Abstract
Despite their essential function in terminating translation, readthrough of stop codons occurs more frequently than previously supposed. However, little is known about the regulation of stop codon readthrough by anatomical site and over the life cycle of animals. Here, we developed a set of reporters to measure readthrough in Drosophila melanogaster. A focused RNAi screen in whole animals identified upf1 as a mediator of readthrough, suggesting that the stop codons in the reporters were recognized as premature termination codons (PTCs). We found readthrough rates of PTCs varied significantly throughout the life cycle of flies, being highest in older adult flies. Furthermore, readthrough rates varied dramatically by tissue and, intriguingly, were highest in fly brains, specifically neurons and not glia. This was not due to differences in reporter abundance or nonsense-mediated mRNA decay (NMD) surveillance between these tissues. Readthrough rates also varied within neurons, with cholinergic neurons having highest readthrough compared with lowest readthrough rates in dopaminergic neurons. Overall, our data reveal temporal and spatial variation of PTC-mediated readthrough in animals, and suggest that readthrough may be a potential rescue mechanism for PTC-harboring transcripts when the NMD surveillance pathway is inhibited.
Subject terms: Translation, Molecular neuroscience, Ageing
Introduction
Fidelity of protein biosynthesis, including termination of the nascent polypeptide chain, is critical for all cellular functions. In eukaryotes, translation termination is a highly conserved process, and ribosomes can terminate protein biosynthesis with remarkable fidelity when encountering a stop codon1,2. Sometimes, however, translation can continue through a stop codon and terminate at the next in-frame stop codon, a mechanism commonly known as stop codon readthrough3.
In RNA viruses, readthrough is extensively utilized to append an extension domain to a proportion of coat proteins4. However, readthrough also occurs in the synthesis of eukaryotic proteins. In yeast, [PSI+] cells which carry the prion form of eRF3 (Sup35p) show a reduced efficiency of translation termination and higher rates of stop codon readthrough3. The [PSI+] strains exhibit phenotypic diversity which is beneficial to adapt to fluctuating environments5,6. In Drosophila, the headcase (hdc) gene encodes two different proteins, one of them produced by translational readthrough, and the readthrough product is necessary for Drosophila tracheal development7. There have also been a number of reports of readthrough in mammals, including in mouse brains8–10.
Validation of eukaryotic readthrough candidates had been confined to relatively small numbers until a comparative genomics methodology was used to analyze nucleotide sequences immediately adjacent to protein-coding regions in 12 Drosophila species. By identifying highly conserved sequences following native stop codons, Kellis and colleagues proposed more than 300 novel readthrough candidates11. Using ribosome profiling, Dunn et al. experimentally validated a large number of these evolutionarily conserved readthrough candidates, as well as identifying more than 300 examples of non-conserved stop codon readthrough events in D. melanogaster embryos and the S2 cell-line12. Although there is some debate about whether stop codon readthrough truly represents a regulatory mechanism13, and there are mechanisms to mitigate canonical readthrough14, these data suggest that stop codon readthrough in eukaryotes is far more pervasive than previously appreciated. We therefore decided to measure readthrough in flies using a set of novel gain-of-function reporter lines that could sensitively detect translation through stop codons in animals throughout their life cycle, as well as in specific tissues. Furthermore, we confirmed that the stop codons in our readthrough reporters are recognized as premature termination codons (PTCs) in flies. We observed that stop codon readthrough frequency in two candidate gene reporters varied widely throughout fly development, and appeared to be highest in Drosophila neurons. High frequency readthrough of PTCs may be an alternative rescue pathway for translation of transcripts with premature termination codons in flies.
Results
An in vivo gain-of-function reporter fly line can sensitively detect translational readthrough
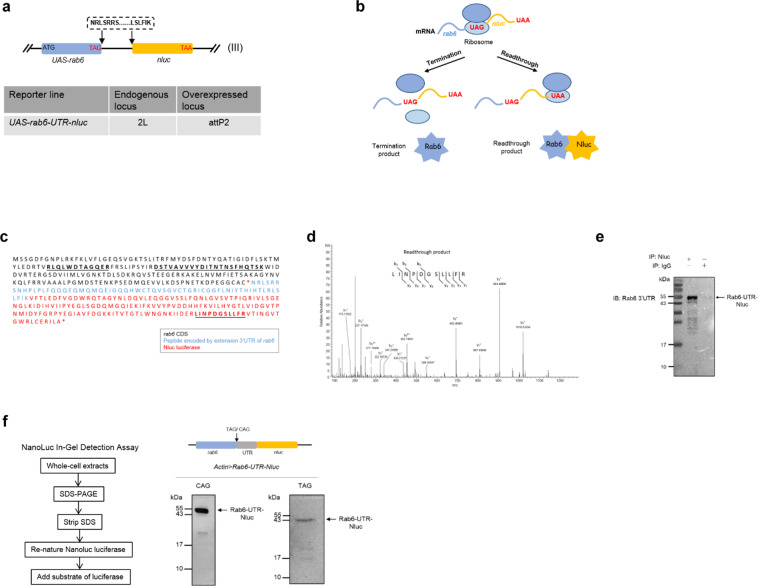
We wished to measure stop codon readthrough in flies across developmental stages of their life cycle. We initially chose to measure readthrough using a candidate gene, rab6, which had been identified as undergoing moderately elevated readthrough rates in a ribosome profiling study of fly embryos12. We decided to use a gain-of-function reporter15,16, since this approach would be able to detect low levels of readthrough. The gene for Nanoluc luciferase – Nluc –17, missing its start codon, was cloned immediately downstream of rab6 complete with its native stop codon and 3′ UTR, but missing the second, in-frame, stop codon (Fig. 1a). In our reporter, translation past the native UAG stop codon would result in functional Nanoluc luciferase enzyme, which could be detected using commercially available reagents (Fig. 1b). We were able to identify by tandem mass spectrometry a peptide derived from Nluc in flies expressing the reporter (Fig. 1c,d). To further confirm that readthrough was occurring, we raised a polyclonal antibody to a peptide coded by the 3′ UTR of rab6, and verified that a protein band of the appropriate size was identified by Western blot (Fig. 1e). To verify Nluc protein was actually the product of translation past the stop codon, not alternative initiation of nluc translation bypassing the stop codon, we performed an “in-gel” Nanoluc luciferase assay capable of detecting functional enzyme on an SDS-PAGE separated whole-cell lysate (see Methods). We could detect functional Nluc as the major band corresponding to the size of the Rab6-translated-3′ UTR-Nluc gene product, comparable to a control Rab6-Nluc fusion protein where the native TAG stop codon of rab6 had been replaced by CAG, corresponding to a glutamine residue (Fig. 1f). Although some smaller bands were visible, they comprised a minority of the total signal, and may have represented alternate initiation or breakdown products. Taken together, these data confirm that Nluc was expressed in reporter flies as a result of stop codon readthrough.
Figure 1.
An in vivo gain-of-function reporter fly line can sensitively detect translational readthrough. (a) Schematic for construction of the in-frame stop codon readthrough reporter. The gene coding for nluc, missing its start codon, was cloned immediately upstream of the second in-frame stop codon following rab6, its native stop codon and 3′ UTR. (b) Schematic for basis of the stop codon readthrough reporter. The expression of UAS-Rab6-UTR-Nluc was driven by Actin-Gal4. Normal termination of translation at the first in-frame stop codon would result in no expression of Nluc. Translational readthrough would result in expression of Rab6-UTR-Nluc, and detection of luciferase activity. (c) Primary sequence of the C-terminal extended polypeptide that would result from translational readthrough of the reporter. Red stars represent the stop codons. Underlined sequences represent peptides detected by mass spectrometry. (d) MS/MS spectrum representing a peptide, LINPDGSLLFR, from Nluc. The spectrum contains a total of eight C-terminal “y” ions and three N-terminal “b” ions consistent with this sequence. (e) Western blot with antibody raised against a 3′ UTR peptide of Rab6 following IP of Nluc from the reporter fly line. (f) Nanoluc luciferase in-gel detection identifies Nluc with migration consistent with translational readthrough. Whole-cell extracts of Actin>Rab6-TAG-Nluc adult flies and the variant with TAG-to-CAG substitution were assayed.
The stop codon of reporter flies is recognized as a premature termination codon
We wished to use our reporter system to measure relative readthrough rates in a semi-quantitative way. To control for differential expression of the reporter, both the UAS-rab6-UTR-STOP-nluc reporter and UAS-gfp were driven by Gal4 (see Methods and Fig. S1a). Comparison of the GFP and Nluc ratios would allow for relative quantitation of stop codon readthrough18,19.
To investigate potential cellular mediators of readthrough in our reporter system, we crossed our reporter fly line with a focused sub-library of UAS-RNAi fly lines from the Harvard Drosophila RNAi Screening Centre (DRSC) Resource20 at Tsinghua. Knock-down was driven via an actin-promoter except where knockdown of the target gene via an actin-driver promoter was lethal, in which case knock-down was driven by BM-40-SPARC-Gal4 and Cg-Gal4. We chose to define “hits” if Nluc/GFP signal was either two-fold decreased or increased compared with control. We screened a total of 615 candidate genes, and identified 90 potential regulators of rab6 readthrough (Fig. S1b). To eliminate rab6-specific hits, we rescreened the hits with a second reporter line encoding a second readthrough candidate rps2012. Testing knock-down of these hits with the rps20 readthrough reporter identified 25 genes, which showed similar readthrough phenotypes when knocked down in both rab6 and rps20 lines (Fig. S1c–f and Dataset S1). Only one candidate, the nonsense-mediated mRNA decay (NMD) gene upf121,22, showed increased Nluc/GFP in rab6 and rps20 reporters when knocked down (Fig. S2a).
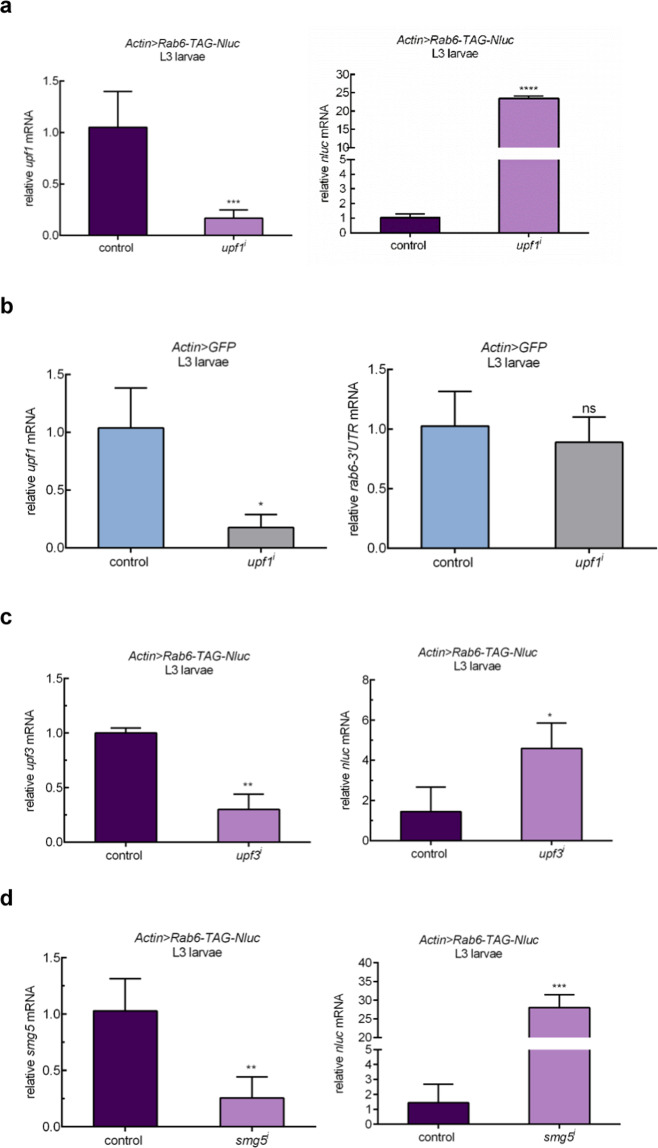
Stop codons can be classified as ‘native’ or ‘premature’, and mechanisms of both termination and readthrough vary and depend on how the stop codon is recognized by the cellular machinery22. Nonsense-mediated mRNA decay, is an important surveillance mechanism for monitoring of transcripts encoding PTCs, and prevents their translation by rapidly degrading such transcripts. Our identification of upf1 as a potential mediator of readthrough in our screen suggested that our reporter constructs were recognized as coding premature termination codons (PTCs). However, systematic investigation of Upf substrates in yeast and mammalian cells suggest that apparently normal mRNAs without the classical features of PTC-containing transcripts may also be targeted for degradation23–25. Was the native rab6 transcript a non-canonical Upf substrate? Knock-down of upf1 led to increased abundance of reporter mRNA (Fig. 2a), but the stability of native rab6 mRNA was not significantly affected by upf1 knockdown (Fig. 2b). These data suggested that rather than representing native gene readthrough, readthrough rates measured in our reporter flies represented variation in PTC suppression. To further confirm whether our reporters measured PTC readthrough, we knocked down two other NMD-associated factors, smg5 and upf326. Knockdown of both factors increased reporter abundance (Fig. 2c,d), as well as Nluc/GFP measurements (Fig. S2b), verifying that the reporter constructs, but not the native genes are subject to NMD, and that relative readthrough rates measured using the reporters would represent PTC readthrough.
Figure 2.
The stop codon of reporter flies is recognized as a premature termination codon. (a) Knockdown of Upfl is associated with increased abundance of rab6-based reporter transcript. Actin>Rab6-TAG-N/uc flies expressed both readthrough reporter and GFP. upfl (THFC_TH02846.N) was knocked down efficiently by Actin promoter (left panel). rab6 reporter mRNA abundance was measured by quantitative RT-PCR with nluc primer. rp49 abundance was used for normalization. (b) Knockdown of Upfl does not affect abundance of endogenous rab6. upfl was knocked down efficiently by Actin promoter (left panel). Native rab6 mRNA abundance was measured by quantitative RT-PCR with rab6-3′UTR primer. rp49 abundance was used for normalization. (c,d) Knockdown of NMD associated factors increases rab6-based reporter transcript. upf3 (BDSC_58181) and smgS (BDSC_62261) were knocked down efficiently by Actin promoter. rp49abundance was used for normalization. Data represent means of three independent experiments±s.d in A-D. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, P >0.05 by Student’s t-test.
PTC readthrough is regulated in a developmental stage-dependent manner
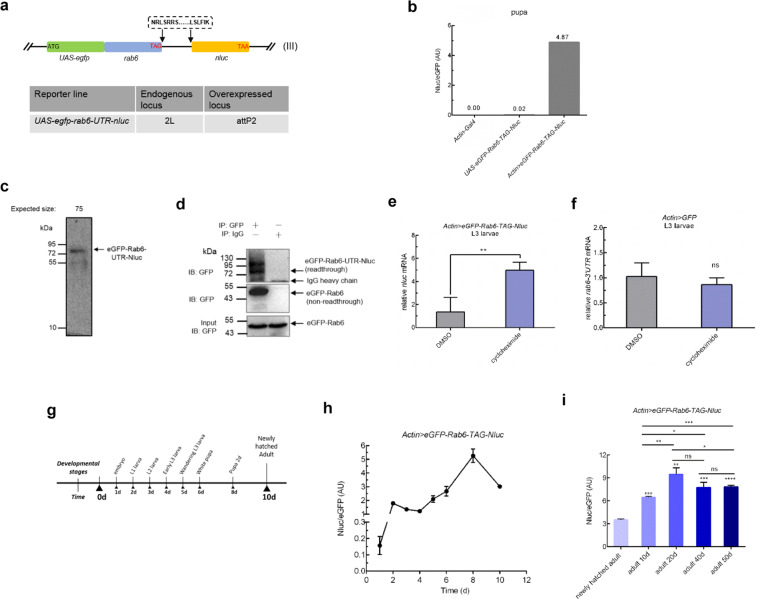
Since the initially-designed reporters were expressed as two constructs, and only one, expressing Nluc, but not the other expressing GFP was subject to NMD, it was possible that our apparent readthrough rates may be skewed, and that some of the hits from the RNAi screen represented interference with NMD and not readthrough per se. We therefore constructed a single-fusion reporter construct, representing egfp-rab6-TAG-UTR-nluc (Fig. 3a). Activity from the reporter was detected only when both Nluc and Gal4 were expressed (Fig. 3b), verifying its specificity and absence of background signal. The Nluc signal from whole-cell extract was derived almost entirely from a protein representing the correct molecular weight for the fusion product, which was strongly supportive that the reporter represented readthrough (Fig. 3c). Furthermore, immunoprecipitating and blotting against GFP identified two proteins with sizes representing normally the terminated translation, as well as a minority product (<1%) representing the extended polypeptide that would result from stop codon readthrough (Fig. 3d). To determine whether the new reporter was also subject to NMD, and therefore whether readthrough represented PTC suppression, we raised flies on cycloheximide or DMSO27. Cycloheximide treatment can stabilize mRNA transcripts subject to NMD28,29. Consistent with our other reporters, the new fusion reporter, but not native rab6 also appeared to be subject to NMD (Fig. 3e,f). Having constructed a reporter that would measure relative PTC readthrough rates, we wanted to determine whether levels of readthrough in our PTC reporter varied by development stage (Fig. S3). We measured Nluc/GFP through larval development (Fig. 3g,h) and also when adults were hatched at day 0, through day 50 of adult life (Fig. 3i). In immature flies, readthrough increased, peaking at the pupa stage (Fig. 3h). In adult flies, readthrough increased from the newly hatched stage through to day 20, but decreased thereafter, although staying higher than the newly hatched adult (Fig. 3i), suggesting that increase in readthrough was not due to aging per se. These results were confirmed with a second fusion egfp-rps20-TAA-UTR-nluc reporter (Fig. S4).
Figure 3.
PTC readthrough is regulated in a developmental stage-dependent manner. (a) Schematic for construction of the in-frame stop codon readthrough reporter. The gene coding for nluc, missing its start codon, was cloned immediately upstream of the second in-frame stop codon following rab6, its native stop codon and 3′ UTR. Here, the peptide sequence NRLSRRS……LSLFIK were encoded by extension 3′ UTR of rab6. To construct the transgene egfp_rab6_UTR_nluc, egfp, missing its stop codon, was fused immediately 5′ to rab6 with its start codon removed. (b) The readthrough reporter sensitively detects translational readthrough with minimal background signal. Nluc activity was only detected when the fly line expressed both the reporter and actin-driven Gal4. (c) Readthrough product from Actin>eGFP-Rab6-TAG-Nluc whole-cell extracts was detected by Nanoluc luciferase in-gel detection assay. (d) Readthrough efficiency was assayed by western blot. Lysates from Actin>eGFP-Rab6-TAG-Nluc adult flies were immunoprecipitated with mouse anti-GFP antibody or normal mouse IgG. eGFP-Rab6 (non-readthrough) and eGFP-Rab6-UTR-Nluc (readthrough) were detected by immunoblot analysis with rat anti-GFP antibody. PVDF membrane was cut at 55 kDa, eGFP-Rab6 was exposure with 10s and eGFP-Rab6-UTR-Nluc with 3600s, respectively. (e,f) The reporter transcript of rab6 is subjected to NMD. (E) Suppressed NMD increased readthrough reporter mRNA abundance. Actin>eGFP-Rab6-TAG-Nluc L3 larvae were transferred to standard fly food containing 500 μg/ml cycloheximide (King-Jones et al., 2005) or DMSO for 24h. Reporter transcript abundance was assessed by real-time RT-PCR. (f) Suppressed NMD does not affect endogenous rab6. Endogenous rab6 mRNA abundance was measured by quantitative RT-PCR with rab6-3′ UTR primer. rp49 abundance was used for normalization. (g) Cartoon representing time-points throughout the fly life-cycle when measurements were taken in (h). (i) Higher readthrough level in old flies is not aging associated. Adult flies at day 10, 20, 40 and 50 of adult life were collected separately for analysis. Relative readthrough rates in Actin>eGFP-Rab6-TAG-Nluc adult flies were measured by normalized Nluc activity (Nluc/eGFP). Data represent means of three independent experiments + s.d. *P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001; ns, P >0.05 by Student’s t-test.
Certain types of neurons undergo higher rates of stop codon readthrough
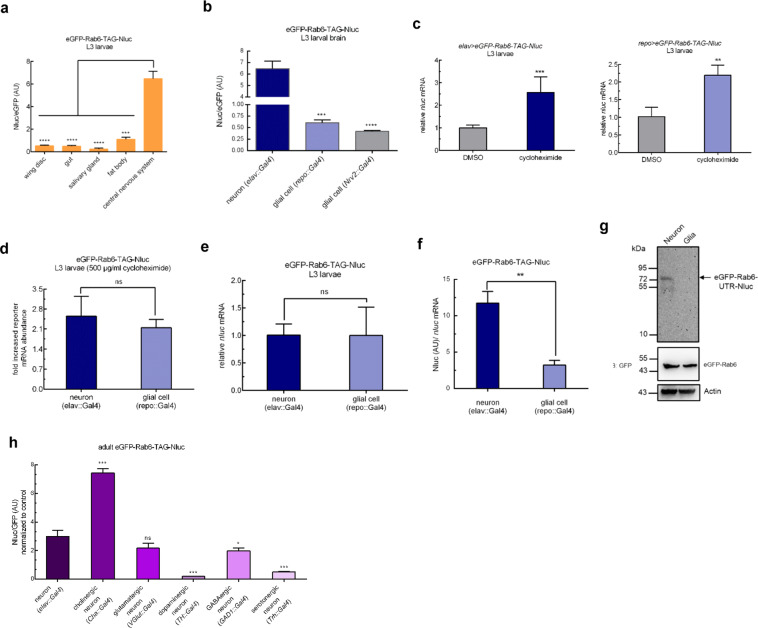
To test the spatial variation of PTC-mediated readthrough, we constructed a series of reporter flies expressing the fusion reporters, driven by tissue-specific promoters. There were clear differences in levels of readthrough by larval tissue, with the highest PTC readthrough in brain tissue (Fig. 4a). To further confirm whether neuronal or glial cells were responsible for the high readthrough rates, we compared stop codon (PTC) readthrough in neurons with those in glia, by expression of the reporters in those tissues specifically. Neurons exhibited higher readthrough than glial cells (Figs. 4b, S5). Could differences in NMD and hence reporter mRNA stability explain this observation? In both neurons and glia, the reporter was subject was to NMD, but to a similar extent (Fig. 4c,d), and nluc transcript abundance was similar in both tissues (Fig. 4e), suggesting that neither tissue variation in NMD or transcript abundance were sufficient to explain the observed differences between glia and neurons. However, measured Nluc activity, corrected by GFP (Fig. 4b) or nluc transcript abundance (Fig. 4f) was higher in neurons than glia. Furthermore, we measured Nluc abundance, as well as the size of the Nluc-containing polypeptide by the Nluc in-gel assay, in reporters expressed in neurons or glia. For similar loading of either total protein or non-extended reporter (as measured by blotting against Actin or GFP respectively), there was substantially more Nluc, at a size corresponding to the readthrough product, in neurons compared with glia (Fig. 4g). These data confirm that PTC-readthrough in fly neurons occurred at higher rates than in fly glia. Finally, we wished to investigate whether PTC readthrough rates varied by type of neuron. Using neurotransmitter-specific expression of the egfp-rab6-TAG-UTR-nluc reporter, we observed that readthrough rates were highest in cholinergic neurons, at more than twice the relative rate seen across neurons as a whole. By contrast, relative readthrough rates in dopaminergic and serotoninergic neurons was significantly lower than the pan-neuronal rates (Fig. 4h), suggesting that even within fly brains, PTC-readthrough rates varied substantially.
Figure 4.
Neurons undergo higher rates of PTC readthrough. (a) Rates of readthrough vary by tissue in wandering L3 larvae. Expression in wing disc was driven by rn-Gal4, gut by crq-Gal4, salivary gland by He-Gal4, fat body by Cg-Gal4, brain by elav-Gal4. Larval tissues were dissected to measure relative readthrough level. (b) Neurons undergo higher rates of stop codon readthrough. Higher rates of translational readthrough in larval neurons compared with glial cells. Neuronal expression was driven by elav-Gal4 and glial expression by repo-Gal4 and Nrv2-Gal4, respectively. Larval brains were dissected and readthrough rates measured in cell-lysates. (c) Reporter transcripts in larval neurons and glial cells are subjected to NMD. L3 larvae were transferred to standard fly food containing 500 μg/ml cycloheximide (King-Jones et al., 2005) or DMSO for 24h. Reporter transcript abundance was assessed by real-time RT-PCR. rp49 abundance was used for normalization. (d) The measured variation in Nluc/eGFP are not due to variation of NMD activity. NMD was inhibited by 500 μg/ml cycloheximide, and increased reporter mRNA in different neural cells were measured by quantitative RT-PCR. rp49 abundance was used for normalization. (e) Reporter transcripts in neural cells are similar. Reporter transcript abundance was assessed by real-time RT-PCR. rp49 abundance was used for normalization. (f) Neurons undergo higher rates of PTC readthrough. Relative readthrough level in neurons and glial cells in eGFP-Rab6-TAG-Nluc larvae. Nluc activity was normalized to the abundance of nluc mRNA, and nluc mRNA expression was determined by qRT-PCR. Data represent means of three independent experiments + s.d. (g) The measured variation in Nluc/eGFP are due to variation of readthrough rates. Neuronal expression was driven by elav-Gal4 and glial expression by repo-Gal4. Readthrough products from Neural cell > eGFP-Rab6-TAG-Nluc whole-cell extracts were detected by Nanoluc luciferase in-gel detection assay. Loading control by Western blot. (h), Rates of readthrough vary by neuron types in adult flies. Expression in neurons by pan-neuronal driver elav-Gal4, cholinergic neurons by Cha-Gal4, glutamatergic neurons by Vglut-Gal4, dopaminergic neurons by TH-Gal4, GABAergic neurons by GAD1-Gal4, serotonergic neurons by Trh-Gal4. Adult flies at day 1 post-hatching were collected to measure relative readthrough rate. Data represent means of three independent experiments + s.d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, P >0.05 by Student’s t-test.
Discussion
Higher readthrough rates in old flies is not associated with aging
Most studies on errors in gene translation have been on single-celled organisms, such as bacteria and yeast18,30–33 and therefore the relationship between translational error and development or anatomy has received limited attention. Mistranslation increases in response to stress, for example viral infection or oxidative damage34, and therefore it has been proposed that increased mistranslation over time may result in “error catastrophe” and may be one of the causes of aging35–37. Experimental evidence for this has been limited. Measuring fidelity of translation of polyU transcripts in vitro in brain homogenates from young and old rats, Filion and Laughrea failed to identify increased error rates with old age38. A more recent study found a strong positive correlation between fidelity of translation of the first and second codon and longevity in rodents39, suggesting that although mistranslation may not increase in old age, getting to old age may require high fidelity translation. However, of note, in that study, there was no correlation between stop codon readthrough and longevity39. In our study, whilst we found that older adult flies had higher rates of readthrough than newly hatched adults (Fig. 3i), this increase in readthrough did not increase further in very old flies (Fig. 3i), suggesting that some other regulatory factor than ‘old age’ may be responsible for the observation.
Specific neuronal types are responsible for higher rate of PTC readthrough in CNS
Studies of tissue-specific translational regulation are still in their infancy40. A recent study measured the translatome by ribosome profiling of fly muscle through tissue-specific expression of tagged ribosomes41. Mutations in Rpl38 resulted in profound patterning defects due to tissue-specific expression of Rpl38 and its role in translation of Homeobox mRNAs42. CNS-specific expression of a mutated tRNA resulted in neurodegeneration in mice43, again verifying that components and regulation of the translation apparatus can be expressed in a tissue-specific manner. A recent study observed high rates of native stop codon readthrough in mouse brains, although differences in readthrough rates between neurons and glia were not apparent10. A recent seminal study examined olfaction in Drosophila44. In D. sechellia, a key olfactory receptor gene, Ir75a has a premature termination codon, and the gene had been classified as a “pseudogene”. However, Prieto-Godino and colleagues elegantly demonstrated substantial translation of full-length, functional Ir75a protein, due to PTC-readthrough. Intriguingly, this phenomenon was also observed in D. melanogaster, in which Ir75a has no PTC, suggesting a conserved, generalizable mechanism of PTC readthrough in olfactory neurons in flies44. Similar to us, they observed that there was no substantial readthrough in fly glia, suggesting that our findings are not restricted to our candidate reporters. Our study extends these findings by measurement of relative PTC-readthrough in several fly tissues, and confirmed that highest readthrough rates were observed in neurons, but more specifically, in a subset of neurons, being highest in cholinergic, followed by glutaminergic neurons, and relatively low rates of readthrough in dopaminergic neurons. The difference in PTC-readthrough between fly glia and neurons was not due to differences in NMD activity. Further mechanistic studies are needed to explain the high readthrough in specific Drosophila neurons, perhaps by differential expression studies in different neuronal subtypes.
Readthrough may represent an alternative mechanism of PTC transcript rescue
Readthrough of the stop codon in our reporters mechanistically represented readthrough of a PTC (Fig. 2) and as such we cannot determine whether canonical readthrough rates also vary by developmental stage and anatomical site. In mammals, a stop codon is usually labelled as premature if it is located more than 50 nucleotides upstream of the last exon-exon junction21,45. However, in Drosophila, as well as yeast, the definition of a PTC occurs independently of exon-exon junctions46, and may include other elements for classification, such as “faux” 3′ UTRs47. Long 3′ UTRs are permissive for targeting of non-PTC-containing transcripts as substrates of Upf1 and NMD48–50. It is therefore likely that by incorporating the nluc gene in the extended 3′ UTRs of our reporter constructs, despite conserving all other elements of the stop codon context, the ‘native’ stop codons of rab6 and rps20 were re-classified as PTCs by the Drosophila cellular machinery.
Despite their potentially disastrous consequences and association with pathology51, premature termination codon-containing transcripts are surprisingly common. Alternative splicing of mRNA can result in a high frequency of PTC-containing transcripts52. And one study estimated that the typical human individual codes for approximately 100 nonsense- (PTC-containing) gene variants, of which 20 would be homozygous53. Since translation of proteins abnormally truncated due to the presence of a premature termination codon could result in protein misfolding, all eukaryotic cells have evolved nonsense-mediated mRNA degradation as a quality control mechanism to target PTC-containing transcripts for destruction22,45. However, NMD is not 100% efficient, a significant proportion of PTC-containing transcripts escape NMD54. Remarkably, a recent comparison of homozygous PTC-containing genes in humans with their homozygous “wild-type” counterparts showed that mRNA and protein abundance were similar in the two groups, suggesting that escape of PTC-containing transcripts from NMD-mediated degradation may be the norm rather than the exception55.
The Upf-mediated NMD pathway has regulatory functions beyond surveillance of PTC-containing mRNA and may itself be repressed in a developmental manner22,56. Bioinformatic predictions have suggested that neural tissue may be particularly susceptible to mistranslation-misfolded protein-induced damage57,58. It was therefore surprising that a brain-specific microRNA, miR-128, actively repressed NMD59, and that this downregulation of NMD represented a switch that triggered neuronal development60. Potential mechanisms for rescue pathways for nonsense-containing transcripts that do not rely on NMD, include RNA editing61, alternative splicing, truncated proteins bypassing the stop codon, and readthrough of the stop codon55. Our data supports a model in which neuronal cells may still be protected from the potentially deleterious effects of translating PTC-containing transcripts, despite downregulation of NMD62 via enhanced readthrough of PTC-containing mRNAs.
Materials and Methods
Fly Genetics
Standard fly husbandry techniques and genetic methodologies were used to assess transgenes in the progeny of crosses, construct intermediate fly lines and obtain flies of the required genotypes for each experiment63. The Gal4-UAS binary expression system was used to drive transgene expression with temporal and spatial control64. Flies were cultured at 25 °C, and anesthetized by CO2 prior to use in experiments. Fly strains used in this study are listed in Table S1. Intermediate strains were constructed using these strains.
Generation of Nluc-tagged readthrough reporter transgenic lines
To obtain UAS-RpS20-TAA-Nluc, UAS-Rab6-TAG-Nluc and UAS-Rab6-CAG-Nluc lines, the relevant rps20-TAA-nluc, rab6-TAG-nluc and rab6-CAG-nluc were separately cloned into vector pVALIUM10-roe using Gateway recombination. RNA extracted from S2 cells (a kind gift from Dr. Gong Cheng) was used to synthesize cDNA (Bio-Rad, cat#1708890). Coding sequences of rps20, rab6 and their following extension 3′ UTR12 were PCR-amplified from cDNA template with primers adding attB site at the 5′ termini of the ORF. In order to increase the expression of the transgenes in Drosophila, a 5′ UTR element Syn2165 and the Kozak sequence CAAAATG (the start codon underlined)66 were added to the coding sequence. nluc was codon optimized by GENEWIZ (Nanjing), and both start codon and stop codon were removed, and this nluc gene was inserted before the second in-frame stop codon of the extension 3′ UTR to acquire a fusion protein. Simultaneously, attB site was added to the 3′ termini of the modified fusion protein for subsquent Gateway cloning. Additionally, rab6-CAG-nluc was constructed by site directed mutagenesis using rab6-TAG-nluc as template. To construct UAS-eGFP-Rab6-TAG-Nluc and UAS-eGFP-RpS20-TAA-Nluc, egfp, missing its stop codon, was PCR-amplified and fused with rab6-TAG-nluc and rps20-TAA-nluc, respectively by overlap PCR.
The PCR products were purified by gel extraction (cwbiotech, cat#CW2302M) and recombined into vector pDONR221 (Life Technologies, cat#12536017) using Gateway BP Clonase (Life Technologies, cat#11789020). Then the entry clones were recombined with destination vector pVALIUM10-roe using Gateway LR clonase (Life Technologies, cat#12538120). The final plasmids UAS-RpS20-TAA-Nluc, UAS-Rab6-TAG-Nluc, UAS-eGFP-Rab6-TAG-Nluc, UAS-eGFP-RpS20-TAA-Nluc and UAS-Rab6-CAG were sent to Tsinghua Fly Center to obtain transgenic fly lines20 by site-directed insertion. To overexpress protein on the second chromosome, entry clone was integrated into attP40 loci, while the overexpressed protein on the third chromosome was obtained by integrating into attP2 loci67. All the primers used in transgenic fly lines construction are listed in Table S2.
Luciferase-GFP assays
Luciferase was measured using the NanoGlo Luciferase Assay Kit (Promega, cat#N1120). Euthanized flies were collected in 200 μl of NB buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 2 mM EDTA, 0.1% NP-40) with addition of protease inhibitor cocktail (Biotool, cat#B14003), and homogenized with a 96-well plate multiple homogenizer (Burkard Scientific, BAMH-96). Homogenized samples were centrifuged at 20,000 rcf (4 °C) to pellet the larval remains. For measuring readthrough level (Nluc/GFP), 30 μl of each sample supernatant was transferred to a white-walled 96-well plate (Costar, cat#3922), an equal volume of Promega Luciferase Reagent was added to each well and incubated in the dark for 5 min. Another 30 μl of each sample supernatant was correspondingly transferred to a black-walled 96-well plate (Corning, cat#3925), and an equal volume of NB buffer was added to each well. Luminescence and fluorescence signal were measured by Fluoroskan Ascent FL (Thermo Scientific).
Immunoprecipitation of Nluc
For western blot (Fig. 1e), Nluc luciferase was immunoprecipitated by addition of 12 μg of rabbit polyclonal anti-Nluc IgG (a generous gift from Lance Encell, Promega) or normal rabbit IgG as a negative control (Cell Signaling Technology, cat#2729) respectively, to concentrated fly lysates, and incubated overnight at 4 °C to form the immune complex. The immune complex was captured by Protein A/G Plus Agarose (Pierce, cat#26146) and eluted by heating the resin with SDS sample buffer.
For mass spectrometry (Fig. 1d), readthrough product of rab6 was enriched by a Pierce Direct IP Kit (Pierce, cat#26148). For each enrichment, 10 μg of rabbit polyclonal anti-Nluc IgG (Promega) was coupled to AminoLink Plus Coupling Resin, and concentrated cell lysate was incubated with the resin overnight at 4 °C to form the immune complex. The enriched readthrough product was eluted by a neutral pH elution buffer (Thermo Scientific, cat#21027).
Immunoprecipitation of eGFP-tagged readthrough reporter
For western blot (Fig. 3d), eGFP-tagged readthrough reporter was immunoprecipitated by addition of 10 μg of mouse monoclonal anti-GFP IgG (Roche, cat#11814460001) or normal mouse IgG as a negative control (Proteintech, cat#66360-3-Ig) respectively, to concentrated fly lysates, and incubated overnight at 4 °C to form the immune complex. The immune complex was captured by Protein A/G Plus Agarose (Pierce, cat#26146) and eluted by heating the resin with SDS sample buffer.
NanoLuc In-Gel Detection Assay
The protocol from the Promega technical manual (https://www.promega.com/-/media/files/resources/protocols/technical-manuals/500/nano-glo-in-gel-detection-system-technical-manual.pdf?la=en) was followed. Briefly, cell lysates were resolved on 15% SDS-PAGE. The gel was extracted from its casing and transferred to an appropriate tray. SDS was stripped from the gel by washing three times with 25% isopropanol in water (20 min each). NanoLuc was re-natured with three water washes (20 min each). The gel was developed with Furimazime (Promega, cat#N1120)/TBST (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.05% Tween 20, 25 uM Furimazine). The gel image was captured on a white reflective background by Chemidoc XRS + (Bio-Rad).
Western blot
Western bolt was performed using standard methods. Rabbit polyclonal antibody to Drosophila Rab6 3′ UTR was acquired by immunizing the New Zealand rabbit with peptide NRLSRRSNHPLPLFC by GenScript company (Nanjing). Readthrough product of rab6 (Fig. 1e) was detected using rabbit anti-Rab6 3′ UTR antibody (2 μg/ml, GenScript) and revealed with Clean-Blot IP Detection Reagent (Thermo Scientific, cat#21230) and ECL Western Blotting Substrate (Pierce, cat#32106). For eGFP-tagged reporter Western blots (Fig. 3d), elution was detected using 1:2500 dilution of monoclonal rat anti-GFP antibody (chromotek, RRID: AB_10773374), followed by a 1:5000 dilution of goat anti-rat IgG-HRP secondary antibody (easybio, cat#BE0109). Expression of reporter from input was detected by mouse anti-GFP IgG (Roche, cat#11814460001), followed by a 1:5000 dilution of goat anti-mouse IgG-HRP secondary antibody (cwbiotech, cat#CW0102). Loading control of neural cells (Fig. 4g) was using 1:2000 dilution of anti-insect beta Actin mouse antibody (cmctag, cat#AT0008), followed by a 1:5000 dilution of goat anti-mouse IgG-HRP secondary antibody, or 1:2500 dilution of monoclonal rat anti-GFP antibody as mentioned before.
Mass Spectrometry
Readthrough product of rab6 was enriched by a developed IP method. After electrophoresis, SDS PAGE was stained by Imperial Protein Stain (Thermo Scientific, cat#24615), and the gel between 43 kDa to 55 kDa were cut, digested with trypsin, analyzed by Tsinghua Protein Chemistry Facility.
Candidate forward genetic screen
Readthrough reporter fly line Actin > Rab6-TAG-Nluc or Actin > RpS20-TAA-Nluc was crossed with a set of UAS-RNAi fly lines (Tsinghua Fly Center) to knockdown target genes. Reporter fly line was crossed with w11,18, and readthrough level (Nluc/GFP) of progeny was as control. BM-40-SPARC > RpS20-TAA-Nluc and Cg > Rab6-TAG-Nluc reporter fly lines were utilized when knockdown target genes by actin > Gal4 resulted in growth deficiency. For each knockdown genotype, 8–12 wandering L3 larvae were collected to measure readthrough level. Readthrough level of control was normalized to 1, and for each knockdown genotype, relative readthrough fold to control was calculated.
Developmental stage assay
Parent transgenic reporter line y v sc; UAS-RpS20-TAA-Nluc (II) or y v sc; UAS-Rab6-TAG-Nluc (III) and an actin driven Gal4 line w; Sp/CyO; act-FO-Gal4 UAS-GFP/TM6B was crossed to express the readthrough reporter ubiquitously in the whole body of progeny. To eliminate the effect by variation in NMD efficiency, reporter line y v sc; UAS-eGFP-RpS20-TAA-Nluc (II) or y v sc; UAS-eGFP-Rab6-TAG-Nluc (III) was crossed with y w; Adv/CyO; actin-Gal4/TM6B. After 6 hours of crossing, parent flies were removed to ensure the majority of the progenies were in the same developmental stage. Cell lysates were flash frozen by liquid nitrogen and stored at −80 °C until all the samples of different developmental stages had been collected. Readthrough value was measured in whole cell lysate as above.
For embryo collection, standard apple juice agar plates were supplemented with fresh baker yeast paste, and collection cages were placed on the plates. Parent flies lay eggs on the plates for 4 hours. 0–4 hr embryos were collected from the agar plate using a small paintbrush.
For aged flies collection, newly hatched male and female flies were sorted into independent groups and cultured them in standard environmental conditions with a 12:12 hr light dark cycle. During the experimental period, flies were transferred to new vials containing fresh food every 2–3 days68. After 40 days of adult life, flies started to die and living flies would be transferred to fresh food every day to avoid the flies sticking to the food in the old vial.
Protein extraction from Drosophila embryos
Collected embryos were washed gently in a collection basket (Corning, cat#352350). The base of the basket was dried by a paper tissue, and the basket was transferred to a container with 50% commercial bleach solution. Incubated for 5 min with gentle, periodic stirring to remove the chorionic membrane of the embryos. The dechorionated embryos became hydrophobic and floated on the surface of the bleach solution. Transferred the basket to a new container with deionized water, washed for 2 min and repeated twice. After wash, about 30 embryos of each independent sample were transferred to a 1.5 ml eppendorf tube containing 200 ul of ice-cold NB lysis buffer (protease inhibitor cocktail was added) and homogenized by a cordless motor (Kimble, cat#749540-0000). Homogenized samples were centrifuged at 20,000 rcf (4 °C) to pellet the larval remains, and supernatant avoiding the upper lipid layer was transferred to a new 1.5 ml eppendorf tube.
Protein extraction from different Drosophila organs or tissues
Expression of readthrough reporter in wing disc was driven by rn-Gal4, gut by crq-Gal4, salivary gland by He-Gal4, fat body by Cg-Gl4, neuron by elav-Gal4, glial cells by repo-Gal4 and Nrv2-Gal4. Different larval organs or tissues were dissected and collected in 100 μl of NB lysis buffer with addition of protease inhibitor cocktail, homogenized by a cordless motor. Homogenized samples were centrifuged at 20,000 rcf (4 °C) to pellet the larval remains, and supernatant avoiding the upper lipid layer was transferred to a new 1.5 ml eppendorf tube. The final cleared cell lysates were flash frozen by liquid nitrogen and stored at −80 °C until all the samples of different Drosophila tissues or organs had been collected.
Antibiotics treatment
L3 larvae were transferred to standard fly food containing 500 μg/ml cycloheximide (MedChemExpress, cat#HY-12320)27 or DMSO for 24 h. Afterwards, total RNA were extracted for real-time RT-PCR.
Real-time quantitative RT-PCR
Approximate 10–20 wandering L3 larvae were collected and RNA was extracted by TransZol Up (TransGen Biotech, cat#ET111-01). DNase I (TransGen Biotech, cat#GD201-01) was used to digest genomic DNA. cDNA was synthesized with iScript Reverse Transcription kit (Bio-Rad, cat#1708890) and iTaq Universal SYBR Green Supermix (Bio-Rad, cat#1725120) was used for quantitative PCR. Analysis was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad). rp49 was used as a reference gene69,70. All PCR reactions were performed in biological triplicate. Primers used were:
rp49-For: 5′-GGCCCAAGATCGTGAAGAAG-3′;
rp49-Rev: 5′-ATTTGTGCGACAGCTTAGCATATC-3′;
upf1-For: 5′-ACTTCCGGTTCGCACATCAT-3′;
upf1-Rev: 5′-CTTCCACTGTTCCTGGTCCC-3′;
upf3-For: 5′- ATGCTCCCTTCCAGTGCTTC-3′;
upf3-Rev: 5′- CCGCTTGATGAACTCCTGGT-3′;
smg5-For: 5′- GCTTTTTGACTGGCTGCGAA-3′;
smg5-Rev: 5′- ACCAGAGAATCACGCACGTT-3′;
nluc-For: 5′-GATCATCCCCTACGAGGGCT-3′;
nluc-Rev: 5′-GTCGATCATGTTGGGGGTCA-3′;
rab6-3′UTR-For: 5′-ATCCAACCATCCTCTCCCCC-3′;
rab6-3′UTR-Rev: 5′-GCAGATCCGGCCAGTACATA-3′;
rps20-3′UTR-For: 5′- ATCATCGACTTGCACTCGCC-3′;
rps20-3′UTR-Rev: 5′- GCACGCCAAACTTTTCGAGG-3′.
Quantitation and statistical analysis
Most of experiments were performed at least three times on separate days (that is, independent experiments). Statistical analysis and graphic representation were performed with Graphpad Prism software. Two-tailed non-parametric Mann-Whitney tests were used in Fig. S2A (data did not pass normality tests), otherwise means between two sample sets were compared by unpaired, two-directional Student’s t-tests in the rest of comparisons. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 were considered statistically significant results.
Supplementary information
Acknowledgements
We thank Dr.Yi Zhong (elav-Gal4 and repo-Gal4) and Dr. Wei Zhang (Cha-Gal4, VGlut-Gal4, TH-Gal4, GAD1-Gal4 and Trh-Gal4) for kindly sharing fly strains; Lance Encell, Paul Otto and Thomas Machleidt from Promega for generously providing the anti-Nluc antibody and the protocol for the NanoLuc luciferase In-Gel Detection Assay; and the Tsinghua Fly Center for constructing transgenic in vivo readthrough reporter fly lines. This work was supported in part by start-up funds from Tsinghua University to BJ; and a grant from the National Science Foundation of China [31771600] to JCPP. BJ is an Investigator of the Wellcome Trust [207487/B/17/Z]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
B.J. conceived the project. Y.N.C. and B.J. designed research. Y.N.C. performed the vast majority of experiments with assistance from T.H.S. and Z.B. J.Q.N. provided the platform for generation of fly lines. J.C.P.P. and B.J. supervised research. Y.N.C. and B.J. wrote the manuscript with input from J.C.P.P. and other authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jose C. Pastor-Pareja, Email: jose.pastor@biomed.tsinghua.edu.cn
Babak Javid, Email: bjavid@gmail.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-65348-8.
References
- 1.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 7.Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2001;2:593–597. doi: 10.1093/embo-reports/kve128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswarappa SM, et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014;157:1605–1618. doi: 10.1016/j.cell.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loughran G, et al. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014;42:8928–8938. doi: 10.1093/nar/gku608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapkota D, et al. Cell-Type-Specific Profiling of Alternative Translation Identifies Regulated Protein Isoform Variation in the Mouse Brain. Cell reports. 2019;26:594–607 e597. doi: 10.1016/j.celrep.2018.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungreis I, et al. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21:2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Zhang J. Stop-codon read-through arises largely from molecular errors and is generally nonadaptive. PLoS Genet. 2019;15:e1008141. doi: 10.1371/journal.pgen.1008141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arribere JA, et al. Translation readthrough mitigation. Nature. 2016;534:719–723. doi: 10.1038/nature18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su HW, et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol. 2016;1:16147. doi: 10.1038/nmicrobiol.2016.147. [DOI] [PubMed] [Google Scholar]
- 16.Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: friend or foe? Trends in biochemical sciences. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Hall MP, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS chemical biology. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javid B, et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci USA. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer, E. B. & Farabaugh, P. J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA13, 87–96, rna.294907 (2007). [DOI] [PMC free article] [PubMed]
- 20.Ni JQ, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta P, Li YR. Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay. Mol Biol Rep. 2018;45:39–55. doi: 10.1007/s11033-017-4139-7. [DOI] [PubMed] [Google Scholar]
- 22.He F, Jacobson A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annual review of genetics. 2015;49:339–366. doi: 10.1146/annurev-genet-112414-054639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tani H, et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012;9:1370–1379. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Q, et al. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avery P, et al. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17:624–638. doi: 10.1261/rna.2404211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King-Jones K, Charles JP, Lam G, Thummel CS. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell. 2005;121:773–784. doi: 10.1016/j.cell.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Colak D, Ji SJ, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–1265. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue K, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 30.Meyerovich, M., Mamou, G. & Ben-Yehuda, S. Visualizing high error levels during gene expression in living bacterial cells. Proc Natl Acad Sci USA107, 11543-11548, doi:0912989107 (2010). [DOI] [PMC free article] [PubMed]
- 31.Miranda, I. et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio4, 10.1128/mBio.00285-13 (2013). [DOI] [PMC free article] [PubMed]
- 32.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz MH, Pan T. Temperature dependent mistranslation in a hyperthermophile adapts proteins to lower temperatures. Nucleic Acids Res. 2016;44:294–303. doi: 10.1093/nar/gkv1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Netzer, N. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature462, 522–526, nature08576 (2009). [DOI] [PMC free article] [PubMed]
- 35.Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proceedings of the National Academy of Sciences. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing: a correction. Proceedings of the National Academy of Sciences. 1970;67:1476–1476. doi: 10.1073/pnas.67.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orgel LE. Ageing of clones of mammalian cells. Nature. 1973;243:441. doi: 10.1038/243441a0. [DOI] [PubMed] [Google Scholar]
- 38.Filion AM, Laughrea M. Translation fidelity in the aging mammal: studies with an accurate in vitro system on aged rats. Mechanisms of ageing and development. 1985;29:125–142. doi: 10.1016/0047-6374(85)90013-2. [DOI] [PubMed] [Google Scholar]
- 39.Ke Z, et al. Translation fidelity coevolves with longevity. Aging cell. 2017;16:988–993. doi: 10.1111/acel.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Z, Barna M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu Rev Cell Dev Biol. 2015;31:31–54. doi: 10.1146/annurev-cellbio-100814-125346. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Dickman D. Development of a tissue-specific ribosome profiling approach in Drosophila enables genome-wide evaluation of translational adaptations. PLoS Genet. 2017;13:e1007117. doi: 10.1371/journal.pgen.1007117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondrashov N, et al. Ribosome-Mediated Specificity in Hox mRNA Translation and Vertebrate Tissue Patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishimura R, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prieto-Godino LL, et al. Olfactory receptor pseudo-pseudogenes. Nature. 2016;539:93–97. doi: 10.1038/nature19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicholson P, Muhlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 46.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 48.Hogg JR, Goff SP. Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci USA. 2013;110:3357–3362. doi: 10.1073/pnas.1219908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 52.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends in biochemical sciences. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 53.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 55.Jagannathan S, Bradley RK. Translational plasticity facilitates the accumulation of nonsense genetic variants in the human population. Genome Res. 2016;26:1639–1650. doi: 10.1101/gr.205070.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng Q, Jagannathan S, Bradley RK. The RNA Surveillance Factor UPF1 Represses Myogenesis via Its E3 Ubiquitin Ligase Activity. Mol Cell. 2017;67:239–251 e236. doi: 10.1016/j.molcel.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond, D. A. & Wilke, C. O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell134, 341–352, 10.1016/j.cell.2008.05.042 S0092-8674(08)00705-8 [pii] (2008). [DOI] [PMC free article] [PubMed]
- 58.Drummond, D. A. & Wilke, C. O. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet10, 715–724, nrg2662 (2009). [DOI] [PMC free article] [PubMed]
- 59.Bruno IG, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lou CH, et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell reports. 2014;6:748–764. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan Y, Dou S, Luo S, Zhang H, Lu J. Adaptation of A-to-I RNA editing in Drosophila. PLoS Genet. 2017;13:e1006648. doi: 10.1371/journal.pgen.1006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaffrey SR, Wilkinson MF. Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci. 2018;19:715–728. doi: 10.1038/s41583-018-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roote J, Prokop A. How to design a genetic mating scheme: a basic training package for Drosophila genetics. G3 (Bethesda) 2013;3:353–358. doi: 10.1534/g3.112.004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 65.Pfeiffer BD, Truman JW, Rubin GM. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:6626–6631. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic acids research. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Linford, N. J., Bilgir, C., Ro, J. & Pletcher, S. D. Measurement of lifespan in Drosophila melanogaster. J Vis Exp, 10.3791/50068 (2013). [DOI] [PMC free article] [PubMed]
- 69.Zang Y, et al. Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. Elife. 2015;4:e07187. doi: 10.7554/eLife.07187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.