Abstract
In the past decade, a large number of studies have detected herpesvirus sequences from many bat species around the world. Nevertheless, the discovery of bat herpesviruses is geographically uneven. Of the various bat species tested to date, only a few were from the New World. Seeking to investigate the distribution and diversity of herpesviruses circulating in neotropical bats, we carried out molecular screening of 195 blood DNA samples from 11 species of three bat families (Phyllostomidae, Mormoopidae, and Molossidae). Using polymerase chain reaction amplification, with degenerate consensus primers targeting highly conserved amino acid motifs of the herpesvirus DNA polymerase and Glycoprotein B genes, we characterized novel viral sequences from all tested species. BLAST searches, pairwise nucleotide and amino acid sequence comparisons, as well as phylogenetic analyses confirmed that they all belonged to the Herpesviridae family, of the Beta- and Gammaherpesvirinae subfamilies. Fourteen partial DNA polymerase gene sequences, of which three beta- and 11 gamma-herpesviruses, were detected. A total of 12 partial Glycoprotein B gene sequences, all gamma-herpesviruses, were characterized. Every sequence was specific to a bat species and in some species (Desmodus rotundus, Carollia perspicillata, and Pteronotus rubiginosus) multiple viruses were found. Phylogenetic analyses of beta- and gammaherpesvirus sequences led to the identification of bat-specific clades. Those composed of sequences obtained from different bat species belonging to distinct subfamilies follow the taxonomy of bats. This study confirms the astonishing diversity of bat herpesviruses and broadens our knowledge of their host range. Nevertheless, it also emphasizes the fact that, to better appreciate the evolutionary history of these viruses, much remains to be done at various taxonomic levels.
Keywords: Chiroptera, Betaherpesvirus, Gammaherpesvirus, DNA polymerase, Glycoprotein B, Evolution, Phylogeny
Highlights
-
•
Molecular screening was carried out on 11 bat species from French Guiana and Martinique (French West Indies).
-
•
Partial DNA polymerase gene sequences of 14 viruses were characterized as well as 12 Glycoprotein B sequences.
-
•
Genetic characterization of these sequences reveals a high degree of genetic divergence.
-
•
Phylogenetically, most of the newly discovered sequences fall within bat-specific clades well correlated with the taxonomy of their hosts.
-
•
This study is the largest conducted to date in terms of species diversity from the New World.
1. Introduction
With over 1200 species, representing more than 20% of all registered mammal species, Chiroptera is the second-largest species-rich mammalian order (Fenton, 1997). Bats are present on all continents except the poles and a few isolated oceanic islands. They are highly diverse in terms of their anatomy and lifestyles, and have different diets (insectivorous, frugivorous, nectarivorous, carnivorous, piscivorous, or hematophagous). Thanks to their biological traits, bats provide key ecosystem services (Kunz et al., 2011; López-Baucells et al., 2018). At the same time, as a natural reservoir of many viruses, their global distribution, abundance, ability to fly and migrate over large distances, and the diversity of their diets and sociality are all factors that favor the acquisition and spread of viruses (Calisher et al., 2006; Drexler et al., 2012; Mackenzie, 2005; Wong et al., 2007). They are thus considered to play a major role in the emergence and transmission of zoonotic viruses such as lyssaviruses, coronaviruses, paramyxoviruses, or filoviruses (Baker et al., 2013b; Calisher et al., 2006; Davis et al., 2006; Freuling et al., 2011; Halpin et al., 2000; Kupferschmidt, 2013; Leroy et al., 2005; Li, 2005; Luby et al., 2009; Smith and Wang, 2013; Towner et al., 2009; Wong et al., 2007; Yob et al., 2001; Zaki et al., 2012). Bats are also carriers of other viruses, including adeno-, astro-, polyoma-, picorna-, or herpesviruses (Drexler et al., 2011; Fagrouch et al., 2012; Jánoska et al., 2011; Lau et al., 2011; Li et al., 2010a, Li et al., 2010b).
Within the Herpesvirales order, viruses of mammals all belong to the Herpesviridae family. Members of this viral family are grouped into three subfamilies (Alpha-, Beta-, and Gammaherpesvirinae) and 13 genera (Davison et al., 2009; Pellett et al., 2011). This repartition into subfamilies and genera reflects the diversity of the different known viral species. This diversity concerns the nature of the host and the cellular tropism as well as the functional organization of the viral genome and the pathogenic properties. In addition, each mammal species can carry multiple herpesvirus species that are host specific. However, there are examples of cross-species transmission (Ehlers et al., 2008). The first description of bat herpesvirus sequences dates back to 2007 (Wibbelt et al., 2007). Over the past decade, dozens of herpesvirus sequences have been described from different bat species on every continent (Anthony et al., 2013; Baker et al., 2013a; Dacheux et al., 2014; Dietrich et al., 2018; Donaldson et al., 2010; Escalera-Zamudio et al., 2016; Ge et al., 2012; Geldenhuys et al., 2018; He et al., 2013; Holz et al., 2018; Hu et al., 2017; Jánoska et al., 2011; Li et al., 2010a, Li et al., 2010b; Molnár et al., 2008; Mühldorfer et al., 2011; Noguchi et al., 2018; Paige Brock et al., 2013; Pozo et al., 2016; Razafindratsimandresy et al., 2009; Salmier et al., 2017; Sano et al., 2015; Sasaki et al., 2014; Shabman et al., 2016; Subudhi et al., 2018; Wada et al., 2018; Watanabe et al., 2010; Watanabe et al., 2009; Wray et al., 2016; Wu et al., 2012; Yang et al., 2013; Zhang et al., 2012; Zheng et al., 2016, Zheng et al., 2018). Most of them were characterized from apparently healthy animals sampled during trapping campaigns in the frame of random surveillance programs. To date, two bat herpesviruses, fruit bat alphaherpesvirus 1 and bat gammaherpesvirus 8 (FBAHV1 and BGHV8, respectively) have been recognized as species by the International Committee for Taxonomy of Viruses (ICTV) according to the latest master species list (MSL# 34) released on March 8, 2019 (https://talk.ictvonline.org/files/master-species-lists/m/msl/8266). Their ICTV official names are Pteropodid alphaherpesvirus 1 (PtAHV1) and Vespertilionid gammaherpesvirus 1 (VeGHV1). They were classified in the Simplexvirus and Percavirus genera, respectively. As of November 2019, when it was last updated, the DBatVir (http://www.mgc.ac.cn/DBatVir/) database reported 233 bat herpesvirus sequences (Chen et al., 2014). A few others have not yet been included (Holz et al., 2018; Noguchi et al., 2018; Wada et al., 2018). They are distributed among the three Herpesvirinae subfamilies, but more than half of them (144 sequences) are unclassified Herpesviridae. In total, they were derived from 58 species of seven bat families (Vespertilionidae, Pteropodidae, Rhinolophidae, Molossidae, Phyllostomidae, Miniopteridae, and Hipposideridae). Nevertheless, the discovery of bat herpesviruses is geographically uneven. Most sequences are from Asian, African, and European bat species. Comparatively few data are available regarding herpesviruses of New World species. Indeed, of the 233 recorded sequences, only eight are from New World bat species, six from North and Central America and two from South America (Donaldson et al., 2010; Escalera-Zamudio et al., 2016; Razafindratsimandresy et al., 2009; Salmier et al., 2017; Shabman et al., 2016; Subudhi et al., 2018). The ones from North and Central America were obtained from two insectivorous and two hematophagous bat species of the Vespertilionidae and Phyllostomidae families, respectively, and corresponded to beta- and gammaherpesvirus sequences (Donaldson et al., 2010; Escalera-Zamudio et al., 2016; Shabman et al., 2016; Subudhi et al., 2018). Those from South America, one alpha- and one gammaherpesvirus, were derived from bats belonging to the Phyllostomidae and Molossidae families, respectively (Razafindratsimandresy et al., 2009; Salmier et al., 2017). In addition, Wray et al. reported the characterization of two other herpesvirus sequences, DrHV-1 and DrHV-2, described as gamma- and betaherpesvirus, respectively, from Desmodus rotundus individuals from Guatemala (Wray et al., 2016). Unfortunately, these last sequences have not been released in the databases.
Strikingly, with the exception of the two aforementioned sequences, there has been no prior organized effort to discover herpesviruses in neotropical bat species from South America and in particular from Amazonia, which hosts one of the highest diversity of bat species in the world (López-Baucells et al., 2016; Paglia et al., 2012). To ascertain the distribution and diversity of herpesviruses circulating in neotropical bats, additional investigations were required. Taking advantage of a unique collection of bat samples collected during the past 10 years in French Guiana (South America) and Martinique (French West Indies), we addressed the presence of herpesviruses in the different bat species collected by analyzing two distinct informative partial genes. The partial characterization of DNA polymerase and Glycoprotein B sequences constitutes a powerful tool in the search for new herpesviruses, which in addition allows unambiguous classification of the newly detected sequences within the different Herpesvirinae subfamilies. Here, we report finding sequences of herpesviruses in all New World bat species tested and describe their phylogenetic relationships.
2. Results
2.1. Screening and characterization of partial sequences of herpesvirus DNA polymerase gene in bat samples
To look for the presence of herpesvirus sequences in our collection of bats, we attempted to amplify a fragment of the highly conserved herpesvirus DNA polymerase (DPol) gene from the PBMC DNA of each wild-caught bat using two different sets of degenerate primers and PCR conditions, as described previously (Ehlers et al., 1999; Rose et al., 1997; VanDevanter et al., 1996). A total of 195 samples from 11 bat species were tested (Table 1 ). DNA samples (39/195 = 20%) from 10 species scored positive after the nested PCRs (nPCRs) (Table 1). No bat belonging to the Pteronotus rubiginosus species scored positive. BLAST searches demonstrated that all of the sequences identified belonged to the Herpesviridae family. In addition, cloning of all PCR products and sequencing of multiple clones of each amplicon allowed detecting different herpesvirus sequences for six of them, for a total of 45 sequences. Then, sequence comparison and phylogenetic analyses revealed the presence of 14 distinct sequences. Three sequences belonged to the Betaherpesvirinae subfamily while the other 11 belonged to the Gammaherpesvirinae. Three distinct sequences, all gamma, were obtained from the D. rotundus individuals as well as two distinct gammaherpesvirus sequences from the Carollia perspicillata individuals. Informal virus names and abbreviations were given to the 14 distinct viruses, as explained in the Materials and Methods section (Table 1).
Table 1.
New World bat species tested for herpesviruses using molecular methods and survey results.
| O | Family | Subfamily | Species | Common name | Origin | N |
DNA Polymerase |
Glycoprotein B |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Informal name | Acronym | n | Informal name | Acronym | n | |||||||
| Chiroptera | Phyllostomidae | Desmodontinae | Desmodus rotundus | Common vampire bat | French Guiana | 22 | Desmodus rotundus Gammaherpesvirus 1 | DrotGHV1 | 10 | Desmodus rotundus Gammaherpesvirus A | DrotGHVA | 5 |
| Desmodus rotundus Gammaherpesvirus 2 | DrotGHV2 | 4 | Desmodus rotundus Gammaherpesvirus B | DrotGHVB | 3 | |||||||
| Desmodus rotundus Gammaherpesvirus 3 | DrotGHV3 | 3 | Desmodus rotundus Gammaherpesvirus C | DrotGHVC | 1 | |||||||
| Diaemus youngi | White-winged vampire bat | French Guiana | 8 | Diaemus youngi Gammaherpesvirus 1 | DyouGHV1 | 5 | Diaemus youngi Gammaherpesvirus A | DyouGHVA | 1 | |||
| Glossophaginae | Anoura geoffroyi | Geoffroy's tailless bat | French Guiana | 20 | Anoura geoffroyi Gammaherpesvirus 1 | AgeoGHV1 | 2 | Anoura geoffroyi Gammaherpesvirus A | AgeoGHVA | 1 | ||
| Carolliinae | Carollia perspicillata | Seba's short-tailed bat | French Guiana | 20 | Carollia perspicillata Gammaherpesvirus 1 | CperGHV1 | 2 | Carollia perspicillata Gammaherpesvirus A | CperGHVA | 2 | ||
| Carollia perspicillata Gammaherpesvirus 2 | CperGHV2 | 2 | ||||||||||
| Stenodermatinae | Artibeus planirostris | Flat-faced fruit-eating bat | French Guiana | 20 | Artibeus planirostris Gammaherpesvirus 1 | AplaGHV1 | 5 | |||||
| Sturnira tildae | Tilda's yellow-shouldered bat | French Guiana | 18 | Sturnira tildae Betaherpesvirus 1 | StilBHV1 | 2 | Sturnira tildae Gammaherpesvirus A | StilGHVA | 1 | |||
| Sturnira angeli | Martinique | 18 | Sturnira angeli Gammaherpesvirus 1 | SangGHV1 | 5 | Sturnira angeli Gammaherpesvirus A | SangGHVA | 4 | ||||
| Mormoopidae | Pteronotus rubiginosus | French Guiana | 15 | Pteronotus rubiginosus Gammaherpesvirus A | PrubGHVA | 1 | ||||||
| Pteronotus rubiginosus Gammaherpesvirus B | PrubGHVB | 2 | ||||||||||
| Pteronotus alitonus | French Guiana | 5 | Pteronotus alitonus Betaherpesvirus 1 | PaliBHV1 | 1 | |||||||
| Molossidae | Molossus coibensis | Coiban mastiff bat | French Guiana | 17 | Molossus coibensis Gammaherpesvirus 1 | McoiGHV1 | 2 | Molossus coibensis Gammaherpesvirus A | McoiGHVA | 1 | ||
| Molossus molossus | Pallas's mastiff bat | French Guiana | 12 | Molossus molossus Gammaherpesvirus 1 | MmolGHV1 | 1 | ||||||
| Molossus molossus | Pallas's mastiff bat | Martinique | 20 | Molossus molossus Betaherpesvrius 1 | MmolBHV1 | 1 | Molossus molossus Gammaherpesvirus A | MmolGHVA | 1 | |||
Abbreviations: O, order; N: number of tested animals; n: number of herpesvirus-positive animals (by PCR, cloning and sequencing) for either DNA polymerase or Glycoprotein B.
All obtained sequences were species-specific and all but the two sequences from Molossus molossus viruses, MmolBHV1 and MmolGHV1, and the one from Pteronotus alitonus, PaliBHV1, were characterized from at least two individuals (Table 1). Thus, DrotGHV1, DrotGHV2, and DrotGHV3 were detected from 10, four, and three out of 22 D. rotundus individuals, respectively while DyouGHV1, AplaGHV1, and SangGHV1 were each identified from five individuals. We also characterized a few cases of co-infections: two D. rotundus individuals were co-infected with DrotGHV1 and DrotGHV2 and three with DrotGHV1 and DrotGHV3 as well as one C. perspicillata individual infected with CperGHV1 and CperGHV2.
We then designed species-specific antisense primers to extend each of the newly identified sequences in upstream direction by about 300 bp with a one round of semi-nested semi-specific PCR (Supplementary Table 1). This resulted in concatenated nucleotide sequences of 439 and 454 bp in size for betaherpesviruses and of 472 bp in size for gammaherpesviruses.
2.2. Screening and characterization of partial sequences of herpesvirus Glycoprotein B gene in bat samples
We then attempted to amplify a 500-bp fragment of the Glycoprotein B (Gb) gene (Table 1). An amplification product of the expected size was obtained from 23 samples (23/195 = 11.8%) belonging to nine different bat species (Table 1). We did not obtain any amplification product from Artibeus planirostris, P. alitonus, and M. molossus (from French Guiana) individuals. BLAST searches of the obtained sequences followed by sequence comparison demonstrated that they all belonged to the Herpesviridae family and revealed the presence of 12 distinct sequences, all from the Gammaherpesvirinae subfamily. Among them, three different sequences were found in D. rotundus (DrotGHVA, DrotGHVB, and DrotGHVC) and two in P. rubiginosus (PrubGHVA and PrubGHVB). Each viral sequence detected was specific to a bat species and, even though some viruses were found in different individuals of the same species, we did not identify any case of co-infection (Table 1).
2.3. Pairwise sequence comparison of DPol sequences
Sequence identity was determined by comparing nucleotide and amino acid sequences of all herpesviruses detected in the different bat species with viruses of the same subfamily described in several mammalian orders (Supplementary Table 2 and Supplementary Table 3). For clarity, comparison of the percentage of identity between the different newly identified bat viruses with those of other bat species is reported by grouping viral sequences at the host family level and with those of other mammals according to the viral genus. All sequences obtained from the various bat species differed from each other at the nucleotide and amino acid levels. Sequences obtained from different specimens of the same species were 100% identical, with the exception of DrotGHV1 sequences showing 99.6–100% identity at the nucleotide level and DrotGHV3 sequences showing 99.2–100% nucleotide identity and 97.5–100% amino acid identity (Supplementary Table 3).
For betaherpesviruses, the three new sequences exhibited among themselves from 53.6% (StilBHV1 vs. PaliBHV1) to 65.4% (StilBHV1 vs. MmolBHV1) nucleotide identity and from 63.7% to 66.9% amino acid identity (Supplementary Table 2). They showed 48.8–67.3% nucleotide identity and 43.5–75% amino acid identity with the other bat betaherpesviruses. The three viruses were more distantly related to betaherpesviruses of Rhinolophidae than to those of Miniopteridae, Molossidae, and Vespertilionidae. Finally, they were more closely related to viruses of the Cytomegalovirus and Muromegalovirus genera than to those of the two other betaherpesvirus genera (Supplementary Table 2).
Considering gammaherpesviruses, the new sequences exhibited among themselves from 48.6% (DrotGHV2 vs. DyouGHV1) to 95.5% (McoiGHV1 vs. MmolGHV1) nucleotide identity and from 43.2% (DrotGHV1 vs. DrotGHV2) to 97.5% (McoiGHV1 vs. MmolGHV1) amino acid identity (Supplementary Table 3). The percentage of identity between the three viruses detected in D. rotundus ranged from 49% to 55.1% at the nucleotide level and from 43.2% to 58% at the amino acid level. The two sequences detected in C. perspicillata showed 62.9% and 64.2% identity in nucleotides and amino acids, respectively. Comparison with the other bat gammaherpesviruses yielded similar results, ranging from 44.1% to 86.9% nucleotide identity and from 40.7% to 91.4% amino acid identity. In addition, the level of nucleotide and amino acid identity with the other mammal gammaherpesviruses ranged from 42% to 69.4% and from 35.8% to 69.1%, respectively.
2.4. Pairwise sequence comparison of Gb sequences
Sequence identity of Gb sequences was determined by comparing sequences of all bat gammaherpesviruses identified as well as with those of other mammal gammaherpesviruses representative of the different gammaherpesvirus genera (Supplementary Table 4). All sequences obtained here from the different bat species differed from each other at the nucleotide and amino acid levels. Sequences obtained from several individuals of the same species were 100% identical with the exception of DrotGHVA sequences, showing 99.1–100% sequence identities at the nucleotide and amino acid levels, and PrubGHVB sequences showing 99.4% nucleotide identity and 98.3% amino acid identity (Supplementary Table 4). The percentage of identity between the three viruses detected in D. rotundus ranged from 62.1% to 85.5% at the nucleotide level and from 59.1% to 93% at the amino acid level, while the two distinct sequences detected in P. rubiginosus showed 56.4% and 54.8–55.7% nucleotide and amino acid identities, respectively.
Overall, the percentages of nucleotide and amino acid identities between the newly obtained sequences with those already published from bats or other mammals were higher or on the same order of magnitude as for DPol sequences (Supplementary Table 3 vs. Supplementary Table 4). Thus, the new sequences exhibited among them from 56.4% (PrubGHVA vs. PrubGHVB) to 92% (McoiGHVA vs. MmolGHVA) nucleotide identity and from 51.3% (CperGHVA vs. PrubGHVB) to 95.7% (McoiGHVA vs. MmolGHVA) amino acid identity (Supplementary Table 4). Comparison with the other chiropteran gammaherpesviruses showed from 48.7% to 93.7% nucleotide identity and from 41.7% to 98.3% amino acid identity. Finally, the level of nucleotide and amino acid identity with the other mammal gammaherpesviruses ranged from 49.3% to 69.8% and from 45.2% to 74.8%, respectively.
2.5. Phylogenetic analyses of betaherpesvirus DPol sequences
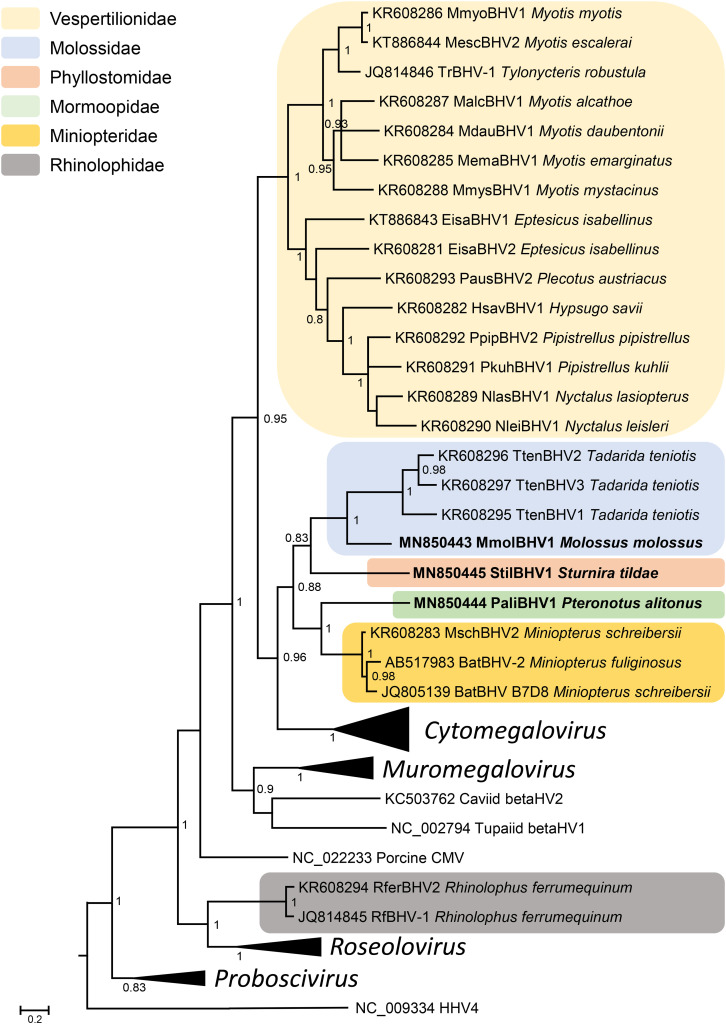
Phylogenetic analyses performed on nucleotide sequences of the newly characterized betaherpesvirus partial DPol sequences with those of other betaherpesviruses available in the databases grouped the bat viral sequences in three well-supported monophyletic clades only composed of bat virus sequences (Fig. 1 ). These monophyletic groups are distinct from the other groups of Betaherpesvirinae corresponding to the genera Cytomegalovirus, Muromegalovirus, Roseolovirus, and Proboscivirus. The first group of bat sequences is composed of two viral sequences detected in Rhinolophus ferrumequinum (Rhinolophidae) (Pozo et al., 2016; Wu et al., 2012). These viruses are closely related to viruses of the Roseolovirus genus. The two other lineages are close to the Cytomegalovirus genus. The first one is only composed of sequences derived from Vespertilionidae bats. In addition, this clade is divided into two different groups, (a) one containing sequences detected in bats belonging to the genus Myotis and the one identified from Tylonycteris robustula and (b) the second one containing viral sequences detected from the other Vespertilionidae genera (Nyctalus, Pipistrellus, Eptesicus, etc.). The third clade is composed of sequences of Molossidae, Phyllostomidae, Mormoopidae, and Miniopteridae. It is further subdivided into four well-supported lineages corresponding to the different families of the bats studied. The three betaherpesvirus sequences we characterized belong to this third clade. The sequence from M. molossus, MmolBHV1, is grouped with sequences from Tadarida in a clade of Molossidae viruses. The two others, StilBHV1 and PaliBHV1, obtained from bats of the Phyllostomidae and Mormoopidae families, respectively, are the only representative viruses of these bat families. StilBHV1 and PaliBHV1 belong to well-supported monophyletic lineages of Molossidae + Phyllostomidae and of Mormoopidae + Miniopteridae viruses, respectively.
Fig. 1.
Phylogenetic tree of betaherpesvirus DNA polymerase sequences. The phylogenetic tree was derived from the partial nucleotide sequences of the DNA polymerase gene (471 bp) of 84 representatives of betaherpesviruses using the Bayesian method with the GTR + I + G model of nucleotide evolution. Human herpesvirus 4 sequence (HHV4 NC_009334) served as outgroup. The tree is shown as a majority rule consensus tree. Support for nodes was provided by the posterior probabilities of the corresponding clades. All resolved nodes have posterior probability greater than 0.75. A scale indicating divergence, as substitutions per site, is at the foot. Sequences generated in this study are in boldface. The virus names are associated with their accession numbers. The Proboscivirus, Roseolovirus, Muromegalovirus, and Cytomegalovirus genera are collapsed for clarity, and the size of the collapsed clade is arbitrary. The collapsed Proboscivirus clade comprises sequences of Elephant endotheliotropic herpesviruses: EEHV1 (NC_020474), EEHV1B (HM568550), EEHV2 (HM568558), EEHV3 (JQ300065), EEHV4 (EU658934), EEHV5 (NC_024696), EEHV6 (HM060765) and EEHV7A (JQ300083). The Roseolovirus clade comprises sequences of MndHVβ (AF282942), PanHV6 (AY359407), HHV6A and HHV6B (NC_001664 and NC_000898, respectively), HHV7 (NC_001716), PtroHV7 (KJ843227 and KJ843228), PpanHV7 (KJ843230), GgorHV7 (KJ843231), and MneHV7 (NC_030200). The Muromegalovirus clade is composed of RatCMV/MuHV2/MuHV8 strains (AY728086, KP967684 and NC_019559), RexuCMV1 (EF125071), BindCMV3 (EF125067), MmusCMV2 (GU017485), MCMV/MuHV1 (AM886412 and NC_004065), MarvCMV1 (EF125059), MglaCMV1 (EF125061), AflaCMV2 (EF125063), and RatCMV Maastricht (NC_002512). Finally, viruses of the Cytomegalovirus collapsed clade are: AoHV1 (FJ483970), PpitCMV1 (KU963229), AsenCMV1 (KU963225), AmacCMV1 (KU963227), ApanCMV1 (KU963228), SalbCMV1 (KU963231), SaHV4 (FJ483967), CapeCMV1 (KU963230), CebHV1 (JQ264772), MndCMV (AY129399), BaCMV (NC_027016), CeHV5 strains (JQ264771 and FJ483968), MfasCMV1 strains (KP796148 and AY728171), McHV3 (AF033184 and DQ120516), CgueCMV1.1 (AY129397), GgorCMV2.1 (FJ538490), PnHV2 (AF480884), and HHV5 strains (M14709, AC146905 and AY315197).
2.6. Phylogenetic analyses of gammaherpesviruses DPol sequences
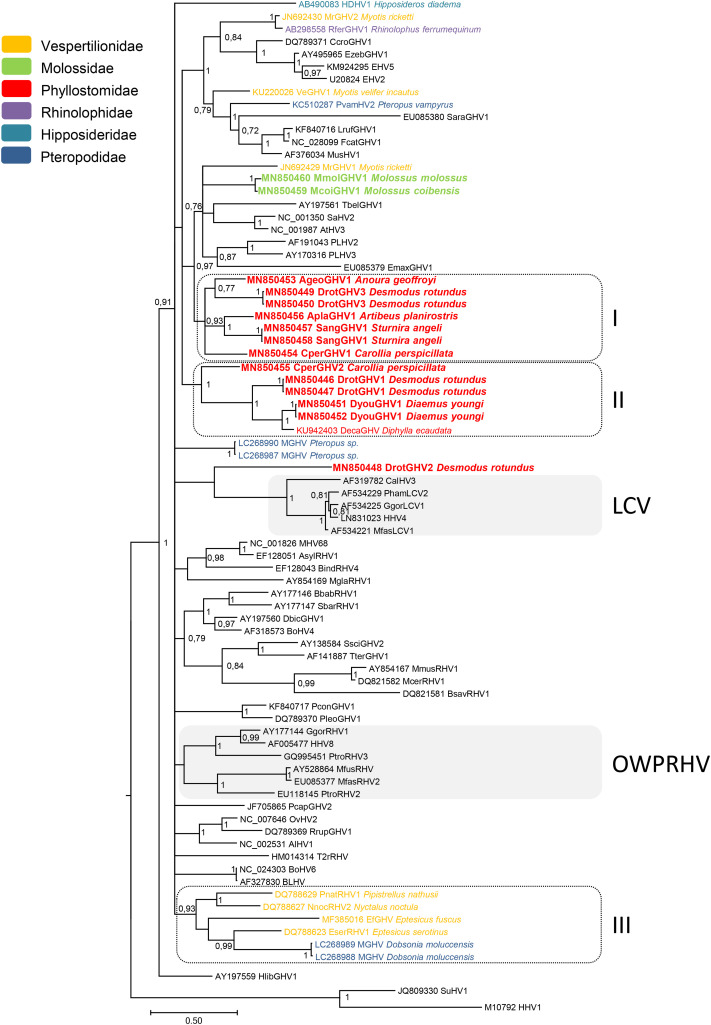
The phylogenetic analysis based on DNA polymerase sequences of Gammaherpesvirinae shows that bat viral sequences are distributed all over the tree (Fig. 2 ). Based on this sequence dataset, all that could be obtained is a multifurcating tree with low support for the deepest nodes. Therefore, resolution of the branching pattern is incomplete and phylogenetic relationships between the different Gammaherpesvirinae genera are not supported. Nevertheless, in the terminal branchings, certain highly supported clades can be recognized. Two of them correspond to the Lymphocryptovirus (LCV) and Old World primate Rhadinovirus (OWPRHV) genera. Three others are bat-specific (clades I, II, and III). Eight of the 11 new sequences reported here fall within two clades (clades I and II). These two clades only comprise viral sequences of Phyllostomidae. Clade I consists of five novel sequences (AgeoGHV1, DrotGHV3, AplaGHV1, SangGHV1, and CperGHV1 from A. geoffroyi, D. rotundus, A. planirostris, S. angeli, and C. perspicillata, respectively) representative of the four different subfamilies of Phyllostomidae tested. Clade II comprises three viral sequences infecting Desmodontinae bats: two new sequences, DrotGHV1 and DyouGHV1, from D. rotundus and Diaemus youngi, as well as the one previously described from Diphylla ecaudata (Diphylla ecaudata gammaherpesvirus SD12, accession number KU942403). Clade II also includes, at its basis, CperGHV2 from C. perspicillata. Clades I and II are highly supported with posterior probability (pp) values of 0.93 and 1, respectively. The third clade, clade III, supported by a pp. of 0.93, is constituted of previously published viral sequences detected from Vespertilionidae of different species and from one Pteropodidae species. Regarding the three other sequences we identified that do not belong to these well-supported clades, the third sequence from D. rotundus, DrotGHV2, clusters with sequences of the Lymphocryptovirus genus, although with a low support, while the two sequences from Molossus, MmolGHV1 and McoiGHV1, are grouped together in a separate lineage.
Fig. 2.
Phylogenetic tree of Gammaherpesvirus DNA polymerase sequences. The phylogenetic tree was derived from the partial nucleotide sequences of the DNA polymerase gene (489 bp) of 80 representatives of gammaherpesviruses using the Bayesian method with the GTR + I + G model of nucleotide evolution. Herpes simplex virus type 1 sequence (HHV1 M10792) served as outgroup. The tree is shown as a majority rule consensus tree. Support for nodes was provided by the posterior probabilities of the corresponding clades. All resolved nodes have posterior probability greater than 0.75. A scale indicating divergence, as substitutions per site, is at the foot. Sequences generated in this study are in boldface. The virus names are associated with their accession numbers. For bat viruses, the host species from which the virus has been detected is indicated after the virus name. In addition, they are color-coded according to the bat families. Abbreviations of virus names use the first letter of the generic host name in uppercase and the first three letters of the specific host name followed by either GHV (Gammaherpesvirus), LCV (Lymphocryptovirus), RHV (Rhadinovirus), or HV (Herpesvirus) and an Arabic numeral (1, 2, 3, 4) depending on the virus. Therefore, Rfer stands for Rhinolophus ferrumequinum, Ccro for Crocuta crocuta, Ezeb for Equus zebra, Pvam for Pteropus vampyrus, Sara for Sorex araneus, Lruf for Lynx rufus, Fcat for Felis catus, Tbel for Tupaia belangeri, Emax for Elephas maximus, Deca for Diphylla ecaudata, Pham for Papio hamadryas, Ggor for Gorilla gorilla, Mfas for Macaca fascicularis, Asyl for Apodemus sylvaticus, Bind for Bandicota indica, Mgla for Myodes glareolus, Bbab for Babyrousa babyrussa, Sbar for Sus barbatus, Dbic for Diceros bicornis, Ssci for Saimiri sciureus, Tter for Tapirus terrestris, Mmus for Mus musculus, Mcer for Mus cervicolor, Bsav for Bandicota savilei, Pcon for Puma concolor, Pleo for Panthera leo, Ptro for Pan troglodytes, Mfus for Macaca fuscata, Pcap for Procavia capensis, Rrup for Rupicapra rupicapra, and Hlib for Hexaprotodon liberiensis. Others that do not exactly follow this nomenclature are listed: HDHV1 stands for Hipposideros diadema herpesvirus 1, MrGHV1 and MrGHV2 for Myotis ricketti gammaherpesvirus 1 and 2, respectively, EHV2 and EHV5 for Equid herpesvirus 2 and 5, respectively, VeGHV1 for Vespertilionid Gammaherpesvirus 1, MusHV1 for Mustelid herpesvirus 1, SaHV2 for Saimiriine herpesvirus 2, AtHV3 for Ateline herpesvirus 3, PLHV2 and 3 for Porcine lymphotropic herpesvirus 2 and 3, respectively, MGHV for Megabat gammaherpesvirus, CalHV3 for Callitrichine herpesvirus 3, HHV4 and 8 for Human herpesvirus 4 and 8, MHV68 for Murine herpesvirus 68, BoHV4 and 6 for Bovine herpesvirus 4 and 6, respectively, OvHV2 for Ovine herpesvirus 2, AlHV1 for Alcelaphine herpesvirus 1, T2rRHV for Type 2 ruminant rhadinovirus, BLHV for Bovine lymphotropic herpesvirus, SuHV1 for Suid herpesvirus 1. Latin numerals I, II, and III on the right-hand side indicate the three well-supported bat gammaherpesvirus clades. LCV and OWPRHV on the right-hand side stand for Lymphocryptovirus and Old World primate Rhadinovirus.
2.7. Phylogenetic analyses of gammaherpesviruses Gb sequences
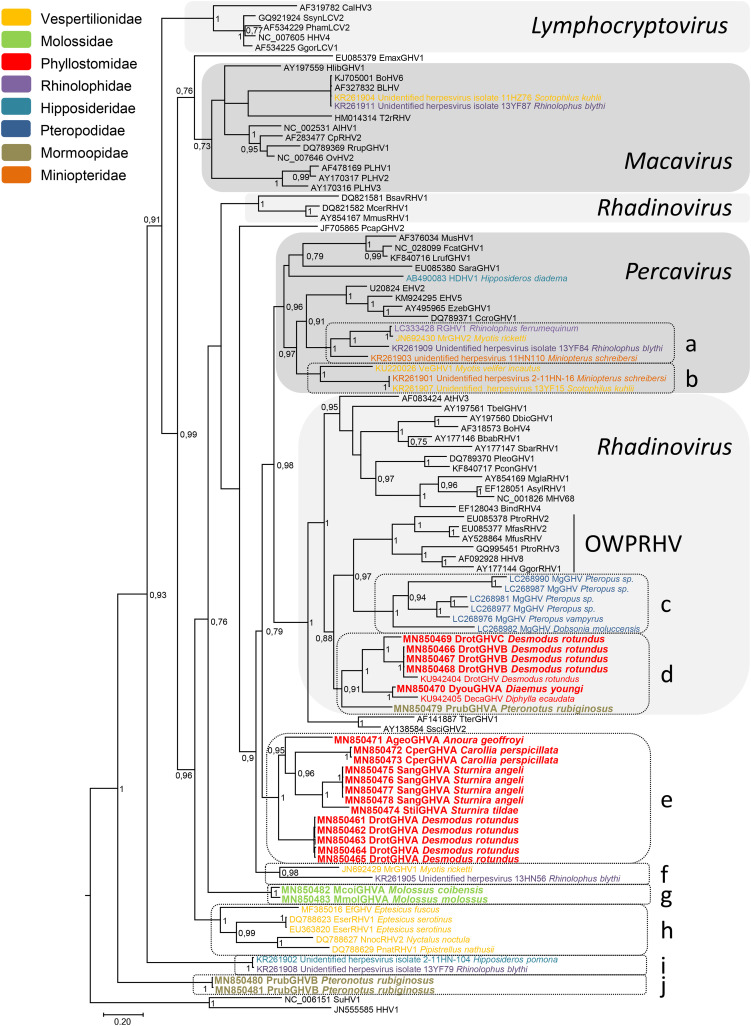
The phylogenetic analysis between the newly characterized bat gammaherpesvirus Gb sequences and that of already published sequences from bats and of representative gammaherpesvirus sequences available in the databases is presented in Fig. 3 . Almost all nodes of the tree are highly supported (pp > 0.7) and the main clades of Lymphocryptovirus, Rhadinovirus, Macavirus, and Percavirus genera can be recognized. This analysis clearly places the bat viral sequences into 10 well-supported (pp over 0.91) and bat-specific clades (named a to j) scattered over the entire tree. Five clades are only composed of sequences obtained from bat species of the same family (clades c, e, g, h, and j) while the other five comprise sequences from bats belonging to different families (clades a, b, d, f, and i). Some sequences do not belong to these clades: They correspond to previously published viral sequences from Scotophilus kuhlii and Rhinolophus blythi closely associated with bovine herpesvirus sequences (Bovine herpesvirus 6 and Bovine lymphotropic herpesvirus) that belong to the Macavirus genus and to the viral sequence derived from Hipposideros pomona that is associated with Sorex araneus gammaherpesvirus (SaraGHV1) and viral sequences from Carnivora (MusHV1, FcatGHV1 and LrufGHV1) but with a pp. value of less than 0.7 (Watanabe et al., 2009; Zheng et al., 2016). This last sequence and those of clades a and b from bat species of the Vespertilionidae, Rhinolophidae, and Miniopteridae families belong to a monophyletic well-supported (pp = 0.96) lineage of Percaviruses.
Fig. 3.
Phylogenetic tree of Gammaherpesvirus Glycoprotein B sequences. The phylogenetic tree was derived from the partial nucleotide sequences of the Glycoprotein B gene (464 bp) of 101 representatives of gammaherpesviruses using the Bayesian method with the GTR + I + G model of nucleotide evolution. Herpes simplex virus type 1 sequence (HHV1 JN555585) served as outgroup. The tree is shown as a majority rule consensus tree. Support for nodes was provided by the posterior probabilities of the corresponding clades. All resolved nodes have posterior probability greater than 0.7. A scale indicating divergence, as substitutions per site, is at the foot. Sequences generated in this study are in boldface. The virus names are associated with their accession numbers. For bat viruses, the host species from which the virus has been detected is indicated after the virus name. They are further color-coded according to bat families. Abbreviations of virus names follow the same system as for Fig. 2. SsynLCV2 stands for Symphalangus syndactylus lymphocryptovirus 2, CpRHV2 for Caprine herpesvirus 2, MusHV1 for Mustelid herpesvirus 1 and RGHV1 for Rhinolophus gammaherpesvirus 1. The different well-supported bat gammaherpesvirus clades are indicated by letters (a, b, …, j) on the right-hand side. OWPRHV on the right-hand side stands for Old World primate Rhadinovirus.
All new sequences detected here fall within four distinct clades (d, e, g, and j). Clade d is composed of sequences of Mormoopidae (PrubGHVA) and of Phyllostomidae of the Desmodontinae subfamily that includes viral sequences from D. rotundus, D. youngi, and D. ecaudata. Two distinct sequences from D. rotundus, DrotGHVB and DrotGHVC, belong to this clade. These two sequences are closely associated with the D. rotundus gammaherpesvirus sequence previously described from Mexico (Escalera-Zamudio et al., 2016). In addition, the sequence from D. youngi, DyouGHVA, is associated with the one from D. ecaudata (Escalera-Zamudio et al., 2016). Within this clade, viruses of Desmodontinae form a highly supported (pp = 1) subclade associated with PrubGHVA from P. rubiginosus. In addition, clade d belongs to a highly supported monophyletic lineage (pp = 0.88) also composed of two other clades, clade c of Pteropodidae bats and the Rhadinovirus clade of Old World primates (OWPRHV). Clade e comprises five newly identified Phyllostomidae viruses, AgeoGHVA from A. geoffroyi, CperGHVA from C. perspicillata, SangGHVA and StilGHVA, from S. angeli and S. tildae, respectively, and DrotGHVA from D. rotundus. The phylogenetic relationships between the different viral sequences of clade e are well correlated with the taxonomy of the bats and the different well-supported subclades are differentiated according to the subfamilies (Glossophaginae, Carolliinae, Stenodermatinae, and Desmodontinae) to which the infected bats belong. Indeed, viruses from Sturnira species (SangGHVA and StilGHVA) group together and are related to CperGVHA from C. perspicillata in a monophyletic clade with a pp. value of 0.96. These two clades are related to AgeoGHVA with a pp. value of 0.95. Finally, these Stenodermatinae, Carolliinae, and Glossophaginae clades group together with DrotGHVA of Desmodontinae in a monophyletic clade (clade e) supported with a pp. value of 1. The same phylogenetic clustering that correlated with the bat subfamilies from which the viruses have been detected is also observed for clade h of Vespertilionidae viruses. Clade e is located at the basis of a highly supported monophyletic lineage comprising Percavirus and Rhadinovirus sequences, with the exception of rhadinoviruses of Muridae (BsavRHV1, McerRHV1 and MmusRHV1), bat sequences of clades a–d, as well as the unclassified Tapirus terrestris and Saimiri sciureus gammaherpesviruses, TterGHV1 and SsciGHV2. From its side, clade g is composed of sequences from the two Molossidae species tested, M. molossus and M. coibensis. It possesses a basal position relative to a large clade comprising all percaviruses and rhadinoviruses including rhadinoviruses of Muridae as well as the unclassified Procavia capensis gammaherpesvirus, PcapGHV2, and clade f. Finally, clade j, composed of two sequences of PrubGHVB from P. rubiginosus, is located at the basis of the Gammaherpesvirinae subfamily.
3. Discussion
This is the largest study conducted to date to assess the occurrence and diversity of herpesviruses in New World bat species. A viral sequence was obtained for every tested species. Each obtained sequence was novel, species-specific and no case of cross-transmission between bat species was identified. Except for P. rubiginosus, for which we did not generate any DPol sequence, and A. planirostris and P. alitonus, for which no Gb sequence was amplified, the two types of sequences were obtained for all other species. Nevertheless, for the species from which both sequences were obtained, PCR targeting Gb was less sensitive than the one targeting DPol with a lower number of positive samples (Table 1). In addition, for Gb, only gammaherpesvirus sequences were obtained. These results prove that, to screen samples, the combinations of primers targeting DPol are a better tool both in terms of sensitivity and diversity of amplified sequences. This also suggests that for Gb other combinations of primers should be used to allow for amplification of sequences of other subfamilies. Nevertheless, the fact that we preferentially amplified gammaherpesvirus sequences is not only a question of primer degeneracy, but also depends on the type of samples tested. Indeed, of all the bat herpesviruses currently described in the literature, most alpha- and betaherpesviruses (globally underrepresented compared with gammaherpesviruses) were amplified from oral swabs (Pozo et al., 2016; Razafindratsimandresy et al., 2009). It will therefore be important for future studies to test not only other bat species but also different types of samples from the same species.
From a phylogenetic perspective, in view of the available data, it appears that differential clustering exists between bat betaherpesvirus sequences according to their host families (Fig. 1). In addition, these clusters are also congruent with the evolutionary relationships of most large groups within families (Hoofer and Bussche, 2003; Simmons, 2005; Teeling, 2005). Nevertheless, the phylogenetic relationships between them are not concordant with the current interpretations of the host pattern of diversification at the family level (Agnarsson et al., 2011). Even though the different clades are all well supported, interpretations are currently limited by the fact that only a few bat betaherpesvirus sequences are available and that only one betaherpesvirus sequence is available for certain families while others are not represented.
Regarding Gammaherpesvirinae, the phylogeny based on Gb sequences is well supported with well-defined clades corresponding to the different known genera. Comparatively, the topology of the DPol tree is less reliable and exhibits poor overall support. The two phylogenies are based on alignments of similar size, and thus the lower support observed for DPol can be attributed to an overall higher level of sequence divergence. Nevertheless, the two phylogenies show that bat gammaherpesviruses are scattered over the entire tree (Fig. 2, Fig. 3). They also demonstrate that viral sequences from Phyllostomidae are distributed in two distinct clades, while those of Molossidae fall together on a separate branch. Based on the Gb phylogeny, which is in agreement with the known phylogeny of Gammaherpesvirinae, we further observe that viruses of Phyllostomidae are close to the Rhadinovirus and Percavirus genera, while those of Molossidae possess a basal position relative to these genera. Finally, within the different clades (c, d, e, and h), composed of viral sequences obtained from bat species belonging to different subfamilies, the phylogenetic relationships of the different viral sequences demonstrate a good correlation with the taxonomy of the host species (Fig. 3) (Agnarsson et al., 2011). These results are in support of a co-evolutionary scenario. Nevertheless, each bat-specific clade is only represented by a few sequences from different species (2 < n < 5). Owing to the paucity of the available data, considering the number of bat species, genera, subfamilies, and families that remain to be tested, this assumption will only be confirmed when more sequences become available.
This study greatly expands our knowledge on the distribution and genetic variation of bat herpesviruses. It adds new insights into the viral diversity hosted by bats from French Guiana and Martinique and, as may be expected, confirms the astonishing diversity of bat herpesviruses. However, we are still far from having deciphered the in-depth details of this diversity. Indeed, the number of New World bat species tested still accounts for only a tiny part. In French Guiana alone, 107 bat species are currently recognized, with the number evolving steadily, mostly due to the splitting of taxa on the basis of new genetic evidence (Catzeflis, 2017; Catzeflis, 2015; Simmons and Voss, 1998). Therefore, the number of tested species accounts for just 10% of the local bats, suggesting a wide diversity of herpesviruses awaiting discovery. These results emphasize the crucial need for a better assessment of herpesvirus distribution in bats. Analysis of other species at a wider geographical scale, as well as of the same species but on other types of samples, should maximize our chances of detecting the whole diversity of bat herpesviruses and thereby expanding our understanding of the diversification processes and evolutionary history of these viruses.
4. Materials and methods
4.1. Ethical and legal statements
All animals were captured, handled, and sampled following ASM guidelines under the supervision of researchers granted the French animal experimentation level 1 diploma (Sikes and the Animal Care and Use Committee of the American Society of Mammalogists, 2016). For bats captured in Martinique, ad hoc authorization was received (# 2014 140–008) delivered by the Préfecture de Martinique. In French Guiana, bats are not protected by law, but captures that occurred within protected areas (nature reserves) received ad hoc authorizations (# 35 from 03/21/2013 and # 59 dated 04/17/2013 delivered by the Préfecture de la Guyane). In compliance with the Access and Benefit Sharing procedure implemented by the Loi pour la Reconquête de la Biodiversité, the use of the genetic resources was declared to the French Ministry of Environment under reference # TSP 128316, and received an internationally recognized certificate of compliance reference # ABSCH-IRCC-FR-246973-1.
4.2. Collection of specimens and biological material and DNA extraction
All bats examined in this study were collected as part of an investigation program on rabies virus circulation. In French Guiana, captures were implemented in bat communities during a 10-year period (de Thoisy et al., 2016). In Martinique, bats were captured in January 2015. All captures were performed at night with Japanese mist nets erected near breeding sites, roosts, at forest edges, around livestock, or through putative foraging courses. Animals were kept in individual bags before being sampled for blood with a sterile needle and capillary at the brachial vein. Before release, external pressure was exerted on the vein with a sterilized absorbent hemostatic sponge to prevent bleeding and facilitate healing. Blood samples were preserved at 4 °C until arrival at the laboratory and centrifuged at 6500g for 10 min to separate sera. Sera and buffy coat samples were stored at −80 °C for later use in the laboratory. The sex and age of all animals were recorded, and the animals were identified morphologically in the field. When possible errors of species identification were suspected, identification was molecularly confirmed by sequencing a fragment of the mitochondrial Cytochrome oxidase I or Cytochrome b genes (Borisenko et al., 2008). Nucleic acids were extracted using the NucliSENS easyMAG® bio-robot (bioMérieux®, Marcy l'Etoile, France). A total of 195 DNA samples from 11 bat species belonging to three families (Phyllostomidae, Mormoopidae, and Molossidae) were obtained (Table 1).
4.3. Screening of samples
Molecular screening was done by semi-nested or nested PCR amplifications with degenerate consensus primers targeting highly conserved amino acid (aa) motifs of the herpesvirus DNA polymerase and Glycoprotein B genes (Supplementary Table 1) (Prepens et al., 2007; Rose et al., 1997; VanDevanter et al., 1996). For the DNA polymerase amplification, two sets of primers targeting the same region of the gene but with different levels of degeneracy were used on each DNA sample in separate reactions for the first-round PCR (DFASA/GDTD1B or DFA + ILK/KG1) and second-round PCR (VYGA/GDTD1B or TGV/IYG) (Supplementary Table 1) (Rose et al., 1997; VanDevanter et al., 1996). For the Glycoprotein B amplification, one set of primers was used: 2759 s/2762as for the first-round PCR and 2760 s/2761as for the second-round PCR (66). All amplicons of approximately the expected size were purified, cloned via TA cloning, and sent for sequencing to Genewiz®, Takeley, UK (https://www.genewiz.com/). To help increase the possibility of identifying different herpesvirus sequences from each amplicon obtained, five to eight clones of the “screening amplicons” were sequenced on both strands.
4.4. Partial DNA polymerase gene amplification
To obtain the nucleotide sequence upstream of the VYGA or TGV motif, species-specific non-degenerate primers were derived from the complementary sequences of the small fragments and used in an nPCR amplification with the DFASA or DFA primer pools using the initial PCR products as templates (Supplementary Table 1). Overlapping amplicons were generated, cloned, and sequenced as described above. Each sequence corresponds to at least three independent clones sequenced on both strands. Contig sequences were then assembled using MEGA 5.05 software (Tamura et al., 2011). The concatenated nucleotide sequences are between 439 and 472 bp in size depending on the strain.
4.5. Phylogenetic analysis
Raw sequences were analyzed and edited in MEGA 5.05 (Tamura et al., 2011). Sequence homology analyses were performed using the BLAST program at the National Center of Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (48). Then, multiple sequence alignments were constructed using ClustalW with all other previously published bat herpesvirus sequences extracted from GenBank (http://www.ncbi.nlm.nih.gov/nucleotide) or DBatVir (http://www.mgc.ac.cn/DBatVir/) as well as representative sequences for each genus or subfamily. Alignments were checked manually. Nucleotide and amino acid sequences were checked for gaps. Pairwise sequence identity (at the nucleotide and amino acid levels) of the partial coding sequences was calculated with MEGA version 5.05 using uncorrected p-distances.
Phylogenetic trees were inferred from the aligned nucleotide sequences. The best-fitted model of nucleotide substitution (GTR + I + G) was determined for each dataset using MEGA 5.05 under corrected Akaike information criteria (AICc) and used for the Bayesian approach with Mr. Bayes 3.2.2 (49, 50). Markov Chain Monte Carlo (MCMC) simulations were run for 10,000,000 generations, with four simultaneous chains, using a sample frequency of 100 and a burn-in of 25,000. Majority rule consensus trees were obtained from the output. Validation of the inference was assessed based on the standard deviation of split frequencies, which was less than the expected threshold value of 0.01. Trees obtained were visualized using the FigTree v1.4 program (http://tree.bio.ed.ac.uk/software/figtree/) .
4.6. Informal naming of novel herpesviruses
For the readability of the paper, the novel viruses were named trinomially according to the host species from which they were detected and the viral subfamily they were assigned to. The first two words designate the name of the bat species in italics, while the third word designates the tentative assignment of the novel virus to a herpesvirus subfamily within the Herpesviridae. Then, a number (for DNA polymerase sequences) or a letter (for Glycoprotein B sequences) was given to differentiate different viruses obtained from the same host species and belonging to the same herpesvirus subfamily. Example: Desmodus rotundus Gammaherpesvirus 1. Abbreviations use the first letter of the generic host name in uppercase and the first three letters of the specific host name, followed by three uppercase letters corresponding to the viral subfamily (BHV or GHV for Betaherpesvirus or Gammaherpesvirus, respectively) to which they were then assigned the Arabic numeral (1, 2, etc.) for the DNA polymerase sequences or a letter in uppercase following alphabetical order (A, B, etc.) for the Glycoprotein B sequences. Example: Desmodus rotundus GammaHerpesVirus 1, DrotGHV1.
4.7. Accession numbers
The sequences reported in this paper were deposited in the GenBank database under accession numbers MN850443 to MN850483.
The following are the supplementary data related to this article.
Primers used for Glycoprotein B and DNA polymerase consensus and specific PCRs.
Nucleotide and amino acid identities between the three novel bat betaherpesviruses identified and other betaherpesviruses on the basis of DNA polymerase partial gene sequences.
Nucleotide and amino acid identities between the novel bat gammaherpesviruses identified and other gammaherpesviruses on the basis of DNA polymerase partial gene sequences.
Nucleotide and amino acid identities between the novel bat gammaherpesviruses identified and other gammaherpesviruses on the basis of Glycoprotein B partial gene sequences.
Funding
This study benefited from the CAROLIA and RESERVOIRS programs supported by European funds (ERDF/FEDER) and assistance from Région Guyane and Direction Régionale pour la Recherche et la Technologie. It also received a European Commission “REGPOT-CT-2011-285837-STRonGer” grant within the FP7 and “Investissement d'Avenir” grants managed by the Agence Nationale de la Recherche (CEBA, Ref. ANR-10-LABX-25-01). S.J. was supported by a grant from the Université de la Guyane, Ecole doctorale 587 “Diversités, Santé et Développement en Amazonie” and by a grant from the Collectivité Territoriale de la Guyane. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Authors' contribution
SJ and VL conceived the project; AL and BdT provided samples; SJ and DD performed the molecular and genetic analyses; SJ, AL, and VL performed the sequence and phylogenetic analyses; SJ, AL, and VL analyzed the data; SJ and VL wrote the paper. All authors read and commented on the manuscript. All authors read and approved the final manuscript.
Data statement
The sequences reported in this paper were deposited in the GenBank database under accession numbers MN850443 to MN850483.
Declaration of Competing Interest
The authors declare that they have no competing financial interests.
Acknowledgments
All field volunteers and owners and/or managers of capture sites are warmly acknowledged for their assistance in captures.
References
- Agnarsson I., Zambrana-Torrelio C.M., Flores-Saldana N.P., May-Collado L.J. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia) PLoS Curr. 2011;3 doi: 10.1371/currents.RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Epstein J.H., Murray K.A., Navarrete-Macias I., Zambrana-Torrelio C.M., Solovyov A., Ojeda-Flores R., Arrigo N.C., Islam A., Ali Khan S., Hosseini P., Bogich T.L., Olival K.J., Sanchez-Leon M.D., Karesh W.B., Goldstein T., Luby S.P., Morse S.S., Mazet J.A.K., Daszak P., Lipkin W.I. A strategy to estimate unknown viral diversity in mammals. mBio. 2013:4. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.S., Leggett R.M., Bexfield N.H., Alston M., Daly G., Todd S., Tachedjian M., Holmes C.E.G., Crameri S., Wang L.-F., Heeney J.L., Suu-Ire R., Kellam P., Cunningham A.A., Wood J.L.N., Caccamo M., Murcia P.R. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.S., Todd S., Marsh G.A., Crameri G., Barr J., Kamins A.O., Peel A.J., Yu M., Hayman D.T.S., Nadjm B., Mtove G., Amos B., Reyburn H., Nyarko E., Suu-Ire R., Murcia P.R., Cunningham A.A., Wood J.L.N., Wang L.-F. Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum. J. Virol. 2013;87:1348–1358. doi: 10.1128/JVI.01202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko A.V., Lim B.K., Ivanova N.V., Hanner R.H., Hebert P.D.N. DNA barcoding in surveys of small mammals communities: a field study in Suriname. Mol. Ecol. Resour. 2008;8:471–479. doi: 10.1111/j.1471-8286.2007.01998.x. [DOI] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catzeflis F. Liste des Mammifères de Guyane française (octobre 2015) 2015. http://www.kwata.net/medias/images/upload/MAMM-GUYANE_Catzeflis-oct2015.pdf
- Catzeflis F. Liste des Mammifères de Guyane française (novembre 2017) 2017. https://www.sfepm.org/pdf/MAMM-GUYANE_Catzeflis_mars2017.pdf
- Chen L., Liu B., Yang J., Jin Q. 2014. DBatVir: the Database of Bat-Associated Viruses. Database 2014, bau021–bau021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux L., Cervantes-Gonzalez M., Guigon G., Thiberge J.-M., Vandenbogaert M., Maufrais C., Caro V., Bourhy H. A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis P.L., Bourhy H., Holmes E.C. The evolutionary history and dynamics of bat rabies virus. Infect. Genet. Evol. 2006;6:464–473. doi: 10.1016/j.meegid.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Davison A.J., Eberle R., Ehlers B., Hayward G.S., McGeoch D.J., Minson A.C., Pellett P.E., Roizman B., Studdert M.J., Thiry E. The order herpesvirales. Arch. Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Thoisy B., Bourhy H., Delaval M., Pontier D., Dacheux L., Darcissac E., Donato D., Guidez A., Larrous F., Lavenir R., Salmier A., Lacoste V., Lavergne A. Bioecological drivers of rabies virus circulation in a neotropical bat community. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M., Kearney T., Seamark E.C.J., Paweska J.T., Markotter W. Synchronized shift of oral, faecal and urinary microbiotas in bats and natural infection dynamics during seasonal reproduction. R. Soc. Open Sci. 2018;5:180041. doi: 10.1098/rsos.180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three north American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Wegner T., Tateno A.F., Zerbinati R.M., Gloza-Rausch F., Seebens A., Müller M.A., Drosten C. Amplification of emerging viruses in a bat Colony. Emerg. Infect. Dis. 2011;17:449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Müller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Cottontail V.M., Rasche A., Yordanov S., Seebens A., Knörnschild M., Oppong S., Adu Sarkodie Y., Pongombo C., Lukashev A.N., Schmidt-Chanasit J., Stöcker A., Carneiro A.J.B., Erbar S., Maisner A., Fronhoffs F., Buettner R., Kalko E.K.V., Kruppa T., Franke C.R., Kallies R., Yandoko E.R.N., Herrler G., Reusken C., Hassanin A., Krüger D.H., Matthee S., Ulrich R.G., Leroy E.M., Drosten C. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B., Borchers K., Grund C., Frolich K., Ludwig H., Buhk H.-J. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes. 1999;18:211–220. doi: 10.1023/A:1008064118057. [DOI] [PubMed] [Google Scholar]
- Ehlers B., Dural G., Yasmum N., Lembo T., de Thoisy B., Ryser-Degiorgis M.P., Ulrich R.G., McGeoch D.J. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J. Virol. 2008;82:3509–3516. doi: 10.1128/JVI.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M., Rojas-Anaya E., Kolokotronis S.-O., Taboada B., Loza-Rubio E., Méndez-Ojeda M.L., Arias C.F., Osterrieder N., Greenwood A.D. Bats, primates, and the evolutionary origins and diversification of mammalian gammaherpesviruses. mBio. 2016;7 doi: 10.1128/mBio.01425-16. e01425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagrouch Z., Sarwari R., Lavergne A., Delaval M., de Thoisy B., Lacoste V., Verschoor E.J. Novel polyomaviruses in south American bats and their relationship to other members of the family Polyomaviridae. J. Gen. Virol. 2012;93:2652–2657. doi: 10.1099/vir.0.044149-0. [DOI] [PubMed] [Google Scholar]
- Fenton M. Science and the conservation of bats. J. Mammal. 1997;78:1–14. [Google Scholar]
- Freuling C.M., Beer M., Conraths F.J., Finke S., Hoffmann B., Keller B., Kliemt J., Mettenleiter T.C., Mühlbach E., Teifke J.P., Wohlsein P., Müller T. Novel lyssavirus in Natterer's bat, Germany. Emerg. Infect. Dis. 2011;17:1519–1522. doi: 10.3201/eid1708.110201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., Shi Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012;86:4620–4630. doi: 10.1128/JVI.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys M., Mortlock M., Weyer J., Bezuidt O., Seamark E.C., Kearney T., Gleasner C., Erkkila T.H., Cui H., Markotter W. A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- He B., Li Z., Yang F., Zheng J., Feng Y., Guo H., Li Y., Wang Y., Su N., Zhang F., Fan Q., Tu C. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz P.H., Lumsden L.F., Druce J., Legione A.R., Vaz P., Devlin J.M., Hufschmid J. Virus survey in populations of two subspecies of bent-winged bats (Miniopterus orianae bassanii and oceanensis) in South-Eastern Australia reveals a high prevalence of diverse herpesviruses. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofer S.R., Bussche R.A.V.D. Molecular phylogenetics of the chiropteran family vespertilionidae. Acta Chiropterologica. 2003;5:1–63. doi: 10.3161/001.005.s101. [DOI] [Google Scholar]
- Hu D., Zhu C., Wang Y., Ai L., Yang L., Ye F., Ding C., Chen J., He B., Zhu J., Qian H., Xu W., Feng Y., Tan W., Wang C. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jánoska M., Vidovszky M., Molnár V., Liptovszky M., Harrach B., Benkő M. Novel adenoviruses and herpesviruses detected in bats. Vet. J. 2011;189:118–121. doi: 10.1016/j.tvjl.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Kunz T.H., Braun de Torrez E., Bauer D., Lobova T., Fleming T.H. Ecosystem services provided by bats. Ann. N. Y. Acad. Sci. 2011;1223:1–38. doi: 10.1111/j.1749-6632.2011.06004.x. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Link to MERS virus underscores Bats' puzzling threat. Science. 2013;341:948–949. doi: 10.1126/science.341.6149.948. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Lai K.K.Y., Huang Y., Yip C.C.Y., Shek C.-T., Lee P., Lam C.S.F., Chan K.-H., Yuen K.-Y. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 2011;85:8819–8828. doi: 10.1128/JVI.02364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Délicat A., Paweska J.T., Gonzalez J.-P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li W. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y., Ge X., Zhang H., Zhou P., Zhu Y., Zhang Y., Yuan J., Wang L.-F., Shi Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H., Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Baucells A., Rocha R., Bobrowiec P., Bernard E., Palmeirim J., Meyer C. 2016. Field Guide to Amazonian Bats. Instituto Nacional de Pesquisas de Amazonia, Manaus, BR. [Google Scholar]
- López-Baucells A., Rocha R., Fernández-Llamazares Á. When bats go viral: negative framings in virological research imperil bat conservation. Mammal Rev. 2018;48:62–66. doi: 10.1111/mam.12110. [DOI] [Google Scholar]
- Luby S.P., Gurley E.S., Hossain M.J. Transmission of human infection with Nipah virus. Clin. Infect. Dis. 2009;49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S. Emerging zoonotic encephalitis viruses: lessons from Southeast Asia and Oceania. J. NeuroVirol. 2005;11:434–440. doi: 10.1080/13550280591002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár V., Jánoska M., Harrach B., Glávits R., Pálmai N., Rigó D., Sós E., Liptovszky M. Detection of a novel bat gammaherpesvirus in Hungary. Acta Vet. Hung. 2008;56:529–538. doi: 10.1556/AVet.56.2008.4.10. [DOI] [PubMed] [Google Scholar]
- Mühldorfer K., Speck S., Kurth A., Lesnik R., Freuling C., Müller T., Kramer-Schadt S., Wibbelt G. Diseases and causes of death in European bats: dynamics in disease susceptibility and infection rates. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K., Kuwata R., Shimoda H., Mizutani T., Hondo E., Maeda K. The complete genomic sequence of Rhinolophus gammaherpesvirus 1 isolated from a greater horseshoe bat. Arch. Virol. 2018 doi: 10.1007/s00705-018-4040-2. [DOI] [PubMed] [Google Scholar]
- Paglia A.P., da Fonseca G.A.B., Rylands A.B., Herrmann G., Aguiar L.M.S., Chiarello A.G., Leite Y.L.R., Costa L.P., Siciliano S., Kierulff M.C.M., Mendes S.L., da Tavares V.C., Mittermeier R.A., Patton J.L. 2nd ed. Occasional Papers in Conservation Biology. Conservation International; Arlington, VA: 2012. Annotated Checklist of Brazilian Mammals. [Google Scholar]
- Paige Brock A., Cortés-Hinojosa G., Plummer C.E., Conway J.A., Roff S.R., Childress A.L., Wellehan J.F., Jr. A novel gammaherpesvirus in a large flying fox (Pteropus vampyrus) with blepharitis. J. Vet. Diagn. Investig. 2013;25:433–437. doi: 10.1177/1040638713486645. [DOI] [PubMed] [Google Scholar]
- Pellett P., Davison A., Eberle R., Ehlers B., Hayward G., Lacoste V., Minson A., Nicholas J., Roizman B., Studdert M., Wang F. Virus Taxonomy; Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press; London, United Kingdom: 2011. Herpesvirales; pp. 99–107. [Google Scholar]
- Pozo F., Juste J., Vázquez-Morón S., Aznar-López C., Ibáñez C., Garin I., Aihartza J., Casas I., Tenorio A., Echevarría J.E. Identification of novel betaherpesviruses in Iberian bats reveals parallel evolution. PLoS One. 2016;11 doi: 10.1371/journal.pone.0169153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prepens S., Kreuzer K.A., Leendertz F., Nitsche A., Ehlers B. Discovery of herpesviruses in multi-infected primates using locked nucleic acids (LNA) and a bigenic PCR approach. Virol. J. 2007;4:84. doi: 10.1186/1743-422X-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafindratsimandresy R., Jeanmaire E.M., Counor D., Vasconcelos P.F., Sall A.A., Reynes J.-M. Partial molecular characterization of alphaherpesviruses isolated from tropical bats. J. Gen. Virol. 2009;90:44–47. doi: 10.1099/vir.0.006825-0. [DOI] [PubMed] [Google Scholar]
- Rose T.M., Strand K.B., Schultz E.R., Schaefer G., Rankin G.W., Thouless M.E., Tsai C.C., Bosch M.L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmier A., Tirera S., de Thoisy B., Franc A., Darcissac E., Donato D., Bouchier C., Lacoste V., Lavergne A. Virome analysis of two sympatric bat species (Desmodus rotundus and Molossus molossus) in French Guiana. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano K., Okazaki S., Taniguchi S., Masangkay J.S., Puentespina R., Eres E., Cosico E., Quibod N., Kondo T., Shimoda H., Hatta Y., Mitomo S., Oba M., Katayama Y., Sassa Y., Furuya T., Nagai M., Une Y., Maeda K., Kyuwa S., Yoshikawa Y., Akashi H., Omatsu T., Mizutani T. Detection of a novel herpesvirus from bats in the Philippines. Virus Genes. 2015;51:136–139. doi: 10.1007/s11262-015-1197-6. [DOI] [PubMed] [Google Scholar]
- Sasaki M., Setiyono A., Handharyani E., Kobayashi S., Rahmadani I., Taha S., Adiani S., Subangkit M., Nakamura I., Sawa H., Kimura T. Isolation and characterization of a novel alphaherpesvirus in fruit bats. J. Virol. 2014;88:9819–9829. doi: 10.1128/JVI.01277-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman R.S., Shrivastava S., Tsibane T., Attie O., Jayaprakash A., Mire C.E., Dilley K.E., Puri V., Stockwell T.B., Geisbert T.W., Sachidanandam R., Basler C.F. Isolation and characterization of a novel gammaherpesvirus from a microbat cell line. mSphere. 2016:1. doi: 10.1128/mSphere.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes R.S., the Animal Care and Use Committee of the American Society of Mammalogists . Vol. 97. 2016. 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education: JMAMMAL; pp. 663–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons N.B. Order chiroptera. In: Wilson D.E., Reeder D.M., editors. Mammal Species of the World: A Taxonomic and Geographic Reference. John Hopkins University Press; Baltimore, MD: 2005. pp. 312–525. [Google Scholar]
- Simmons N.B., Voss R.S. The mammals of Paracou, French Guiana, a Neotropical lowland rainforest fauna. Part 1, Bats. Bull. AMNH. 1998;237 (Paracou bats) [Google Scholar]
- Smith I., Wang L.-F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi S., Rapin N., Dorville N., Hill J.E., Town J., Willis C.K.R., Bollinger T.K., Misra V. Isolation, characterization and prevalence of a novel Gammaherpesvirus in Eptesicus fuscus , the North American big brown bat. Virology. 2018;516:227–238. doi: 10.1016/j.virol.2018.01.024. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Towner J.S., Amman B.R., Sealy T.K., Carroll S.A.R., Comer J.A., Kemp A., Swanepoel R., Paddock C.D., Balinandi S., Khristova M.L., Formenty P.B.H., Albarino C.G., Miller D.M., Reed Z.D., Kayiwa J.T., Mills J.N., Cannon D.L., Greer P.W., Byaruhanga E., Farnon E.C., Atimnedi P., Okware S., Katongole-Mbidde E., Downing R., Tappero J.W., Zaki S.R., Ksiazek T.G., Nichol S.T., Rollin P.E. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDevanter D.R., Warrener P., Bennett L., Schultz E.R., Coulter S., Garber R.L., Rose T.M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y., Sasaki M., Setiyono A., Handharyani E., Rahmadani I., Taha S., Adiani S., Latief M., Kholilullah Z.A., Subangkit M., Kobayashi S., Nakamura I., Kimura T., Orba Y., Sawa H. Detection of novel gammaherpesviruses from fruit bats in Indonesia. J. Med. Microbiol. 2018;67:415–422. doi: 10.1099/jmm.0.000689. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Ueda N., Iha K., Masangkay J.S., Fujii H., Alviola P., Mizutani T., Maeda K., Yamane D., Walid A., Kato K., Kyuwa S., Tohya Y., Yoshikawa Y., Akashi H. Detection of a new bat gammaherpesvirus in the Philippines. Virus Genes. 2009;39:90–93. doi: 10.1007/s11262-009-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Maeda K., Suzuki K., Ueda N., Iha K., Taniguchi S., Shimoda H., Kato K., Yoshikawa Y., Morikawa S., Kurane I., Akashi H., Mizutani T. Novel betaherpesvirus in bats. Emerg. Infect. Dis. 2010;16:986–988. doi: 10.3201/eid1606.091567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibbelt G., Kurth A., Yasmum N., Bannert M., Nagel S., Nitsche A., Ehlers B. Discovery of herpesviruses in bats. J. Gen. Virol. 2007;88:2651–2655. doi: 10.1099/vir.0.83045-0. [DOI] [PubMed] [Google Scholar]
- Wong S., Lau S., Woo P., Yuen K.-Y. Bats as a continuing source of emerging infections in humans. Rev. Med. Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray A.K., Olival K.J., Morán D., Lopez M.R., Alvarez D., Navarrete-Macias I., Liang E., Simmons N.B., Lipkin W.I., Daszak P., Anthony S.J. Viral diversity, prey preference, and bartonella prevalence in desmodus rotundus in Guatemala. EcoHealth. 2016;13:761–774. doi: 10.1007/s10393-016-1183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., Liu L., Xue Y., Wang J., Yang F., Zhang S., Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang Y., Zheng W., He B., Jiang T., Li Y., Xia L., Feng Y., Fan Q., Tu C. Metagenomic analysis of bat virome in several Chinese regions. Sheng wu gong cheng xue bao. 2013;29:586–600. [PubMed] [Google Scholar]
- Yob J.M., Field H., Rashdi A.M., Morrissy C., van der Heide B., Rota P., Bin Adzhar A., White J., Daniels P., Jamaluddin A., Ksiazek T. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001;7:439–441. doi: 10.3201/eid0703.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang H., Todd S., Tachedjian M., Barr J.A., Luo M., Yu M., Marsh G.A., Crameri G., Wang L.-F. A novel bat herpesvirus encodes homologues of major histocompatibility complex classes I and II, C-type lectin, and a unique family of immune-related genes. J. Virol. 2012;86:8014–8030. doi: 10.1128/JVI.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Qiu M., Chen S., Xiao J., Ma L., Liu S., Zhou J., Zhang Q., Li X., Chen Z., Wu Y., Chen H., Jiang L., Xiong Y., Ma S., Zhong X., Huo S., Ge J., Cen S., Chen Q. High prevalence and diversity of viruses of the subfamily Gammaherpesvirinae, family Herpesviridae, in fecal specimens from bats of different species in southern China. Arch. Virol. 2016;161:135–140. doi: 10.1007/s00705-015-2614-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.Y., Qiu M., Guan W.J., Li J.M., Chen S.W., Cheng M.J., Huo S.T., Chen Z., Wu Y., Jiang L.N., Chen Q. Viral metagenomics of six bat species in close contact with humans in southern China. Arch. Virol. 2018;163:73–88. doi: 10.1007/s00705-017-3570-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for Glycoprotein B and DNA polymerase consensus and specific PCRs.
Nucleotide and amino acid identities between the three novel bat betaherpesviruses identified and other betaherpesviruses on the basis of DNA polymerase partial gene sequences.
Nucleotide and amino acid identities between the novel bat gammaherpesviruses identified and other gammaherpesviruses on the basis of DNA polymerase partial gene sequences.
Nucleotide and amino acid identities between the novel bat gammaherpesviruses identified and other gammaherpesviruses on the basis of Glycoprotein B partial gene sequences.