Abstract
When it is desired to identify infectious agents involved in an outbreak of bovine respiratory disease, a variety of possible sampling methods may be used. For field use, the deep nasopharyngeal swab, transtracheal wash, and nonendoscopic bronchoalveolar lavage are most feasible. At present, bacterial culture and polymerase chain reaction testing are most commonly used to identify infectious agents. Interpretation of test results can be challenging, particularly for opportunistic pathogens. Evidence-based guidelines for precise interpretation of microbiologic tests results are lacking; however, approaches that have been practically useful for the management of bovine respiratory disease outbreaks are presented.
Keywords: Bronchoalveolar lavage, Calves, MALDI-TOF, PCR, Next-generation sequencing
Key points
-
•
Diagnostic tests to identify pathogens involved in respiratory diseases of cattle are increasingly used, predominantly driven by the need to rationalize antimicrobial use.
-
•
Several methods to sample the respiratory tract are available, of which a deep nasopharyngeal swab, transtracheal wash, and nonendoscopic bronchoalveolar lavage best fit practice. Each technique has advantages and disadvantages, and consensus regarding the best choice is not yet reached.
-
•
Next to microbial culture, for microbiologic diagnosis polymerase chain reaction is especially popular. Promising techniques for a rapid diagnosis are matrix-assisted laser desorption/ionization time of flight mass spectrometry and next-generation sequencing, which will become widely available in coming years.
-
•
It is still difficult to interpret identification of opportunistic pathogens, both at the individual and herd level. More research is needed before evidence-based guidelines can be developed.
Introduction
In 2012, Fulton and Confer1 warned that the speed of development of new laboratory diagnostics has outpaced clinicians’ ability to properly interpret test results. A range of diagnostic tests is now available, ranging from culture to polymerase chain reaction (PCR) to next-generation sequencing (NGS). However, there is still doubt regarding the clinical significance of some pathogens detected, and how to interpret a diagnostic test result depending on what sample was tested. In contrast to this is the increasing pressure on antimicrobial use and the need for veterinarians and farmers to use antimicrobials more rationally.2 The use of diagnostic support by laboratory analysis is one of the frequently mentioned cornerstones of antimicrobial stewardship programs.3 However, scientifically reasoned, the evidence that systematic use of laboratory diagnostics, especially the antibiogram, would result in selection of a different first-choice therapy compared with an empiric decision preferably following (evidence-based) guidelines, is limited, especially in cattle. Antimicrobial resistance in respiratory tract bacteria from cattle is present and varies highly between systems. Resistance levels are generally lower in closed dairy and beef herds, substantially higher in feedlots, and most worrisome in veal calf operations, where oral mass medication is frequently used.4 , 5 Although there is no doubt of the presence of resistance, and multiresistance, in respiratory bacteria from cattle, to what extent this results in therapy failure when following guidelines for antimicrobial therapy is poorly documented. In recent years, guidelines specifying first-line, second-line, and third-line antimicrobial choices for the different cattle diseases have been initiated in several European Union countries, including the Netherlands, Belgium, Denmark, Sweden, and Germany.2 , 6 , 7 However, the amount of literature reporting the clinical benefit of every antimicrobial-bacteria combination in highly variable field settings is currently very limited. Therefore, these guidelines mainly include the spectrum of the antimicrobial, pharmaceutical leaflet recommendations and follow classification of the importance of antimicrobials for human medicine of the World Health Organization.8 In contrast with human medicine, to the authors’ knowledge there are no extensive, sufficiently detailed, and large-scale studies available on therapy failure caused by antimicrobial resistance in cattle.
Regardless of the limitations mentioned earlier, diagnostics are more and more frequently used when addressing bovine respiratory disease (BRD). This increased use is understandable, because antimicrobial decision making for BRD still often involves a decision to use group therapy, and, in the current climate, mass medicating without any evidence of the need for this therapy will increasingly be criticized. The authors fully acknowledge the complexity of advising on the implementation and interpretation of diagnostic tests for BRD, given the huge gaps in the current knowledge.9 However, the need is urgent, and therefore this article provides a framework to assist practitioners and clinicians in their everyday decision-making process. This reasoning may not withstand time; this article does not represent a consensus of all leading experts, nor is the objective to provide a complete literature overview. This discussion reflects on the body site sampled, the test used, and the pathogen detected.
Types of samples and sampling procedures
The selected sampling site of the respiratory tract is of great importance for interpretation of the test result. Table 1 provides an overview of available sampling techniques, with their advantages and drawbacks. Descriptions of these techniques are available elsewhere (eg, deep nasopharyngeal swabs [DNSs] and nonendoscopic bronchoalveolar lavage [nBAL],10 transtracheal aspiration [TTA]/transtracheal wash [TTW],11 and endoscopic bronchoalveolar lavage [BAL]12; Fig. 1 ).
Table 1.
Overview of available sampling techniques of the respiratory tract of calves and cattle, with advantages and disadvantages
| Nasopharyngeal Swab | Transtracheal Wash or Transtracheal Aspirate | Nonendoscopic Bronchoalveolar Lavage | Endoscopic Bronchoalveolar Lavage | |
|---|---|---|---|---|
| Sampling Site | Nasopharyngeal mucosa | Tracheal bifurcation | Individual random lung lobe | Individual (or multiple) targeted lung lobes |
| Use | Single use, disposable | Single use, disposable, or multiple use, sterilizable | Multiple use, sterilizable | Multiple use, sterilizable |
| Representative for Lower Airways | ± | Yes | Yes, but controversial | Yes |
| Sampled Surface | <0.5 cm2 | 5–10 cm2 | >10 cm2 | >10 cm2 |
| Procedure Costs | − | ++ | + | ++++ |
| Estimated Procedure Time per Animal, Including Preparation (min) | < 1 | 10 | 1–10 | 10 |
| Contamination Risk from Nasal Passage | High | Absent | Moderate (protective sleeve or agar plug possible) | Low (protective sleeve or agar plug possible) |
| Difficulty of the Technique | − | + | + | ++ |
| Possible Complications |
|
|
|
|
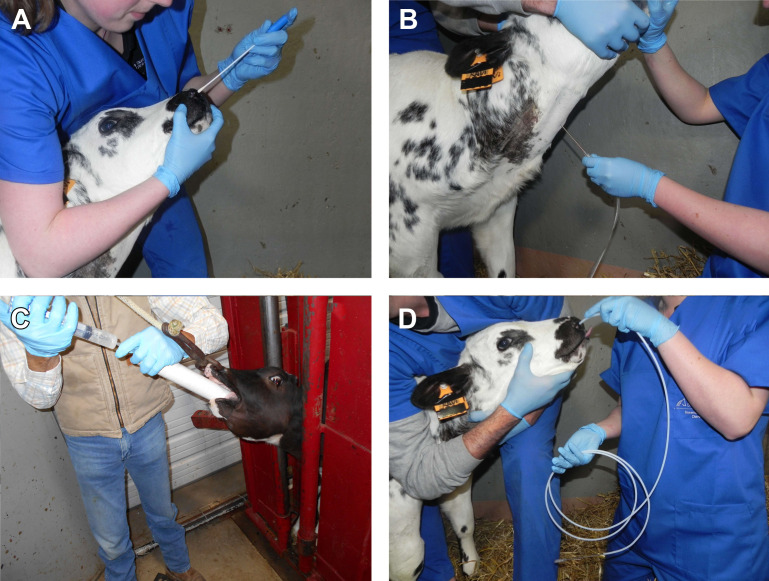
Fig. 1.
Overview of accessible sampling methods of the airways in cattle. (A) DNS, (B) TTW, (C) nBAL through the mouth under visual control; (D) nBAL performed blindly through the nose.
Nasal swabs, predominantly sampling the cutaneous part of the nose, are generally considered of limited value for infectious diagnostics. In contrast, DNSs sample the respiratory and associated lymphoid epithelium of the nasopharynx and return more meaningful samples. However, the biggest issue with nasopharyngeal swabs is the large number of polymicrobial samples recovered (>80%),10 which heavily compromises clinical interpretation when only opportunistic pathogens are retrieved. Contamination can be reduced by rinsing the nares (with a single-use paper towel or a gauze with alcohol) or by using a guarded DNS. However, studies specifically focusing on the effect of guarded swabs to reduce nasal contamination are, to the authors’ knowledge, not available in cattle. Recent reports on the respiratory microbiome in cattle also put the idea of contamination at that sampling site into another perspective, given the large variation in bacterial species normally present.13 , 14 The largest disadvantage of DNSs is that they do not directly sample the lower respiratory tract. Despite some conflicting results, previous studies overall showed that, for most pathogens, an association between DNS results and TTW or BAL is present.10 , 15, 16, 17 In addition to cotton swabs, brush swabs also exist, which cause more intensive swabbing of the mucosa (although possibly also blood staining of the sample), presumably with higher detection rates. No evidence on their benefit for use in cattle is currently available. Complications of DNS are rare and included nasal hemorrhage and fracture of the shaft of the swab. The latter is without any harmful consequence because the animal evacuates the remaining part of the swab either by sneezing or swallowing.
To overcome the issue of nasal contamination, transtracheal sampling techniques relying on perforation of the trachea with a needle or catheter after surgical preparation of the skin have been developed. Historically, transtracheal swabs have been used,18 but the transtracheal aspirate and wash are now common. Although an aspirate (TTA) only involves aspiration of mucus present in the respiratory tract, a wash (TTW) requires fluid instilment and immediate aspiration. Despite the terminology TTA being frequently used in the field, the technique usually used is a TTW. Most frequently, for TTW in cattle, TTW kits (Large Animal Trans-Tracheal Wash Kit, MILA International, Inc, Florence, KY), or human central venous catheters (eg. Centracath 75, Vygon, Ecouen, France) are used, which are commercially available and sterile packed for single use. Alternatively, a male dog urinary catheter can be used in combination with a 12-G catheter/needle to perforate the trachea in between 2 tracheal rings.19
In veterinary medicine, the common thinking is that the TTW is preferred for bacteriology and BAL to study inflammation (cytology). However, this recommendation generally comes from horse medicine, and seems to be expert opinion rather than supported by substantial peer-reviewed studies.20 In humans, TTW is generally not used for ethical reasons. The general idea is that the bronchial bifurcation is the site where the efflux of the mucociliary system of the whole of the lung comes together. Hence sampling there would be representative for the whole of the lung.20 However, there are some counterarguments for this reasoning. First, the mucociliary system can be heavily impaired by pneumonia. Second, microbial aspiration from the nasopharynx into the upper trachea is likely frequent. Third, normal pathogenesis involves gradual descent of bacteria down the respiratory tree toward the lung. Taking the second and third arguments into account, a positive TTW culture might equally represent a bacterial tracheitis or even an insignificant colonization or upper airway contamination, resulting in false positive diagnosis of infectious bronchopneumonia. Advantages are that a new disposable catheter can easily be used for each animal, and sampling is theoretically achievable within a predictable time frame given that no active cooperation of the animal is required, in contrast with BAL. However, sedation of the animal and local anesthesia of the puncture site can be done to improve animal comfort during the procedure.
In a BAL procedure, a BAL catheter or flexible endoscope is introduced through the nose and trachea into the lower airways until it wedges into a larger (or smaller depending on catheter diameter) bronchus. Next, while holding in this wedged position, a volume (usually 60 mL in calves, if necessary followed by a second or third injection) of sterile saline is injected and immediately aspirated. Classically, as in human medicine, a BAL is performed by endoscopy. The major advantage is that a specific lung lobe, previously shown to be affected on radiology or ultrasonography, can be sampled. Also, protective sheets or agar plugs can be used to reduce the risk of nasal contamination. The major disadvantage of the endoscope is the high operating costs and risk for equipment damage in the farm setting. Also, sampling multiple animals becomes difficult because time to resterilize the endoscope between animals is needed (15–20 minutes minimum).
To overcome the cost and risks of endoscopic BAL, nBAL techniques have been developed. In nBAL, a BAL catheter is blindly introduced through the nose, larynx, and trachea until the wedged position in a large bronchus is reached. Next, a volume of saline is injected and gently aspirated. The volume used varies substantially between studies (30–250 mL16 , 17), but a trend to reduce the volume for welfare/comfort reasons is present.21 On average, 33.5% of the volume (12.0%–73.8%) can be recovered in nBAL, which is substantially larger than in a TTW procedure.21 It is important to realize that sedation not only suppresses the required responses (coughing, curving of the nose, and extroversion of the tongue) to ensure an intratracheal position but also causes systematic sampling of the diaphragmatic lung lobes, which are less likely to be affected.22 Good restraint of the calf with the head fixed with the nose pointing upward as much as possible is advisable to ease blind introduction of the tube into the trachea. Alternatively, the calf might be surprised into allowing the tube to be advanced into the airways by placing the head in a horizontal position, and introducing the catheter on inspiration visible by the opening of the nostrils. Overall, in 80% of animals, nBAL sampling can be completed within minutes. For the remaining 20%, the practical advice is to select another animal to sample when undertaking group diagnosis, rather than spending excessive time and causing prolonged irritation to a reluctant animal. Alternatively, a technique where a double-guarded BAL catheter is orally introduced into the larynx through a PVC (polyvinyl chloride) speculum has been described for calves that are at least 3 to 4 months old, where the guarded catheter is inserted through the larynx under visual control.23
The use of BAL samples (especially nBAL) for bacteriology is still highly controversial, mainly because of the risk of nasal contamination. Although contamination is far less than with DNS, 20.8% of nBAL samples were still polymicrobial.10 A large influence of the sampler seems to be present,10 likely depending not only on differences in hygienic sample handling but also on skills to swiftly introduce the catheter without touching too much of the nasopharynx. However, it is important to realize that hard evidence on substantial nasal contamination by using nBAL catheters is currently not available in any species. The only available study on this matter showed pure culture and negative results in 29.2% and 40.3% of the nBAL samples, even though DNS samples of the same animals were polymicrobial.10 Further, the currently most extensive study on sample method comparison showed very good agreement for bacteriology between DNS, TTW, and nBAL.17 Interestingly, in human medicine, there are growing efforts toward the use of a mini-BAL procedure for bacterial diagnosis in ventilator-assisted pneumonia.24 Overall, sample contamination should be avoided and, in the case of nBAL, this can be done by adequate training or visualization of the larynx by a video speculum (Ivetscope, Dairymac Limited, Hampshire, United Kingdom) or endoscopic cameras intended for plumbers or auto mechanics. These devices are available at much lower prices than traditional endoscopes.
Next to the site of the respiratory tract sampled (upper or lower airway), the cultural perception of the effect of the sampling technique on animal welfare also plays an important role in what technique is currently preferred in a given country/region. No studies on the effect of respiratory tract sampling on stress or pain have yet been conducted in calves. A Master of Science thesis showed that both animals sampled by DNS or nBAL spent less time walking compared with the unsampled control group, whereas lying or eating were unaffected.25 For TTW as well as nBAL, the required volume of saline to be instilled is unclear; it is also unclear whether the volume instilled influences bacteriology results, as it does for cytology.26
In summary, sampling techniques for the field need to be economically feasible both in terms of equipment/disposables cost and also invested time. The DNS, TTW, and nBAL best suit this profile and are currently most frequently used in the field. Differences in use exist between countries, which mainly originate from historical or cultural preference.
Available diagnostic tests for the causal diagnosis of bovine respiratory disease
An overview of available diagnostic tests for pathogen identification in respiratory diseases in cattle is shown in Table 2 . It is beyond the scope of this article to provide a complete overview of all tests possible. The focus is on the most frequently used tests and the most promising future tests likely to become widely available for practice within the next 5 years. In the current international context, the pressure to reduce antimicrobial use has become the main driver of diagnostic test performance for causal diagnosis of BRD. A crucial aspect for field efficacy is a short turnaround time (TAT), the time between sampling and availability of the test result. In order to be able to use the diagnostic test result to target therapy or initiate control measures TAT needs to be as short as possible, ideally less than a day. However, having test results the next morning might also be workable for most outbreaks. Also, the use of cow-side testing for a causal diagnosis of BRD has great potential to reduce TAT. However, to the authors’ knowledge, no such tests are currently commercially available. Hence, attention should be given to ensuring proper and timely transport to the laboratory. At refrigerator temperature (4°C–8°C), the isolation rate of Mannheimia haemolytica and Pasteurella multocida was not reduced for 24 hours, whereas a transport temperature of more than 30°C resulted in reduced isolation as soon as 2 hours later.27
Table 2.
Advantages and disadvantages of available diagnostic tests to detect bacterial pathogens involved in respiratory disease in live cattle
| Use | Turnaround Time∗ | Advantages | Disadvantages | |
|---|---|---|---|---|
| Microbial Culture | Live bacteria detection |
|
|
|
| PCR | DNA detection (specific genomic region) |
|
|
|
| Serology (Antibody ELISA) | Antibody detection |
|
|
|
| Culture-enriched Direct MALDI-TOF | Live bacteria detection |
|
|
|
| Nanosequencing | DNA detection (whole genome) |
|
|
|
∗ Turnaround time is the time between arrival in the laboratory and availability of the test result. Reported times are in optimal conditions.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; MALDI-TOF, matrix-assisted laser desorption/ionization time of flight; MBT-ASTRA, MALDI Biotyper Antibiotic Susceptibility Test Rapid Assay; qPCR, quantitative PCR.
Serology
Serologic tests are useful to target vaccination programs, to determine protective status, and to evaluate infection dynamics at larger scale. However, they are not suitable to direct immediate therapy because they have a TAT of 3 weeks (required time for seroconversion) and only provide indirect evidence of infection. Also, for the opportunistic Pasteurellaceae family, maternal immunity smoothly shifts to acquired immunity, without any signs of disease or seroconversion.28 Another important issue is that sensitivity and specificity can be highly variable between different antibody enzyme-linked immunosorbent assays (ELISAs), hampering clinical interpretation and their use for individual animal decisions (eg, culling or purchase).29 For targeting therapy, direct identification of the pathogen is needed, and this can be achieved by microbial culture, matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (MS), PCR, or NGS/third-generation sequencing.
Microbial Culture
Microbial culture is most frequently used for identification of bacteria. Next to low operating costs, the possibility of antimicrobial susceptibility testing is an important advantage of culture. For mycoplasmata, specific media are required,29 and fastidious growers, especially Histophilus somni, are easily overgrown, resulting in false-negative results.10 , 30 Sensitivity and specificity of microbial culture have not been determined for most of the bacteria involved in BRD. A recent study using bayesian latent class analysis showed that Mycoplasma bovis culture on solid medium containing Tween 80 is 70.7% (95% bayesian credible intervals [BCI], 52.1 to 87.1) sensitive and 93.9% (95% BCI, 85.9–98.4) specific.31
Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
In the last decade, MALDI-TOF MS, which identifies bacteria by their unique protein profiles, has revolutionized routine diagnostics. It is primarily used for identification of bacteria after culturing, including Mycoplasma species.32 However, MALDI-TOF MS can also be applied directly on the sample after a very short period of enhanced growth in a liquid medium. Relative to classic microbial culture, these culture-enriched direct MALDI-TOF MS techniques allow correct bacterial identification in 73% of the samples (sensitivity = 59.1%; 95% confidence interval [CI], 47.2–71.0; specificity = 100% [100–100]) within 6 hours.33 The technique performed less well in polymicrobial samples and in samples with mixed infection. Also for M bovis, a culture-enriched direct MALDI-TOF MS technique was developed, which was 86.6% (95% BCI, 54–99) sensitive and 86.4% (95% BCI, 80–96) specific in a bayesian latent class model including PCR and microbial culture on solid agar.31 TAT was reduced from more than 5 days to less than 3 days.31 In addition, different MALDI-TOF MS methods are available for antimicrobial susceptibility testing. By means of MBT-ASTRA (MALDI Biotyper Antibiotic Susceptibility Test Rapid Assay), oxytetracycline resistance in P multocida could be identified with high accuracy (Se = 95.7%; 95% CI, 86.3–100.6; Sp = 100%; [95% CI, 100–100]) in as little as 3 hours, outperforming the disc diffusion antibiogram.34 The MBT-ASTRA technique can be designed for every bacterium-antibiotic combination, but logistical changes are needed to create a good intralaboratory workflow. The costs of MALDI-TOF procedures are generally low, in line with microbial culture.
Polymerase Chain Reaction
PCR for the causal diagnosis of BRD is now very popular. The main reasons are that multiplex PCR or multiple single PCRs allow detection of multiple bacteria and viruses, providing practitioners with a more extended view of the pathogens involved and hence more options to better target therapy, control, and prevention. Fastidious and metabolically active viable but unculturable viruses and bacteria can be detected, in contrast with standard microbial culture.35 However, in contrast with sequencing techniques, specific primers are needed, and the pathogen of interest needs to be determined beforehand. In this way, the diagnostics are potentially biased and possibly lead to false-negative results. Another problem is that viral genomes evolve rapidly and primers might become outdated, limiting the efficient detection of the pathogens of interest. PCR is generally not cheap, but, by pooling samples (DNS, TTW, or BALs), a group diagnosis can be reached and costs are decreased. In available studies, pools of samples from 5 animals were shown to improve diagnostic accuracy at the group level.36 , 37 The largest disadvantage of PCR is interpretative difficulty, because PCR can identify dead pathogens, opportunists currently not involved in infection, and contaminants, none of which signify a clinically meaningful test result. This disadvantage was shown, for example, for H somni.19 The use of quantitative PCR is more informative because the pathogen load, especially in the respiratory disease complex, is important to consider. For this reason, quantitative PCR is increasingly used in veterinary laboratories, although interpretative questions remain. Especially, when multiple pathogens are detected, determining the attributable fraction of each pathogen to the clinical presentation remains very difficult.38
Next-Generation and Third-Generation Sequencing
NGS technologies are now becoming more widely available because of the democratization of the technologies, and because platforms such as MinION (Oxford Nanopore Technologies, Oxford, UK) allow decentralized sequencing experiments. The first studies using NGS (metagenomics) to detect viruses involved in BRD in feedlots are already reported; these detected known pathogenic viruses as well as previously unknown or incompletely understood viruses (eg, influenza D virus).39 , 40 Hence, the advantage of NGS is that all pathogens can be simultaneously detected, without prior selection of which pathogens to test for. Also, semiquantitative results can be reported because, for most viruses, the number of reads corresponds to the initial load of the pathogen present.41 Not only viruses but also whole genomes can be recovered at a scale that is constantly increasing. Eventually, direct sequencing of bacteria will allow detection of virulence genes, phylogenetic clustering of strains during outbreaks, and ultimately prediction of antimicrobial resistance based on single nucleotide polymorphisms or resistance genes. A high total bacterial burden and low bacterial community diversity were associated with positive culture results in classic microbial culture.35 NGS is the basis for microbiome studies, which are discussed elsewhere in this issue.42 Disadvantages are that NGS is costly and requires a long TAT under the current conditions using the most accurate devices (eg, Illumina, San Diego, CA). However, with nanopore sequencing platforms (eg, MinION) a higher throughput and shorter TAT can be achieved. This long-read technology has been commercially available since 2014 and has made tremendous improvements in output and accuracy. In humans and pigs, MinION has been used to characterize pathogens in different types of samples, even at the site of disease outbreaks, because data analysis can be done in the field on portable hardware.43 , 44 On human lower respiratory tract samples, within 6 hours of sampling, a result was given at a sensitivity of 96.6%.45 However, in order to achieve wide implementation in veterinary practice, the cost, the ability to correctly interpret, and setup of an actionable logistic chain will be essential. Therefore, this technology is another case in which analysis of pooled samples from multiple animals to obtain a group diagnosis of primary pathogens will be the most likely application.
Interpretation of diagnostic test results and sampling strategy
Clinical interpretation of a diagnostic test result to determine the infectious cause of a respiratory tract disease requires information on the pathogen identified, the site of the respiratory tract sampled, the diagnostic test used, the clinical condition of the animal, and whether the sample originates from a single animal or is pooled. There is no current consensus on the way to sample the respiratory tract or to interpret diagnostic test results in humans and many other species. Based on the available research, it is unlikely that an evidence-based consensus on respiratory tract sampling method, diagnostic testing, and interpretation of results in cattle can be reached. Hence, this article assists readers to properly interpret results of testing by providing information not only on current recommendations but especially on the drawbacks and research gaps.
Detection of Primary Pathogens
The first point to consider is the nature of the pathogen retrieved, whether it is a primary or secondary pathogen. A primary or obligate pathogen (when present), per definition, induces damage to the respiratory tract, mostly followed by an inflammatory response. However, depending on the infectious dose and host immunity, infection might result in clinical disease or not. Also, certain primary pathogens can chronically and even asymptomatically infect animals, resulting in carriers: for example, Salmonella spp or M bovis. Most primary pathogens weaken innate immunity of the airways, facilitating superinfection by opportunistic bacteria. Some, such as bovine respiratory syncytial virus, bovine herpesvirus type 1 (BHV-1), and potentially also others (Table 3 ), are able to induce life-threatening disease without bacterial superinfection. Despite still being controversial in some scientific communities or countries, M bovis is generally considered a primary pathogen.46 Detection of a primary pathogen can, with some caution, be interpreted straightforwardly. The primary pathogen should normally not be present, and, depending on its virulence, it can, either as a sole agent or in combination with other agents, be held responsible for the clinical picture. Also, detection from any site of the respiratory tract is meaningful. For animal welfare reasons and following the pathogenesis, which starts with nasal infection, DNS samples might be sufficient and even most appropriate to detect primary pathogens. However, detection rates at the different sites of the respiratory tract differ between pathogens. For bovine coronavirus, DNS was more frequently positive than samples from the lower respiratory tract, whereas the inverse was true for bovine respiratory syncytial virus.17 For BHV-1, DNS is recommended given that the infection most frequently remains limited to the upper airways. The true interest of any diagnostic effort lies in extrapolation of test results from the sampled animals to the whole group. Detection of primary pathogens in some animals makes involvement of the same pathogen in the cohoused animals very likely.47 Hence, to improve sensitivity of the group diagnosis, the use of PCR on a pooled sample (up to 5 animals) can be considered.36 , 37 A pitfall when working with PCR to detect primary pathogens is that vaccine antigen can be detected up to 14 days after intranasal vaccination with a live vaccine, resulting in false-positives.48
Table 3.
Overview of viruses and bacteria commonly isolated from samples of the respiratory tract in cattle
| Pathogen | Primary or Secondary Pathogen | Remarks | Reference |
|---|---|---|---|
| Bovine adenovirus | Primary, but controversial | Widespread, but generally mild disease, except immunocompromised calves (types 3, 4, and 7) Type 10 associated with lethal enteritis |
58, 59, 60, 61 |
| Bovine coronavirus | Primary, but controversial | As a sole agent, experimentally only able to induce mild disease. Outbreaks with single viral infection resulting in severe morbidity and mortality described in calves and adult cattle | 62,63 |
| BHV-1 | Primary | Limited to the nasal cavity, pharynx, and trachea. Immunosuppression by hampering function and number of white blood cells. Potentially lethal as a single agent | 62 |
| Bovine rhinitis virus A and B | Likely apathogenic | — | 39 |
| Bovine respiratory syncytial virus | Primary | As a single viral agent, able to cause lethal bronchopneumonia. In older animals frequently subclinical | 52,62 |
| Bovine viral diarrhea virus | Primary | Mainly immunosuppression by hampering function and number of white blood cells. Potentially lethal as a single agent | 62,64 |
| Parainfluenza virus type 3 | Primary | As a single agent, generally mild disease | 52,62 |
| Influenza D virus | Controversial, likely primary | As a sole agent, experimentally only able to induce mild disease. Epidemiologically linked with disease | 39,65 |
| Bibersteinia trehalosi | Secondary | Occasionally isolated from cattle. More pathogenic role attributed to this bacterium in sheep | 66 |
| Histophilus somni | Controversial, likely secondary | Part of the resident flora. Septicemia is a lethal complication resulting in myocarditis, polyserositis, and thrombotic meningoencephalitis. Risk factors of septicemia unclear | 52 |
| Mannheimia haemolytica | Controversial, likely secondary | Part of the resident flora, differences in strain virulence described possibly resulting in some primary pathogenic strains. Other studies show cattle to become ill from their own resident strain on exposure to other pathogens and/or risk factors | 11,49,52 |
| Chlamydia psittaci | Controversial, likely primary | Natural infections result in mild or subclinical disease | 75 |
| M bovis | Primary | Extended immunosuppressive effect on white blood cells combined with immune-evasive mechanisms resulting in chronicity. Clonal spread of a strain limited in time and space is the general rule | 46,47 |
| Mycoplasma bovirhinis | Apathogenic | — | 19 |
| Mycoplasma dispar | Controversial, likely apathogenic | Recently shown to be more part of the microbiome of feedlot cattle classified as healthy | 14,19,67 |
| Moraxella bovis/ovis | Secondary | Primary eye pathogen, occasionally isolated in pure culture from animals with bronchopneumonia | 68 |
| Pasteurella multocida | Secondary | Part of the resident flora. Strain virulence differences exist, and some disease presentations (eg, septicemia or peritonitis) have been linked to certain strains | 52,69 |
| Salmonella spp | Primary | Primary site of infection of most Salmonella spp is the gastrointestinal tract. Localization in the respiratory tract is possible, most likely after septicemic spread | 70 |
| Trueperella pyogenes | Secondary | Involved in purulent processes. Often regarded as characteristic for chronicity. However, naturally resistant to fluoroquinolones | 71 |
| Escherichia coli, Gallibacterium anatis, Enterobacter hormaechei, staphylococci, streptococci, fungi | Secondary | Single reports on cattle-specific strains isolated in pure culture in an outbreak of pneumonia in calves | 52,72, 73, 74 |
Multiple other bacterial species can be detected in the bovine respiratory tract. This table is limited to either known primary pathogens or frequently isolated pathogens, currently assumed to have a pathogenic significance.
Detection of Secondary Pathogens
A secondary or opportunistic pathogen can be part of the normal respiratory microbiome, without inducing inflammation. In general, breaching of innate immunity, either by another pathogen or a noninfectious cause, is needed before the opportunistic pathogen invades tissues and induces inflammation. Interpretation of detection of an opportunistic pathogen is more difficult, given that they can be present in healthy animals.10 , 16 , 19 Therefore, simply detecting the pathogen cannot be seen as evidence of its involvement. The Pasteurellaceae family and a range of other bacteria (eg, Streptococcus spp and Trueperella pyogenes) are generally considered secondary pathogens. Although there seems to be little discussion of P multocida, scientific opinions on the potential primary role and differences in strain virulence of M haemolytica and H somni vary greatly.11 , 49 It is outside the scope of this article to review or take a position on this matter. Similarly, in other species, including humans, this issue of opportunistic pathogens exists. When interpreting a positive culture result, a differentiation between contamination, colonization, and infection needs to be made. Contamination is defined as the presence, usually in low numbers, of bacteria in a sample that are not expected to be present in the sampled site. Colonization can be defined as the presence of a micro-organism in a host, with growth and multiplication of the organism, but without interaction between host and organisms, hence no inflammatory reaction, immune response, or clinical expression occurs.50 Similarly, infection is isolation of a high number of bacteria from a site of the respiratory tract, but in the presence of inflammation of the mucosa, presenting either clinically or subclinically.50 Hence, simply picking a suspected colony from an agar plate, to confirm the cause of the respiratory disease, and subsequently using an antibiogram based on this single colony may be misleading. More information can be derived from culture results if quantitative descriptions and at least the degree of contamination are described. A possible way to better describe culture results, previously used for research purposes,10 , 33 is presented in Table 4 . It is also important to realize that using selective media for Pasteurellaceae, as, for example, by adding bacitracin, ensures better growth and detection of these opportunistic pathogens, but information on the amount of pathogens and degree of contamination of the sample will be lost.5 , 51
Table 4.
Overview of possible culture results for respiratory samples from cattle
| Observation | Interpretation | Explanation |
|---|---|---|
 |
Negative culture | No growth |
 |
Pure culture | Abundant growth of a single bacterial species |
 |
Dominant culture | Abundant growth of 1 bacterial species combined with a limited number of colonies from other bacteria (contaminants) |
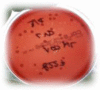 |
Mixed culture | Equal growth of 2 bacterial species (primary or secondary pathogens) |
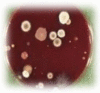 |
Polymicrobial culture | Growth of multiple bacterial species (and possibly molds), of which the most dominant ones are considered apathogenic for the host (contaminants or apathogenic flora) |
All cultures are on Columbia blood agar and derived from nonendoscopic bronchoalveolar lavage samples.
Images courtesy of Dr. L. Van Driessche, PhD, Merelbeke, Belgium.
Pasteurellaceae are part of the normal respiratory flora, and can even be abundantly present in the nasal cavity of healthy animals.52 An association between the presence of a Pasteurella species in the nose and its presence in the lower respiratory tract is described.10 , 15 , 16 However, interpretation of DNS results for opportunistic pathogens remains very difficult, especially because the composition of the nasopharyngeal microbiota seems to be heavily influenced by bioaerosols from the agricultural environment.53 Loss of biodiversity and overgrowth of opportunistic pathogens occurs in the pathogenesis of BRD, resulting in higher odds that Pasteurellaceae can be cultured from nasal swabs in larger quantities in ill animals.10 , 35 , 54 However, with current knowledge on the interpretation of DNS results at the individual or group level, samples of the lower respiratory tract are likely a better option to evaluate potential involvement of opportunistic pathogens. Interpretation of detection of opportunists in lower respiratory tract samples remains difficult, even in ill animals, because, even with very strict clinical case definitions, Pasteurellaceae can also be cultured from the lower airways in healthy animals.10 , 14 Previously explored ways to overcome the issue of interpreting detection of opportunistic pathogens in humans and other species are the use of quantitative cultures or cytologic evidence of inflammation. Quantitative culture is derived from the assumption that, in case of a severe infection, the opportunistic pathogen will be present in larger numbers.50 Cutoffs such as greater than 103 colony-forming units per milliliter of BAL fluid have been suggested in dogs and horses.20 , 55 However, pathogen burden builds up, and sampling early in the disease process could mean that much lower numbers are detected despite the pathogen being involved in the inflammatory process.56 Another option is derived from the assumption that a bacterial infection will result in a massive airway neutrophilia.55 However, clear cutoff percentages for neutrophilia differentiating a bacterial infection from a viral one or a strictly noninfectious airway inflammation have not been determined in calves. Given that some BAL techniques result in a larger contribution of the bronchial component in the total BAL fluid, the neutrophil percentage is increased compared with a larger volume of mainly alveolar lavage fluid.21 Primary insights in calf BAL fluid analysis show that cytologic parameters coincide poorly with clinical or ultrasonographical findings or culture results for opportunistic pathogens, at least when using nBAL.57 The presence of phagocytosis by bacteria in neutrophils or macrophages may be helpful to differentiate active infection versus simple presence of the bacteria.57 Although interpreting culture results for opportunistic pathogens is already difficult, interpretation of PCR results is even more so. Not only can insignificant quantities or even dead bacteria result in a positive PCR, the nasal passages also pick up bacterial DNA, making the lower respiratory tract sample positive. Quantitative PCR might overcome this issue, but, to the authors’ knowledge, no guidelines on how to interpret these results are currently available in the bovine species.
Sampling Strategy
Although laboratory costs are fixed, the return on investment of an analysis greatly depends on selection of appropriate animals to sample and on the technical sampling skills of the veterinarian. An animal in the first days of the disease, not previously treated with antimicrobials and not displaying severe respiratory signs, is first choice. By sampling in the acute phase of the disease, the odds of detecting the viral component are higher. By avoiding previous antimicrobial treatment and by sampling early in the disease course, the probability that the antibiogram derived is useful for empiric therapy increases. By avoiding sampling animals in heavy respiratory distress, the odds of aggravation of disease or even mortality can be decreased.
In addition, in spite of all reasoning made earlier, veterinarians still make decisions at an individual animal level. When sampling an individual, the main interest is usually to make a decision representing the group, and to judge the utility of the diagnostic test to support this group decision. Different epidemiologic approaches are possible to determine an appropriate sample size. The goal is more a detection of disease approach (being 95% confident that the pathogen was detected when present) rather than determining the prevalence of the pathogen in a group of animals. In the field, sample size is currently more driven by practical reasons such as available time to sample or maximum samples allowed to pool for economic reasons. For example, performing PCR on pools of 5 animals increases sensitivity without diluting the sample too much. Fig. 2 provides an overview on the risk of not finding a positive animal in 2 scenarios, 1 related to 100% prevalence of the pathogen in the diseased population and 1 scenario with a pretest probability that 70% of sick calves are affected by the pathogen (ie, where multiple pathogens are involved and can cause the same clinical disease). Results of a test with 70% sensitivity (ie, detects the pathogen in 7 of 10 infected calves) and 100% specificity (no false-positive calves) are presented. Using this test in a scenario with 70% of the affected animals being positive for the pathogen, after 5 calves not finding a positive, will misclassify only 3.5% of the herds (the 2 scenarios assume that the pooled tests’ accuracy is the same as the individual test). In the case of opportunistic pathogens, given that they can be found in healthy or subclinical animals, at this time it might be most prudent to only sample animals with evidence of clinical bronchopneumonia by using a combination of clinical scoring and thoracic ultrasonography.57 Focusing on bacterial isolation to direct intervention strategies, without taking the clinical status into account, holds great danger for overtreatment with antimicrobials.
Fig. 2.
Risk (probability ranging between 0 and 1) of not finding a positive animal for a given pathogen according to sample size (x axis) in a scenario where 100% (solid line) and 70% (dashed line) of affected animals are positive for a given pathogen. The graph represents a test with 70% sensitivity and 100% specificity. It assumes that there are no false-positive results (ie, when the test indicates that the pathogen is present, this is a true-positive result). In the example where the pathogen is causing the disease in 100% of affected calves, the risk of not finding an infected animal after sampling n cases is (1-Se)ˆn, where Se is the test sensitivity. In the alternative scenario where only 70% of cases are caused by the pathogen (ie, in 30% of cases, this is another cause), the probability of not finding a case is (1–0.7∗Se)ˆn.
Concluding remarks
Knowledge of respiratory health is rapidly evolving in animals, following new developments in humans. In particular, better insights into the role of the respiratory microbiome and the interaction of the airway inflammatory response with different organisms and air pollutants are likely to change how the diagnostic tests discussed in this article are interpreted. The authors hope that the information, tools, and provisional advice provided can aid the large group of cattle veterinarians, already having to make rational treatment decisions today.
Acknowledgments
Disclosure
B. Pardon has received honoraria for acting as speaker or consultant for pharmaceutical (Zoetis, MSD, Vetoquinol, Dopharma, Boehringer Ingelheim, Dechra, Hipra, Ceva, Merial, and Elanco), agricultural (Algoet nutrition), and chemical (Proviron) companies and nonprofit organizations (Boerenbond, AMCRA, DGZ-Vlaanderen). S. Buczinski has received honoraria for acting as speaker or consultant as well as research grants for pharmaceutical companies (Zoetis, MSD, Hipra, and Ceva) and companies involved in commercialization of ancillary tests used in respiratory diseases (EI Medical Imaging, Geissler Corp.).
References
- 1.Fulton R.W., Confer A.W. Laboratory test descriptions for bovine respiratory disease diagnosis and their strengths and weaknesses: gold standards for diagnosis, do they exist? Can Vet J. 2012;53:754–761. [PMC free article] [PubMed] [Google Scholar]
- 2.FVE (Federation of Veterinarians of Europe) FVE guidelines for responsible use of antibiotics. https://wwwfveorg/cms/wp-content/uploads/FVE_sheet_vet_doctor_dentist_EN_november_2014_webpdf 2019 Available at:
- 3.de With K., Allerberger F., Amann S. Strategies to enhance rational use of antibiotics in hospital: a guideline by the German Society for Infectious Diseases. Infection. 2016;44:395–439. doi: 10.1007/s15010-016-0885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catry B., Dewulf J., Maes D. Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS One. 2016;11:e0146488. doi: 10.1371/journal.pone.0146488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timsit E., Hallewell J., Booker C. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet Microbiol. 2017;208:118–125. doi: 10.1016/j.vetmic.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 6.AMCRA Classification of antimicrobials: methods. 2019. https://formularium.amcra.be/classification.php Available at:
- 7.KNMVD Procedures for the development of formularies for responsible antimicrobial use. 2015. https://www.knmvd.nl/app/uploads/sites/4/2018/09/150209-procedure-opstellen-formularia-definitief.pdf Available at:
- 8.WHO (World Health Organization) Critically important antimicrobials for human medicine, 6th revision. 2019. https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf?ua=1 Available at:
- 9.Buczinski S., Pardon B. BRD diagnosis: what progress have we made in clinical diagnosis? Vet Clin North Am Food Anim Pract. 2020;36(2):399–423. doi: 10.1016/j.cvfa.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Van Driessche L., Valgaeren B.R., Gille L. A deep nasopharyngeal swab versus nonendoscopic bronchoalveolar lavage for isolation of bacterial pathogens from preweaned calves with respiratory disease. J Vet Intern Med. 2017;31:946–953. doi: 10.1111/jvim.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timsit E., Christensen H., Bareille N. Transmission dynamics of Mannheimia haemolytica in newly-received beef bulls at fattening operations. Vet Microbiol. 2013;161:295–304. doi: 10.1016/j.vetmic.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 12.Viso M., el Jaraki M.R., Espinasse J. A sequential broncho-alveolar washing in non-anaesthetized normal bovines: method and preliminary results. Vet Res Commun. 1985;9:213–219. doi: 10.1007/BF02215144. [DOI] [PubMed] [Google Scholar]
- 13.McMullen C., Orsel K., Alexander T.W. Comparison of the nasopharyngeal bacterial microbiota of beef calves raised without the use of antimicrobials between healthy calves and those diagnosed with bovine respiratory disease. Vet Microbiol. 2019;231:56–62. doi: 10.1016/j.vetmic.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 14.Timsit E., Workentine M., van der Meer F. Distinct bacterial metacommunities inhabit the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bronchopneumonia. Vet Microbiol. 2018;221:105–113. doi: 10.1016/j.vetmic.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Godinho K.S., Sarasola P., Renoult E. Use of deep nasopharyngeal swabs as a predictive diagnostic method for natural respiratory infections in calves. Vet Rec. 2007;160:22–25. doi: 10.1136/vr.160.1.22. [DOI] [PubMed] [Google Scholar]
- 16.Allen J.W., Viel L., Bateman K.G. The microbial flora of the respiratory tract in feedlot calves: associations between nasopharyngeal and bronchoalveolar lavage cultures. Can J Vet Res. 1991;55:341–346. [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle D., Credille B., Lehenbauer T.W. Agreement among 4 sampling methods to identify respiratory pathogens in dairy calves with acute bovine respiratory disease. J Vet Intern Med. 2017;31:954–959. doi: 10.1111/jvim.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heckert H.P., Rohn M., Hofmann W. Sample collection for diagnostics of bovine respiratory diseases. Prakt Tierarzt. 1997;78:1056. [Google Scholar]
- 19.Angen O., Thomsen J., Larsen L.E. Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet Microbiol. 2009;137:165–171. doi: 10.1016/j.vetmic.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgson JL. Collection and interpretation of tracheal wash and bronchoalveolar lavage for diagnosis of infectious and non-infectious lower airway disorders. In: Proceedings of the 9th International Congress of WEVA. Marrakech, Morocco, 2006. p. 71–7.
- 21.van Leenen K., Van Driessche L., De Cremer L. Factors associated with lung cytology as obtained by non-endoscopic broncho-alveolar lavage in group-housed calves. BMC Vet Res. 2019;15:167. doi: 10.1186/s12917-019-1921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Driessche L., Valgaeren B., De Schutter P. Effect of sedation on the intrapulmonary position of a bronchoalveolar lavage catheter in calves. Vet Rec. 2016;179 doi: 10.1136/vr.103676. [DOI] [PubMed] [Google Scholar]
- 23.Allen T.H., Johnson E.G., Edmonds M.D. Influence of tilmicosin on quantified pulmonary concentrations of three bacterial pathogens in calves with naturally-occurring bovine respiratory disease. Bovine Pract. 2013;47:65–72. [Google Scholar]
- 24.Lavigne M.C. Nonbronchoscopic methods [Nonbronchoscopic Bronchoalveolar Lavage (BAL), Mini-BAL, Blinded Bronchial Sampling, Blinded Protected Specimen Brush] to investigate for pulmonary infections, inflammation, and cellular and molecular markers: a narrative review. Clin Pulm Med. 2017;24:13–25. [Google Scholar]
- 25.Roelants B. Ghent University; 2018. Comparison of deep nasopharyngeal swab, bronchoalveolar lavage and transtracheal wash for the diagnosis of infectious bronchopneumonia in calves: which one is most animal friendly? Master students thesis.https://lib.ugent.be/catalog/rug01:002481486 Available at: [Google Scholar]
- 26.Orard M., Depecker M., Hue E. Influence of bronchoalveolar lavage volume on cytological profiles and subsequent diagnosis of inflammatory airway disease in horses. Vet J. 2016;207:193–195. doi: 10.1016/j.tvjl.2015.09.027. [DOI] [PubMed] [Google Scholar]
- 27.Van Driessche L. Ghent University; Merelbeke (Belgium): 2019. It’s about time. Rapid detection and susceptibility testing of Pasteurellaceae causing respiratory disease in cattle by MALDI-TOF MS [PhD thesis] pp. 1–279. [Google Scholar]
- 28.Prado M.E., Prado T.M., Payton M. Maternally and naturally acquired antibodies to Mannheimia haemolytica and Pasteurella multocida in beef calves. Vet Immunol Immunopathol. 2006;111:301–307. doi: 10.1016/j.vetimm.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Parker A.M., Sheehy P.A., Hazelton M.S. A review of mycoplasma diagnostics in cattle. J Vet Intern Med. 2018;32:1241–1252. doi: 10.1111/jvim.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn P.F., Carter M.E., Markey B. Haemophilus species. In: Quinn P.J., editor. Clinical veterinary microbiology. Vol. 35. Mosby International Limited; London: 1994. pp. 121–128. [Google Scholar]
- 31.Bokma J, Van Driessche L, Deprez P, et al. Rapid identification of mycoplasma bovis from bovine bronchoalveolar lavage fluid with MALDI-TOF MS after enrichment procedure. J Clin Microbiol 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 32.Spergser J., Hess C., Loncaric I. Matrix-assisted laser desorption ionization-time of flight mass spectrometry is a superior diagnostic tool for the identification and differentiation of Mycoplasmas isolated from animals. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.00316-19. [pii:e00316-19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Driessche L., Bokma J., Deprez P. Rapid identification of respiratory bacterial pathogens from bronchoalveolar lavage fluid in cattle by MALDI-TOF MS. Sci Rep. 2019;9:18381. doi: 10.1038/s41598-019-54599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Driessche L., Bokma J., Gille L. Rapid detection of tetracycline resistance in bovine Pasteurella multocida isolates by MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA) Sci Rep. 2018;8:13599. doi: 10.1038/s41598-018-31562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson R.P., Erb-Downward J.R., Prescott H.C. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52:3605–3613. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Neill R., Mooney J., Connaghan E. Patterns of detection of respiratory viruses in nasal swabs from calves in Ireland: a retrospective study. Vet Rec. 2014;175:351. doi: 10.1136/vr.102574. [DOI] [PubMed] [Google Scholar]
- 37.Pardon B., Callens J., Maris J. Pathogen-specific risk factors in acute outbreaks of respiratory disease in calves. J Dairy Sci. 2020:2556–2566. doi: 10.3168/jds.2019-17486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deloria Knoll M., Fu W., Shi Q. Bayesian estimation of pneumonia etiology: epidemiologic considerations and applications to the pneumonia etiology research for child health study. Clin Infect Dis. 2017;64:S213–S227. doi: 10.1093/cid/cix144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra N., Cernicchiaro N., Torres S. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J Gen Virol. 2016;97:1771–1784. doi: 10.1099/jgv.0.000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M., Hill J.E., Fernando C. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound Emerg Dis. 2019;66:1379–1386. doi: 10.1111/tbed.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conceicao-Neto N., Zeller M., Lefrere H. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci Rep. 2015;5:16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timsit E., McMullen C., Amat S. Respiratory bacterial microbiota in cattle: from development to modulation to enhance respiratory health. Vet Clin North Am Food Anim Pract. 2020;36(2):297–320. doi: 10.1016/j.cvfa.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Quick J., Grubaugh N.D., Pullan S.T. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theuns S., Vanmechelen B., Bernaert Q. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Sci Rep. 2018;8:9830. doi: 10.1038/s41598-018-28180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charalampous T., Kay G.L., Richardson H. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 46.Maunsell F.P., Woolums A.R., Francoz D. Mycoplasma bovis infections in cattle. J Vet Intern Med. 2011;25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 47.Timsit E., Arcangioli M.A., Bareille N. Transmission dynamics of Mycoplasma bovis in newly received beef bulls at fattening operations. J Vet Diagn Invest. 2012;24:1172–1176. doi: 10.1177/1040638712463211. [DOI] [PubMed] [Google Scholar]
- 48.Timsit E., Le Drean E., Maingourd C. Detection by real-time RT-PCR of a bovine respiratory syncytial virus vaccine in calves vaccinated intranasally. Vet Rec. 2009;165:230–233. doi: 10.1136/vr.165.8.230. [DOI] [PubMed] [Google Scholar]
- 49.Klima C.L., Alexander T.W., Hendrick S. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can J Vet Res. 2014;78:38–45. [PMC free article] [PubMed] [Google Scholar]
- 50.Dani A. Colonization and infection. Cent European J Urol. 2014;67:86–87. doi: 10.5173/ceju.2014.01.art19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catry B., Haesebrouck F., Vliegher S.D. Variability in acquired resistance of Pasteurella and Mannheimia isolates from the nasopharynx of calves, with particular reference to different herd types. Microb Drug Resist. 2005;11:387–394. doi: 10.1089/mdr.2005.11.387. [DOI] [PubMed] [Google Scholar]
- 52.Griffin D., Chengappa M.M., Kuszak J. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract. 2010;26:381–394. doi: 10.1016/j.cvfa.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Mbareche H., Veillette M., Pilote J. Bioaerosols play a major role in the nasopharyngeal microbiota content in agricultural environment. Int J Environ Res Public Health. 2019;16 doi: 10.3390/ijerph16081375. [pii:E1375] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stroebel C., Alexander T., Workentine M.L. Effects of transportation to and co-mingling at an auction market on nasopharyngeal and tracheal bacterial communities of recently weaned beef cattle. Vet Microbiol. 2018;223:126–133. doi: 10.1016/j.vetmic.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Peeters D.E., McKiernan B.C., Weisiger R.M. Quantitative bacterial cultures and cytological examination of bronchoalveolar lavage specimens in dogs. J Vet Intern Med. 2000;14:534–541. doi: 10.1892/0891-6640(2000)014<0534:qbcace>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 56.Ackermann M.R., Gallup J.M., Zabner J. Differential expression of sheep beta-defensin-1 and -2 and interleukin 8 during acute Mannheimia haemolytica pneumonia. Microb Pathog. 2004;37:21–27. doi: 10.1016/j.micpath.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 57.van Leenen K., Van Driessche L., De Cremer L. Comparison of bronchoalveolar lavage fluid bacteriology and cytology in calves classified based on combined clinical scoring and lung ultrasonography. Prev Vet Med. 2020;176:104901. doi: 10.1016/j.prevetmed.2020.104901. [DOI] [PubMed] [Google Scholar]
- 58.Vaatstra B.L., Tisdall D.J., Blackwood M. Clinicopathological features of 11 suspected outbreaks of bovine adenovirus infection and development of a real-time quantitative PCR to detect bovine adenovirus type 10. N Z Vet J. 2016;64:308–313. doi: 10.1080/00480169.2016.1198280. [DOI] [PubMed] [Google Scholar]
- 59.Pardon B., De Bleecker K., Dewulf J. Prevalence of respiratory pathogens in diseased, non-vaccinated, routinely medicated veal calves. Vet Rec. 2011;169:278. doi: 10.1136/vr.d4406. [DOI] [PubMed] [Google Scholar]
- 60.Narita M., Yamada M., Tsuboi T. Bovine adenovirus type 3 pneumonia in dexamethasone-treated calves. Vet Pathol. 2003;40:128–135. doi: 10.1354/vp.40-2-128. [DOI] [PubMed] [Google Scholar]
- 61.Yamada M., Narita M., Nakamura K. Apoptosis in calf pneumonia induced by endobronchial inoculation with bovine adenovirus type 3 (BAV-3) J Comp Pathol. 2003;128:140–145. doi: 10.1053/jcpa.2002.0618. [DOI] [PubMed] [Google Scholar]
- 62.Grissett G.P., White B.J., Larson R.L. Structured literature review of responses of cattle to viral and bacterial pathogens causing bovine respiratory disease complex. J Vet Intern Med. 2015;29:770–780. doi: 10.1111/jvim.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellis J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can Vet J. 2019;60:147–152. [PMC free article] [PubMed] [Google Scholar]
- 64.Larson R.L. Bovine viral diarrhea virus-associated disease in feedlot cattle. Vet Clin North Am Food Anim Pract. 2015;31:367–380. doi: 10.1016/j.cvfa.2015.05.007. vi. [DOI] [PubMed] [Google Scholar]
- 65.Ferguson L., Olivier A.K., Genova S. Pathogenesis of Influenza D virus in cattle. J Virol. 2016;90:5636–5642. doi: 10.1128/JVI.03122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanthorn C.J., Dewell R.D., Cooper V.L. Randomized clinical trial to evaluate the pathogenicity of Bibersteinia trehalosi in respiratory disease among calves. BMC Vet Res. 2014;10:89. doi: 10.1186/1746-6148-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McMullen C., Orsel K., Alexander T.W. Evolution of the nasopharyngeal bacterial microbiota of beef calves from spring processing to 40 days after feedlot arrival. Vet Microbiol. 2018;225:139–148. doi: 10.1016/j.vetmic.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 68.Catry B., Boyen F., Baele M. Recovery of Moraxella ovis from the bovine respiratory tract and differentiation of Moraxella species by tDNA-intergenic spacer PCR. Vet Microbiol. 2007;120:375–380. doi: 10.1016/j.vetmic.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 69.Catry B., Chiers K., Schwarz S. Fatal peritonitis caused by Pasteurella multocida capsular type F in calves. J Clin Microbiol. 2005;43:1480–1483. doi: 10.1128/JCM.43.3.1480-1483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rings D.M. Salmonellosis in calves. Vet Clin North Am Food Anim Pract. 1985;1:529–539. doi: 10.1016/s0749-0720(15)31301-3. [DOI] [PubMed] [Google Scholar]
- 71.Catry B., Croubels S., Schwarz S. Influence of systemic fluoroquinolone administration on the presence of Pasteurella multocida in the upper respiratory tract of clinically healthy calves. Acta Vet Scand. 2008;50:36. doi: 10.1186/1751-0147-50-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z., Duan L., Liu F. First report of Enterobacter hormaechei with respiratory disease in calves. BMC Vet Res. 2020;16:1. doi: 10.1186/s12917-019-2207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Contrepois M., Dubourguier H.C., Parodi A.L. Septicaemic Escherichia coli and experimental infection of calves. Vet Microbiol. 1986;12:109–118. doi: 10.1016/0378-1135(86)90073-8. [DOI] [PubMed] [Google Scholar]
- 74.Van Driessche L., Vanneste K., Bogaerts B. Isolation of drug-resistant Gallibacterium anatis from calves with unresponsive bronchopneumonia, Belgium. Emerg Infect Dis. 2020;26(4) doi: 10.3201/eid2604.190962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osterman C., Rüttger A., Schubert E. Infection, disease, and transmission dynamics in calves after experimental and natural challenge with a Bovine Chlamydia psitacci isolate. Plos One. 2013;8(5):e64066. doi: 10.1371/journal.pone.0064066. [DOI] [PMC free article] [PubMed] [Google Scholar]