Abstract
Clostridioides difficile is the leading cause of hospital-acquired gastrointestinal infections and a major public health burden in the United States. C. difficile infection causes a spectrum of disease from mild diarrhea to severe complications such as pseudomembranous colitis, toxic megacolon and death. This broad range of disease is only partially explained by bacterial genetic factors, host genetics, comorbidities and previous drug exposures. Another important factor is the gut microbiome, the disruption of which results in a loss of colonization resistance to C. difficile. Here, we review how gut microbiota and their metabolites impact C. difficile virulence and influence disease.
Keywords: Microbiome, Clostridium difficile, Microbial ecology, interspecies interactions, Microbiology, Metabolism
Introduction
The vast collection of bacteria, archaea, fungi and viruses that inhabit the gastrointestinal tract is important for human health. One area under continued research is what role this microbial community, termed the gut “microbiome”, plays during infection with gastrointestinal pathogens and how these interactions influence disease. In this review, we will focus on the impact of the gut microbiome on Clostridioides difficile (also known as Clostridium difficile) [1]. C. difficile is a Gram-positive, anaerobic, spore-producing bacterium responsible for 73% of all hospital-care associated gastrointestinal infections (GI) [2]. C. difficile infection (CDI) can cause a spectrum of disease from mild diarrhea to severe complications such as pseudomembranous colitis, toxic megacolon and death (reviewed in [3]). This broad spectrum of C. difficile-associated disease may be explained in part by bacterial genetic factors such as variation in the pathogenicity locus [4] and increased accessory gene content [5,6]. There is also likely a strong role for exogenous factors such as host genetics, comorbidities, treatment modalities and previous drug exposures. Here we focus on how resident microbiota can manipulate pathogen behavior and virulence. We also forecast the impact of uncovering the molecular mechanisms underlying these interactions.
The estimated number of CDIs in the United States in 2015 was 453,000, which were associated with approximately 29,000 deaths [7]. Worldwide, the estimated incidence of C. difficile cases is about 50 cases per 100,000 people per year [8]. Historically thought of as a nosocomial infection, CDI cases have been arising from the community [9] and a potential origin of these infections is from domesticated animals [10,11]. Altogether, CDI is not just a threat to certain vulnerable hospitalized populations, but is a larger public health concern involving both humans and animals. Since the United States Centers for Disease Control aims to reduce CDIs by 30% by 2020 [12], fully understanding C. difficile pathogenesis with the goal of preventing, treating, and reducing disease and disease recurrence is crucial to this endeavor.
A commonality in known risk factors for CDI, which include antibiotic usage, advanced age, inflammatory bowel disease, and immunosuppression, is disruption of the intestinal microbial ecosystem or “dysbiosis” [3]. While exposure to antibiotics is the primary risk factor for CDI, the increased prevalence of cases in the absence of antibiotics [9] suggests that other environmental factors, such as diet, or drug usage, may play a role in modulating the microbiome. Regardless of the cause of microbial community disruption, the result is a decrease in microbial diversity, alterations in the abundances of several important bacterial taxa, and a loss of colonization resistance. Colonization resistance encompasses numerous mechanisms by which the indigenous microbiota impedes exogenous pathogens from establishing infection. These mechanisms include competition for essential nutrients, limiting access to mucosal surfaces, direct production of antimicrobial molecules, modulating the intestinal metabolome, and activating the host immune system against the pathogen of interest [13]. Differences in the host microbiome and metabolome have long been observed to be associated with development, resolution and recurrence of CDI [14–21]. Recent work has begun to investigate the mechanisms by which these specific bacteria and metabolites impact development of CDI and disease manifestations (Figure 1).
Figure 1: Impact of Gut Microbiota and Intestinal Metabolites on C. difficile Infection.
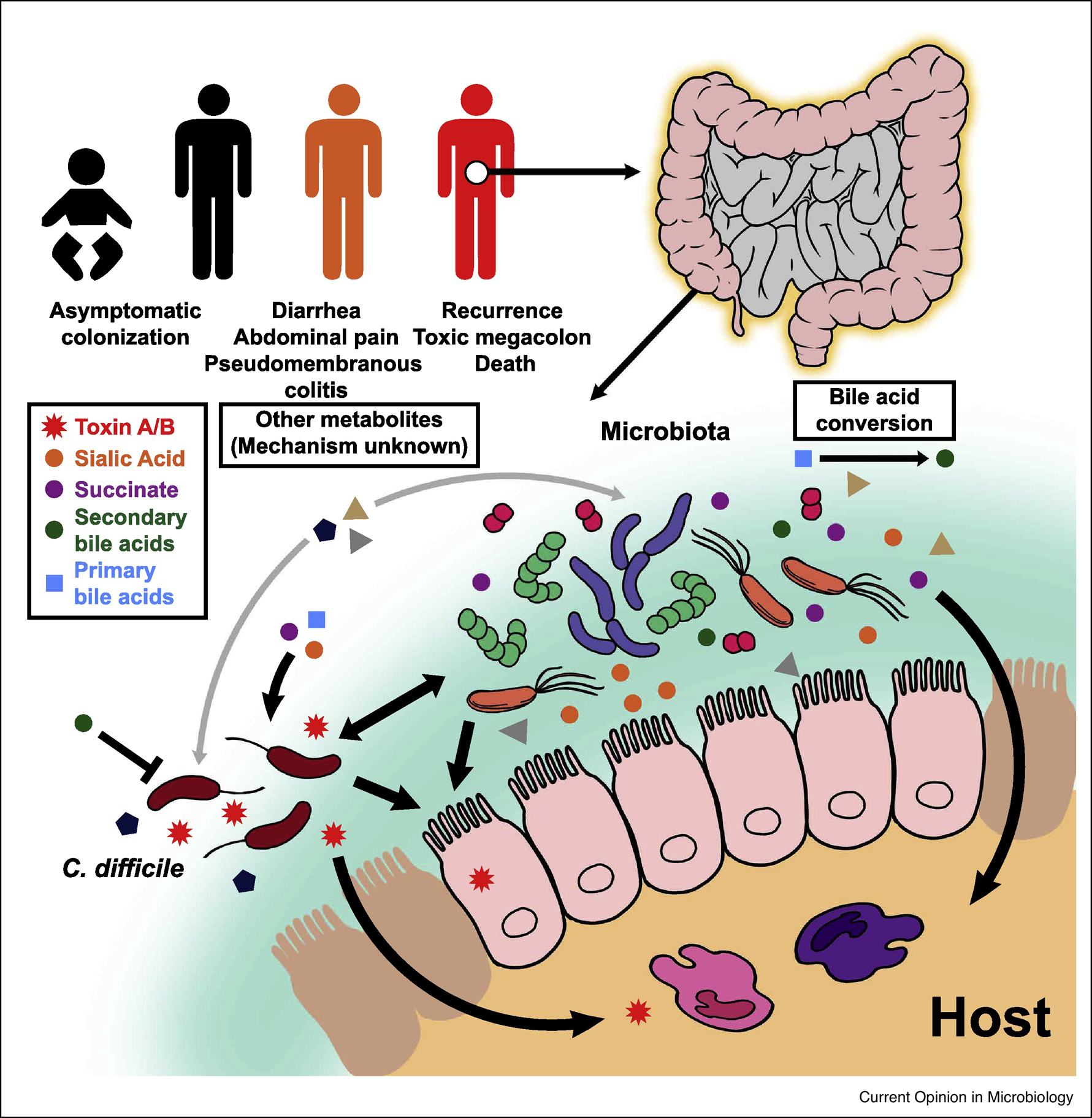
Exposure to C. difficile can cause a spectrum of disease ranging from asymptomatic colonization to mild infection treatable with antibiotics to severe intestinal pathologies. A disturbed gut microbiota usually precedes C. difficile infection as the normal enteric microbial flora provide colonization resistance against the pathogen. This is accomplished by their conversion of primary bile acids to secondary bile acids, which generally inhibit the growth of C. difficile. In contrast, other bacterial metabolic products, such as sialic acid and succinate, promote C. difficile growth. Intestinal epithelial cells and resident innate immune cells are affected by C. difficile toxin, which ultimately leads to disruption of the epithelial layer and development of a pro-inflammatory environment. Several other metabolites and bacteria are under consideration for their role in C. difficile disease (discussed in the text) by either directly impacting the pathogen or indirectly influencing the host immune response to infection.
Intestinal Metabolites Modulate C. difficile Behavior
Metabolism forms a common foundation for all cellular processes for the host, its resident microbiota, and invading pathogens. Metabolic crosstalk between bacteria is already known to profoundly impact the behavior of pathogens in various settings. For example, microbial synergy in the form of polymicrobial biofilms results in pathologic colonization of the oral cavity, the middle ear, chronic ulcerating wounds and the lung [22]. Open questions include which microorganisms synergize with C. difficile and how might metabolites be sensed and integrated by each partner in these relationships to impact their behavior, lifestyle, and potential virulence.
One well-established example of metabolic interactions in CDI is how microbial modulation of host bile salts impacts C. difficile colonization of the host (reviewed in [23]). In order to germinate, C. difficile spores sense primary bile acids [24], such as cholate and taurocholate [25,26], which are produced by the liver and secreted into the intestinal lumen. Numerous gut microbiota metabolize primary bile acids, using bile salt hydrolases and bile acid-inducible enzymes [27,28], to generate both unconjugated primary bile acids (such as cholate and chenodeoxycholate) and secondary bile acids (SBAs), some of which can inhibit C. difficile growth [29–32]. The secondary bile acids lithocholate and deoxycholate are significantly elevated in healthy subjects compared to those with either primary or recurrent CDI [18]. These observations are consistent with the fact that lithocholate inhibits C. difficile spore germination [33] and that deoxycholate inhibits growth of vegetative C. difficile cells [25]. Furthermore, bile-salt hydrolases and SBAs generally increase after fecal microbiota transplantation [34–36]. The functionality of this restored bile acid pool from FMT-treated CDI subjects has been demonstrated by successful inhibition of C. difficile germination and growth in vitro [37]. Future research focused on utilizing rationally designed microbial consortia and probiotic organisms to manipulate the bile acid pool could show promise for treatment of primary and recurrent CDI.
Bile acids may also impact other aspects of C. difficile virulence. C. difficile produces two enterotoxins, TcdA and TcdB, which are the primary drivers of pathogenesis by causing intestinal epithelial cell damage leading to a robust inflammatory response by the host [38]. Recent work investigated the ability of microbial-derived bile acids found in humans (deoxycholate, isodeoxycholate, lithocholate, isolithocholate, and ursodeoxycholate) to impact toxin production by clinically relevant C. difficile strains [32]. Exposure to low concentrations of deoxycholate, one of the most abundant cecal bile acids [39,40], reduced toxin production by most strains, without a concomitant reduction in general vegetative cell growth [32]. Furthermore, sub-lethal concentrations of deoxycholate stimulate antibiotic-resistant C. difficile biofilm formation in vitro [48]. Together, these studies show that a shifting composition of intestinal bile acids can either promote or halt successful colonization, growth, persistence and virulence by C. difficile. A crucial next step should focus on understanding where and when C. difficile is exposed to specific bile acids during colonization, outgrowth, and persistence in the dynamic and volatile environment of the infected intestinal tract.
Beyond bile acids, there is rich metabolic potential in the microbiota that can likely impact C. difficile behavior and virulence, and the outcome of infection. Previous work has shown the importance of microbially-derived sialic acid and succinate in CDI utilizing mice mono-colonized with Bacteroides thetaiotaomicron, a model gut commensal [41,42]. B. thetatiotaomicron encodes sialidases which cleave and release the terminal sugar sialic acid from mucosal glycoconjugates, but does not possess the catabolic enzymes required to actually consume it. In the first study, it was demonstrated that B. thetatiotaomicron cross-feeds sialic acid to C. difficile, and that utilization of sialic acid improves C. difficile expansion in the gut [41]. In the subsequent study, analysis of C. difficile gene expression when infecting a B. thetaiotaomicron mono-colonized mouse revealed the importance of carbohydrate transport and metabolism, and specifically in conversion of succinate to butyrate [42]. It was observed that succinate levels were elevated in cecal contents after antibiotic treatment and experimentally-induced diarrhea. Similar to sialic acid, succinate appears necessary for C. difficile expansion in the gut. The authors posit that B. thetaiotaomicron produces high levels of succinate during its fermentation of dietary carbohydrates, and that C. difficile reduces succinate to butyrate, regenerating the electron acceptor NAD+, to support fermentation of other energy sources. These interactions exemplify how the enzymatic potential of a bacterial community can impact CDI.
Numerous metabolites change in abundance during CDI. For example, proline, branched-chain amino acids, and carbohydrates decrease in abundance as C. difficile colonizes the mouse cecum [21]. Additionally, end products of Stickland fermentation, a process used by C. difficile to metabolize amino acids, are found to increase [20]. In humans, it has been observed that low levels of cholesterol and high levels of coprostanol, a microbially-derived byproduct of cholesterol metabolism in the gut, discriminate between a CDI-associated and healthy gut microbiome [19]. Yet the role of these sterols in disease is still not understood. These shifts in the metabolome may not simply be a hallmark of toxin-induced disruptions in intestinal physiology. Colonization with non-toxigenic C. difficile results in an altered gut metabolome which is different from both healthy and active disease states [16], as might be expected when an invasive organism establishes itself in the gut. While these initial studies were instrumental in highlighting some of the metabolic changes during CDI, gross measures of intestinal metabolites preclude direct implication of the types of bacteria producing and using each compound.
Interspecies Interactions during C. difficile Infection
Which bacteria are primarily responsible for manipulating these key metabolites, and can they can be harnessed to alter the metabolic milieu as a therapeutic intervention in CDI? Attention has been given to Clostridium scindens as analyses of both mouse models and hospitalized patients previously determined that C. scindens is associated with resistance to CDI [29]. C. scindens is one member of the gut microbiome that can convert the primary unconjugated bile acid cholate into deoxycholate by 7α-dehydroxylation [43]. Generally, the prevalence of the baiCD gene cluster, encoding a key enzyme of this biotransformation, is higher in fecal samples of C. difficile negative hospitalized patients than those with active CDI [44]. Furthermore, in vitro co-culture of C. difficile and C. scindens in the presence of cholate leads to inhibition of C. difficile growth [45]. Similarly, administration of a microbiota consortia that included C. scindens to antibiotic-treated or gnotobiotic mice enhanced resistance to subsequent CDI and restored the abundances of the C. difficile inhibitory bile acids, deoxycholate and lithocholate [29,30].
Besides transforming primary bile acids to SBAs and limiting C. difficile germination and growth, C. scindens may impact C. difficile viability by producing antibiotics. It has recently been observed that C. scindens and other bile acid 7α-dehydroxylating human gut bacteria inhibit C. difficile growth by secreting antibiotic compounds. These compounds were subsequently determined to be the indole-derived turbomycin A and 1,1,1-tris(3-indolyl)-methane [45]. Interestingly, the presence of deoxycholate and cholate enhanced the antimicrobial activity of these compounds against C. difficile through a currently unknown mechanism. Given these data, C. scindens is being pursued as a “probiotic” to protect or treat CDI [46]. However, it should be noted that presence of C. scindens in the gut may not protect against or resolve CDI on its own [47], as its probiotic activity might be modulated by other members of the gut microbiota. Furthermore, co-culture of C. scindens with C. difficile in vitro enhances biofilm formation [48] and biofilms are likely to enhance resistance of C. difficile to antibiotics.
Other commensal bacterial species have been targeted as potential probiotics. Lactobacillus is a genus of Gram-positive, facultative anaerobes that are nearly synonymous with the term “probiotic”. Several studies have looked at the effects of various Lactobacillus species (including L. acidophilus, L. delbrueckii, L. fermentum, L. gasseri and L. plantarum) on C. difficile virulence. Generally, in vitro co-cultures of Lactobacillus species and C. difficile lead to inhibition of the latter’s growth, which is likely due to acidification of the environment by the former [49–51]. However, when co-cultures control for changes in pH and in tests of C. difficile growth in cell-free Lactobacillus conditioned media, it has been observed that several species decrease quorum sensing, expression of pathogenicity locus genes and subsequent toxin production by C. difficile [49,50,52]. The protective effect of Lactobacillus given as a monoculture or consortia-based probiotic for CDI has been shown in mice [49,52,53] and in adults and children taking antibiotics [54]. Metabolites and other molecules produced by Lactobacillus may also influence C. difficile’s interaction with the host, as Lactobacillus cell-free supernatants decrease the pathogen’s ability to adhere to epithelial cells in vitro [55,56].
Evidently, different commensal bacterial species and strains impact C. difficile behavior. What, then, of the interactions between C. difficile and enteric bacteria whose expansion is characteristic of the dysbiosis occurring before and during CDI? The broad decreases in gut microbiota diversity, but not necessarily bacterial load, observed after antibiotic treatment are often characterized by expansion of specific bacteria, such as Enterobacteriaceae and Enterococcus [57,58]. These same bacteria are known to thrive during intestinal inflammation (reviewed in [59])) and during CDI [17,60]. Further understanding is needed on whether and how these organisms take advantage of the environment prior to and during CDI and alter the clinical course of infection.
C. difficile contributes to changing the intestinal metabolite milieu that is already departed from homeostasis in the dysbiotic state preceding CDI. For example, high levels of indole were detected in a recent screen of fecal metabolites from CDI patients [61]. It was suggested that C. difficile induces indole production by Escherichia coli. This cross-talk involves the accessory gene regulator (Agr) 1 quorum sensing system of C. difficile and the tryptophanase gene of E. coli, although the precise mechanism is still unknown. Furthermore, this study showed that indole directly limited the growth of many anaerobic gut bacteria in vitro, notably Bifidobacterium longum, other Clostridium species, and Flavobacterium sp [61]. Another mechanism by which C. difficile inhibits specific members of the gut microbiota is through its fermentation of tyrosine to produce para-cresol [62]. This ability was recently shown to grant C. difficile a competitive growth advantage in vitro, specifically over Gram-negative bacteria of the Bacteroidaceae and Enterobacteriaceae families [63]. Additionally, C. difficile deficient in para-cresol production showed a modest reduction in microbial burden in a recurrent mouse model of C. difficile infection, although there was no difference in initial colonization. C. difficile can also secrete proline-based cyclic dipeptides that can inhibit gut bacteria, including commensal Clostridium species [45]. Whether these antibacterial peptides are produced by C. difficile in vivo, and what role they play in contributing to infection-associated dysbiosis remains to be uncovered.
Conclusions
Significant gains in our understanding of CDI have been achieved with high-throughput metagenomic and metabolomic surveys of the gut before, during and after CDI coupled with mechanistic insights from molecular microbiology experiments. Of course, it is inherently difficult to predict what occurs in the complex environment of the human gastrointestinal tract during the dynamism of an ongoing infection. Therefore, gnotobiotic mouse models and bioreactor systems will be instrumental in singling out the importance of a limited number of polymicrobial interactions and metabolic pathways of interest during pathogenesis. Additional understanding will be gained by incorporating advanced imaging technologies to interrogate the spatial dynamics of the microbial and metabolic environment during CDI. Beyond the scope of this review is the adjacent, yet important, topic concerning how microbial metabolites can modulate the host innate and adaptive immune response to C. difficile. Evolving in vitro models, such as the newly developed anaerobic intestine-on-a-chip [64], will allow study of how the microbiota and their metabolites impact the host epithelia and vice versa during CDI and various other gastrointestinal diseases. With the technological advances available at present and those that will undoubtedly be developed in the future, along with classic microbiological techniques, untangling the network of host-microbiota-pathogen interactions to more comprehensively understand CDI is within grasp.
Acknowledgements
J.P.Z is supported by (K22AI7220). A.A. is supported by the Postdoctoral Fellowship for Academic Diversity at the Children’s Hospital of Philadelphia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have no relevant conflict of interests to disclose.
References
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM: Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 2016, 40:95–99. [DOI] [PubMed] [Google Scholar]
- 2.Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, Consortium HAI: Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe 2019. [DOI] [PubMed] [Google Scholar]
- 3.Rupnik M, Wilcox MH, Microbiology G-DN: Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature Reviews Microbiology 2009. [DOI] [PubMed] [Google Scholar]
- 4.Rupnik M: Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS microbiology reviews 2008, 32:541–555. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BB, Carter RA, Ling L, Leiner I, Taur Y, Kamboj M, Dubberke ER, Xavier J, Pamer EG: Pathogenicity Locus, Core Genome, and Accessory Gene Contributions to Clostridium difficile Virulence. mBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, et al. : Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome biology 2009, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, et al. : Burden of Clostridium difficile infection in the United States. N. Engl. J. Med 2015, 372:825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balsells E, Shi T, Leese C, Lyell I, Burrows J, Wiuff C, Campbell H, Kyaw MH, Nair H: Global burden of Clostridium difficile infections: a systematic review and meta-analysis. Journal of global health 2019, 9:10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JS, Monaghan TM, Wilcox MH: Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nature reviews. Gastroenterology & hepatology 2016, 13:206–216. [DOI] [PubMed] [Google Scholar]
- 10.Moono P, Foster NF, Hampson DJ, Knight DR, Bloomfield LE, Riley TV: Clostridium difficile Infection in Production Animals and Avian Species: A Review. Foodborne pathogens and disease 2016, 13:647–655. [DOI] [PubMed] [Google Scholar]
- 11.Knetsch CW, Kumar N, Forster SC, Connor TR, Browne HP, Harmanus C, Sanders IM, Harris SR, Turner L, Morris T, et al. : Zoonotic Transfer of Clostridium difficile Harboring Antimicrobial Resistance between Farm Animals and Humans. Journal of clinical microbiology 2018, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC: Clostridioides difficile. Edited by: Centers for Disease Control and Prevention; 2018. vol 2019.] [Google Scholar]
- 13.Rosa R, Donskey CJ, Munoz-Price LS: The Intersection Between Colonization Resistance, Antimicrobial Stewardship, and Clostridium difficile. Current infectious disease reports 2018, 20:27. [DOI] [PubMed] [Google Scholar]
- 14.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP: Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. Journal of clinical microbiology 2013, 51:2884–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB: Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nature communications 2014, 5:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rojo D, Gosalbes MJJ, Ferrari R, Pérez-Cobas AE, Hernández E, Oltra R, Buesa J, Latorre A, Barbas C, Ferrer M, et al. : Clostridium difficile heterogeneously impacts intestinal community architecture but drives stable metabolome responses. The ISME journal 2015, 9:2206–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent C, Miller MA, Edens TJ, Mehrotra S, Dewar K, Manges AR: Bloom and bust: intestinal microbiota dynamics in response to hospital exposures and Clostridium difficile colonization or infection. Microbiome 2016, 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, Korzenik JR: Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Alimentary pharmacology & therapeutics 2016, 43:1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antharam VC, McEwen DC, Garrett TJ, Dossey AT, Li EC, Kozlov AN, Mesbah Z, Wang GP: An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium difficile Infection. PloS one 2016, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenior ML, Leslie JL, Young VB, Schloss PD: Clostridium difficile Alters the Structure and Metabolism of Distinct Cecal Microbiomes during Initial Infection To Promote Sustained Colonization. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors elucidate the global changes in transcription and metabolism of gut microbiome in mice treated with several different antibiotics and who varied in their resolution of C. difficile infection. This study is one of the first to use paired metatranscriptomics and metabolomics during enteric infection and demonstrates how C. diffiicle modulates the metabolism of the gut microbiome for its own benefit.
- 21.Fletcher JR, Erwin S, Lanzas C, Theriot CM: Shifts in the Gut Metabolome and Clostridium difficile Transcriptome throughout Colonization and Infection in a Mouse Model. mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME: Polymicrobial interactions: impact on pathogenesis and human disease. Clinical microbiology reviews 2012, 25:193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen A: A Gut Odyssey: The Impact of the Microbiota on Clostridium difficile Spore Formation and Germination. PLoS pathogens 2015, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis MB, Allen CA, Shrestha R, Sorg JA: Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS pathogens 2013, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorg JA, Sonenshein AL: Bile salts and glycine as cogerminants for Clostridium difficile spores. Journal of bacteriology 2008, 190:2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heeg D, Burns DA, Cartman ST, Minton NP: Spores of Clostridium difficile Clinical Isolates Display a Diverse Germination Response to Bile Salts. PLoS ONE 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long SL, Gahan CGMGM, Joyce SA: Interactions between gut bacteria and bile in health and disease. Molecular aspects of medicine 2017, 56:54–65. [DOI] [PubMed] [Google Scholar]
- 28.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ: Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Applied microbiology and biotechnology 2017, 101:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. : Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517:205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schürch CM, McCoy KD, Kuehne SA, Minton NP, et al. : Functional Intestinal Bile Acid 7α-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Frontiers in cellular and infection microbiology 2016, 6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon S, Yu J, McDowell A, Kim SH, You HJ, Ko G: Bile salt hydrolase-mediated inhibitory effect of Bacteroides ovatus on growth of Clostridium difficile. Journal of microbiology (Seoul, Korea) 2017, 55:892–899. [DOI] [PubMed] [Google Scholar]
- 32.Thanissery R, Winston JA, Theriot CM: Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 2017, 45:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors surveyed a range of human and mouse microbially-derived secondary bile acids on their ability to impact various aspects of the C. diffiicle life cycle. Their results showed that C. difficile strains were variably impacted by the inhibitory effects of secondary bile acids, highlighting the nuances in susceptibility to C. difficile colonization observed in vivo.
- 33.Sorg JA, Sonenshein AL: Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. Journal of bacteriology 2010, 192:4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A: Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. American journal of physiology. Gastrointestinal and liver physiology 2014, 306:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seekatz AM, Theriot CM, Rao K, Chang Y-MM, Freeman AE, Kao JY, Young VB: Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 2018, 53:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]; A longitudinal analysis of patients treated with FMT for recurrent CDI revealed consistent increases in short chain fatty acids and variable increases in secondary bile acids post-FMT. By modeling metabolite concentrations and presence of specific bacteria, Lachnospiraceae, Ruminococcaceae, and unclassified Clostridiales were found to be associated with these restored metabolites.
- 36.Mullish BH, McDonald JAKAK, Pechlivanis A, Allegretti JR, Kao D, Barker GF, Kapila D, Petrof EO, Joyce SA, Gahan CGMGM, et al. : Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 2019, 68:1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]; By analyzing the microbiome and metabolome of patients with recurrent CDI before and after FMT, this study determined that successful treatment was associated with a rapid and sustained enrichment in bile salt hydrolase-producing bacteria. The sufficiency of bile salt hydrolases in resolving recurrent CDI was further demonstrated in a mouse model.
- 37.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, Khoruts A, Sadowsky MJ: Changes in Colonic Bile Acid Composition following Fecal Microbiota Transplantation Are Sufficient to Control Clostridium difficile Germination and Growth. PloS one 2016, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrasekaran R, Lacy DB: The role of toxins in Clostridium difficile infection. FEMS microbiology reviews 2017, 41:723–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann AF: The continuing importance of bile acids in liver and intestinal disease. Archives of internal medicine 1999, 159:2647–2658. [DOI] [PubMed] [Google Scholar]
- 40.Hamilton JP, Xie G, Raufman J-PP, Hogan S, Griffin TL, Packard CA, Chatfield DA, Hagey LR, Steinbach JH, Hofmann AF: Human cecal bile acids: concentration and spectrum. American journal of physiology. Gastrointestinal and liver physiology 2007, 293:63. [DOI] [PubMed] [Google Scholar]
- 41.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. : Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL: Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell host & microbe 2014, 16:770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallonee DH, White WB, of bacteriology H PB: Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. Journal of bacteriology 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solbach P, Chhatwal P, Woltemate S, Tacconelli E, Buhl M, Gerhard M, Thoeringer CK, Vehreschild MJGTJGT, Jazmati N, Rupp J, et al. : BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PloS one 2018, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee I-KK, Yun B-SS, Matsuzaki K, Furukawa M, Min H-KK, Bajaj JS, et al. : Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell chemical biology 2019, 26:27–340000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pamer E, Buffie C, McKenney P: Methods and compositions for reducing Clostridium difficile infection. US Patent 2019.
- 47.Amrane S, Bachar D, Lagier JC, Raoult D: Clostridium scindens Is Present in the Gut Microbiota during Clostridium difficile Infection: a Metagenomic and Culturomic Analysis. Journal of clinical microbiology 2018, 56. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Dubois T, Tremblay YDNDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, Briandet R, Dupuy B: A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ biofilms and microbiomes 2019, 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yun B, Oh S, Griffiths MW: Lactobacillus acidophilus modulates the virulence of Clostridium difficile. Journal of dairy science 2014, 97:4745–4758. [DOI] [PubMed] [Google Scholar]
- 50.Yong CC, Lim J, Kim BKK, Park DJJ, Oh S: Suppressive effect of Lactobacillus fermentum Lim2 on Clostridioides difficile 027 toxin production. Letters in applied microbiology 2019, 68:386–393. [DOI] [PubMed] [Google Scholar]
- 51.Monteiro CRAVRAV, do Carmo MS, Melo BO, Alves MS, Dos Santos CI, Monteiro SGG, Bomfim MRQRQ, Fernandes ES, Monteiro-Neto V: In Vitro Antimicrobial Activity and Probiotic Potential of Bifidobacterium and Lactobacillus against Species of Clostridium. Nutrients 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quigley L, Coakley M, Alemayehu D, Rea MC, Casey PG, O’Sullivan Ó, Murphy E, Kiely B, Cotter PD, Hill C, et al. : Lactobacillus gasseri APC 678 Reduces Shedding of the Pathogen Clostridium difficile in a Murine Model. Frontiers in microbiology 2019, 10:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. : Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS pathogens 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldenberg JZ, Yap C, Lytvyn L, Lo CK, Beardsley J, Mertz D, Johnston BC: Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. The Cochrane database of systematic reviews 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banerjee P, Merkel GJ, Bhunia AK: Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut pathogens 2009, 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Najarian A, Sharif S, Griffiths MW: Evaluation of protective effect of Lactobacillus acidophilus La-5 on toxicity and colonization of Clostridium difficile in human epithelial cells in vitro. Anaerobe 2019, 55:142–151. [DOI] [PubMed] [Google Scholar]
- 57.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG: Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity 2012, 80:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, et al. : Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of clinical investigation 2010, 120:4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winter SE, Lopez CA, Bäumler AJ: The dynamics of gut-associated microbial communities during inflammation. EMBO reports 2013, 14:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pérez-Cobas AE, Artacho A, Ott SJ, Moya A, Gosalbes MJJ, Latorre A: Structural and functional changes in the gut microbiota associated to Clostridium difficile infection. Frontiers in microbiology 2014, 5:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darkoh C, Plants-Paris K, Bishoff D, DuPont HL: Clostridium difficile Modulates the Gut Microbiota by Inducing the Production of Indole, an Interkingdom Signaling and Antimicrobial Molecule. mSystems 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elsden SR, Hilton MG, of microbiology W JM: The end products of the metabolism of aromatic amino acids by Clostridia. Archives of microbiology 1976. [DOI] [PubMed] [Google Scholar]
- 63.Passmore IJ, Letertre MPMPM, Preston MD, Bianconi I, Harrison MA, Nasher F, Kaur H, Hong HA, Baines SD, Cutting SM, et al. : Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS pathogens 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]; Production of a unique bacteriostic compound was determined to give C. difficile a competitive advantage in vivo against other enteric bacteria, primarily Gram-negative organisms. Furthermore, production of para-cresol during CDI changes the intestinal metabolic milieu, implicating it as a compound with wide-reaching effects.
- 64.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, et al. : A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature biomedical engineering 2019, 3:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]; A microfluidic device with biophysical and biochemical properties of the mammalian intestine enables extended co-culture of epithelial cells and anaerobic prokaryotes. Importantly, the diversity and composition of the human gut microbiome is recapitulated in this tool.


