing factor to extensive drug resistance, and their recent acquisition and dissemination likely forebodes pan-drug resistance in the near future [21, 22]. Across Europe, resistance rates differ between countries, which may be explained in part by different levels of antibiotic use. However, critical knowledge gaps remain relating to how the characteristics of the pathogen population also contribute to the epidemic. Numerous studies have highlighted the role of animals and environmental reservoirs as important evolutionary drivers in the mobilisation of resistance genes within and between species and have argued that increasing selective pressure in animals and the environment warrants a ‘one-health’ surveillance approach [23, 24]. However, this study re-asserts the central role of intra-and inter-hospital transmission for the most burdensome carbapenemase-positive clones that are already well-established within the human population. Indeed, strong links between hospital care and the acquisition of carbapenem-non-susceptible bacteria have previously been noted [14, 25–28], and the impact of community [29] or animal and environmental reservoirs [10] on clinical cases remains circumstantial. In this context, we note that a large faecal resistome analysis from slaughter pigs and broilers failed to identify carbapenemase genes in 181 pig and 178 poultry farms from nine European countries [30].
Here we infer the epidemiology of carbapenemase-positive K. pneumoniae in Europe using genome data for 1717 carbapenem-non-susceptible and -susceptible isolates obtained from consecutive clinical samples in 244 hospitals in 32 countries over a six-month prevalence survey (EuSCAPE) [14]. Most (99.5%) of the 944 isolates submitted as carbapenem-non-susceptible belonged to the species, K. pneumoniae sensu stricto. Furthermore, the majority (69.6%) of isolates submitted as carbapenem-non-susceptible possessed one or more carbapenemase genes, which included the five most frequently reported carbapenemase genes worldwide: blaKPC-like, blaOXA-48-like, blaNDM-like, blaVIM-like, and blaIMP-like. Carbapenemase-positive isolates were concentrated among four major clonal lineages of K. pneumoniae sensu stricto comprising STs 11, 15, 101, and 258/512. All have recently emerged, gained abundance and, for the most part, expanded in southern or eastern European countries. Hence, these lineages bear the hallmarks of so-called “high-risk” clones, which commonly share a recent ancestor, epidemic success, and a defined geographic distribution [31]. They are also often associated with outbreaks [32–35] and likely possess particular characteristics that increase their tenacity, transmissibility and population size, providing greater opportunity for acquisition of antibiotic resistance genes [36].
We demonstrated a strong relationship between genetic and geographic distance among carbapenemase-positive isolates, which cluster at country-level and, even more strikingly, at hospital-level. More than half of the carbapenemase-positive isolates had the genetically nearest neighbour (gNN) sampled from the same hospital and likewise the majority of hospitals that contributed carbapenemase-positive K. pneumoniae had at least one pair of gNNs isolated from patients treated within a six month period at the same hospital. This is suggestive of frequent within-hospital transmission. At the same time, the proportion of gNNs sampled from the same hospital or same country incrementally declined with a decreasing spectrum of beta-lactam resistance determinants. This suggests that, in hospitals, ecological constraints exerted by antibiotic exposure obstruct the spread of isolates with a lower capacity for expressing resistance.
In contrast to our finding that carbapenemase-positive K. pneumoniae are transmitted predominantly via nosocomial routes, a study also using genomic data found that CPE (including those of the K. pneumoniae species complex), collected both retrospectively and prospectively in four US hospitals, could mostly not be linked to transmissions [37]. We cannot explain this difference but suggest that the lack of a consistent and purpose-designed sampling frame, ensuring that isolates from consecutive clinical samples were collected, may have obscured transmission events in these US hospitals. This study did, however, find geographical structuring of ST258 isolates, demonstrating local spread of this particular lineage.
We also noted that the distributions of core genome diversity differentiated same hospital vs. different hospital and different country isolates. This epidemiological signature provided the means to determine an optimal cut-off value for SNP differences that could be used to aid epidemiological investigations of putative institutional outbreaks of ST258/512. In addition to this, we could also provide predictions of the sensitivity and specificity of using a cut-off at different SNP thresholds (Fig. 4f). However, one major limitation of our approach is that we had no detailed epidemiological information to help confirm whether pairs of patient isolates were linked at ward level and by overlapping treatment intervals. Nonetheless, our analysis of a large number of contemporaneous genomes sampled over different geographical scales should improve the external validity over casuistic findings that report epidemiological cut-off values by discriminating single hospital outbreaks from a convenience sample of unrelated isolates. Furthermore, limiting epidemiologic conclusions to the number of core genome differences will also fail to detect possible plasmid transmission events. However, we argue that plasmid spread between ST258/512 isolates likely plays a relatively minor role compared with clonal spread considering the strong relationship between the core genome diversity and geographic distance observed.
It has been previously shown that ST258 emerged from an ST11 ancestor via a large recombination event with an ST442-like strain [19]. It has been divided into two sub-lineages, “clade I” and “clade II”, which are differentiated by a region of recombination affecting the capsular loci [19]. ST258 is predicted to have emerged in the 1990s in the USA where it remains endemic [38]. ST258 and its derivative, ST512, are now also endemic in Israel as well as in some southern European countries, most notably Italy and Greece, and have caused outbreaks elsewhere [39, 40]. However, the ancestral relationships between the dispersed populations of this clone were previously unclear. Here, we observed strong geographical structuring by analysing 236 ST258/512 genomes together with 415 publicly available genomes from diverse international origins. The tree structure (Fig. 5) shows a clear country-specific partitioning with US isolates at the tips of long basal branches and monophyletic short-branched expansions in different countries, suggesting that ST258/512 originated in the USA and has successively spread to Mediterranean countries. Most of the high incidence countries that reported a rapid rise in carbapenem resistance over the past 15 years in Europe witnessed epidemic expansion of ST258/512 as illustrated in Fig. 5 [8, 11, 41].
In summary, we have shown that the epidemic of carbapenem-non-susceptible K. pneumoniae in Europe is driven by the expansion of a small number of clonal carbapenemase-positive lineages that propagate along nosocomial transmission routes and that antibiotic use serves as a major effect modifier. Introductions of progenitor carbapenem resistance genes are rare events in the natural history of the current epidemic and, although there is no denial that gene flow between the “one-health” compartments exist [42], this phenomenon has had no substantial impact on the course of the epidemiology of the major carbapenemase-positive clones of K. pneumoniae in Europe in the period up to 2014. Public health efforts should therefore focus on genomic pathogen surveillance, identifying introductions of high-risk clones and their expansion early in the course of an incipient epidemic, and reinforcing the resilience of national hospital referral networks by a no-tolerance infection control policy. All data from this publication are publicly available (see Data Availability) and can be used as contextual information for the public health decision-making involving investigations of outbreaks and/or isolate collections. It may also serve as a benchmark of the status of carbapenem-non-susceptible K. pneumoniae in Europe in 2013-14, to which future survey data can be compared, facilitating critical surveillance efforts.
Methods
Culture, DNA extraction and Illumina sequencing
2162/2301 (94.0%) samples submitted as K. pneumoniae during the EuSCAPE survey [14] were prepared for whole genome sequencing. The remaining 139/2301 (6.0%) were discarded due to a lack of growth, contamination of the culture plates, or having multiple phenotypes suggestive of contamination with different species.
470/2162 samples were plated on MacConkey agar (Sigma Aldrich) overnight at 37°C and a single colony representing each sample was picked into phosphate buffered saline (PBS) solution. For the remaining 1692/2162 samples, a storage bead was picked into 1.5mL of low salt Luria-Bertani (LB) broth in a well of a 96-well S-block (Qiagen), and incubated at 37°C overnight with shaking. The bacteria were pelleted and re-suspended in 1xPBS solution. Lysozyme (Sigma Aldrich), RNase A (Invitrogen) and Proteinase K (Qiagen) were added to all samples, which were incubated for 1h at 37°C. DNA was extracted using the QIAamp 96-well kit on the QIAcube HT system (Qiagen). Isolates were sequenced using the Illumina HiSeq platform with 125bp paired-end reads.
Quality control (QC) analysis of sequence data
Trimmomatic v0.33 [43] was used to trim the Illumina sequence reads. SPAdes v3.9.0 [44] was used to generate de novo assemblies from the trimmed sequence reads using kmer sizes of 41, 49, 57, 65, 77, 85 and 93 and the –cov-cutoff flag set to “auto”. QUAST v4.6.0 [45] was used to generate assembly statistics. Isolates were discarded from future analysis if the size of the de novo assembly was outside of 5-7Mb, the total number of contigs that were over 1kb was greater than 1000, and if <90% of the assembly was comprised of contigs greater than 1kb.
MASH v2.0 [46] was used to determine the similarity of each isolate to the genomes in the RefSeq bacterial database, using the raw sequence reads, the full de novo assembly, and each individual contig from the de novo assembly. Isolates were discarded from further analysis if the top species matches using either the raw reads and full assembly were not Klebsiella spp., and if the combined percentage length of the individual contigs with a top match to Klebsiella spp. was <70.
For QC purposes only, all raw sequence reads were mapped to the NTUH K2044 reference genome using Burrows Wheeler Aligner [47]. The resulting BAM files were used to calculate the number of heterozygous SNP positions per sample using a Perl script (available at https://github.com/sanger-pathogens/vr-codebase/blob/master/scripts/het_snp_calculator.pl), and the mean depth of coverage. Isolates were discarded from future analysis if >1000 heterozygous SNPs were identified, or if the mean depth of coverage was <20x.
The QC results for all 2162 sequenced isolates are available in Supplementary Tables 1, 2, 3 and Supplementary Fig. 1.
Phylogenetic analysis of all isolates
Prokka v1.5 [48] was used to annotate the de novo assemblies with predicted genes. The annotated assemblies were used as input for Roary v3.11.3 [49], which determined the “core” and “accessory” genes of the 1717 isolates that passed the QC analysis using an identity threshold of 95% (blastp). An alignment of 2539 core genes comprising 2,436,120bp was generated, representing genes that are present in >=95% of isolates from each of the four species (K. pneumoniae sensu stricto, K. quasipneumoniae, K. variicola and K. quasivariicola). Due to the high diversity, it was not feasible to remove recombined regions from the alignment prior to phylogenetic analysis. The variable positions were extracted from the alignment and used to generate a maximum likelihood tree with RAxML v7.0.4 using a general time reversible (GTR) model with a gamma correction for site rate variation [50]. 100 random bootstrap replicates were also performed to assess the support for nodes. 57.6% and 66.7% of nodes have bootstrap support of over 0.9 and 0.75, respectively. The phylogenetic tree was visualized together with associated metadata using Microreact [51]. The average nucleotide identity (ANI) between pairs of isolates was also determined from the alignment of 2539 core genes using Panito (available at https://github.com/sanger-pathogens/panito).
Identification of sequence types and K-type (capsular type)
The seven-gene sequence type [17] of all isolates was determined using the multi-locus sequence typing (MLST) calling function of Ariba v.2.6.1 [52] and the “Klebsiella pneumoniae” database from PubMLST [17]. The K-type (capsular type) of each isolate was determined from the de novo assembly using Kaptive [53].
Identification of carbapenemase genes
Carbapenemase genes (blaKPC-like, blaNDM-like, blaVIM-like, blaOXA-48-like, blaIMP-like, blaGES-like, blaGIM-like, blaAIM-like, blaBIC-like, blaDIM-like, blaIMI-like, blaNMC-A-like, blaSIM-like, blaSPM-like, blaLMB-like, blaFRI-like, blaSME-like, blaKHM-like, blaBKC-like, blaOXA-23-like, blaOXA-51-like and blaOXA-58-like) and their variants were detected using Ariba [52] with a custom resistance gene database available at https://figshare.com/s/94437a301288969109c2. A carbapenemase was identified as present only if the gene coverage was at least 0.2x the minimum coverage of the seven MLST genes in each genome, in order to reduce false positive calls. This cut-off allowed for variation in sequencing coverage between samples and provided maximum concordance with the previously obtained PCR results [14].
Identification of ESBL genes, AmpC genes and porin alterations
As with the carbapenemase genes, the presence of ESBL and AmpC genes was detected using Ariba. Porin genes, ompK35 and ompK36, were also detected with Ariba and checked for completeness. Since Ariba relies upon the raw reads, the de novo assemblies were also cross-checked for the presence of these omp genes, both using BLASTn [54] and by extraction from the annotation files generated using Prokka [48]. Genes were considered defective if the gene was found to be absent, fragmented or interrupted, or if frameshift or stop codon mutations were visible.
Phenotypic re-testing
Antimicrobial susceptibility testing were performed by broth microdilution [55] on 1574/1717 (91.7%) isolates including in the genomic analyses using lyophilized custom plates (Thermofisher Scientific).
Determination of the genetically nearest neighbours (gNNs) and permutation analysis
The genetically nearest neighbour (gNN) to each isolate was defined as that with the highest ANI, as calculated from the alignment of 2539 core genes. The expected proportions of gNNs sampled from different geographic contexts, given an absence of geographic structure, were calculated by performing 100 random permutations of the hospital codes. The gNNs were re-calculated based on each permutation, and the mean and 95% confidence intervals were calculated from the distribution of results.
Phylogenetic analysis of ST258/512
Sequence reads of isolates belonging to STs 258 and 512 (and other nested STs) were mapped to the reference genome, NJST258_1 [56]. Recombined regions were identified and removed from the pseudo-genome alignment using Gubbins v1.4.10 [57], and the resulting pairwise SNP differences were calculated. A phylogenetic tree of the ST258/512 lineage (comprising 236 genomes from EuSCAPE and 415 publicly available genomes) was constructed using only the vertically-inherited SNPs with RAxML v7.0.4 [50], and rooted using an ST11 outgroup isolate (EuSCAPE_PL007). 52.9% and 65.7% of nodes have bootstrap support over 0.9 and 0.75, respectively. The phylogenetic tree was visualized together with associated metadata using Microreact [51] and with predicted recombined regions using Phandango [58].
Predictions of intra- versus inter-hospital transmission in the ST258/512 lineage
A statistical method described by Aspbury et al. [18] was used to predict the hospital origin of each ST258/512 isolate in the EuSCAPE collection from the pseudo-genome alignment (excluding recombined regions). The code is available at: https://github.com/marianne-aspbury/hospital-origin. However, we deviated from the approach described by Aspbury et al. in which a set of new isolate genomes are queried one at a time in chronological order against a genome database of older isolates and subsequently added to the database. Instead, we used all ST258/512 genomes from the EuSCAPE collection (n=236) in the database, and removed one isolate at a time to be the query, regardless of its isolation date. Depending on whether the isolate was predicted to be most likely from the same or different hospital as the actual submitting hospital, we inferred that the isolate has been involved in intra- or inter-hospital transmission.
Statistical tests
A chi-square test for trend was used to test for a significant downward trend in the proportion of genetically nearest neighbours (gNNs) obtained from the same hospital across the beta-lactome resistance groups (i-v). The test was implemented in R (https://www.r-project.org/).
Supplementary Material
Acknowledgements
We would like to thank the Pathogen Informatics group and Core Sequencing facility at the Wellcome Sanger Institute for their contribution to the study. We would also like to thank Fenna Bosma and Marion Zigterman from the Medical Microbiology and Infection Prevention department of the University Medical Center Groningen for their support in assembling the isolate collection.
Source of Funding
This work was funded by The Centre for Genomic Pathogen Surveillance, Wellcome Genome Campus, Wellcome (grants 098051 and 099202) and The NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance (NIHR 16/136/111). The EuSCAPE project was funded by ECDC through a specific framework contract (ECDC/2012/055) following an open call for tender (OJ/25/04/2012-PROC/2012/036).
Footnotes
Data availability
All raw and assembled Illumina sequence data is available from the European Nucleotide Archive (ENA) under the study accession number, PRJEB10018/ERP011196. Individual accession numbers for raw sequence data and de novo assemblies are also available in Supplementary Table 4. Phylogenetic analysis of the 1717 K. pneumoniae isolates (EuSCAPE only) and the ST258/512 isolates (EuSCAPE and public data) together with all metadata, and links to raw sequence data are available at the project URLs https://microreact.org/project/EuSCAPE_Kp and https://microreact.org/project/EuSCAPE_ST258 within Microreact [51]. Phylogenetic analyses and metadata of isolates are also available separately for each of the contributing countries in Microreact (see Supplementary Table 7 for project URLs).
Author Contributions
HG and DMA conceived the study. The EuSCAPE working group collected the bacterial isolates and epidemiological data, and performed preliminary laboratory analyses. The ESGEM facilitated the training and capacity building for the collection of bacterial isolates and preliminary analyses. SD, SR, SRH, CG, TF, SA, KA, RG, TG, GE, MA, SS, EJF, GMR, HG and DMA performed the data analysis. SD, SR, EJF, GMR, HG and DMA wrote the manuscript. All authors read and approved the manuscript.
Competing Interests
The authors declare no competing interests.
The EuSCAPE working group
Andi Koraqi 9, Denada Lacej 9, Petra Apfalter 10, Rainer Hartl 10, Youri Glupczynski 11, Te-Din Huang 11, Tanya Strateva 12, Yuliya Marteva-Proevska 13, Arjana Tambic Andrasevic 14, Iva Butic 14, Despo Pieridou-Bagatzouni 15, Panagiota Maikanti-Charalampous 15, Jaroslav Hrabak 16, Helena Zemlickova 17, Anette Hammerum 18, Lotte Jakobsen 18, Marina Ivanova 19, Anastasia Pavelkovich 19, Jari Jalava 20, Monica Österblad 20, Laurent Dortet 21, Sophie Vaux 22, Martin Kaase 23, Sören G. Gatermann 24, Alkiviadis Vatopoulos 25, Kyriaki Tryfinopoulou 25, Ákos Tóth 26, Laura Jánvári 26, Teck Wee Boo 27, Elaine McGrath 27, Yehuda Carmeli 28, Amos Adler 28, Annalisa Pantosti 29, Monica Monaco 29, Lul Raka 30, Arsim Kurti 30, Arta Balode 31, Mara Saule 31, Jolanta Miciuleviciene 32, Aiste Mierauskaite 32, Monique Perrin-Weniger 33, Paul REICHERT 33, Nina Nestorova 34, Sonia Debattista 34, Gordana Mijovic 35, Milena Lopicic 35, Ørjan Samuelsen 36, Bjørg Haldorsen 36, Dorota Żabicka 37, Elżbieta Literacka 38, Manuela Caniça 39, Vera Manageiro 39, Ana Kaftandzieva 40, Elena Trajkovska-Dokic 40, Maria Damian 41, Brandusa Lixandru 41, Zora Jelesic 42, Anika Trudic 43, Milan Niks 44, Eva Schreterova 45, Mateja Pirs 46, Tjasa Cerar 46, Jesús Oteo 47, Belén Aracil 47, Christian Giske 48, Karin Sjöström 49, Deniz Gür 50, Ası Cakar 50, Neil Woodford 51, Katie Hopkins 51, Camilla Wiuff 52, Derek J. Brown 53.
9 University Hospital Center "Mother Theresa", Tirana, Albania. 10 Elisabethinen Hospital Linz, Linz, Austria. 11 CHU Dinant-Godinne UCL Namur, Namur, Belgium. 12 Faculty of Medicine, Medical University of Sofia, Sofia, Bulgaria. 13 Alexandrovska University Hospital, Sofia, Bulgaria. 14 University Hospital for Infectious Diseases, Zagreb, Croatia. 15 Nicosia General Hospital, Nicosia, Cyprus. 16 Faculty of Medicine in Plzen, Charles University in Prague, Plzen, Czech Republic. 17 National Institute of Public Health, Praha, Czech Republic. 18 Statens Serum Institut, Copenhagen, Denmark. 19 East Tallinn Central Hospital, Tallinn, Estonia. 20 National Institute for Health and Welfare, Turku, Finland. 21 Bicêtre Hospital, Le Kremlin-Bicêtre, France. 22 Institut de Veille Sanitaire, Saint-Maurice, France. 23 Universitätsmedizin Göttingen, Göttingen, Germany. 24 Ruhr-University Bochum, Bochum, Germany. 25 National School of Public Health, Athens, Greece. 26 National Center for Epidemiology, Budapest, Hungary. 27 Galway University Hospitals, Galway, Ireland. 28 Ministry of Health, Tel-Aviv, Israel. 29 Istituto Superiore di Sanità, Rome, Italy. 30 National Institute of Public Health of Kosovo, Prishtina, Kosovo. 31 Pauls Stradins Clinical University Hospital, Riga, Latvia. 32 National Public Health Surveillance Laboratory, Vilnius, Lithuania. 33 Laboratoire National De Sante, Düdelingen, Luxembourg. 34 Mater Dei Hospital, Msida, Malta. 35 Institute of Public Health, Podgorica, Montenegro. 36 University Hospital of North Norway, Tromsø, Norway. 37 Narodowy Instytut Lekow, Warsaw, Poland. 38 National Medicines Institute, Warsaw, Poland. 39 National Institute of Health Dr. Ricardo Jorge, Lisbon, Portugal. 40 Institute of Microbiology and Parasitology, Medical Faculty, Skopje, Republic of Macedonia. 41 Cantacuzino National Research Institute, Bucharest, Romania. 42 Institute for Public Health of Vojvodina, Novi Sad, Serbia. 43 Institute for Pulmonary Diseases of Vojvodina, Sremska Kamenica, Serbia. 44 Public Health Authority of the Slovak Republic, Bratislava, Slovakia. 45 University Hospital of P.J.Safarik, Kosice, Slovakia. 46 Institute of Microbiology and Immunology, Ljubljana, Slovenia. 47 Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain. 48 Karolinska Institutet, Stockholm, Sweden. 49 Public Health Agency of Sweden, Stockholm, Sweden. 50 Hacettepe University, Ankara, Turkey. 51 Reference Microbiology Services, Public Health England, London, United Kingdom - England and Northern Ireland. 52 Sydvestjysk Hospital, Esbjerg, Denmark. 53 Scottish Microbiology Reference Laboratories, Glasgow, United Kingdom - Scotland.
References
- 1.World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organisation; 2017. [Google Scholar]
- 2.Cassini A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infectious Diseases. 2019;19:56–66. doi: 10.1016/S1473-3099(18)306054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathers AJ, et al. Molecular Dissection of an Outbreak of Carbapenem-Resistant Enterobacteriaceae Reveals Intergenus KPC Carbapenemase Transmission through a Promiscuous Plasmid. MBio. 2011;2 doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin J, et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. Journal of Antimicrobial Chemotherapy. 2017;72:3025–3034. doi: 10.1093/jac/dkx264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yigit H, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2004;48:15–22. doi: 10.1128/aac.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yong D, et al. Characterization of a New Metallo-beta-Lactamase Gene, bla(NDM-1), and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrobial Agents and Chemotherapy. 2009;53:5046–5054. doi: 10.1128/aac.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz-Price LS, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infectious Diseases. 2013;13:785–796. doi: 10.1016/s1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2016.260. 16260. [DOI] [PubMed] [Google Scholar]
- 11.Albiger B, et al. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Eurosurveillance. 2015;20:17–34. doi: 10.2807/1560-7917.es.2015.20.45.30062. [DOI] [PubMed] [Google Scholar]
- 12.Kazi M, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis. 2015;34:467–472. doi: 10.1007/s10096-014-2249-x. [DOI] [PubMed] [Google Scholar]
- 13.Singh-Moodley A, Perovic O. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis. 2016;16:536. doi: 10.1186/s12879-016-1858-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundmann H, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infectious Diseases. 2017;17:153–163. doi: 10.1016/s1473-3099(16)30267-2. [DOI] [PubMed] [Google Scholar]
- 15.Holt KE. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long SW, et al. Whole-Genome Sequencing of a Human Clinical Isolate of the Novel Species Klebsiella quasivariicola sp. nov. Genome Announc. 2017;5 doi: 10.1128/genomeA.01057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aspbury M, et al. Rapid statistical methods for inferring intra- and inter-hospital transmission of nosocomial pathogens from whole genome sequence data. biorXiv. 2018 [Google Scholar]
- 19.Chen L, Mathema B, Pitout JDD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 Is a Hybrid Strain. MBio. 2014;5 doi: 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: Past, Present, and Future. Antimicrobial Agents and Chemotherapy. 2011;55:4943–4960. doi: 10.1128/aac.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Man TJB, et al. Genomic Analysis of a Pan-Resistant Isolate of Klebsiella pneumoniae, United States 2016. MBio. 2018;9 doi: 10.1128/mBio.00440-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Therapeutic Advances in Infectious Disease. 2016;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush K, et al. Tackling antibiotic resistance. Nature Reviews Microbiology. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray AK, et al. Novel Insights into Selection for Antibiotic Resistance in Complex Microbial Communities. MBio. 9 doi: 10.1128/mBio.00969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwaber MJ, et al. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis. 2011;52:848–855. doi: 10.1093/cid/cir025. [DOI] [PubMed] [Google Scholar]
- 26.Falagas ME, et al. Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case-control study. Journal of Antimicrobial Chemotherapy. 2007;60:1124–1130. doi: 10.1093/jac/dkm356. [DOI] [PubMed] [Google Scholar]
- 27.Mohan B, et al. Fecal Carriage of Carbapenem-Resistant Enterobacteriaceae and Risk Factor Analysis in Hospitalised Patients: A Single Centre Study from India. Indian Journal of Medical Microbiology. 2017;35:555–562. doi: 10.4103/ijmm.IJMM_17_144. [DOI] [PubMed] [Google Scholar]
- 28.Bartolini A, et al. Prevalence, molecular epidemiology and intra-hospital acquisition of Klebsiella pneumoniae strains producing carbapenemases in an Italian teaching hospital from January 2015 to September 2016. International Journal of Infectious Diseases. 2017;59:103–109. doi: 10.1016/j.ijid.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Yusuf E, et al. OXA-48 Producing Klebsiella pneumoniae in a Household Contact of a Previously Infected Patient: Person-to-Person Transmission or Coincidental Community Acquisition? Microbial Drug Resistance. 2016;22:134–136. doi: 10.1089/mdr.2015.0179. [DOI] [PubMed] [Google Scholar]
- 30.Munk P, et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol. 3:898–908. doi: 10.1038/s41564-018-0192-9. [DOI] [PubMed] [Google Scholar]
- 31.Aanensen DM, et al. Whole-Genome Sequencing for Routine Pathogen Surveillance in Public Health: a Population Snapshot of Invasive Staphylococcus aureus in Europe. MBio. 2016;7 doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitart C, et al. First Outbreak of a Plasmid-Mediated Carbapenem-Hydrolyzing OXA-48 beta-Lactamase in Klebsiella pneumoniae in Spain. Antimicrobial Agents and Chemotherapy. 2011;55:4398–4401. doi: 10.1128/aac.00329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snitkin ES, et al. Tracking a Hospital Outbreak of Carbapenem-Resistant Klebsiella pneumoniae with Whole-Genome Sequencing. Science Translational Medicine. 2012;4 doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou K, et al. Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum beta-lactamase producing ST15 Klebsiella pneumoniae. Scientific Reports. 2016;6 doi: 10.1038/srep20840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu D, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 36.Mathers AJ, Peirano G, Pitout JDD. The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clinical Microbiology Reviews. 2015;28:565–591. doi: 10.1128/cmr.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerqueira GC, et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowers JR, et al. Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic. Plos One. 2015;10 doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Zuluet P, et al. First Outbreak of KPC-3-Producing Klebsiella pneumoniae (ST258) Clinical Isolates in a Mexican Medical Center. Antimicrobial Agents and Chemotherapy. 2013;57:4086–4088. doi: 10.1128/aac.02530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Cerero L, et al. Characterisation of the first ongoing outbreak due to KPC-3-producing Klebsiella pneumoniae (ST512) in Spain. International Journal of Antimicrobial Agents. 2014;44:538–540. doi: 10.1016/j.ijantimicag.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Glasner C, et al. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Eurosurveillance. 2013;18:9–15. doi: 10.2807/1560-7917.es2013.18.28.20525. [DOI] [PubMed] [Google Scholar]
- 42.Kock R, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clinical Microbiology and Infection. 2018;24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ondov BD, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 51.Argimón S, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunt M, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyres KL, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Tenth Edition. CLSI; 2015. [Google Scholar]
- 56.Deleo FR, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hadfield J, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.