Abstract
Breast cancer (BC) represents the most commonly diagnosed cancer among females worldwide. Although targeted therapy has greatly improved the efficacy of treating BC, a large proportion of BC patients eventually develop recurrence or metastasis. Traditional invasive tumor tissue biopsy is short of comprehensiveness in tumor assessment due to heterogeneity. Liquid biopsy, an attractive non-invasive approach mainly including circulating tumor cell and circulating tumor DNA (ctDNA), has been widely utilized in a variety of cancers with the advances of sequencing technologies in recent years. The ctDNA that is found circulating in body fluids refers to DNA released from tumor cells and has shown clinical utility in metastatic breast cancer (MBC). With the results of genomic variants detection, ctDNA could be used to predict clinical outcomes, monitor disease progression, and guide treatment for patients with MBC. Moreover, the drug resistance problem may be addressed by ctDNA detection. In this review, we summarized the technological developments and clinical applications of ctDNA in MBC.
Keywords: circulating tumor DNA, metastatic breast cancer, genomic variants, drug resistance
Introduction
Breast cancer (BC) represents the most commonly diagnosed cancer and the leading cause of cancer death among females worldwide.1 In China, 272.4 thousand women were newly diagnosed as BC each year, bringing heavy medical and financial burden to the whole society.2 The 2011 St. Gallen conference categorized BC into four subtypes based on the molecular landscape of the tumor, including luminal A, luminal B, human epidermal growth factor receptor-positive (HER2+), and triple-negative (ductal) disease.3 This surrogate classification for BC subtypes has been shown to successfully predict tumor presenting features, nodal involvement, recurrence patterns, and disease-free survival.4 Even though the 5-year mortality rate of BC has dropped by 34% since 1990, a number of patients eventually developed resistance to the therapy.5 For instance, the drug resistance rate of estrogen receptor-positive (ER+)/progesterone receptor-positive BC patients could reach 70%.6 Meanwhile, the treatment of advanced BC (ABC) remains a challenge due to the common drug resistance.
Tumor tissue biopsy is the gold standard specimen for tumor genotyping. However, tumor tissue biopsy is invasive, painful, and potentially risky, and performing biopsy in some tumor sites may not be convenient.7 Moreover, in metastatic settings it may be short of comprehensiveness due to tumor heterogeneity.8 Therefore, alternative less-invasive detection methods are desperately required to capture the entire genetic map of the tumor and consequently improve the current treatment options. Liquid biopsy is an attractive non-invasive approach that utilizes readily available body fluids to provide a real-time comprehensive assessment of the tumor, through the analysis of genomic variants in circulating tumor DNA (ctDNA) and circulating tumor cell (CTC).9 In cancer patients, ctDNA is a small proportion of circulating cell-free DNA (cfDNA) in the bloodstream, which was first described by Mandel et al in 1948.10 With the rapid developments of sequencing technologies in recent years, the clinical applications of ctDNA in various tumors such as lung cancer, BC, and gastric cancer have been extensively explored.9,11,12 For BC, ctDNA was shown to provide supplementary information for tumor tissue biopsy in terms of prognosis assessment, drug resistance test, and individualized treatment.13 Importantly, additional mutations detected by ctDNA could reflect the utility of liquid biopsy to capture heterogeneity of metastatic sites in ABC.14
To offer an update of current literature, this review will discuss the recent developments of ctDNA in metastatic BC (MBC) from these aspects: main detection technologies, concordance between ctDNA and tumor tissue biopsy, pre-analytical factors, tumor burden, genomic variants, specific gene mutations, drug resistance, and tumor heterogeneity and sub-clone.
Main Detection Technologies for ctDNA in MBC
It has been demonstrated that cfDNA can be released from apoptosis, necrosis, and macrophage phagocytosis of the cells, whereas the concentration of cfDNA in the bloodstream of healthy individuals is low (approximately 10 ng/mL).15,16 The concentration of cfDNA can be increased by certain events like exercise, cancer, and autoimmune disease.17–19 In patients with BC, the concentration of cfDNA may increase to 0.5, 235, 422, and 1280 ng/mL classified by tumor stage I, II, III, and IV, respectively.20 However, the concentration of ctDNA or the concentration ratio of ctDNA: cfDNA can be very low given that the majority of cfDNA consists of background germline DNA.21 Therefore, highly sensitive approaches capable of detecting the genomic profile of limited available amounts of ctDNA are demanded. So far, diverse detection methods mainly based on Polymerase Chain Reaction (PCR) and Next-Generation Sequencing (NGS) have been applied, while each has its own strength and weakness.22
PCR-Based Technologies
The development of traditional simple PCR has shown some limitations in liquid biopsy such as its low detection sensitivity.23 Currently, digital PCR (dPCR) is frequently used for the detection of ctDNA because of its high sensitivity and ease of use. Two common dPCR methods, the droplet digital PCR (ddPCR) and BEAMing [beads, emulsification, amplification, and magnetics] assay, can accurately detect point mutations present at frequencies ≤ 0.1%, nevertheless restricted to the analysis of a small number of genomic loci and the need of “a priori” knowledge of the investigated genomic aberrations.24 A workflow by means of uniplex or multiplex pre-amplification combined with dPCR was successfully used to investigate the estrogen receptor 1 (ESR1) mutation status of the samples of MBC patients who progressed on endocrine treatment.25 The combination of allele-specific, asymmetric rapid PCR and melting analysis assay was used to detect the phosphoinositide 3-kinase catalytic subunit alpha (PIK3CA) hotspot mutations (E545K, H1047R) and showed a high concordance (81.1%, P = 0.001) with ddPCR.26 Gyanchandani et al utilized a novel whole-genome amplification of cfDNA via rolling-circle amplification before ddPCR and provided physiological evidence that ctDNA was accessible by fingerstick and sustained presence/absence of mutation detection after whole-genome amplification.27
NGS-Based Technologies
NGS, compared with PCR, can interrogate larger regions such as gene panels without prior knowledge of mutations.28 The main methods of NGS consist of targeted sequencing, whole-exome sequencing, and whole-genome sequencing.8 Li et al performed targeted deep sequencing on 17 blood samples from 17 Chinese MBC patients and identified 60 somatic mutations in the blood samples of all patients.7 MammaSeq, a breast cancer-specific NGS panel, was shown to detect 43 protein-coding mutations in 14 ctDNA samples from 7 patients with ABC, targeting 79 genes and 1369 mutations.29 Despite of the comprehensive sequencing ability, the error rate of NGS complicates the calling of mutations with variant allele frequencies (VAFs) < 5%.28 Error correction through random molecular barcodes has been incorporated into NGS ctDNA assays to reduce this error rate, enabling mutation calling with VAFs < 0.1%.30 Masunaga et al analyzed the cfDNA samples from 34 MBC patients and revealed a higher sensitivity (0.1%) of the molecular barcodes-NGS than NGS without barcode (1%) by reducing background errors.31 Nevertheless, increasing proportions of false positives and negatives have been reported at low mutant allele frequencies, which brings the issue of detection reliability.32 Importantly, reproducibility is one of the main caveats of high throughput technologies. The study by Stetson et al suggested that most NGS assay discordance is a result of technical variations.33 Gerratana et al confirmed the detection performance of a novel NGS platform, PredicinePLUSTM, when compared with Guardant360TM and that different NGS platforms could be potentially compared provided that certain sample management and analytical requirements are met.32
Hybrid Techniques
Hybrid techniques such as the combination of PCR and NGS allow to circumvent both the sensitivity limits of NGS (1–2%) and the inherent need for “a priori” knowledge of the target of PCR.22 Studies using hybrid techniques such as synchronous coefficient of drag alteration, tagged-amplicon deep sequencing, and safe sequencing system have been summarized in a previous review.9 Recently, Keup et al analyzed 17 genes in the cfDNA of 44 MBC patients by a targeted PCR-based NGS approach. This method constructed a library using a customized targeted DNA panel with integrated unique molecular indices before sequencing. The comprehensive analysis of cfDNA variants not only confirmed the benefit of unique molecular indices in the variant verification, but also identified new promising variants with clinical relevance.34
The Concordance Between ctDNA and Tumor Tissue Biopsy
The ctDNA cannot be used to replace the need for metastatic tissue biopsy of patients until it is proven to represent the genomic information detected in solid tissue.35 A number of studies have investigated the difference between ctDNA and tumor tissue biopsy in MBC and some of them have revealed high concordances (77~89%).36–40 The remarkable concordance suggests that ctDNA assays can be confidently used to molecularly stratify patients for prognostic and predictive purposes. Furthermore, compared to tumor tissue biopsy, ctDNA may have the superiority of obtaining more comprehensive mutation information.41 For example, Maxwell et al found that the proportion of genomic alteration in MBC patients was lower in tumor than in blood (69% vs 91%; P = 0.06).42 Even so, several studies reported significant differences between the genomic results of tumor tissue biopsy and cfDNA in MBC patients.43–46 Diverse analysis tools, temporal spacing between ctDNA analysis and tumor biopsy, and tumor heterogeneity due to the substantial selection pressure from therapies perhaps contribute to this inconsistency.43,46,47
Pre-Analytical Factors
The quality of ctDNA analysis was affected by a lot of pre-analytical factors.9 Hakan et al reported a significant increase in isolation efficiency with very fresh blood samples of at least 15 milliliters which were processed in minutes.48 Risberg et al investigated the potential pre-analytical influential factors of ctDNA levels. The results revealed that using cell-stabilization tubes, storing at 4 °C after collection for a short period, and performing the centrifugation in different types of centrifuges greatly improved the feasibility of collecting high-quality specimens.49 What’s more, higher frequencies of ESR1 and PIK3CA mutations in the plasma than in the serum were revealed, indicating that plasma samples might be the preferred source of cfDNA.50–52 Possible influential factors in each step of the ctDNA detection from sample collection to sequencing are shown in Figure 1.
Figure 1.
The whole process of ctDNA analysis and possible influencing factors.
Notes: The concentration of ctDNA and mutation frequency are significantly higher in ABC than early BC. Using cell-stabilization tubes, storing at 4°C after collection for a short period, and performing the centrifugation in different types of centrifuges greatly improved the feasibility of collecting high-quality specimens. Plasma samples may be the preferred source of ctDNA than serum. The type of detection technology also affects the results of ctDNA analysis. EMV, extracellular micro vesicle; ctDNA, circulating tumor DNA; ABC, advanced breast cancer; NGS, next-generation sequencing; PCR, polymerase chain reaction.
Assessing Tumor Burden According to the Quantity and Quality of ctDNA/cfDNA
Tumor burden provides key information about objective response and disease progression.53 MBC patients with high panel-specific mutational load had a significantly shorter time to progression than those with a low mutational load (P = 0.0112).42 The molecular tumor burden index, a measure of the percentage of ctDNA in samples, was positively correlated with tumor size as evaluated by computed tomography (P < 0.0001) and detected disease progression 8–16 weeks earlier.54
The ctDNA Concentration
Dynamic changes in ctDNA levels closely reflect changes in tumor burden and increases in ctDNA levels may predict the progressive disease (PD) several months before the standard imaging.55 In recent years, several methods were used to determine the ctDNA concentration in MBC and investigate its relationship with clinical outcomes. Cheng et al evaluated the ctDNA concentration of MBC patients by measuring the short and long fragments of two repetitive DNA elements, ALU and LINE1, and found that ctDNA concentration could serve as an independent prognostic marker.56 Li et al used the VAF of mutations from the major mutated clones to access the ctDNA levels of MBC patients. The results showed that a higher ctDNA level in plasma was associated with a shorter progression-free survival (PFS).7
The Cutoff of ctDNA Concentration
A number of studies have demonstrated the prognostic value of a CTC enumeration > 5 at baseline and follow-up in patients with MBC.55,57,58 Recently, Cristofanilli et al used a CTC threshold of 5 cells per 7.5 mL for the stratification of 2346 MBC patients in a retrospective and pooled analysis of individual patient data from 18 cohorts and confirmed the identification of two subgroups of MBC (Stage IVindolent and Stage IVaggressive), independent of clinical and molecular variables.59 Nevertheless, a cutoff of ctDNA concentration that correlates with a worse prognosis has not been identified yet. A recent retrospective study revealed a statistically significant difference in PFS and overall survival (OS) for baseline %ctDNA (the maximum mutant allele fraction in each sample) < 0.5 vs. ≥ 0.5 (P = 0.003 and P = 0.012).55 Stover et al presented the genomic characterization of a large metastatic triple-negative BC (MTNBC) cohort. A cfDNA tumor fraction threshold of ≥ 10% was associated with significantly worse metastatic survival.60 In one study by Shatsky et al, ABC patients with at least one alteration with 5% ctDNA had poorer median OS (6.7 months) vs. patients with less than 5% ctDNA (17.9 months) (P = 0.01).43
The DNA Integrity
The cfDNA concentration directly reflects the quantity of cfDNA, while the cfDNA integrity (cfDI) represents the quality of cfDNA, which has been investigated as a diagnostic or prognostic marker in multiple cancers.61–63 In general, cfDI is calculated as a ratio of longer DNA to shorter DNA for ALU repeats (247 and 115 bp). Plasma ALU-247, ALU-115, and DNA integrity may be proven to have clinical utility in BC diagnosis.64 In metastatic settings, Cheng et al showed a significantly increased cfDI (P = 1.21E-7 for ALU and P = 1.87E-3 for LINE1) in both repetitive DNA elements after one cycle of therapy. In addition, the multiple cox regression model indicated an independent prognostic role of cfDI.56
Detecting Genomic Variants
Besides the ctDNA concentration, the mutation frequency of ctDNA in ABC is significantly higher than that in early BC.20,23 Analyzing the genomic variants including somatic mutations, somatic copy number alterations (SCNAs), and DNA methylation (DNAme) may have important prognostic significance.65
Somatic Mutations
Somatic mutations identified in ctDNA are widely representative of the tumor genome as they can provide the spectrum of mutations not limited to a single region of the tumor.66 In recent years, several studies identified the somatic mutation profiles of ABC.40,67-69 Chung et al sequenced 62 genes in ctDNA from patients with ER+ MBC and found that the most frequently altered genes were TP53 (38%), ESR1 (31%), and PIK3CA (31%).40 Ding et al identified HER2 mutations in both the tyrosine kinase (n = 14) and extracellular (n = 4) domains in MBC patients.67 In a retrospective study by Yi et al, authors determined the frequency and spectrum of common cancer-related gene mutations in ctDNA among different ABC subtypes. 96.0% (96/100) female ABC patients had somatic genomic alterations including copy number variants (CNVs) and point mutations and different subtypes of ABC had distinct features in terms of genetic alterations.68 In one study by Zill et al, the somatic mutation landscape of 70 cancer genes from cfDNA deep sequencing analysis of 21,807 patients with advanced cancers was depicted. The results revealed the most commonly amplified genes (MYC and FGFR1) and the most common resistance alterations (ESR1 L536/Y537/D538) in ABC.69
Somatic Copy Number Alterations
A study by Stover et al presented the largest genomic characterization of MTNBC to date. They found that certain SCNAs were more frequent in MTNBC patients relative to primary TNBC including copy number gains in drivers NOTCH2, AKT2, and AKT3.60 Theoretically, all HER2+ patients should show a copy number gain of erb-b2 receptor tyrosine kinase 2 (ERBB2). However, in a study by Yi et al, only 8.8% patients in the HER2+ group showed an ERBB2 copy number gain. The reason might be that the ctDNA assay was not sufficiently sensitive to accurately detect CNVs.68 This viewpoint was supported by a recent meta-analysis of Yang et al The pooled results showed that single nucleotide variants detection through cfDNA from MBC patients had higher sensitivity (0.78 [0.71–0.84]) and specificity (0.92 [0.87–0.95]), when compared to tissue DNA detection. However, the cfDNA-based detection of CNV was shown to have no diagnostic value due to the low sensitivity (0.42 [0.24–0.62]) in the cases of CNV.70
DNA Methylation
DNAme has been considered as a hallmark of cancer over the last decade.71 DNAme means the covalent addition of a methyl group to cytosine within the context of CpG dinucleotide and is chemically and biologically stable.72,73 Le et al examined the promoter methylation status of a panel of markers in a range of BC cell lines spanning the epithelial-mesenchymal spectrum with methylation-sensitive high-resolution melting. A distinction in profiling DNA methylation was revealed between epithelial and mesenchymal phenotypes.74 This study provided important insight into selecting an optimal panel of methylation markers to assess minimal residual disease, which is a major determinant of the initiation of metastatic tumors.75 Panagopoulou et al assessed the methylation status of a panel of cancer-related genes in BC. The methylation of WNT5A was statistically significantly correlated to greater tumor size and poorer prognosis characteristics and in advanced stage disease with shorter OS. In the metastatic group, SOX17 methylation was significantly correlated to shorter PFS and OS.76 These studies highlighted that DNA methylation should be further evaluated as a potential liquid biopsy-based biomarker in metastatic settings.
The Significance of Detecting Specific Gene Mutations
The identification of specific point mutations such as PIK3CA and ESR1 in BC may provide key information for the selection of treatment regimens.77 In MBC, the mutation profile of specific genes could be utilized to predict clinical outcomes. Importantly, crucial adjustments of treatment regimens could be made corresponding to the changes of genetic information.78 The detection sites and clinical relevance of detecting specific gene mutations in the ctDNA of ABC patients from studies in recent years are summarized in Table 1.
Table 1.
Selected Studies Detecting Specific Gene Mutations in Recent Years
| Mutant Gene | Detection Sites | Researcher | Study Type | Patients Classification | Clinical Relevance |
|---|---|---|---|---|---|
| PIK3CA | Not specific | Ma et al45 | Phase I study | HER2+ MBC | Predicting the efficacy of pyrotinib |
| R88Q, R93Q/W, K111E/N, G118D, E365K, C420R, E542K, E545G/K, Q546K, and H1047L/R/Y | Baselga et al36 | Phase III study | HR+HER2- ABC | Associated with longer PFS in patients with PIK3CA mutation than patients without PIK3CA mutation | |
| E542K, E545K, and H1047L/R | O’Leary et al82 | Phase III study | HR+HER2- ABC | Predicting PFS of ABC patients on palbociclib and fulvestrant. | |
| E542K, E545K, and H1047R | Park et al81 | Phase II study | MTNBC | Revealing PIK3CA mutational status in cfDNA from blood samples of MTNBC | |
| E542K, E545K, and H1047R | Takano et al65 | Phase II study | HER2+ MBC | Exploring the predictive value of the differential benefit of anti-HER2 drugs | |
| E542K, E545G/K, Q546K, M1043I, and H1047 L/R/Y | Di Leo et al37 | Phase III study | HR+HER2- ABC | PFS was in favour of buparlisib plus fulvestrant versus fulvestrant alone in patients with PIK3CA mutations | |
| E545K, and H1047R | Li et al84 | Formal-prospective/retrospective study | ABC | Associated with shorter median PFS and worse OS | |
| E542K, E545K, and H1047L/R | Kodahl et al38 | Observational study | ABC | Changes in PIK3CA ctDNA level correlated with treatment response | |
| E545K, and H1047R | Tzanikou et al26 | Observational study | MBC | Concordance of PIK3CA hotspot mutations between ctDNA and CTCs | |
| E542K, E545K, and H1047R | Jacot et al83 | Observational study | ER+/HER2−ABC | Four-week persistence of cfDNA PIK3CA mutation appears highly correlated with PFS | |
| ESR1 | E380Q, D538G, and Y537S/N/C | Takeshita et al94 | Retrospective study | ER+ MBC | Prevalence of ESR1 mutation in Japanese MBC patients |
| E380Q, D538G, I298N, V560E, and Y537S/N | Yi et al68 | Retrospective study | ABC | Positively related to the line of endocrine therapy | |
| D538G, and Y537S/N/C | Beije et al93 | Observational study | MBC | Conferring endocrine resistance | |
| D538G, and Y537S/N | Takeshita et al46 | Retrospective study | MBC | Independent distribution of ESR1 mutations between plasma and tumor tissue | |
| E380Q, D538G, S463P, L536R, and Y537S/N/C | O’Leary et al82 | Phase III study | HR+HER2- ABC | ESR1 ctDNA dynamics offering limited prediction of clinical outcome. | |
| D538G, and Y537S/N/C | Allouchery et al86 | Retrospective study | HR+ MBC | ESR1 mutation frequency increased with the development of relapse and progression | |
| E380Q, D538G, Y537S/N/C, V534E, L536Q/R | Paoletti et al88 | Phase I study | ER+HER2- MBC | Elevated ESR1mutation associated with worse PFS | |
| HER2 | Not specific | Ma et al95 | Single-arm phase II study | HER2mut, non-amplified MBC | As a screening tool to identify future trial patients |
| Not specific | Sakai et al101 | Retrospective study | HER2+ MBC | Predicting primary resistance to T-DM1 | |
| Not specific | Li et al102 | Phase I study | HER2+ MBC | Multiple genetic alterations significantly associated with shorter PFS compared to none or one genetic alteration | |
| RB1 | Not specific | Condorelli et al96 | Observational study | HER2+ MBC | Conferring therapeutic resistance of CDK4/6 inhibitor |
| BCL2 | Not specific | Lok et al97 | Phase Ib study | BCL2+ MBC | Combination therapy for patients with BCL2-positive tumors |
Notes: PIK3CA, phosphoinositide 3-kinase catalytic subunit alpha; ESR1, estrogen receptor 1; HER2, human epidermal growth factor receptor 2; RB1, retinoblastoma 1; BCL2, B-cell lymphoma 2; MBC, metastatic breast cancer; HR, hormone receptor; ABC, advanced breast cancer; MTNBC, metastatic triple-negative breast cancer; ER, estrogen receptor; PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; TDM-1, trastuzumab emtansine; CDK4/6, cyclin-dependent kinase 4 and 6; +, positive; −, negative.
PIK3CA
PIK3CA, an oncogene encoding the p110α component of the phosphoinositide 3-kinase (PI3K), represents one of the most frequent driver mutations observed in BC.38 As shown in Figure 2, the four most frequent hotspot mutations of PIK3CA are located within two exons (exon 9: E545K and E542K; exon 20: H1047R and H1047L), accounting for 80–90% of all PIK3CA mutations in human malignancies.79,80 High-frequency (35/73 (47.9%)) PIK3CA hotspot mutations were present in the plasma-ctDNA of MBC patients.26 Park et al reported that more than 70% of the cfDNA samples of patients with MTNBC had PIK3CA hotspot mutations and the mutation frequency increased after the treatment.81
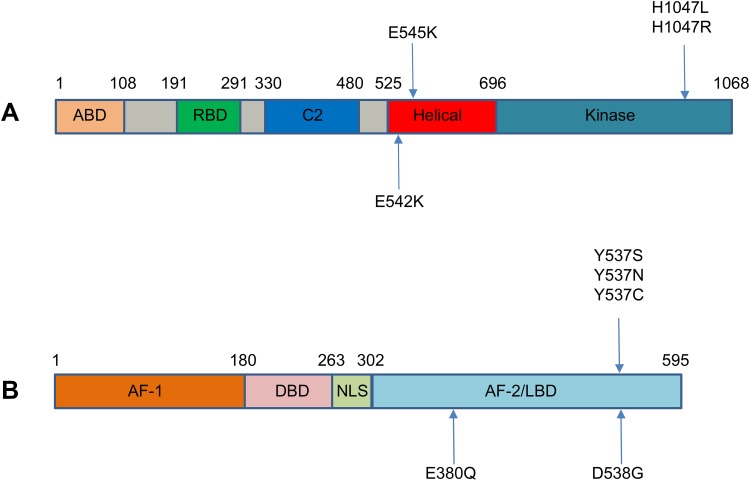
Figure 2.
Major mutation sites of (A) PIK3CA and (B) ESR1.
Notes: Most of the reported mutations in PIK3CA cluster in conserved regions within the region coding for the helical and kinase domains of p110α, including E542K, E545K, H1047L, and H1047R. ESR1 point mutations mainly localized in the ligand-binding domain, including 3 hotspot mutations in codons 380, 537, and 538. ABD, adaptor binding domain; RBD, ras binding domain; AF, activation function; DBD, DNA binding domain; NLS, nuclear localizing signal; LBD, ligand binding domain.
In recent years, a number of studies supporting the prognostic role of PIK3CA in ABC have been carried out.36–38,45,65,82-84 O’Leary et al found that PIK3CA ctDNA level after 15 days’ treatment strongly predicted the PFS of ABC patients on palbociclib and fulvestrant (hazard ratio 3.94, log-rank P = 0.0013).82 In a study by Jacot et al, a detectable circulating PIK3CA mutation after 4 weeks’ hormone therapy was highly correlated with a worse PFS.83 A retrospective study by Li et al revealed that PIK3CA p.H1047R mutation was more frequent in HER2+ disease and associated with worse OS. Moreover, this was the only mutation associated with shorter PFS on multivariate analysis in HER2+ patients who were treated with trastuzumab, suggesting that trastuzumab had lower activity in these patients.84 Takano et al found that in patients without PIK3CA mutations, lapatinib plus capecitabine (LX) yielded relatively longer PFS (P = 0.035) and OS (P = 0.025) than trastuzumab plus capecitabine (HX), indicating that these patients might benefit from LX rather than HX.65 In a Phase I study by Ma et al, all patients with PIK3CA or TP53 mutations were relatively insensitive to pyrotinib, whereas a significantly higher response rate was revealed in patients with wild-type PIK3CA and TP53 (P = 0.013).45 The BELLE-2 study was the first randomized Phase 3 clinical trial to include a prospective analysis of PFS in patients with ctDNA PIK3CA mutations. A significant difference in PFS in the buparlisib group vs. the placebo group was recorded in patients with ctDNA PIK3CA mutations, but not in patients with non-mutant ctDNA.36 On this basis, Di Leo et al demonstrated that PFS was in favour of combination therapy (buparlisib plus fulvestrant) vs. fulvestrant alone in patients with PIK3CA mutations in the BELLE-3 study.37 Overall, the ctDNA was analyzed in 1698 ABC patients of these studies and 552 (31.5%) of them had PIK3CA mutations. Of note, 1429 of these patients had HR+ ABC and 31.1% (445/1429) had PIK3CA mutations. Available data indicates that H1047R (118/256) and E545K (94/256) are the two most frequent mutation sites in PIK3CA (Table 2). These studies suggest that PIK3CA may be a powerful target to predict the clinical outcomes and guide treatment decisions especially for HR+ or HER2+ MBC.
Table 2.
The Distribution of PIK3CA Mutation Sites
ESR1
The activation of ESR1 is a key feature of 70–80% BC patients in whom ER expression is detected.85 As shown in Figure 2, ESR1 point mutations mainly localized in the ligand-binding domain (LBD), including 3 hotspot mutations in codons 380, 537, and 538. Allouchery et al found that the mutation frequency of ESR1 increased with the development of relapse and progression (5.3% and 33%, respectively), suggesting that detecting circulating ESR1 mutations should be focused on the metastatic setting.86 A secondary analysis of the BOLERO-2 clinical trial by Chandarlapaty et al demonstrated that each of the ESR1 mutations was associated with shorter OS.87 Paoletti et al found that patients with persistently elevated ESR1LBDm+ ctDNA had worse PFS than patients who did not (P = 0.0007).88 However, an analysis of the data from PALOMA-3 study indicated that ESR1 mutations selected by prior hormone therapy were frequently sub clonal, with ESR1 ctDNA dynamics offering limited prediction of clinical outcomes.82
Endocrine therapy is the basis of treatment for ER+ MBC patients. However, 40% of these patients obtain no clinical benefit from first-line endocrine therapy, and virtually all of the patients in whom the tumor initially responds eventually develop resistance.89 ESR1 mutation emergence has been recently highlighted as a frequent mechanism of acquired aromatase inhibitor (AI) resistance in the metastatic setting.90,91 It was found that the mutation frequency of ESR1 was significantly higher in HR+ patients than in HR- patients, especially in the HR+/HER2- group. In addition, ESR1 mutations were positively related to the line of endocrine therapy. These results support the viewpoint that ESR1 mutations are acquired from endocrine therapy.68,92 Beije et al reported that the ESR1 mutations of cfDNA were more prevalent in the progressing cohort (42%) than in the baseline cohort (11%) (P = 0.04), which strongly suggested a role in conferring endocrine resistance in MBC.93 Takeshita et al showed that there were distinct populations of ESR1 mutations in metastatic tissue and plasma.94 Each ESR1 mutation might have different clinical significance, and it would be necessary to investigate them all. This viewpoint is consistent with Chandarlapaty et al In their analysis, the ESR1 D538G mutation group derived a large benefit from treatment with a mechanistic target of rapamycin kinase (mTOR) inhibitor, whereas the ESR1 Y537S mutation group did not.87 Altogether, the ctDNA was analyzed in 685 ABC patients and 169 (24.7%) of them had ESR1 mutations in these studies. Notably, 524 of these patients had HR+ ABC and 26.9% (141/524) had ESR1 mutations. D538G (23/56) and Y537S (18/56) are the two most frequent mutation sites in ESR1 (Table 3). Selected by the treatment pressure, ESR1 mutations in ABC often evolve into polyclone, which may weaken its prognostic value. Still, detecting the mutation profile of ESR1 may assist in knowing the resistance mechanism of endocrine therapy and guiding clinical treatment (for example, the use of fulvestrant in ABC patients after AI treatment).
Table 3.
The Distribution of ESR1 Mutation Sites
Other Gene Mutations
Besides ESR1 and PIK3CA, the clinical impacts of other gene mutations such as HER2, retinoblastoma 1 (RB1), and B-cell lymphoma 2 (BCL2) have been investigated in ABC.95–97 HER2 is a well-established therapeutic target in BC.98–100 HER2 gene amplification in ctDNA and ER/PR status may predict primary resistance to T-DM1 (trastuzumab emtansine), which is an antibody-drug conjugate with high efficacy approved for the treatment of patients with HER2+ ABC.101 In a phase I study by Li et al, HER2 bypass signaling pathway along with PI3K/AKT/mTOR pathway and TP53 was shown to be significantly associated with poorer PFS in patients with HER2+ MBC treated with the combination of pyrotinib and capecitabine (median, 16.8 vs. 29.9 months, P = 0.006).102 Theoretically, HER2- (non-amplified) BC patients do not benefit from HER2-directed drugs.103 However, recent identification of recurrent HER2 mutations (HER2mut) in a subset of HER2 gene non-amplified BC suggested an additional HER2 targeting opportunity.95,104-106 RB1 is a tumor suppressor protein that is dysfunctional in several major cancers.107 Deregulation of the cyclin D1-CDK4/6-RB pathway is implicated in endocrine treatment resistance.108 Condorelli et al described the acquisition of de novo RB1 mutations in three patients with MBC treated with CDK4/6 inhibitors. None of the RB1 mutations was present in the pre-CDK4/6 specimen, highlighting that these molecular alterations likely emerged under selective pressure from the CDK4/6 inhibitor potentially conferring therapeutic resistance.96 Further research is needed to identify the frequencies of these mutations and how these mutations temporally emerge under the selective pressure of CDK4/6 treatment. BCL2, a key member of the BCL2 pro-survival family, is often expressed at high levels in poorer-prognosis luminal B tumors and in good-prognosis luminal A tumors.109,110 Lok et al demonstrated that combining venetoclax with endocrine therapy had a tolerable safety profile and elicited notable activity in ER+BCL2+ MBC. Their findings warrant further investigation of combination therapy for patients with BCL2+ tumors.97
Drug Resistance
Targeted therapies have remarkably changed the treatment of cancer over the last decade. However, almost all tumors acquire resistance to systemic treatment as a result of tumor heterogeneity and clonal evolution.111 Multiple mechanisms involved in signal transduction molecules have been demonstrated to be associated with the primary or acquired resistance in ABC, including activation of various intracellular pathways such as the PI3K/AKT/mTOR and CDK4/6/RB pathways.83,102
Resistance to Endocrine Therapy in ER+ BC
ESR1 mutation is not only associated with AI treatment failure in ER+ BC patients, but also confers ligand independent cell growth and relative resistance to tamoxifen and fulvestrant.112,113 Moreover, the crosstalk between PI3K and ER pathway and between PI3K/ERBB2 pathway also cause patients’ resistance to endocrine therapy.114 Zhou et al evaluated the genetic response of ctDNA to chemotherapy in ER+ MBC patients. The results showed that more patients with PD exhibited increased PIK3CA mutation frequency than non-PD patients (56.25% vs 0%, P = 0.002). Furthermore, ESR1 and GATA3 mutations were shown to correlate with PIK3CA mutation, suggesting that the accumulation of PIK3CA, ESR1 and GATA3 mutations might significantly increase the risk of endocrine therapy resistance.115
Resistance to CDK4/6 Inhibitors
Selective CDK4/6 inhibitors have become the standard of care for ER+HER2- MBC patients.116 These agents prevent cell cycle progression from G0/G1-phase to S-phase by directly blocking the activity of the cyclin D-CDK4/6 holoenzyme, leading to the proliferation inhibition of sensitive tumor cells.117 The potent anticancer activity of CDK4/6 inhibitors has been demonstrated in pre-clinical and clinical studies.118,119 However, little is known about the mechanisms of resistance to these agents. Therefore, there is a pressing need to develop biomarker approaches to identify which ER+ BC patients will derive long-term benefit from CDK4/6 inhibition. O’ Leary et al conducted ctDNA sequencing to 195 patients from the PALOMA-3 study. The evolution of ESR1 mutations was observed, with selection of ESR1 Y537S as the variant most likely promoting resistance to fulvestrant in the combination.120 Formisano et al identified FGFR1/2 amplification or activating mutations in 14/34 (41%) post-progression specimens by sequencing the ctDNA in 34 patients after progression on CDK4/6 inhibitors. In addition, the presence of FGFR1 alterations in baseline plasma tumor DNA was associated with a shorter PFS in the MONALEESA-2 study, suggesting that aberrant FGFR signaling was a potential mechanism of escape from endocrine therapy plus CDK4/6 inhibitors.121
Tumor Heterogeneity and Sub-Clone
Intra-tumoral genetic heterogeneity presents a challenge to targeted therapy response, as sub-clones may have divergent response to therapy.82 A study by Ma et al provided evidence that tumor heterogeneity present in ctDNA could predict therapeutic outcome. The results indicated that patients with higher tumor heterogeneity had significantly worse PFS outcomes, with a median PFS of 30.0 weeks vs. 60.0 weeks for patients with low tumor heterogeneity (P = 0.02).54 Sub-clonal mutations are reported most commonly in ESR1. Multiple concurrent ESR1 genomic alterations including mutation, rearrangement, and amplification were observed in 40% of ESR1-altered cases, suggesting polyclonal origin.40 Paoletti et al proposed that integrating multiple biomarkers (ctDNA ESR1LBDm, CTC enumeration, and CTC-ER status) could improve outcome prediction and endocrine treatment resistance mechanisms’ identification over a single biomarker.88 Li et al explored potential predictive biomarkers of HER2+ MBC patients and found that compared to none or one genetic alteration, multiple genetic alterations in HER2 bypass signaling pathway, PI3K/AKT/mTOR pathway, and TP53 were significantly associated with poorer PFS (median, 16.8 vs 29.9 months, P = 0.006).102
Summary and Conclusions
Compared to tumor tissue biopsy, ctDNA can non-invasively detect more comprehensive genomic information from limited amount of plasma. In recent years, great progress has been made in the detection technologies of liquid biopsy from PCR to NGS, providing strong technical support for ctDNA analysis. A lot of efforts were made to increase the sensitivity and reduce the error rate of ctDNA sequencing, leading to an improvement of the quality control of ctDNA analysis. Recent studies have revealed high concordances (77–89%) between ctDNA and tumor tissue biopsy, which confirmed the detection capability of ctDNA and its role in replacing tumor tissue biopsy. In addition, a series of pre-analytical factors such as sample freshness, storage tube and temperature, and centrifugation speed could affect the analysis results of ctDNA and should be controlled appropriately. Tumor burden, providing key information about objective response and disease progression, could be reflected through the evaluation of ctDNA concentration and integrity. The prognostic value of specific gene mutations such as PIK3CA in MBC has been confirmed in numerous studies. Hopefully, by longitudinally monitoring the treatment response and ctDNA variants of MBC patients, the drug resistance may be addressed, however, which needs to be further validated in large-scale prospective trials. Moreover, given that the previous ctDNA tests are mainly for patients with advanced cancers, the applications of ctDNA in early-stage cancers, such as early diagnosis, relapse monitoring, and screening may be well explored. To conclude, ctDNA has promising prospects in cancer research, especially in promoting the realization of personalized medicine, and perhaps it will completely change the existing patterns of cancer treatment in the near future.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflict of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes – dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22(8):1736–1747. doi: 10.1093/annonc/mdr304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasconcelos I, Hussainzada A, Berger S, et al. The St. Gallen surrogate classification for breast cancer subtypes successfully predicts tumor presenting features, nodal involvement, recurrence patterns and disease free survival. Breast. 2016;29:181–185. doi: 10.1016/j.breast.2016.07.016 [DOI] [PubMed] [Google Scholar]
- 5.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203 [DOI] [PubMed] [Google Scholar]
- 6.Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. doi: 10.1038/ng.2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Wang X, Li Y, Lai H, Liu Y, Jin L. Non-invasive analysis of tumor mutation profiles and druggable mutations by sequencing of cell free DNA of Chinese metastatic breast cancer patients. Thorac Cancer. 2019;10(4):807–814. doi: 10.1111/1759-7714.13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appierto V, Di Cosimo S, Reduzzi C, Pala V, Cappelletti V, Daidone MG. How to study and overcome tumor heterogeneity with circulating biomarkers: the breast cancer case. Semin Cancer Biol. 2017;44:106–116. doi: 10.1016/j.semcancer.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 9.Buono G, Gerratana L, Bulfoni M, et al. Circulating tumor DNA analysis in breast cancer: is it ready for prime-time? Cancer Treat Rev. 2019;73:73–83. doi: 10.1016/j.ctrv.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 10.Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l’homme (Nucleic acids in human blood plasma). C R Seances Soc Biol Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 11.Lim M, Kim CJ, Sunkara V, Kim MH, Cho YK. Liquid biopsy in lung cancer: clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines (Basel). 2018;9(3):100. doi: 10.3390/mi9030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Zhang K, Xi H, et al. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget. 2017;8(4):6330–6340. doi: 10.18632/oncotarget.14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Ju S, Wang X, Cong H. Advances in liquid biopsy using circulating tumor cells and circulating cell-free tumor DNA for detection and monitoring of breast cancer. Clin Exp Med. 2019;19(3):271–279. doi: 10.1007/s10238-019-00563-w [DOI] [PubMed] [Google Scholar]
- 14.Savas P, Teo ZL, Lefevre C, et al. The subclonal architecture of metastatic breast cancer: results from a prospective community-based rapid autopsy program “CASCADE”. PLoS Med. 2016;13(12):e1002204. doi: 10.1371/journal.pmed.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cappelletti V, Appierto V, Tiberio P, Fina E, Callari M, Daidone MG. Circulating biomarkers for prediction of treatment response. J Natl Cancer Inst Monogr. 2015;2015(51):60–63. doi: 10.1093/jncimonographs/lgv006 [DOI] [PubMed] [Google Scholar]
- 16.Fournie GJ, Gayral-Taminh M, Bouche JP, Conte JJ. Recovery of nanogram quantities of DNA from plasma and quantitative measurement using labeling by nick translation. Anal Biochem. 1986;158(2):250–256. doi: 10.1016/0003-2697(86)90545-2 [DOI] [PubMed] [Google Scholar]
- 17.Kohler C, Radpour R, Barekati Z, et al. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer. 2009;8(1):105. doi: 10.1186/1476-4598-8-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tug S, Helmig S, Menke J, et al. Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell Immunol. 2014;292(1–2):32–39. doi: 10.1016/j.cellimm.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 19.Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med. 2012;42(7):565–586. doi: 10.2165/11631380-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 20.Tangvarasittichai O, Jaiwang W, Tangvarasittichai S. The plasma DNA concentration as a potential breast cancer screening marker. Indian J Clin Biochem. 2015;30(1):55–58. doi: 10.1007/s12291-013-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Board RE, Williams VS, Knight L, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137(1):98–107. doi: 10.1196/annals.1448.020 [DOI] [PubMed] [Google Scholar]
- 22.Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
- 25.Vitale SR, Sieuwerts AM, Beije N, et al. An optimized workflow to evaluate estrogen receptor gene mutations in small amounts of cell-free DNA. J Mol Diagn. 2019;21(1):123–137. doi: 10.1016/j.jmoldx.2018.08.010 [DOI] [PubMed] [Google Scholar]
- 26.Tzanikou E, Markou A, Politaki E, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. 2019;13(12):2515–2530. doi: 10.1002/1878-0261.12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyanchandani R, Kvam E, Heller R, et al. Whole genome amplification of cell-free DNA enables detection of circulating tumor DNA mutations from fingerstick capillary blood. Sci Rep. 2018;8(1):17313. doi: 10.1038/s41598-018-35470-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15(1):75. doi: 10.1186/s12916-017-0840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith NG, Gyanchandani R, Shah OS, et al. Targeted mutation detection in breast cancer using MammaSeq. Breast Cancer Res. 2019;21(1):22. doi: 10.1186/s13058-019-1102-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masunaga N, Kagara N, Motooka D, et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Treat. 2018;167(1):49–58. doi: 10.1007/s10549-017-4487-y [DOI] [PubMed] [Google Scholar]
- 32.Gerratana L, Zhang Q, Shah AN, et al. Performance of a novel next generation sequencing circulating tumor DNA (ctDNA) platform for the evaluation of samples from patients with metastatic breast cancer (MBC). Crit Rev Oncol Hemat. 2020;145:102856. doi: 10.1016/j.critrevonc.2019.102856 [DOI] [PubMed] [Google Scholar]
- 33.Stetson D, Ahmed A, Xu X, et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis Oncol. 2019;3:1–9. [DOI] [PubMed] [Google Scholar]
- 34.Keup C, Benyaa K, Hauch S, et al. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell Mol Life Sci. 2020;77(3):497–509. doi: 10.1007/s00018-019-03189-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017;109(12):12. doi: 10.1093/jnci/djx118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baselga J, Im SA, Iwata H, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(7):904–916. doi: 10.1016/S1470-2045(17)30376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Leo A, Johnston S, Lee KS, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19(1):87–100. doi: 10.1016/S1470-2045(17)30688-5 [DOI] [PubMed] [Google Scholar]
- 38.Kodahl AR, Ehmsen S, Pallisgaard N, et al. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol Oncol. 2018;12(6):925–935. doi: 10.1002/1878-0261.12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. doi: 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung JH, Pavlick D, Hartmaier R, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol. 2017;28(11):2866–2873. doi: 10.1093/annonc/mdx490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 42.Maxwell KN, Soucier-Ernst D, Tahirovic E, et al. Comparative clinical utility of tumor genomic testing and cell-free DNA in metastatic breast cancer. Breast Cancer Res Treat. 2017;164(3):627–638. doi: 10.1007/s10549-017-4257-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shatsky R, Parker BA, Bui NQ, et al. Next-generation sequencing of tissue and circulating tumor DNA: the UC San Diego Moores center for personalized cancer therapy experience with breast malignancies. Mol Cancer Ther. 2019;18(5):1001–1011. doi: 10.1158/1535-7163.MCT-17-1038 [DOI] [PubMed] [Google Scholar]
- 44.Goetz MP, Suman VJ, Reid JM, et al. First-in-human phase i study of the tamoxifen metabolite Z-Endoxifen in women with endocrine-refractory metastatic breast cancer. J Clin Oncol. 2017;35(30):3391–3400. doi: 10.1200/JCO.2017.73.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma F, Li Q, Chen S, et al. Phase I study and biomarker analysis of pyrotinib, a novel irreversible Pan-ErbB receptor tyrosine kinase inhibitor, in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer. J Clin Oncol. 2017;35(27):3105–3112. doi: 10.1200/JCO.2016.69.6179 [DOI] [PubMed] [Google Scholar]
- 46.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast cancer patients. Transl Oncol. 2017;10(5):766–771. doi: 10.1016/j.tranon.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savli H, Sertdemir N, Aydin D, et al. TP53, EGFR and PIK3CA gene variations observed as prominent biomarkers in breast and lung cancer by plasma cell-free DNA genomic testing. J Biotechnol. 2019;300:87–93. doi: 10.1016/j.jbiotec.2019.05.005 [DOI] [PubMed] [Google Scholar]
- 49.Risberg B, Tsui DWY, Biggs H, et al. Effects of collection and processing procedures on plasma circulating cell-free DNA from cancer patients. J Mol Diagn. 2018;20(6):883–892. doi: 10.1016/j.jmoldx.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Iwase H. ESR1 and PIK3CA mutational status in serum and plasma from metastatic breast cancer patients: a comparative study. Cancer Biomark. 2018;22(2):345–350. doi: 10.3233/CBM-171161 [DOI] [PubMed] [Google Scholar]
- 51.Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer Treat Rev. 2002;28(5):255–271. doi: 10.1016/S0305-7372(02)00077-4 [DOI] [PubMed] [Google Scholar]
- 52.Lam NYL, Rainer TH, Chiu RWK, Lo YM. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem. 2004;50(1):256–257. doi: 10.1373/clinchem.2003.026013 [DOI] [PubMed] [Google Scholar]
- 53.Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18(3):e143–e152. doi: 10.1016/S1470-2045(17)30074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma F, Guan Y, Yi Z, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2020;146(5):1359–1368. doi: 10.1002/ijc.32536 [DOI] [PubMed] [Google Scholar]
- 55.Rossi G, Mu Z, Rademaker AW, et al. Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin Cancer Res. 2018;24(3):560–568. doi: 10.1158/1078-0432.CCR-17-2092 [DOI] [PubMed] [Google Scholar]
- 56.Cheng J, Holland-Letz T, Wallwiener M, et al. Circulating free DNA integrity and concentration as independent prognostic markers in metastatic breast cancer. Breast Cancer Res Treat. 2018;169(1):69–82. doi: 10.1007/s10549-018-4666-5 [DOI] [PubMed] [Google Scholar]
- 57.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 58.Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12(14 Pt 1):4218–4224. doi: 10.1158/1078-0432.CCR-05-2821 [DOI] [PubMed] [Google Scholar]
- 59.Cristofanilli M, Pierga JY, Reuben J, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Crit Rev Oncol Hemat. 2019;134:39–45. doi: 10.1016/j.critrevonc.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 60.Stover DG, Parsons HA, Ha G, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol. 2018;36(6):543–553. doi: 10.1200/JCO.2017.76.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang WW, Zahurak M, Goldenberg D, et al. Increased plasma DNA integrity index in head and neck cancer patients. Int J Cancer. 2006;119(11):2673–2676. doi: 10.1002/ijc.22250 [DOI] [PubMed] [Google Scholar]
- 62.Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63(14):3966–3968. [PubMed] [Google Scholar]
- 63.El-Shazly SF, Eid MA, El-Sourogy HA, Attia GF, Ezzat SA. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis C virus-related hepatocellular carcinoma. Int J Biol Markers. 2010;25(2):79–86. doi: 10.1177/172460081002500204 [DOI] [PubMed] [Google Scholar]
- 64.Hussein NA, Mohamed SN, Ahmed MA. Plasma ALU-247, ALU-115, and cfDNA integrity as diagnostic and prognostic biomarkers for breast cancer. Appl Biochem Biotechnol. 2019;187(3):1028–1045. doi: 10.1007/s12010-018-2858-4 [DOI] [PubMed] [Google Scholar]
- 65.Takano T, Tsurutani J, Takahashi M, et al. A randomized Phase II trial of trastuzumab plus capecitabine versus lapatinib plus capecitabine in patients with HER2-positive metastatic breast cancer previously treated with trastuzumab and taxanes: WJOG6110B/ELTOP. Breast. 2018;40:67–75. doi: 10.1016/j.breast.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 66.Rothe F, Laes JF, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol. 2014;25(10):1959–1965. doi: 10.1093/annonc/mdu288 [DOI] [PubMed] [Google Scholar]
- 67.Ding Q, Chen H, Lim B, et al. HER2 somatic mutation analysis in breast cancer: correlation with clinicopathological features. Hum Pathol. 2019;92:32–38. doi: 10.1016/j.humpath.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 68.Yi Z, Ma F, Li C, et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci Rep. 2017;7(1):5995. doi: 10.1038/s41598-017-06327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zill OA, Banks KC, Fairclough SR, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24(15):3528–3538. doi: 10.1158/1078-0432.CCR-17-3837 [DOI] [PubMed] [Google Scholar]
- 70.Yang X, Zhang K, Zhang C, Peng R, Sun C. Accuracy of analysis of cfDNA for detection of single nucleotide variants and copy number variants in breast cancer. BMC Cancer. 2019;19(1):465. doi: 10.1186/s12885-019-5698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Green BB, Karagas MR, Punshon T, et al. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the new hampshire birth cohort study (USA). Environ Health Perspect. 2016;124(8):1253–1260. doi: 10.1289/ehp.1510437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fackler MJ, Lopez BZ, Umbricht C, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Res. 2014;74(8):2160–2170. doi: 10.1158/0008-5472.CAN-13-3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer. 2016;23(3):R157–R171. doi: 10.1530/ERC-15-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le AV, Szaumkessel M, Tan TZ, Thiery JP, Thompson EW, Dobrovic A. DNA methylation profiling of breast cancer cell lines along the epithelial mesenchymal spectrum-implications for the choice of circulating tumour DNA methylation markers. Int J Mol Sci. 2018;19(9). doi: 10.3390/ijms19092553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson JR, Kim Y, Seeger RC, Kim ES. A novel minimal residual disease model of neuroblastoma in mice. J Pediatr Surg. 2016;51(6):991–994. doi: 10.1016/j.jpedsurg.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Panagopoulou M, Karaglani M, Balgkouranidou I, et al. Circulating cell-free DNA in breast cancer: size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38(18):3387–3401. doi: 10.1038/s41388-018-0660-y [DOI] [PubMed] [Google Scholar]
- 77.Gradishar WJ, Anderson BO, Balassanian R, et al. Breast cancer, version 4.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;13(12):310–320. doi: 10.6004/jnccn.2018.0012 [DOI] [PubMed] [Google Scholar]
- 78.Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95(6):643–660. doi: 10.1111/cge.13514 [DOI] [PubMed] [Google Scholar]
- 79.Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15(16):5049–5059. doi: 10.1158/1078-0432.CCR-09-0632 [DOI] [PubMed] [Google Scholar]
- 80.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94(4):455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park IH, Kong SY, Kwon Y, et al. Phase I/II clinical trial of everolimus combined with gemcitabine/cisplatin for metastatic triple-negative breast cancer. J Cancer. 2018;9(7):1145–1151. doi: 10.7150/jca.24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9(1):896. doi: 10.1038/s41467-018-03215-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacot W, Dalenc F, Lopez-Crapez E, et al. PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res Treat. 2019;177(3):659–667. doi: 10.1007/s10549-019-05349-y [DOI] [PubMed] [Google Scholar]
- 84.Li H, Xu Y, Zhao F, et al. Plasma PIK3CA ctDNA specific mutation detected by next generation sequencing is associated with clinical outcome in advanced breast cancer. Am J Cancer Res. 2018;8(9):1873–1886. [PMC free article] [PubMed] [Google Scholar]
- 85.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- 86.Allouchery V, Beaussire L, Perdrix A, et al. Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Cancer Res. 2018;20(1):40. doi: 10.1186/s13058-018-0968-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paoletti C, Schiavon G, Dolce EM, et al. Circulating biomarkers and resistance to endocrine therapy in metastatic breast cancers: correlative results from AZD9496 oral SERD Phase I trial. Clin Cancer Res. 2018;24(23):5860–5872. doi: 10.1158/1078-0432.CCR-18-1569 [DOI] [PubMed] [Google Scholar]
- 89.Pritchard KI. Endocrine therapy: is the first generation of targeted drugs the last? J Intern Med. 2013;274(2):144–152. doi: 10.1111/joim.12065 [DOI] [PubMed] [Google Scholar]
- 90.Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 mutations and the treatment of estrogen receptor–positive advanced breast cancer. J Clin Oncol. 2016;34(25):2961–2968. doi: 10.1200/JCO.2016.67.3061 [DOI] [PubMed] [Google Scholar]
- 91.Clatot F, Perdrix A, Augusto L, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget. 2016;7(46):74448–74459. doi: 10.18632/oncotarget.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schiavon G, Hrebien S, Garcia-Murillas I, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beije N, Sieuwerts AM, Kraan J, et al. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol Oncol. 2018;12(1):48–57. doi: 10.1002/1878-0261.12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Prevalence of ESR1 E380Q mutation in tumor tissue and plasma from Japanese breast cancer patients. BMC Cancer. 2017;17(1):786. doi: 10.1186/s12885-017-3779-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma CX, Bose R, Gao F, et al. Neratinib efficacy and circulating tumor DNA detection of HER2 mutations in HER2 nonamplified metastatic breast cancer. Clin Cancer Res. 2017;23(19):5687–5695. doi: 10.1158/1078-0432.CCR-17-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Condorelli R, Spring L, O’Shaughnessy J, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29(3):640–645. doi: 10.1093/annonc/mdx784 [DOI] [PubMed] [Google Scholar]
- 97.Lok SW, Whittle JR, Vaillant F, et al. A Phase Ib dose-escalation and expansion study of the BCL2 inhibitor venetoclax combined with tamoxifen in ER and BCL2-positive metastatic breast cancer. Cancer Discov. 2019;9(3):354–369. doi: 10.1158/2159-8290.CD-18-1151 [DOI] [PubMed] [Google Scholar]
- 98.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152 [DOI] [PubMed] [Google Scholar]
- 99.Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353(16):1652–1654. doi: 10.1056/NEJMp058197 [DOI] [PubMed] [Google Scholar]
- 100.Abramson V, Arteaga CL. New strategies in HER2-overexpressing breast cancer: many combinations of targeted drugs available. Clin Cancer Res. 2011;17(5):952–958. doi: 10.1158/1078-0432.CCR-09-1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakai H, Tsurutani J, Iwasa T, et al. HER2 genomic amplification in circulating tumor DNA and estrogen receptor positivity predict primary resistance to trastuzumab emtansine (T-DM1) in patients with HER2-positive metastatic breast cancer. Breast Cancer. 2018;25(5):605–613. doi: 10.1007/s12282-018-0861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Q, Guan X, Chen S, et al. Safety, efficacy, and biomarker analysis of pyrotinib in combination with capecitabine in HER2-positive metastatic breast cancer patients: a Phase I clinical trial. Clin Cancer Res. 2019;25(17):5212–5220. doi: 10.1158/1078-0432.CCR-18-4173 [DOI] [PubMed] [Google Scholar]
- 103.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–726. doi: 10.1200/JCO.2002.20.3.719 [DOI] [PubMed] [Google Scholar]
- 104.Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224–237. doi: 10.1158/2159-8290.CD-12-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grellety T, Soubeyran I, Robert J, Bonnefoi H, Italiano A. A clinical case of invasive lobular breast carcinoma with ERBB2 and CDH1 mutations presenting a dramatic response to anti-HER2-directed therapy. Ann Oncol. 2016;27(1):199–200. doi: 10.1093/annonc/mdv496 [DOI] [PubMed] [Google Scholar]
- 106.Ross JS, Gay LM, Nozad S, et al. Clinically advanced and metastatic pure mucinous carcinoma of the breast: a comprehensive genomic profiling study. Breast Cancer Res Treat. 2016;155(2):405–413. doi: 10.1007/s10549-016-3682-6 [DOI] [PubMed] [Google Scholar]
- 107.Murphree AL, Benedict WF. Retinoblastoma: clues to human oncogenesis. Science. 1984;223(4640):1028–1033. doi: 10.1126/science.6320372 [DOI] [PubMed] [Google Scholar]
- 108.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perillo B, Sasso A, Abbondanza C, Palumbo G. 17beta-estradiol inhibits apoptosis in MCF-7 cells, inducing bcl-2 expression via two estrogen-responsive elements present in the coding sequence. Mol Cell Biol. 2000;20(8):2890–2901. doi: 10.1128/MCB.20.8.2890-2901.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaillant F, Merino D, Lee L, et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell. 2013;24(1):120–129. doi: 10.1016/j.ccr.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 111.Mastoraki S, Strati A, Tzanikou E, et al. ESR1 methylation: a liquid biopsy-based epigenetic assay for the follow-up of patients with metastatic breast cancer receiving endocrine treatment. Clin Cancer Res. 2018;24(6):1500–1510. doi: 10.1158/1078-0432.CCR-17-1181 [DOI] [PubMed] [Google Scholar]
- 112.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12(10):573–583. doi: 10.1038/nrclinonc.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuang Y, Siddiqui B, Hu J, et al. Unraveling the clinicopathological features driving the emergence of ESR1 mutations in metastatic breast cancer. NPJ Breast Cancer. 2018;4:22. doi: 10.1038/s41523-018-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ribas R, Pancholi S, Rani A, et al. Targeting tumour re-wiring by triple blockade of mTORC1, epidermal growth factor, and oestrogen receptor signalling pathways in endocrine-resistant breast cancer. Breast Cancer Res. 2018;20(1):44. doi: 10.1186/s13058-018-0983-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou D, Ouyang Q, Liu L, et al. Chemotherapy modulates endocrine therapy-related resistance mutations in metastatic breast cancer. Transl Oncol. 2019;12(5):764–774. doi: 10.1016/j.tranon.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Turner NC, Neven P, Loibl S, Andre F. Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet. 2017;389(10087):2403–2414. doi: 10.1016/S0140-6736(16)32419-9 [DOI] [PubMed] [Google Scholar]
- 117.Pernas S, Tolaney SM, Winer EP, Goel S. CDK4/6 inhibition in breast cancer: current practice and future directions. Ther Adv Med Oncol. 2018;10:1758835918786451. doi: 10.1177/1758835918786451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malorni L, Curigliano G, Minisini AM, et al. Palbociclib as single agent or in combination with the endocrine therapy received before disease progression for estrogen receptor-positive, HER2-negative metastatic breast cancer: tREnd trial. Ann Oncol. 2018;29(8):1748–1754. doi: 10.1093/annonc/mdy214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Leary B, Cutts RJ, Liu Y, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8(11):1390–1403. doi: 10.1158/2159-8290.CD-18-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Formisano L, Lu Y, Servetto A, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10(1):1373. doi: 10.1038/s41467-019-09068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]