Abstract
Aberrant Src kinase activity is known to be involved in a variety of human malignancies, whereas the regulatory mechanism of Src has not been completely clarified. Here, we demonstrated that tripartite motif containing 7 (TRIM7) directly interacted with Src, induced Lys48-linked polyubiquitination of Src and reduced the abundance of Src protein in hepatocellular carcinoma (HCC) cells. We further identified TRIM7 as a tumor suppressor in HCC cells through its negative modulation of the Src-mTORC1-S6K1 axis in vivo and in vitro in several HCC models. Moreover, we verified the dysregulated expression of TRIM7 in clinical liver cancer tissues and its negative correlation with Src protein in clinical HCC specimens. Overall, we demonstrated that TRIM7 suppressed HCC progression through its direct negative regulation of Src and modulation of the Src-mTORC1-S6K1 axis; thus, we provided a novel insight into the development of HCC and defined a promising therapeutic strategy for cancers with overactive Src by modulating TRIM7.
Subject terms: Tumour-suppressor proteins, Tumour-suppressor proteins, Diagnostic markers
Introduction
The proto-oncogene tyrosine kinase Src is the prototype of the membrane-bound, nonreceptor tyrosine kinase family that includes Fyn, Yes, Lyn, Hck, Yrk, Lck, and Blk [1–3]. The Src protein is composed of several functional domains including a membrane-targeting myristoylated or palmitoylated SH4 domain, an SH3 domain, an SH2 domain, a protein tyrosine kinase domain (SH1) and a regulatory tail in the C-terminus [4]. Intermolecular autophosphorylation of Tyr419, which is located in the activation loop of the kinase domain, significantly promotes Src kinase activity [5]. Src kinase functionally interacts and cooperates with several receptor and nonreceptor protein tyrosine kinases to provide a diversity of signals for the regulation of cellular functions [6].
In the past decade, remarkable progress has been achieved in understanding the diverse functions of Src. As a nonreceptor protein tyrosine kinase, Src plays a broad role in cellular biological processes including proliferation, differentiation, survival, and metabolism. Activation of Src kinase promotes multiple downstream transduction cascades including phosphatidylinositol 3-kinase (PI3K) and protein kinase B (PKB), mitogen-activated protein kinase, and signal transducer and activator of transcription 3, which are vital regulators of survival, proliferation, and gene expression [4].
There is a large body of evidence showing that aberrant expression and activation of Src is involved in a variety of human malignancies and that Src dramatically facilitates cancer cell division, proliferation, and invasion [7, 8]. It has been reported that cytoplasmic Src family kinase activity is associated with more aggressive disease and shorter patient survival in several types of cancers [9–11]. The oncogenic role of Src in cancers has been well recognized whereas the regulatory mode of Src in human malignancies is far from being clarified.
In this study, we identified tripartite motif containing (TRIM) 7 as a novel negative regulator of Src protein. TRIM7 is a new member of TRIM family proteins, and it was first identified as glycogenin-interacting protein in consideration of its role in binding and activating glycogen proteins [12–14]. TRIM7 possesses an N-terminal RING (really interesting new gene) domain, a B-box domain (zinc-finger motif), a coiled-coil domain, and a C-terminal B30.2/SPRY domain, which indicates its role as a RING-type E3 ubiquitin ligase to directly bind with target proteins for ubiquitination [15].
Here we showed for the first time that TRIM7 directly interacted with Src and acted as a negative regulator of Src protein in hepatocellular carcinoma (HCC) cells. We demonstrated that TRIM7 suppressed oncogenesis and progression of HCC by modulating the Src-mTORC1-S6K1 axis. Moreover, experiments including in vivo xenograft tumor models and clinical liver cancer specimens further validated the tumor suppressor role of TRIM7 via its negative regulation of Src. Altogether, this study demonstrated a novel regulatory effect of TRIM7 on Src and provided a therapeutic strategy to manipulate Src-overactive cancers by regulating TRIM7.
Results
TRIM7 directly bound to the Src protein
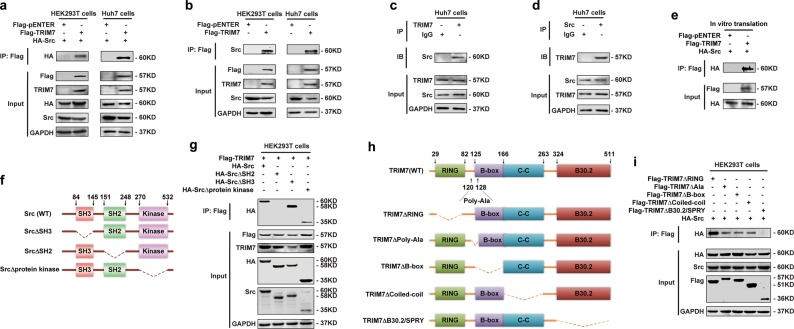
To investigate whether TRIM7 has a potential regulatory effect on Src, we first performed endogenous and exogenous co-immunoprecipitation (co-IP) assays to determine whether TRIM7 could interact with Src. Our data showed that TRIM7 interacted with both exogenous and endogenous Src protein (Fig. 1a, b). Moreover, endogenous TRIM7 could bind to endogenous Src (Fig. 1c, d), which indicated their interaction in natural conditions. Then, we obtained TRIM7 and Src proteins by an in vitro transcription and translation system and further demonstrated that TRIM7 could directly interact with Src protein in vitro (Fig. 1e).
Fig. 1.
TRIM7 directly bound to Src protein. a Co-IP analysis of the binding between exogenous TRIM7 and exogenous Src in HEK293T and Huh7 cells co-transfected with Flag-TRIM7 plasmid and HA-Src plasmid. b Co-IP analysis of the binding between exogenous TRIM7 and endogenous Src in HEK293T and Huh7 cells transfected with Flag-TRIM7 plasmid or mock control plasmid. c Co-IP analysis of the binding between endogenous TRIM7 and endogenous Src in Huh7 cells. d Co-IP analysis of the binding between endogenous TRIM7 and endogenous Src in Huh7 cells. e Co-IP analysis of the direct binding between Flag-TRIM7 and HA-Src proteins obtained from an in vitro transcription and translation system. f Schematic illustration of Src, showing wild-type and truncation mutants of Src. g Co-IP analysis of the interaction between TRIM7 and Src or Src truncation mutants in HEK293T cells co-transfected with HA-Src plasmid or HA-Src truncation mutant plasmids together with Flag-TRIM7 plasmid. h Schematic illustration of TRIM7, showing the wild-type and truncation mutants of TRIM7. i Co-IP analysis of the interaction between Src and TRIM7 truncation mutants in HEK293T cells co-transfected with Flag-TRIM7 truncation mutant plasmids together with HA-Src plasmid. The presented figures are the representative of data from at least three independent experiments
To investigate the molecular basis of the interaction between TRIM7 and Src, we generated a series of domain-deleted mutants of both TRIM7 and Src, and tried to define their interacting domains (Fig. 1f, h). Our data showed that the SH2 domain of Src (Fig. 1g) and the B30.2/SPRY domain of TRIM7 (Fig. 1i) were required for the interaction between TRIM7 and Src. Thus, these data demonstrated that the binding between the SH2 domain of Src and the B30.2/SPRY domain of TRIM7 was the molecular basis for the direct interaction between TRIM7 and Src.
TRIM7 reduced the abundance of Src protein
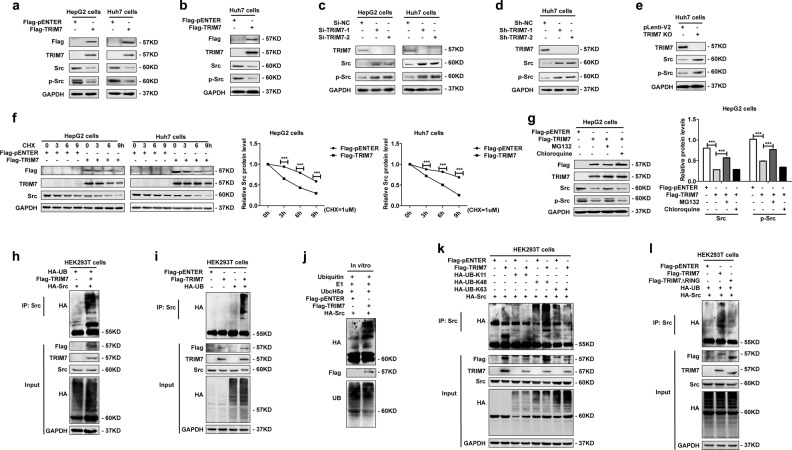
Since we identified a direct interaction between TRIM7 and Src, we were further interested in defining whether TRIM7 regulated the abundance and stability of Src protein. First, we constructed gain-of-function models by transient and stable transfection of a TRIM7 expression plasmid into HCC cells (Fig. 2a, b, Supplementary Fig. S1a, b). Our data showed that after successful overexpression of TRIM7 by both transient and stable transfection of the TRIM7 plasmid (Fig. 2a, b, Supplementary Fig. S1a, b), the protein levels of both Src and p-Src were significantly reduced (Fig. 2a, b). Second, we constructed three different loss-of-function models including Si-TRIM7 transiently transfected cells (Fig. 2c, Supplementary Fig. S1c), lentivirus-mediated Sh-TRIM7 stably transfected cells (Fig. 2d), and TRIM7-knockout cells based on the CRISPR-Cas9 system (Fig. 2e). Our data showed that in all of these cell models of TRIM7 inhibition, the protein levels of both Src and p-Src were significantly increased (Fig. 2c, d, e). All these data indicated that TRIM7 inhibited the proto-form as well as the activated form of Src.
Fig. 2.
TRIM7 reduced the abundance of Src protein and directly induced K48-linked polyubiquitination. a Western blot analysis of Src and p-Src protein levels in HepG2 and Huh7 cells transiently transfected with Flag-TRIM7 plasmid or mock control plasmid. b Western blot analysis of Src and p-Src protein levels in Huh7 cells stably transfected with Flag-TRIM7 plasmid or mock control plasmid. c Western blot analysis of Src and p-Src protein levels in HepG2 and Huh7 cells transiently transfected with Si-NC, Si-TRIM7-1, or Si-TRIM7-2. d Western blot analysis of Src and p-Src protein levels in Huh7 cells stably transfected with lentivirus-mediated Sh-NC, Sh-TRIM7-1, or Sh-TRIM7-2. e Western blot analysis of Src and p-Src protein levels in TRIM7-knockout Huh7 cells. f HepG2 and Huh7 cells were transfected with Flag-TRIM7 plasmid or mock control plasmid and were cultured for 24 h before being further incubated with CHX for 0, 3, 6, and 9 h. The Src protein level of the transfected cells was detected by western blot. g HepG2 cells were transfected with Flag-TRIM7 plasmid or mock control plasmid and were cultured for 24 h before being further incubated with MG132 (10 μM) for 4 h or chloroquine (25 μM) for 6 h. The Src and p-Src protein levels of the transfected cells were detected by western blot. h Co-IP analysis of the ubiquitination of Src in HEK293T cells co-transfected with Flag-TRIM7 plasmid, HA-UB plasmid and HA-Src plasmid. i Co-IP analysis of ubiquitination of endogenous Src in HEK293T cells co-transfected with Flag-TRIM7 plasmid and HA-UB plasmid. j In vitro ubiquitination analysis of ubiquitination of Src in the presence of in vitro-translated Flag-TRIM7 and HA-Src, E1, UbcH5a, and ubiquitin. k Co-IP analysis of ubiquitination of Src in HEK293T cells co-transfected with Flag-TRIM7 plasmid, HA-Src plasmid, and HA-UB-K11, HA-UB-K48, or HA-UB-K63 plasmid. l Co-IP analysis of ubiquitination of Src in HEK293T cells co-transfected with Flag-TRIM7 plasmid or Flag-TRIM7 truncation mutant (TRIM7ΔRING) plasmid as well as HA-Src plasmid and HA-UB plasmid. The band intensities of the key proteins were further quantitatively analyzed in the indicated groups and were presented in the right panel. ***P < 0.001 for statistical analysis of the indicated groups. The presented figures are representative of data from at least three independent experiments
Our data further showed that the TRIM7-mediated negative regulation of Src occurred at the translational level but not the transcription level of Src (Supplementary Fig. S1a, b, c). After blocking de novo protein synthesis in the TRIM7-overexpressing cells by cycloheximide, we found that TRIM7 significantly facilitated the decrease of Src protein level (Fig. 2f). Further investigation suggested that the TRIM7-mediated negative regulation of Src could be significantly rescued by the proteasome inhibitor MG132 (Fig. 2g), which indicated that Src was degraded by TRIM7 through proteasome-mediated degradation. Altogether, these data indicated that TRIM7 significantly reduced the abundance of Src protein via proteasome-mediated degradation.
TRIM7 induced Lys48-linked polyubiquitination of Src
Considering that TRIM7 belongs to the TRIM family, most of which are RING-type E3 ligases, we tried to define whether TRIM7 negatively regulated Src protein by its E3 ligase activity. Our data showed that TRIM7 was able to attach a polyubiquitin chain to Src protein (Fig. 2h, i), and further investigation confirmed the polyubiquitin chain could be attached to Src protein by TRIM7 in an in vitro ubiquitination system (Fig. 2j), which suggested that TRIM7 directly degraded Src protein through polyubiquitination.
Further investigation revealed that TRIM7 could attach a Lys48 (K48)-linked but not Lys11 (K11)-linked or Lys63 (K63)-linked polyubiquitin chain to Src protein, which implied that TRIM7 could induce Lys48-linked polyubiquitination of Src (Fig. 2k). When we transfected HCC cells with the RING domain-deleted TRIM7 mutant plasmid, the polyubiquitination of Src was significantly reduced in comparison with that of the wild-type TRIM7 plasmid-transfected group (Fig. 2l). Thus it indicated that TRIM7 induced ubiquitination of Src via its RING domain. Taken together, these data demonstrated that TRIM7 directly interacted with Src protein and induced Lys48-linked polyubiquitination of Src via its RING domain.
TRIM7 suppressed the Src-mTORC1-S6K1 axis
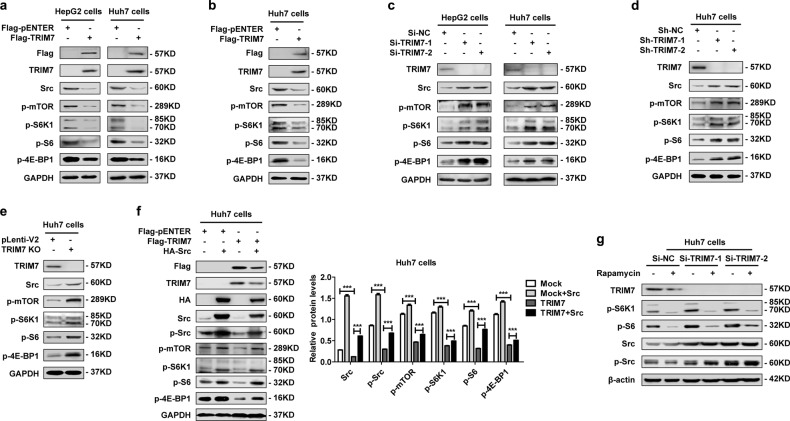
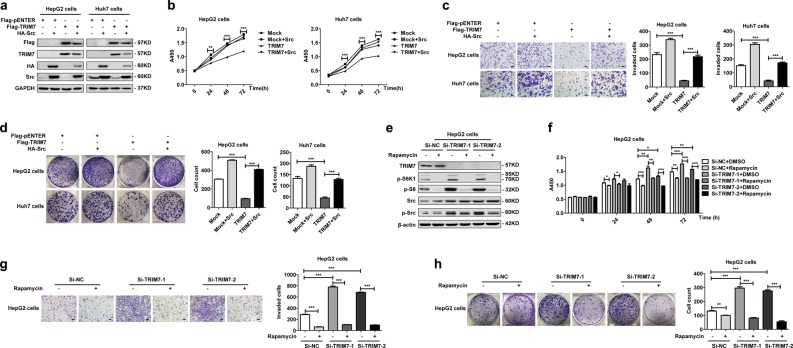
We determined that TRIM7 directly bound to Src and mediated Lys48-linked polyubiquitination of Src protein, so we were interested in defining the downstream molecular pathway of the TRIM7-Src axis. Our data showed that both transient and stable overexpression of TRIM7 efficiently reduced the protein levels of p-mTOR, p-S6K1, p-S6, and p-4E-BP1, which indicated its inhibitory effect on the mTORC1 pathway (Fig. 3a, b). In contrast, in TRIM7-knockdown and -knockout cells, the protein levels of p-mTOR, p-S6K1, p-S6, and p-4E-BP1 were significantly upregulated (Fig. 3c, d, e). Our data further showed that overexpression of Src could rescue the TRIM7-mediated suppression of the mTORC1-S6K1 signaling pathway (Fig. 3f), while the mTORC1 pathway inhibitor rapamycin did not change the protein levels of either Src or p-Src (Fig. 3g), which indicated that mTORC1-S6K1 signaling was downstream of the TRIM7-mediated negative regulation of Src. In summary, our data demonstrated that TRIM7 inhibited Src protein as well as its downstream mTORC1-S6K1 signaling in HCC cells.
Fig. 3.
TRIM7 modulated the Src-mTORC1-S6K1 axis. a Western blot analysis of Src, p-mTOR, p-S6K1, p-S6, and p-4E-BP1 protein levels in HepG2 and Huh7 cells transiently transfected with Flag-TRIM7 plasmid or mock control plasmid. b Western blot analysis of Src, p-mTOR, p-S6K1, p-S6, and p-4E-BP1 protein levels in Huh7 cells stably transfected with Flag-TRIM7 plasmid or mock control plasmid. c Western blot analysis of Src, p-mTOR, p-S6K1, p-S6, and p-4E-BP1 protein levels in HepG2 cells and Huh7 cells transiently transfected with Si-NC, Si-TRIM7-1, or Si-TRIM7-2. d Western blot analysis of Src, p-mTOR, p-S6K1, p-S6, and p-4E-BP1 protein levels in Huh7 cells stably transfected with lentivirus-mediated Sh-NC, Sh-TRIM7-1 or Sh-TRIM7-2. e Western blot analysis of Src, p-mTOR, p-S6K1, p-S6, and p-4E-BP1 protein levels in TRIM7-knockout Huh7 cells. f Western blot analysis of Src-mTORC1-S6K1 signaling in Huh7 cells transiently co-transfected with Flag-TRIM7 plasmid and HA-Src plasmid. g Western blot analysis of Src-mTORC1-S6K1 signaling in Huh7 cells transiently transfected with Si-NC, Si-TRIM7-1, or Si-TRIM7-2 before treatment with rapamycin (100 nM) for 24 h. ***P < 0.001 for statistical analysis of the indicated groups. The presented figures are representative of data from at least three independent experiments
TRIM7 acted as a tumor suppressor through its suppression of the Src-mTORC1-S6K1 axis in HCC cells
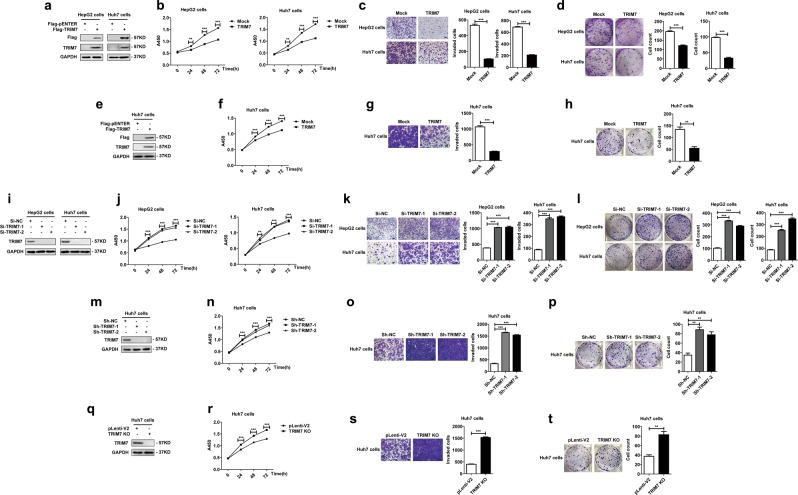
Since we defined the TRIM7-mediated negative regulation of Src, we were further interested in investigating whether TRIM7 had any effect on HCC cells. In HCC cells transiently or stably overexpressing TRIM7 (Fig. 4a, e), the proliferation (Fig. 4b, f), invasion (Fig. 4c, g), and colony formation (Fig. 4d, h) were significantly inhibited. In contrast, in the TRIM7-knockdown and -knockout HCC cells (Fig. 4i, m, q), the proliferation (Fig. 4j, n, r), invasion (Fig. 4k, o, s) and colony formation (Fig. 4l, p, t) were all significantly promoted. Thus, these data suggested that TRIM7 acted as a tumor suppressor and inhibited the malignant behaviors of HCC cells.
Fig. 4.
TRIM7 inhibited the malignant behaviors of HCC cells. a–d HepG2 and Huh7 cells were transiently transfected with Flag-TRIM7 plasmid or mock control plasmid, and successful overexpression of TRIM7 in the transfected cells was detected by western blot (a). The malignant behaviors of the transfected cells, including the proliferation (b), invasion (c), and colony formation (d) were further detected and analyzed. e–h Huh7 cells were transfected with Flag-TRIM7 plasmid or mock control plasmid, and the stably transfected cells were selected and further cultured. Effective stable overexpression of TRIM7 in these transfected cells was verified by western blot (e). The proliferation (f), invasion (g), and colony formation (h) of HCC cells stably overexpressing TRIM7 were further detected and analyzed. i–l HepG2 and Huh7 cells were transiently transfected with Si-TRIM7-1, Si-TRIM7-2, or Si-NC, and the successful knockdown of TRIM7 in the transfected cells was verified by western blot (i). The proliferation (j), invasion (k), and colony formation (l) of the TRIM7-knockdown cells were further detected and analyzed. m–p Huh7 cells were transfected with lentivirus-mediated Sh-TRIM7-1, Sh-TRIM7-2, or Sh-NC, and the stably transfected cells were selected and further analyzed. The successful knockdown of TRIM7 in the transfected cells was verified by western blot (m). The proliferation (n), invasion (o), and colony formation (p) of the TRIM7-knockdown Huh7 cells were further detected and analyzed. q–t TRIM7-knockout Huh7 cells were generated by the CRISPR/Cas9 system, and stable TRIM7-knockout clones were selected and further analyzed. The successful deletion of TRIM7 in these cells was verified by western blot (q). The proliferation (r), invasion (s), and colony formation (t) of the TRIM7-knockout cells were further detected and analyzed. **P < 0.01 and ***P < 0.001 for statistical analysis of the indicated groups. Scale bar in c, g, k, o, s: 100 μm. The presented figures are representative of data from at least three independent experiments
To define whether TRIM7-mediated degradation of Src protein was responsible for its antitumor effect, we co-transfected TRIM7 and Src plasmids into HCC cells and further detected the malignant behaviors of HCC cells. After successful overexpression of both TRIM7 and Src proteins (Fig. 5a), the results showed that exogenous overexpression of Src significantly rescued the TRIM7-mediated suppression of the proliferation (Fig. 5b), invasion (Fig. 5c), and colony formation (Fig. 5d) of HCC cells. These data verified that TRIM7 exerted its antitumor effect through its negative regulation of Src.
Fig. 5.
TRIM7 exerted its antitumor effect through modulation of the Src-mTORC1-S6K1 axis. a–d HepG2 and Huh7 cells were co-transfected with Flag-TRIM7 plasmid and HA-Src plasmid, and successful overexpression of TRIM7 and Src in the transfected cells was detected by western blot (a). The proliferation (b), invasion (c), and colony formation (d) of the transfected HCC cells were further detected and analyzed. e–h HepG2 cells were transfected with Si-NC, Si-TRIM7-1, or Si-TRIM7-2 before treatment with rapamycin (100 nM) for 48 h, and the protein levels of p-S6K1, p-S6, Src, and p-Src in the transfected cells were detected by western blot (e). The proliferation (f), invasion (g), and colony formation (h) of the transfected HepG2 cells were further detected and analyzed. *P < 0.05, **P < 0.01, and ***P < 0.001 for statistical analysis of the indicated groups. Scale bar in c, g: 100 μm. The presented figures are representative of data from at least three independent experiments
Next, we attempted to define whether TRIM7 acted as a tumor suppressor through its suppression of the mTORC1-S6K1 signaling pathway. Our data showed that treatment with the mTORC1 inhibitor rapamycin significantly abolished the increased protein levels of p-S6K1 and p-S6 but not Src and p-Src in Si-TRIM7-transfected HCC cells (Fig. 5e), which indicated that TRIM7 induced suppression of the mTORC1 pathway downstream of its negative regulation of the Src protein. Moreover, the proliferation (Fig. 5f), invasion (Fig. 5g), and colony formation (Fig. 5h) of Si-TRIM7-transfected cells were strikingly suppressed by rapamycin. Altogether, these data confirmed that TRIM7 exerted its antitumor effect through its suppression of the Src-mTORC1-S6K1 axis.
TRIM7 played a tumor suppressor role in HCC through its functional domains
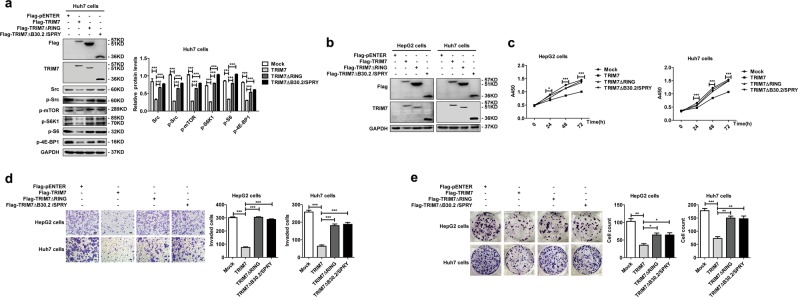
We identified the B30.2/SPRY domain as the binding domain of TRIM7 with Src, while the RING domain was necessary for the TRIM7 ubiquitination function. Thus, we further examined whether TRIM7 exerted its antitumor effect on HCC cells through these two domains. Our data showed that deletion of either the B30.2/SPRY domain or the RING domain in TRIM7 efficiently rescued the TRIM7-mediated inhibition of the Src-mTORC1-S6K1 axis (Fig. 6a), which indicated that TRIM7 suppressed the Src-mTORC1-S6K1 axis through these two domains. We further transfected these domain-deleted mutant plasmids into HCC cells and verified the successful overexpression of these two proteins in HCC cells by western blot (Fig. 6b). Our data showed that the overexpression of either of these two mutants could significantly abolish the TRIM7-mediated suppression of the proliferation (Fig. 6c), invasion (Fig. 6d), and colony formation (Fig. 6e) of HCC cells. Altogether, these data verified that both the B30.2/SPRY and RING domains were necessary for the tumor suppressor role of TRIM7 in HCC cells.
Fig. 6.
The RING domain and the B30.2/SPRY domain of TRIM7 were critical for the suppressor role of TRIM7. a Western blot analysis of Src-mTORC1-S6K1 signaling in Huh7 cells transfected with Flag-TRIM7 plasmid or TRIM7 truncation mutant (TRIM7ΔRING or TRIM7ΔB30.2/SPRY) plasmids. b–e HepG2 and Huh7 cells were transfected with Flag-TRIM7 plasmid or TRIM7 truncation mutant (TRIM7ΔRING or TRIM7ΔB30.2/SPRY) plasmids, and the successful overexpression of TRIM7 or its truncation mutants (TRIM7ΔRING or TRIM7ΔB30.2/SPRY) in the transfected cells was detected by western blot (b). The proliferation (c), invasion (d), and colony formation (e) of these transfected HCC cells were further detected and analyzed. *P < 0.05, **P < 0.01, and ***P < 0.001 for statistical analysis of the indicated groups. Scale bar in d: 100 μm. The presented figures are the representative of data from at least three independent experiments
Exogenous overexpression of TRIM7 suppressed tumorigenesis in xenograft tumor models
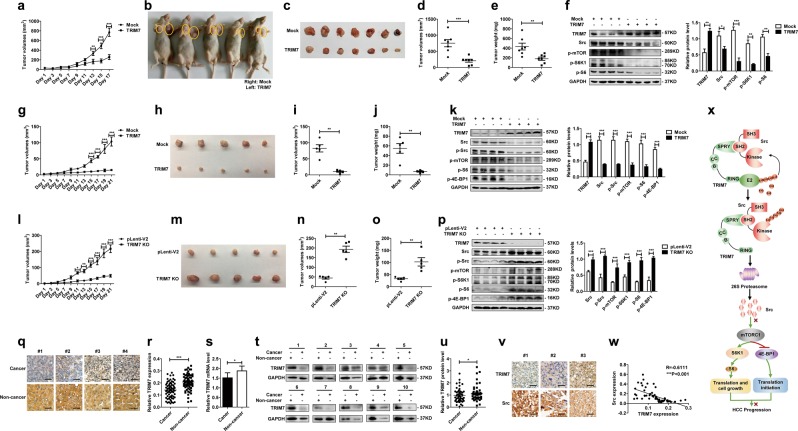
To investigate the effect of the TRIM7-Src-mTORC1 axis in an animal model, we constructed xenograft tumors in nude mice as described before [16, 17]. After visible tumors appeared, we injected Flag-TRIM7 plasmid into tumors in the left flanks and mock control plasmid into tumors in the right flanks of the mice every other day before the mice were sacrificed for analysis. Growth kinetics of the formed tumors showed that exogenous overexpression of TRIM7 significantly suppressed the growth of xenograft tumors overexpressing TRIM7 compared with the mock control tumors (Fig. 7a). The excised tumors from each group were compared, and the results indicated that tumors with TRIM7 overexpression were much smaller than those of the mock group (Fig. 7b, c). In addition, detailed analysis of the excised tumors showed that the exogenous overexpression of TRIM7 significantly reduced the volume and weight of xenograft tumors (Fig. 7d, e). The successful overexpression of TRIM7 in the TRIM7 plasmid-transfected group was verified by analyzing the extracted tumors from each group by western blot (Fig. 7f). Moreover, the Src level in the TRIM7-overexpressing tumors was significantly decreased, and the activation of the mTORC1-S6K1 pathway was also significantly inhibited (Fig. 7f), which further verified the TRIM7-mediated negative regulation of the Src-mTORC1 axis in vivo.
Fig. 7.
Xenograft tumor models and clinical HCC tissues validated the tumor suppressor role of TRIM7. a–f The Xenograft tumor model with TRIM7 plasmid injection was constructed as described before. When visible tumors appeared, 30 μg of TRIM7 plasmid or equal amount of mock control plasmid were injected into tumors in either flank once every other day. Tumors were measured every other day before the mice were sacrificed (a). Images were presented to show the representative of the mice with subcutaneous xenograft tumors in the TRIM7-transfected group and mock control group (b). Images were presented to show formed tumors isolated from sacrificed mice in the TRIM7-overexpressing group and mock control group (c). Statistical analysis of the volume (d) and weight (e) of formed tumors in the TRIM7-overexpressing group or mock control group. The protein levels of TRIM7 and the Src-mTORC1-S6K1 signaling in the TRIM7-overexpressing tumors and mock control tumors were detected by western blot (f). Band intensities of the key proteins were further quantitatively analyzed in the indicated groups and were presented in the right panel. g–k The TRIM7 stably overexpressing xenograft tumor model was constructed by subcutaneously injecting 1 × 107 TRIM7 stably overexpressing Huh7 cells into one flank of athymic nude mice, and injecting equal amount of mock control cells into the other flank of the athymic nude mice. Growth curves of the TRIM7 stably overexpressing tumors and mock control tumors were monitored every other day before the mice were sacrificed (g). Images were presented to show the formed tumors isolated from the TRIM7 stably overexpressing group and mock group (h). The volume (i) and weight (j) of the formed tumors in the TRIM7 stably overexpressing group and mock control group were statistically analyzed. The protein levels of TRIM7 and the Src-mTORC1-S6K1 signaling in the TRIM7 stably overexpressing tumors and mock control tumors were analyzed by western blot, and intensities of the key proteins were further quantitatively analyzed and were presented in the right panel (k). l–p The TRIM7-knockout xenograft tumor model was constructed by injecting 1 × 107 TRIM7-knockout cells into one flank of the nude mice and equal amount of control cells into the other side of the mice. The formed tumors were measured every other day before the mice were sacrificed (l). Images were presented to show the formed tumors isolated from the sacrificed mice in the TRIM7-knockout tumors and mock control tumors (m). The volume (n) and weight (o) of formed tumors in the TRIM7-knockout group and mock control group were statistically analyzed. The protein levels of TRIM7 and the Src-mTORC1-S6K1 signaling in the TRIM7-knockout tumors and mock control tumors were analyzed by western blot (p). Band intensities of the key proteins were further quantitatively analyzed in the indicated groups and were presented in the right panel. q IHC staining of TRIM7 in HCC tissues and corresponding noncancerous liver tissues from clinical HCC patients. Presented images are representative of figures from the investigated HCC patients. r Statistical analysis of TRIM7 expression by IHC assay in HCC tissues and corresponding noncancerous liver tissues from the investigated HCC patients. s Real-time PCR analysis of TRIM7 mRNA level in HCC tissues and corresponding noncancerous liver tissues from clinical HCC patients. t Western blot analysis of TRIM7 protein level in HCC tissues and corresponding noncancerous liver tissues from clinical HCC patients. Presented images are the representative of blots from the investigated HCC patients. u Statistical analysis of TRIM7 expression by western blot assay in HCC tissues and corresponding noncancerous liver tissues from the investigated HCC patients. v IHC staining of Src expression in HCC tissues from clinical HCC patients. Presented images are representative of figures from the investigated HCC patients. w Correlations analysis of TRIM7 and Src expression by IHC assay in HCC tissues from the investigated HCC patients. x Schematic illustration of the interaction between TRIM7 and Src proteins, as well as the related signaling pathway. *P < 0.05, **P < 0.01, and ***P < 0.001 for statistical analysis of the indicated groups. Scale bar in q, v: 50 μm
To further verify the tumor suppressor role of TRIM7 and its inhibitory effect on the Src-mTORC1 axis in vivo, we constructed another xenograft tumor model by subcutaneously injecting TRIM7 stably overexpressing Huh7 cells into one flank and mock control cells into the other flank of nude mice. The growth status of the formed tumors was monitored every other day before the mice were sacrificed. Our data showed that the stable overexpression of TRIM7 significantly suppressed tumor growth and that TRIM7 stably overexpressing tumors were significantly smaller than those of the mock group (Fig. 7g, h). Statistical analysis showed that both the volume and weight of the TRIM7 stably overexpressing tumors were significantly decreased compared with those of the mock group (Fig. 7i, j). Western blot assay verified the successful overexpression of TRIM7 and its significant suppressive effect on Src protein as well as its downstream mTORC1 pathway in TRIM7 stably overexpressing tumors (Fig. 7k).
To further confirm the effect of TRIM7 in vivo, we performed another set of xenograft tumor model by injecting TRIM7-knockout Huh7 cells generated by the CRISPR/Cas9 system into one flank of nude mice and the mock control cells into the other flank of nude mice. The growth status of the formed tumors was monitored before the tumors were excised. Our data showed that growth of the TRIM7-knockout tumors was significantly promoted (Fig. 7l, m). Statistical analysis demonstrated that the volume and weight of the TRIM7-knockout tumors were significantly increased compared with those of the mock group (Fig. 7n, o). Western blot analysis of these excised tumors demonstrated successful knockout of TRIM7 and subsequent activation of the Src-mTORC1 axis in response to TRIM7 knockout (Fig. 7p).
Altogether, these in vivo data verified that TRIM7 acted as a Src inhibitor and inhibited HCC tumor growth in xenograft tumor models through its suppression of the Src-mTORC1-S6K1 axis.
Loss of TRIM7 expression contributed to hepatocarcinogenesis in clinical HCC tissues
We determined that TRIM7 exerted antitumor effect through its negative regulation of Src in vitro and in vivo, and we were further interested in investigating whether abnormal TRIM7 expression contributed to tumorigenesis in clinical HCC patients. Thus, we assessed the expression level of TRIM7 in HCC tissues and corresponding distal noncancerous liver tissues by immunohistochemistry (IHC), real-time PCR, and western blot. First, the location and the expression of TRIM7 were detected by IHC in paired tissues from 80 HCC patients. The IHC data showed that TRIM7 was mainly expressed in the cytoplasm of HCC cells and hepatocytes, and the expression level of TRIM7 in HCC tissues was significantly downregulated compared with that in the corresponding distal noncancerous liver tissues (Fig. 7q, r, Supplementary Table S1). Next, we measured TRIM7 mRNA and protein levels in paired tissues from one cohort of 64 HCC patients and another cohort of 52 HCC patients, respectively. Similarly, both real-time PCR and western blot assays confirmed the IHC data, showing that both TRIM7 mRNA and protein levels in HCC tissues were significantly decreased compared with those in the corresponding distal noncancerous liver tissues (Fig. 7s, t, u). To verify the TRIM7-mediated negative regulation of Src in clinical HCC specimens, we further detected Src expression in HCC tissues. Our data showed that Src protein accumulated significantly in HCC tissues with the lower expression of TRIM7 (Fig. 7v), and statistical analysis showed that TRIM7 expression was significantly inversely correlated with the abundance of Src (Fig. 7w), which verified the TRIM7-mediated negative regulation of Src in clinical HCC patients. Altogether, the clinical investigations demonstrated that the loss of TRIM7 expression in HCC tissues might be involved in HCC progression and further verified the TRIM7-mediated negative regulation of Src in clinical specimens.
Altogether, we demonstrated that TRIM7 acted as an important negative regulator of Src as well as its downstream mTORC1-S6K1 signaling, thus suppressing HCC progression (Fig. 7x).
Discussion
Src interacts with several types of tyrosine kinase receptors, and it has been defined as a key molecule in tumor progression that can provide oncogenic signals for mitogenesis, cell survival, invasion, and metastasis [18]. Emerging evidence has shown that Src is hyperactive in the development of several types of cancers, including HCC [4, 19–22]. It has been reported that increased Src expression and activation are detected in more than 60% of HCC patients and is involved in disease progression [23, 24]. Due to the positive correlation between increased Src activity and cancer progression, targeting Src is emerging as a promising attractive strategy for improving the clinical therapeutic effects for cancer patients. Bosutinib, dasatinib, ponatinib, and vandetanib have been approved by the Food and Drug Administration as Src inhibitors for drug therapy of malignant diseases [4], which indicates the great potential of targeting Src in clinical treatment. Furthermore, Src mutants are very uncommon in tumors [4], thus discovering novel regulators of Src provides great therapeutic potential for cancers.
In this study, we demonstrated that TRIM7, a newly identified TRIM family member, is a novel negative regulator of Src. To date, ~100 human TRIM genes have been identified, and studies by us and others have indicated that alterations of TRIM proteins are involved in diverse pathological conditions, including carcinogenesis [16, 17, 25–27]. Many TRIM proteins act as E3 ubiquitin ligases and induce ubiquitination by directly interacting with their substrates and further regulating multiple biological processes, including protein stability, transcriptional control, signaling transduction, and cell cycle progression [14, 28, 29]. TRIM7 belongs to class IV of the TRIM family proteins, and the exact role of TRIM7 in physiological and pathological conditions remains largely unknown. In this study, we verified that TRIM7 directly interacted with Src protein both in vivo and in vitro, and the B30.2/SPRY domain of TRIM7 and the SH2 domain of Src protein were the molecular basis for the direct binding between TRIM7 and Src. We further showed that TRIM7 induced Lys48-linked polyubiquitination of Src via its RING domain, which ultimately led to the significant inhibition of Src activity. Thus, we have identified TRIM7 as a novel inhibitor of Src protein and defined the direct binding domains as well as the functional domains of TRIM7 and Src, which provided the molecular basis for further development of targeted therapy for the treatment of cancers.
Based on the critical involvement of Src activity in multiple cancers and the role of TRIM7 as a negative regulator of Src, we expected an antitumor effect of TRIM7 on cancers. Therefore, we tested the effect of TRIM7 on malignant behaviors of HCC cells, and our data demonstrated that TRIM7 suppressed the proliferation, invasion, and colony formation of HCC cells via its direct inhibition of Src protein in several cellular and animal models. We further demonstrated abnormal loss of TRIM7 expression in clinical HCC tissues and verified the negative correlation between TRIM7 and Src protein in clinical specimens, which indicated the involvement of the TRIM7-Src axis in HCC progression.
The contribution of Src activity in cancers is mainly attributable to its powerful activation of multiple downstream oncogenic signaling pathways; thus it has been recognized that targeting Src together with its downstream signaling is an attractive therapeutic strategy for cancer patients. In this study, we demonstrated that TRIM7 directly targeted Src for degradation and thus further led to efficient suppression of its downstream mTORC1-S6K1 signaling. Over the past decade, the critical role of mTORC1 signaling in HCC progression has been widely accepted. It has been reported that the mTORC1 pathway is frequently activated in up to half of HCCs and is highly associated with poor prognosis in HCC [30]. Thus, targeting the mTORC1 pathway provides an attractive strategy for improving clinical therapeutic effects for HCC patients. The critical role of Src in activating mTORC1 signaling has been demonstrated in several recent studies [31, 32], whereas the TRIM7-mediated regulation of the Src-mTORC1 signaling axis in HCC cells is reported here for the first time. Thus, this study provided evidence to define TRIM7 as a novel inhibitor of Src and opened a new avenue for treating mTORC1-overactive cancers by modulating TRIM7.
In conclusion, we identified TRIM7 as a novel negative regulator of Src. TRIM7 induced Lys48-linked, RING domain-dependent polyubiquitination of Src protein, which further led to the suppression of its downstream mTORC1-S6K1 signaling pathway, and acted as a tumor suppressor in HCC. Thus, this study identified a novel TRIM7-mediated regulatory mechanism of Src, and it also has clinical significance by providing a novel therapeutic strategy for Src-overactive cancers by modulating TRIM7.
Materials and methods
Cell culture and transfection
Human HepG2 and Huh7 HCC cells and human embryonic kidney 293T cells were obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China). All mycoplasma-free cells were cultured in DMEM-High Glucose medium (HyClone, Massachusetts, USA) supplemented with 10% fetal bovine serum and were authenticated by STR profiling by HKGENE (HKGENE Genetechnology, Beijing, China). The small interfering RNA targeting TRIM7 was synthesized by RIBOBIO (RIBOBIO, Guangzhou, China). TRIM7 and Src plasmids were synthesized by OriGene (OriGene Technologies, Maryland, USA) and Vigene (Vigene Biosciences, Rockville, USA), respectively. Truncation mutants of TRIM7 were generated using a KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. All of the transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Co-IP, western blot assay, immunofluorescence, and IHC
Co-IP, western blot assay, immunofluorescence, and IHC were performed as described previously [17, 33, 34]. Immunofluorescence and IHC assays were conducted in a manner blinded to the sample identity. The specific primary antibodies used in these assays included antibodies against TRIM7 (#ab170538, Abcam, Cambridge, MA, USA; and #HPA039213, Sigma-Aldrich, Saint Louis, MO, USA), Src (#2109, Cell Signaling Technology, Beverly, USA; and #11097-1-AP, Proteintech, Chicago, USA), Flag (#F1804, Sigma-Aldrich, Saint Louis, MO, USA), HA (#51064-2-AP, Proteintech, Chicago, USA), ubiquitin (#L1714, Santa Cruz Biotechnology, Dallas, USA), GAPDH (#6004-1-Ig, Proteintech, Chicago, USA), and β-actin (#TA-09, ZSGB-BIO, Beijing, China). The antibodies against the phosphorylation form of proteins used in western blot including antibodies against p-Src (#6943), p-mTOR (#2971), p-S6K1 (#9234), p-S6 (#4858), and p-4E-BP1 (#2855) were all bought from Cell Signaling Technology (Cell Signaling Technology, Beverly, USA).
Cell proliferation, invasion, and colony formation assays
HepG2 and Huh7 cells were plated in 96-well plates at a density of 104 cells/well, and the proliferation of HCC cells was detected by a CCK-8 Kit (Dojindo, Kumamoto, Japan) at 0, 24, 48, and 72 h according to the manufacturer’s instructions. Invasion assays and colony formation assays were performed as previously described [35, 36].
Real-time PCR assay
Total RNA was isolated from HCC cells or liver cancer tissues, and real-time PCR was performed as described before [17]. Primers for the human TRIM7 gene were forward: 5′-GCTCGGGGTTGAGATCACC-3′, reverse: 5′-CCAGGCACATTGCTACACCT-3′. Primers for the human Src gene were forward: 5′-GAGCGGCTCCAGATTGTCAA-3′, reverse: 5′-CTGGGGATGTAGCCTGTCTGT-3′. Relative mRNA levels of the genes were normalized to β-actin. Primers for the β-actin gene were forward: 5′-GGCACCACACCTTCTACAATG-3′, reverse: 5′-TAGCACAGCCTGGATAGCAAC-3′. The relative mRNA levels of target genes were obtained by using the 2−ΔΔCt method, and all assays were performed in triplicate.
In vitro binding and ubiquitination assay
The direct interaction between TRIM7 and Src proteins was assessed by a TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The Flag-TRIM7 and HA-Src proteins were expressed in vitro, mixed together, and analyzed by co-IP assay with anti-Flag antibody, followed by western blot assay with anti-HA antibody to determine the direct binding between TRIM7 and Src proteins. In vitro ubiquitination was analyzed with a ubiquitination kit (Boston Biochem, Cambridge, MA, USA) following the protocol recommended by the manufacturer. Briefly, Flag-TRIM7, and HA-Src proteins were expressed in vitro and further incubated with ubiquitin conjugation mixture at 30 °C for 60 min, followed by subsequent ubiquitination analysis of the Src protein by co-IP assay.
Generation of genetically modified stable HCC cell lines
To construct stable TRIM7-overexpressing HCC cells, a TRIM7 plasmid was transfected into Huh7 cells using Lipofectamine 2000. The successfully transfected cells were selected by complete medium containing 2 μg/mL of puromycin at 48 h after the transfection. The surviving colonies of transfected cells were further amplified, followed by validation by western blot. To generate a stable TRIM7-knockdown cell line, lentivirus carrying TRIM7-RNA interference sequence (Sh-TRIM7-1 and Sh-TRIM7-2) or its mock control (Sh-NC) were transfected into Huh7 cells using lentivirus (GeneChem Co., Ltd., Shanghai, China) according to the manufacturer’s protocol. The transfected cells were further selected with puromycin (2 μg/ml) and stable TRIM7-knockdown clones were validated by western blot. TRIM7-knockout cells were constructed with the CRISPR/Cas9 system according to the procedure described in the reference [37]. Briefly, a single-guide RNA (sgRNA) targeting TRIM7 (forward: 5′-CACCGAGAGAGGATGAGGCGCGGGT-3′, reverse: 5′-AAACACCCGCGCCTCATCCTCTCTC-3′) was transfected into Huh7 cells by using the pLentiCRISPRv2 system (Addgene). The sgRNA-transfected cells and the pLenti-V2-transfected mock control cells were further selected with puromycin (2 μg/ml) and the selected positive clones were cultured for further experiments.
In vivo tumor growth assay
All of the xenograft tumor models were constructed with 5-week-old immunodeficient BALB/c athymic male nude mice (Huafukang Biotechnology Ltd, Beijing, China). The xenograft tumor model with TRIM7 plasmid injection was constructed and analyzed in seven nude mice as described before [16, 17]. To construct the xenograft tumor model with TRIM7 stably overexpressing cells, 1 × 107 TRIM7 stably overexpressing Huh7 cells were injected into one flank, and equal amount of cells stably overexpressing mock plasmid were injected into the other flank of five athymic male nude mice. To construct the xenograft tumor model with the TRIM7-knockout cells, we injected 1 × 107 CRISPR/Cas9-based TRIM7-knockout Huh7 cells into one flank of five athymic male nude mice, and injected equal amount of mock control cells into the other flank. The formed tumors were measured and analyzed as previously described [33]. All mice for xenograft tumor models were randomly assigned. All of the protocols dealing with the animals were in accordance with guidelines of the Institutional Animal Care and Use Committee and were approved by the Medical Ethics Committee of Shandong University.
Clinical liver cancer specimens
Paired samples of HCC tissues and corresponding noncancerous liver tissues were collected from the Department of Hepatobiliary Surgery, Shandong Provincial Hospital Affiliated to Shandong University. Among these samples, 80 pairs of HCC tissues and corresponding noncancerous tissues were used for the IHC assay, 52 pairs of matched specimens were used for the western blot assay to detect the protein level of TRIM7, and 64 pairs of matched specimens were used for real-time PCR to determine the mRNA level of TRIM7. Details of the clinicopathologic characteristics of the recruited HCC patients were shown in Table 1. Written informed consents were obtained from all patients before the study was initiated. All of the protocols were in accordance with the Helsinki Declaration and approved by the Medical Ethics Committee of Shandong University.
Table 1.
Clinicopathologic characteristics of HCC patients in cohorts for IHC, real-time PCR, or western blot
| Cohort 1 (n = 80, for IHC) | Cohort 2 (n = 64, for real-time PCR) | Cohort 3 (n = 52, for western blot) | |
|---|---|---|---|
| Characteristics | No. of patients (%) | No. of patients (%) | No. of patients (%) |
| Gender | |||
| Male | 72 (90.0%) | 54 (84.4%) | 36 (69.2%) |
| Female | 8 (10.0%) | 10 (15.6%) | 16 (30.8%) |
| Age | |||
| <54 | 38 (47.5%) | 28 (43.8%) | 26 (50.0%) |
| ≥54 | 42 (52.5%) | 36 (56.2%) | 26 (50.0%) |
| HBV infection | |||
| Yes | 14 (17.5%) | 20 (31.3%) | 20 (38.5%) |
| No | 66 (82.5%) | 44 (68.7%) | 32 (61.5%) |
| Liver cirrhosis history | |||
| Yes | 75 (93.8%) | 48 (75.0%) | 39 (75.0%) |
| No | 5 (6.2%) | 16 (25.0%) | 13 (25.0%) |
| Tumor size | |||
| <5 cm | 39 (48.8%) | 34 (53.1%) | 35 (67.3%) |
| ≥5 cm | 41 (51.2%) | 30 (46.9%) | 17 (32.7%) |
| TNM stage | |||
| I | 36 (45.0%) | 31 (48.4%) | 33 (63.5%) |
| II | 3 (3.8%) | 3 (4.7%) | 2 (3.8%) |
| III | 37 (46.2%) | 21 (32.8%) | 11 (21.2%) |
| IV | 4 (5.0%) | 9 (14.1%) | 6 (11.5%) |
| BCLC stage | |||
| 0 | 2 (2.5%) | 14 (21.9%) | 1 (1.9%) |
| A | 39 (48.8%) | 19 (29.7%) | 29 (55.8%) |
| B | 13 (16.3%) | 8 (12.5%) | 12 (23.1%) |
| C | 25 (31.2%) | 23 (35.9%) | 8 (15.4%) |
| D | 1 (1.2%) | 0 (0.0%) | 2 (3.8%) |
Statistical analysis
Data were statistically analyzed using SPSS 22.0 software (SPSS, IL, USA) and GraphPad Prism 5 software (GraphPad, CA, USA). Quantitative variables were evaluated with two-tailed Student’s t test or one-way analysis of variance, while categorical variables were analyzed by χ2-tests. Correlations for categorical and continuous variables were evaluated by Spearman’s rank correlation test and Pearson’s correlation test, respectively. Data were presented as mean ± SEM. A P value < 0.05 was considered statistically significant, and the P value used in all analyses was two-tailed.
Supplementary information
Supplementary materials-The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein
Acknowledgements
This study was supported by the National Natural Science Foundation of China (no. 81672391, no.81972275, no.81472269, no.81602550, and no.81772626) and the Major Innovation Project of Shandong Province (no. 2018CXGC1217).
Data availability
All relevant data that support the findings of this study are available from the corresponding author upon request. Supplementary information is available at Cell Death & Differentiation’s website.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by V. D’Angiolella
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-019-0464-9) contains supplementary material, which is available to authorized users.
References
- 1.Engen JR, Wales TE, Hochrein JM, Meyn MA, 3rd, Banu Ozkan S, Bahar I, et al. Structure and dynamic regulation of Src-family kinases. Cell Mol life Sci. 2008;65:3058–73. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chojnacka K, Mruk DD. The Src non-receptor tyrosine kinase paradigm: new insights into mammalian Sertoli cell biology. Mol Cell Endocrinol. 2015;415:133–42. doi: 10.1016/j.mce.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–27. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 4.Roskoski R., Jr Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9–25. doi: 10.1016/j.phrs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–49. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 6.Espada J, Martin-Perez J. An update on Src family of nonreceptor tyrosine kinases biology. Int Rev cell Mol Biol. 2017;331:83–122. doi: 10.1016/bs.ircmb.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. J Signal Transduct. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–58. doi: 10.1023/A:1023772912750. [DOI] [PubMed] [Google Scholar]
- 9.Elsberger B, Paravasthu DM, Tovey SM, Edwards J. Shorter disease-specific survival of ER-positive breast cancer patients with high cytoplasmic Src kinase expression after tamoxifen treatment. J Cancer Res Clin Oncol. 2012;138:327–32. doi: 10.1007/s00432-011-1096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE. Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis. Cancer. 2002;94:344–51. doi: 10.1002/cncr.10221. [DOI] [PubMed] [Google Scholar]
- 11.He P, Wu W, Wang H, Liao K, Zhang W, Xiong G, et al. Co-expression of Rho guanine nucleotide exchange factor 5 and Src associates with poor prognosis of patients with resected non-small cell lung cancer. Oncol Rep. 2013;30:2864–70. doi: 10.3892/or.2013.2797. [DOI] [PubMed] [Google Scholar]
- 12.Skurat AV, Dietrich AD, Zhai L, Roach PJ. GNIP, a novel protein that binds and activates glycogenin, the self-glucosylating initiator of glycogen biosynthesis. J Biol Chem. 2002;277:19331–8. doi: 10.1074/jbc.M201190200. [DOI] [PubMed] [Google Scholar]
- 13.Zhai L, Dietrich A, Skurat AV, Roach PJ. Structure-function analysis of GNIP, the glycogenin-interacting protein. Arch Biochem Biophys. 2004;421:236–42. doi: 10.1016/j.abb.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Montori-Grau M, Pedreira-Casahuga R, Boyer-Diaz Z, Lassot I, Garcia-Martinez C, Orozco A, et al. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism. 2018;83:177–87. doi: 10.1016/j.metabol.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y, et al. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24:762–5. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Ma X, Qiu Y, Zhu L, Lin Y, You Y, et al. TRIM50 suppressed hepatocarcinoma progression through directly targeting SNAIL for ubiquitous degradation. Cell Death Dis. 2018;9:608. doi: 10.1038/s41419-018-0644-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo P, Ma X, Zhao W, Huai W, Li T, Qiu Y, et al. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1-TSC2 complex. Oncogene. 2018;37:478–88. doi: 10.1038/onc.2017.349. [DOI] [PubMed] [Google Scholar]
- 18.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 19.Ku M, Wall M, MacKinnon RN, Walkley CR, Purton LE, Tam C, et al. Src family kinases and their role in hematological malignancies. Leuk Lymphoma. 2015;56:577–86. doi: 10.3109/10428194.2014.907897. [DOI] [PubMed] [Google Scholar]
- 20.Varkaris A, Katsiampoura AD, Araujo JC, Gallick GE, Corn PG. Src signaling pathways in prostate cancer. Cancer Metastasis Rev. 2014;33:595–606. doi: 10.1007/s10555-013-9481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu ST, Pham H, Pandol SJ, Ptasznik A. Src as the link between inflammation and cancer. Front Physiol. 2013;4:416. doi: 10.3389/fphys.2013.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Sayed I, Helmy MW, El-Abhar HS. Inhibition of SRC/FAK cue: a novel pathway for the synergistic effect of rosuvastatin on the anti-cancer effect of dasatinib in hepatocellular carcinoma. Life Sci. 2018;213:248–57. doi: 10.1016/j.lfs.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Lau GM, Lau GM, Yu GL, Gelman IH, Gutowski A, Hangauer D, et al. Expression of Src and FAK in hepatocellular carcinoma and the effect of Src inhibitors on hepatocellular carcinoma in vitro. Digestive Dis Sci. 2009;54:1465–74. doi: 10.1007/s10620-008-0519-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Guo TF, Jing ZT, Yang Z, Liu L, Yang YP, et al. Hepatitis B virus core protein promotes hepatocarcinogenesis by enhancing Src expression and activating the Src/PI3K/Akt pathway. FASEB J. 2018;32:3033–46. doi: 10.1096/fj.201701144R. [DOI] [PubMed] [Google Scholar]
- 25.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Zhang Y, Huang J, Wong CC, Zhai J, Li C, et al. TRIM67 activates p53 to suppress colorectal cancer initiation and progression. Cancer Res. 2019;79:4086–98. doi: 10.1158/0008-5472.CAN-18-3614. [DOI] [PubMed] [Google Scholar]
- 27.Tan P, Ye Y, He L, Xie J, Jing J, Ma G, et al. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018;16:e3000051. doi: 10.1371/journal.pbio.3000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty A, Diefenbacher ME, Mylona A, Kassel O, Behrens A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat Commun. 2015;6:6782. doi: 10.1038/ncomms7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, et al. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in arabidopsis. Dev Cell. 2017;41:47–58 e44. doi: 10.1016/j.devcel.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhat M, Sonenberg N, Gores GJ. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810–8. doi: 10.1002/hep.26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ondrusova L, Reda J, Zakova P, Tuhackova Z. Inhibition of mTORC1 by SU6656, the selective Src kinase inhibitor, is not accompanied by activation of Akt/PKB signalling in melanoma cells. Folia Biologica. 2013;59:162–7. [PubMed] [Google Scholar]
- 32.Vojtechova M, Tureckova J, Kucerova D, Sloncova E, Vachtenheim J, Tuhackova Z. Regulation of mTORC1 signaling by Src kinase activity is Akt1-independent in RSV-transformed cells. Neoplasia. 2008;10:99–107. doi: 10.1593/neo.07905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia X, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. doi: 10.1038/srep06248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X, et al. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Investig. 2014;94:52–62. doi: 10.1038/labinvest.2013.126. [DOI] [PubMed] [Google Scholar]
- 35.Cao L, Han L, Zhang Z, Li J, Qu Z, Du J, et al. Involvement of anoikis-resistance in the metastasis of hepatoma cells. Exp Cell Res. 2009;315:1148–56. doi: 10.1016/j.yexcr.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Wei Q, Guo P, Mu K, Zhang Y, Zhao W, Huai W, et al. Estrogen suppresses hepatocellular carcinoma cells through ER beta-mediated upregulation of the NLRP3 inflammasome. Lab Investig. 2015;95:804–16. doi: 10.1038/labinvest.2015.63. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Lun M, Liu Y, Wang H, Yan Y, Sun J. CRISPR/Cas9-mediated chicken TBK1 gene knockout and its essential role in STING-mediated IFN-beta induction in chicken cells. Front Immunol. 2018;9:3010. doi: 10.3389/fimmu.2018.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials-The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein
Data Availability Statement
All relevant data that support the findings of this study are available from the corresponding author upon request. Supplementary information is available at Cell Death & Differentiation’s website.