Abstract
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) pose a serious threat to clinical patient management and public health, as they are generally resistant to most antibiotics and cause infections with high mortality rates. Klebsiella pneumoniae ranks second among Enterobacteriaceae species that cause nosocomial infections. In this study, we investigated the epidemic characteristics of carbapenem-resistant K. pneumoniae (CRKP) in the pediatric intensive care unit (PICU) of Yanbian University Hospital.
Materials and Methods
A total of 14 non-duplicate CRKP strains, collected from March 2015 to November 2019, were subjected to automated microbial identification and antimicrobial susceptibility tests using the Phoenix-100 ID/AST system. The strains were also subjected to genotypic resistance testing, polymerase chain reaction assays to detect genes encoding carbapenemases and other β-lactamases, multi-locus sequence typing (MLST), and pulsed-field gel electrophoresis (PFGE)-based homology analysis.
Results
Two carbapenemase genes, KPC-2 and NDM-1 (in eight and six strains, respectively), were detected. MLST enabled the division of the strains into two sequence types, ST11 and ST1224 (containing eight and six strains, respectively). PFGE results classified the 14 strains into clonotypes A–D, of which clonotypes A and B belonged to ST11, while clonotypes C and D belonged to ST1224.
Conclusion
Our study reveals that epidemics of the KPC-2-ST11 and NDM-1-ST1224 strains occurred in the PICU of Yanbian University Hospital. Surveillance and strict implementation of prevention and control measures are crucial to prevent the occurrence and rapid spread of nosocomial infections.
Keywords: Klebsiella pneumoniae, carbapenemase, sequence type 1224, pediatric intensive care unit
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that mainly causes pneumonia, bloodstream infections, and urinary tract infections.1 It produces extended-spectrum β-lactamases (ESBL) that confer resistance to multiple antibiotics. Carbapenem antibiotics are the last line of defense against multidrug-resistant K. pneumoniae. Recent years have witnessed the emergence of carbapenem-resistant K. pneumoniae (CRKP), with a gradual increase in cases following the widespread use of carbapenem antibiotics. CRKP is also resistant to other antibiotics, making its treatment a great challenge. Carbapenemase production is the predominant mechanism of antibiotic resistance in CRKP, followed by high production of ESBL and AmpC β-lactamases, coupled with reduced membrane permeability. Carbapenemases are mainly divided into class A (K. pneumoniae carbapenemase, KPC), class B (IMP, New Delhi metallo-β-lactamases (NDM), and Verona integron encoded metallo-β-lactamase (VIM)), and class D (OXA-48).2,3 In China, the main carbapenemases in CRKP include KPC and NDM enzymes. KPC-2 is a common KPC, while NDM-1 is the main NDM subtype.4 KPC-2 and NDM-1 were first discovered in carbapenemase-producing K. pneumoniae in 2002 and 2009, respectively.5,6 They were subsequently found in other Enterobacteriaceae, such as Escherichia coli and Enterobacter cloacae. KPC-2 and NDM-1 expression in carbapenemase-producing K. pneumoniae has been widely reported worldwide,7 but there are only a limited number of studies on newborns. In this study,8 we have retrospectively investigated the epidemic characteristics of CRKP in the pediatric intensive care unit (PICU) of Yanbian University Hospital.
Materials and Methods
Collection of Bacterial Strains and Clinical Information
All clinical strains of carbapenem-resistant Enterobacteriaceae (CRE) (resistant to imipenem or meropenem) isolated from blood and sputum samples in the PICU of Yanbian University Hospital from March 2015 to November 2019 were non-duplicate strains. Strain identification and antimicrobial sensitivity testing (AST) were performed using the Phoenix-100 ID/AST System (Becton, Dickinson and Co., USA). The quality control strain was E. coli ATCC25922. Collected clinical data included patients age, sex, underlying diseases, antibiotic usage 30 days prior to strain isolation, invasive procedures, and treatment outcomes.
Phenotypic Screening for Carbapenemases and Detection of Antibiotic Resistance Genes
Phenotypic screening for antibiotic resistance genes was performed in accordance with the modified Carbapenem Inactivation Method (mCIM) recommended by the Clinical and Laboratory Standards Institute Guidelines (2017). A 10-uL loopfull bacterial in 2 mL TSB emulsified, supplemented with a 10-μg meropenem disk and incubated at 35°C for 4 hours. Prior to removing the meropenem disk, a 0.5 McFarland suspension of E. coli ATCC25922 was prepared using saline and spread over the surface of Mueller Hinton (MH) agar plates using the K-B method. The meropenem disk was transferred onto the surface of MH agar plates, followed by incubation at 35°C for 18–24 hours. DNA was extracted using the boiling method. Briefly, 3–5 colonies were suspended in 400 μL of double-distilled water. The suspension was boiled for 10 minutes and centrifuged at 12,000 rpm for 3 minutes. The resulting supernatant was collected and used as the DNA template. Carbapenemase genes (blaNDM, blaKPC, blaIMP, blaVIM, and blaOXA48) and other β-lactamase genes (blaCTX-M, blaACT, blaDHA, and blaCMY) were detected by polymerase chain reaction (PCR).9,10 The products were sequenced by Sanger sequencing on an ABI3730xl Sequencer (BeijingTsingke Biotechnology Co., Ltd,Chian). Primer sequences, PCR cycling conditions, and PCR product sizes are listed in Table 1.
Table 1.
Sequences of the Primers (Carbapenemase Genes and Other β-Lactamase Genes)
| Target Gene | Primer | Primer Sequence (5′–3′) | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| KPC | KPC-F | TGTCACTGTATCGCCGTC | 1010 | 58 |
| KPC-R | CTCAGTGCTCTACAGAAAACC | |||
| IMP1 | IMP1-full-F | TGAGCAAGTTATCTGTATTC | 740 | 55 |
| IMP1-full-R | TTAGTTGCTTGGTTTTGATG | |||
| IMP2 | IMP2-full-F | GGCAGTCGCCCTAAAACAAA | 737 | 55 |
| IMP2-full-R | TAGTTACTTGGCTGTGATGG | |||
| NDM | NDM-full F | ATGGAATTGCCCAATATTATGCAC | 816 | 61 |
| NDM-full R | TCAGCGCAGCTTGTCGGC | |||
| VIM1 | VIM1-full-F | TTATGGAGCAGCAACCGATGT | 920 | 55 |
| VIM1-full-R | CAAAAGTCCCGCTCCAACGA | |||
| VIM2 | VIM2-full-F | AAAGTTATGCCGCACTCACC | 865 | 55 |
| VIM2-full-R | TGCAACTTCATGTTATGCCG | |||
| OXA-48 | OXA-48-F | GCGTGGTTAAGGATGAACAC | 438 | 52 |
| OXA-48-R | CATCAAGTTCAACCCAACCG | |||
| CTX-M1 group | CM3-full-F | TTTCGGAAGCATAAAATCGG | 1021 | 56 |
| CM3-full-R | GGCGATAAACAAAAACGGAA | |||
| CTX-M2 group | P3-F | ATGATGACTCAGAGCATTCG | 832 | 65 |
| P2b-R | TCCCGACGGCTTTCCGCCTT | |||
| CTX-M3 group | YW3459-F | AAAAATGATTGAAAGGTGGT | 1242 | 56 |
| YW3460-R | GTGAAGAAGGTGTTGCTGAC | |||
| DHA | DHA1-F | CTGATGAAAAAATCGTTATC | 1141 | 56 |
| DHA1-R | ATTCCAGTGCACTCAAAATA | |||
| CMY | CMY-F | TGTCAACACGGTGCAAATCA | 1346 | 56 |
| CMY-R | AGCAACGACGGGCAAAATG | |||
| ACT | ACT-F | CGAACGAATCATTATTCAGCACCG | 1518 | 56 |
| ACT-R | CGGCAATGTTTACTACACAGCG |
Multi-Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
All K. pneumoniae strains were subjected to MLST of several housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tnoB) as described on the MLST website (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html).4 K. pneumoniae strains were characterized by PFGE according to the previously published protocol by Ribot et al11 and previously described by Hu et al.12 Salmonella enterica serotype H9812 was used as a marker, and PFGE was established using XbaI digestion and agarose gel electrophoresis for 19 h at 14°C, with switch times of 6 and 36 s at 6 V/cm on a Bio-Rad CHEF Mapper Pulsed Field Electrophoresis System. Comparison of the PFGE patterns was performed in BioNumerics 7.6 using the Dice Similarity coefficient.
Results
Strain Isolation and AST
A total of 429 bacterial strains were isolated in the PICU from March 2015 to November 2019, including 57 Enterobacteriaceae strains, which mainly comprised K. pneumoniae and E. coli (36 and 17 strains, respectively). Of these, 14 strains (all K. pneumoniae) displayed resistance to carbapenems and the resistance rates to meropenem and imipenem were 24.6%. Eleven strains of CRKP were isolated from blood samples, while three were isolated from sputum samples. AST revealed that all 14 strains were resistant to imipenem, meropenem, and other β-lactams. KPC-2-producing strains were susceptible to trimethoprim/sulfamethoxazole, tetracycline, and polymyxin, but displayed resistance to other antibiotics. NDM-1-producing strains were susceptible to gentamicin, amikacin, ciprofloxacin, levofloxacin, polymyxin, and chloramphenicol, but showed resistance to other antibiotics (Table 2).
Table 2.
Microbiological and Molecular Characteristics, Antibiotic Susceptibilities (mg/L) of Carbapenem-Resistant K. pneumoniae
| Isolate Number | Specimen | Gender | Age | Date of Isolation (y) | mCIM (mm) | Carbapenemase | CTX–M | DHA | ST | PFGE Pattern | MEM | IPM | ATM | FEP | CAZ | CTX | TZP | AMK | CIP | LVX | POL | GEN | SXT | TCY | CHL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Blood | M | 7m | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 2 | Sputum | F | 16d | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 3 | Blood | M | 9d | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 4 | Blood | F | 9d | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 5 | Blood | M | 11d | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 6 | Blood | M | 22d | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 7 | Sputum | M | 1m | 2015 | 6 | KPC-2 | CTX-M-65 | – | 11 | A | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | ≤0.5 | >8 | ≤0.5 | 4 | >16 |
| 10 | Blood | F | 9d | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | C | >8 | >8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 11 | Sputum | M | 1m | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | C | >8 | >8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 12 | Blood | M | 12d | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | C | >8 | >8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 13 | Blood | M | 8d | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | C | 8 | 8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 14 | Blood | M | 8d | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | D | >8 | >8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 15 | Blood | F | 9d | 2017 | 6 | NDM-1 | – | DHA1 | 1224 | D | >8 | >8 | >16 | >16 | >16 | >32 | >64 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤2 | >2 | >8 | ≤4 |
| 77 | Blood | M | 10d | 2018 | 6 | KPC-2 | CTX-M-65 | – | 11 | B | >8 | >8 | >16 | >16 | >16 | >32 | >64 | >32 | >2 | >8 | 1 | >8 | ≤0.5 | 4 | 8 |
Abbreviations: MEM, meropenem; IPM, imipenem; ATM, aztreonam; FEP, cefepime; CAZ, ceftazidime; CTX, cefotaxime; TZP, piperacillin/tazobactam; AMK, amikacin; CIP, ciprofloxacin; LVX, levofloxacin; POL, polymyxin; GEN, Gentamicin; SXT, trimethoprim/sulfamethoxazole; TCY, tetracycline; CHL, chloramphenicol.
Phenotypic Screening and Genotyping of Carbapenemase Genes
All strains yielded positive mCIM results. Two types of carbapenemases, KPC-2 and NDM-1, were detected, in eight and six K. pneumoniae strains, respectively. All KPC-2-producing strains were also found to produce CTX-M-65, while all NDM-1-producing strains were also found to produce the AmpC enzyme DHA-1 (Table 2).
MLST and PFGE
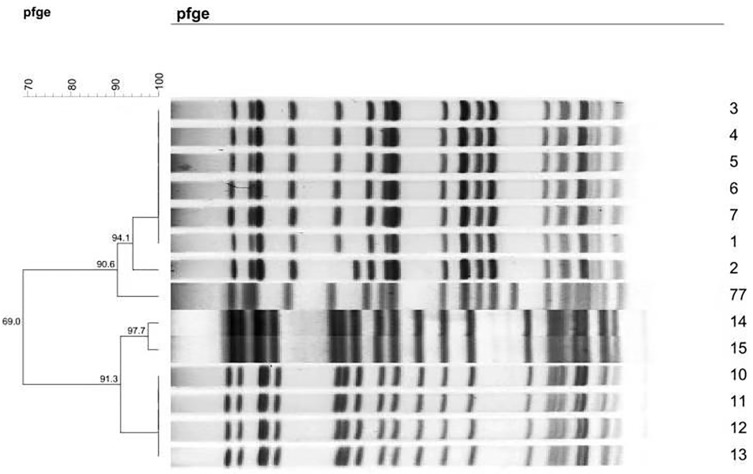
MLST divided the K. pneumoniae strains into two sequence types, ST11 (allelic profile: 3-3-1-1-1-1-4) and ST1224 (allelic profile: 18-67-26-63-142-38-169), which comprised eight and six strains, respectively. PFGE classified the 14 strains into four clonotypes. Six strains (1, 3, 4, 5, 6, and 7) belonged to the same PFGE clonotype A (with 100% similarity) and four strains (10, 11, 12, and 13) belonged to clonotype C (with 100% similarity) (Figure 1).
Figure 1.
Dendrogram of patterns for carbapenem-resistant K. pneumonia isolates obtained by PFGE.
Discussion
The clinical impact of carbapenem resistance has become an important global public health issue. The K. pneumoniae detection rate in clinical settings has gradually increased. Data from the China Antimicrobial Resistance Surveillance System showed that K. pneumoniae ranks second among Gram-negative bacteria (after E. coli) in terms of the number of detected isolates. Additionally, the system revealed increased resistance to carbapenems over time. To date, there have been numerous reports of CRKP. However, there are only a limited number of studies focusing on CRKP infections in children. All strains in this study were isolated from patients in the PICU, from patients aged less than one year.
In this study, a total of 14 CRKP strains were collected over the past four years, while no other CRE were detected.Some studies documented that Carbapenemase production is one of the main mechanisms of antibiotic resistance in CRE. The detection of carbapenemases, such as OXA-48, IMP-4, VIM-1, NDM-1, and KPC-2, has been reported in CRKP isolated from newborn patients.13–17 NDM-1-producing strains predominate in China.8 KPC-2 and NDM-1 were identified in 57.1% and 42.9% of the isolated K. pneumoniae strains, respectively, while other carbapenemases were not. In addition, none of the strains carried two or more carbapenemases. KPC-2-producing strains were prevalent in 2015, while NDM-1-producing strains were prevalent in 2017. However, there was only one KPC-2-producing strain detected in 2018. We also found that KPC-2-producing strains contained CTX-M-65, while NDM-1-producing strains also contained the AmpC enzyme DHA-1. Studies on the production of KPC-2 in carbapenem-resistant K. pneumoniae strains isolated from newborns are rarely reported in China. KPC-2-producing strains were also found to produce CTX-M-14 or CTX-M-15 β-lactamases,18,19 inconsistent with our findings.
AST demonstrated that all strains were resistant to β-lactams. KPC-2-producing strains were susceptible to trimethoprim/sulfamethoxazole, tetracycline, and polymyxin, and NDM-1-producing strains were susceptible to gentamicin, amikacin, ciprofloxacin, levofloxacin, polymyxin, and chloramphenicol. however, both strains were resistant to other antibiotics. These findings indicate that polymyxin has excellent antimicrobial activity, and thus can be administered empirically, whereas other antibiotics should be administered based on AST results. There are limited antibiotic options available for the treatment of infections caused by carbapenemase-producing Enterobacteriaceae in newborn patients,20 rendering the prevention and control of infections caused by these bacteria particularly important. Physicians, veterinarians, pharmacists, and nurses all need to be aware of the risks of drug resistance, especially carbapenem-resistance.21–23
MLST revealed two sequence types of CRKP, ST11 and ST1224. The former was isolated in 2015 (seven strains) and 2018 (one strain), while the latter was isolated in 2017 (six strains). PFGE identified six strains that could be classified into the same clonotype (with 100% similarity), and exhibited homology with strains 2 and 77 (94.1% and 90.6% similarities, respectively). ST1224 could be further divided into two PFGE clonotypes: clonotype C, comprised of strains 10, 11, 12, and 13 with 100% similarity, and clonotype D, comprised of strains 14 and 15. Both clonotypes exhibited homology with one another with 91.3% similarity. ST1224 and ST11 lacked homology with one another. There have been no large-scale epidemics of CRKP over the past four years, but changes have occurred in the epidemic strains.
The KPC2-ST11 strain is the predominant epidemic strain in China,4 and deserves great attention as there was a small outbreak of the KPC2-CTX-M-65-ST-11 strain in 2015, as well as an infection case reported in 2018 during the course of this study. NDM-1-producing K. pneumoniae has diverse MLST sequence types, such as ST14, ST37, ST105, and ST20.3,17,24,25 ST1224 is a relatively rare sequence type, and has been reported previously in newborns in India.26 In 2016, detection of the KPC-2-ST1224 K. pneumoniae strain in an adult patient was also reported in China.12 In 2017, the NDM-1-ST1224 strain was detected in a blood sample of a patient with leukemia. The NDM-1 gene is carried by a transferable plasmid of approximately 55 kb.27 Six NDM-1-DHA1-ST1224 strains were detected in this study. PFGE analysis showed that those six strains exhibited homology and were epidemic strains in 2017. Attention should be paid to the large-scale transfer of NDM-1 gene within and between departments, as it is a plasmid-harbored gene.
Children, and particularly infants, are a vulnerable population with varying risks of infection, depending on the degree of immune maturation, the presence of comorbidities, the use of invasive medical devices, and the use of antibiotics. The immature immune system of infants renders them more vulnerable to infections.17 CRE colonization is a high-risk factor in infections. Previous studies have demonstrated that the administration of carbapenems for more than four days is an independent risk factor for colonization of KPC-producing K. pneumoniae,28 with an infection rate of 9% among CRKP-colonized patients.29 Clinical data revealed that there were ten males and 13 newborns among the 14 patients, who all shared a common underlying disease (neonatal pneumonia), and have been administered antibiotics prior to the isolation of bacterial strains. There were 1 patient administered cefepime and 13 patients administered cefuroxime at the beginning, Later 4 of 13 patients were changed to cefepime.The use of second- and fourth-generation cephalosporins may exert antibiotic selective pressures that lead to the emergence of CRKP. In addition, 9/14 patients were diagnosed with neonatal sepsis. The mortality rates from lower respiratory tract infections and bacteremia caused by CRE are 34.8% and 43.1%, respectively.30 All patients in this study have fully recovered after treatment and have been discharged.
The presence of antibiotic-resistant strains in the environment is an important route for infections. These bacteria, which contaminate hospital environments via infected patients, can be directly or indirectly disseminated through medical personnel. Thus, it is essential to determine whether CRE colonization or infection in a patient originates from other patients or microbial communities that are difficult to eradicate via cleaning or disinfection in ward environments. Previous studies have found that CRE can be detected on the surface of beds, infusion pumps, and personal tables. The detection rate of environmental CRE diminishes with increasing distance from carriers, with the surface of beds being the most contaminated sites. The reduction in detection with distance may be attributed to the fact that medical equipment and items at a distance from patients are less exposed to hand contact or the body secretions of CRE carriers.31 It has also been demonstrated that outbreaks of carbapenem-resistant K. oxytoca are associated with damp environments, such as sink drainpipes in patient wards or sinks in intensive care units.32 Furthermore, CRE was found on the hands of medical personnel and incubators in neonatal intensive care units.14 Mother-to-child transmission may also serve as a source of bacteria that lead to infections in other patients sharing the same ward.33 After antibiotic resistant strains were detected in Yanbian University Hospital, we dedicated substantial attention to the issue and conducted screening tests on the air, incubators, bed stalls, and the hands of medical personnel. However, CRKP was not detected in the environment. Nevertheless, since then, we have enhanced the surveillance and control of infections, and there have been no large-scale nosocomial outbreaks.
Conclusions
There are extremely limited therapeutic options available for infections caused by carbapenemase-producing K. pneumoniae. Therefore, the prevention and control of infections caused by these bacteria are particularly important, and combination therapies can effectively prevent the emergence of new antibiotic resistant strains. Our results suggest that epidemics of KPC-2-ST11 and NDM-1-ST1224 strains occurred in the PICU of Yanbian University Hospital. When this occurs, hand hygiene, patient isolation, regular disinfection, and reasonable use of antibiotics should be emphasized as epidemic controls. Surveillance and strict implementation of prevention and control measures are crucial to prevent the occurrence and rapid spread of nosocomial infections.
Acknowledgments
The authors thank the excellent technical assistance provided by Hua Zong.
Ethics Statement
This study was approved by the research ethics board at Yanbian University Hospital.
Author Contributions
All authors contributed to data analysis, drafting, or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gajdacs M, Abrok M, Lazar A, Burian K. Comparative epidemiology and resistance trends of common urinary pathogens in a Tertiary-Care Hospital: a 10-year surveillance study. Medicina (Kaunas). 2019;55(7):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerdsin A, Deekae S, Chayangsu S, et al. Genomic characterization of an emerging blaKPC-2 carrying Enterobacteriaceae clinical isolates in Thailand. Sci Rep. 2019;9(1):18521. doi: 10.1038/s41598-019-55008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y, Shao C, Li J, Fan H, Bai Y, Wang Y. Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS One. 2015;10(3):e0119571. doi: 10.1371/journal.pone.0119571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Wang X, Wang J, et al. Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: data From a Longitudinal Large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi: 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- 5.Bradford PA, Bratu S, Urban C, et al. Emergence of Carbapenem-Resistant Klebsiella Species Possessing the Class A Carbapenem-Hydrolyzing KPC-2 and Inhibitor-Resistant TEM-30 β-Lactamases in New York City. Clin Infect Dis. 2004;39(1):55–60. doi: 10.1086/421495 [DOI] [PubMed] [Google Scholar]
- 6.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Y, Wang Y, Hsia Y, Sharland M, Heath PT. Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China. Ann Clin Microbiol Antimicrob. 2019;18(1):36. doi: 10.1186/s12941-019-0334-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54(1):573–577. doi: 10.1128/AAC.01099-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis JS 2nd, Herrera M, Wickes B, Patterson JE, Jorgensen JH. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother. 2007;51(11):4015–4021. doi: 10.1128/AAC.00576-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribot EM, Wierzba RK, Angulo FJ, Barrett TJ. Salmonella enterica serotype Typhimurium DT104 isolated from Humans, United States, 1985, 1990, and 1996. Emerg Infect Dis. 2002;8(4):387–391. doi: 10.3201/eid0804.010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu L, Liu Y, Deng L, et al. Outbreak by ventilator-associated ST11 K. pneumoniae with Co-production of CTX-M-24 and KPC-2 in a SICU of a Tertiary Teaching Hospital in Central China. Front Microbiol. 2016;7:1190. doi: 10.3389/fmicb.2016.01190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karaaslan A, Soysal A, Altinkanat Gelmez G, Kepenekli Kadayifci E, Soyletir G, Bakir M. Molecular characterization and risk factors for carbapenem-resistant Gram-negative bacilli colonization in children: emergence of NDM-producing Acinetobacter baumannii in a newborn intensive care unit in Turkey. J Hosp Infect. 2016;92(1):67–72. doi: 10.1016/j.jhin.2015.09.011 [DOI] [PubMed] [Google Scholar]
- 14.Yu F, Ying Q, Chen C, et al. Outbreak of pulmonary infection caused by Klebsiella pneumoniae isolates harbouring blaIMP-4 and blaDHA-1 in a neonatal intensive care unit in China. J Med Microbiol. 2012;61(Pt 7):984–989. doi: 10.1099/jmm.0.043000-0 [DOI] [PubMed] [Google Scholar]
- 15.Esposito EP, Gaiarsa S, Del Franco M, et al. A novel IncA/C1 Group Conjugative Plasmid, encoding VIM-1 metallo-beta-lactamase, mediates the acquisition of carbapenem resistance in ST104 klebsiella pneumoniae isolates from neonates in the intensive care unit of V. Monaldi Hospital in Naples. Front Microbiol. 2017;8:2135. doi: 10.3389/fmicb.2017.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad N, Ali SM, Khan AU. Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int J Antimicrob Agents. 2019;53(4):525–529. doi: 10.1016/j.ijantimicag.2018.12.005 [DOI] [PubMed] [Google Scholar]
- 17.Vanegas JM, Parra OL, Jimenez JN. Molecular epidemiology of carbapenem resistant gram-negative bacilli from infected pediatric population in tertiary - care hospitals in Medellin, Colombia: an increasing problem. BMC Infect Dis. 2016;16:463. doi: 10.1186/s12879-016-1805-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang M, Qin X. Investigation on β-lactam resistance gene of Klebsiella pneumoniae with decreased sensitivity to carbapenem antibiotics. Chin J Exp Clin Infect Dis. 2012;6(04):327–332. [Google Scholar]
- 19.Liu Y, Li XY, Wan LG, Jiang WY, Yang JH, Li FQ. Acquisition of carbapenem resistance in multiresistant Klebsiella pneumoniae isolates of sequence type 11 at a university hospital in China. Diagn Microbiol Infect Dis. 2013;76(2):241–243. doi: 10.1016/j.diagmicrobio.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 20.Gajdacs M. Intravenous or oral antibiotic therapy: sophie’s choice? Gen Internal Med Clinl Innovations. 2019;4(2). doi: 10.15761/GIMCI.1000176 [DOI] [Google Scholar]
- 21.Rabano-Blanco A, Dominguez-Martis EM, Mosteiro-Miguens DG, Freire-Garabal M, Novio S. Nursing students’ knowledge and awareness of antibiotic use, resistance and stewardship: a descriptive cross-sectional study. Antibiotics (Basel). 2019;8(4):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odetokun IA, Akpabio U, Alhaji NB, et al. Knowledge of antimicrobial resistance among veterinary students and their personal antibiotic use practices: a national cross-sectional survey. Antibiotics (Basel). 2019;8(4):243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajdacs M, Paulik E, Szabo A. Knowledge, attitude and practice of community pharmacists regarding antibiotic use and infectious diseases: a cross-sectional survey in hungary (KAPPhA-HU). Antibiotics (Basel). 2020;9(2). doi: 10.3390/antibiotics9020090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Hu X, Chen F, et al. Co-outbreak of multidrug resistance and a novel ST3006 Klebsiella pneumoniae in a neonatal intensive care unit: A retrospective study. Medicine (Baltimore). 2019;98(4):e14285. doi: 10.1097/MD.0000000000014285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng R, Zhang Q, Guo Y, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016;15:10. doi: 10.1186/s12941-016-0124-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S, Bhattacharjee A, Naha S, et al. Molecular characterization of NDM-1-producing Klebsiella pneumoniae ST29, ST347, ST1224, and ST2558 causing sepsis in neonates in a tertiary care hospital of North-East India. Infect Genet Evol. 2019;69:166–175. doi: 10.1016/j.meegid.2019.01.024 [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Du J, Zhou C, et al. An uncommon ST1224 NDM-1-producing klebsiella pneumoniae isolated from the bloodstream of a leukemia patient in China. Chemotherapy. 2017;62(4):262–268. doi: 10.1159/000469699 [DOI] [PubMed] [Google Scholar]
- 28.Ma MS, Wang DH, Sun XJ, Li ZH, Wang C. Risk factors for Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae colonization in neonates. Chin J Contemp Pediatr. 2014;16(10):970–974. Chinese. [PubMed] [Google Scholar]
- 29.Borer A, Saidel-Odes L, Eskira S, et al. Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K pneumoniae. Am J Infect Control. 2012;40(5):421–425. doi: 10.1016/j.ajic.2011.05.022 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistantEnterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62:e01882–17. doi: 10.1128/AAC.01882-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerner A, Adler A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental contamination by carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2013;51(1):177–181. doi: 10.1128/JCM.01992-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herruzo R, Ruiz G, Gallego S, Diez J, Sarria A, Omeñaca F. VIM-Klebsiella oxytoca outbreak in a neonatal intensive care unit. This time it wasn’t the drain. J Prev Med Hyg. 2017;58:E302–E307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonfanti P, Bellu R, Principe L, et al. Mother-to-child transmission of KPC carbapenemase-producing klebsiella pneumoniae at birth. Pediatr Infect Dis J. 2017;36(2):228–229. doi: 10.1097/INF.0000000000001403 [DOI] [PubMed] [Google Scholar]