Abstract
Early diagnosis of Ehrlichia ruminantium in cattle is a recipe for effective control of heartwater in ruminants. Hence, we assessed the presence of E. ruminantium in the blood of cattle and the engorged Amblyomma variegatum by nested PCR. The electrophoresed PCR products obtained after primary and secondary amplifications revealed amplicon sizes of about350 bp and 280 bp respectively, which corresponded with the partial region of pSC20 gene amplified. Sequences obtained had 95–99% homology with those sequences available in GenBank. The prevalence of the E. ruminantium in ticks (50%; 126/252) was significantly (p < 0.05) higher than that in cattle blood 23.55% (61/259). The prevalence was significantly (p < 0.05) higher in ticks from adult cattle 51.47% (133/259) than those from the young cattle 44.86% (116/259) and in tick from females 54.55% (141/259) than in ticks from the males 41.38% (107/259). Alignment of autochthonous sequences revealed that the three sequences were polymorphic with two sequences showing similar nucleotides deletion at points 87–91 and 107–108. The phylogenetic trees inferred by ML showed topologies with two autochthonous sequences, one each from cattle blood and tick, clustering together in one clade and the other clustering within those sequences from South Africa and Zimbabwe in another clade. In conclusion, this study revealed a higher prevalence of E. ruminantium in engorged A. variegatum than in the blood of infected cattle. Hence, it is suggested that the amplification that targets the pCS20 gene in engorged ticks may be more suitable to determine the E. ruminantium carrier status of cattle.
Keywords: Cattle, Ehlichia ruminantium, Amblyomma variegatum, pCS20 gene, Nigeria
Introduction
Cowdriosis, also known as heartwater, is a rickettsial disease of ruminants caused by Ehrlichia ruminantium. It is transmitted by the hard ticks of the genus Amblyomma (Allsopp 2010; Kasari et al. 2010). The organism is pleomorphic in nature but the coccoid shape is usually encountered in naturally infected ruminants (Hart et al. 1991). Though the organisms are found in the granulocytes of the infected animals, the replicating stage is found within the endothelial cells of the blood vessels (Kasari et al. 2010). The occurrence and distribution of heartwater is well documented with its animal reservoir poorly understood (Deem et al. 1996) but infections in ruminants have been reported where the tick vectors are present (Deem et al. 1996; Yunker 1996). Heartwater is an important disease that could result in the death or survival of the infected ruminants and in the recent past, it has been listed among the emerging zoonoses (Louw et al. 2006; Esemu et al. 2011; Chitanga et al. 2014) as well as a potent agricultural biothreat (Esemu et al. 2011).
The clinical manifestation of heartwater depends on the strain of the infecting parasite, the breed of animal infected and its immune system. The infection may be peracute (characterized by sudden death of the infected animal) or acute (in which the infected animal has fever, anorexia, listlessness, congested mucous membranes and respiratory signs). In these two forms of heartwater, nervous signs characterized by chewing movements, protrusion of the tongue, twitching of the eyelids and walking in circles with a high-stepping gait are observed. Terminally, the animal often goes into lateral recumbency with paddling movements before death (Van de Pypekamp and Prozesky 1987).
Detection and diagnosis of heartwater can be achieved by xenodiagnosis (Peter et al. 2000), a method with its own limitations. Aside the fact that it is time consuming, cumbersome and expensive, it has been shown to have low sensitivity in mice infected with stabilate from infected ticks (Peter et al. 2000). However, the definitive diagnosis of cowdriosis has been achieved by examination of capillaries in brain smears. These two methods are not suitable for wide scale epidemiological study of heartwater. Hence, more sensitive PCR-based assay methods for the detection of Ehrlichia ruminantium in blood sample or ticks have been developed and applied worldwide (Peter et al. 2000; Martinez et al. 2004; Sayler et al. 2016).
In Nigeria, information on the prevalence of heartwater in cattle and E. ruminantum in Ambyomma ticks is scanty due to the difficulty in the diagnosis of the infection in ruminants and ticks. Two studies in the Northern part of the country attempted to characterize the strains of E. ruminantum by xenodiagnostic method (Leeflang and Ilemobade 1977) and shed light on the prevalence of E. ruminantium in cattle using generic PCR assay and sequencing (Lorusso et al. 2016) but to the best of our knowledge, there is no report on the prevalence and molecular detection of the parasite in the blood of cattle and engorged ticks infesting them using species specific PCR assay. Hence, this study attempted to shed light on the E. ruminantium carrier status of Amblyomma variegatum ticks and cattle that were under semi-intensive management in Abeokuta, in southern part of Nigeria using species specific nested PCR, sequencing and sequence analysis in an effort to add to the existing body of knowledge on epidemiology and genetic diversity of Ehrlichia ruminantium.
Materials and methods
Study area
The animals sampled were cattle under semi-intensive system of management in Abeokuta, Ogun State. The State is very close to Lagos, the major entry point into Nigeria. Ogun State is bordered by Oyo and Osun States to the north, Lagos State to the south, Ondo State to the east and Republic of Benin to the west. It is located between Longitude 3.0° E and 5.0° E and Latitude 6.2° N and 7.8° N. The weather in the state is favorable for tick development all year round with average temperature of 27.2 °C.
Animals, sample collection and preservation
Semi-intensively managed cattle of different breeds and ages were randomly sampled from Abeokuta and environs. For the purpose of analysis, the cattle (259) were grouped into two, the Bos indicus (218) and Bos taurus (41). Blood samples were collected from the jugular vein into 5 ml tubes containing Disodium ethylenediaminetetraacetic acid (EDTA) as anticoagulant. The samples were transported on ice packs to the laboratory. Ticks were collected manually from various body regions of the sampled cattle as described by Okello-Onen and Hassan (2006). The ticks collected from each cattle were kept in separate bottles containing 70% ethanol, labelled appropriately and transported to the laboratory. The ticks were identified as described by Okello-Onen and Hassan (2006). 252 engorged Amblyomma ticks, one each from a cattle, was collected and checked for presence of E. ruminantium DNA.
DNA extraction
Genomic DNA was extracted from the EDTA anti-coagulated blood samples using Quick-gDNA™ BloodPrep (Zymo Research Corporation, Irvine, CA 92614, U.S.A) as described by Takeet et al. (2013). Genomic DNA was extracted from the ticks using the Zymo kit above but with slight modification. Briefly: one engorged tick collected from an animal was thoroughly washed in sterile water and dried. The ticks were crushed using clean sterilized pestle and mortar. About 400 µl of nuclease free water was added to the crushed ticks and thoroughly mixed to form a homogenous mixture. 200 µl of Genomic Lysis Buffer and 15 µl of proteinase K was added to 50 µl of the mixture, vortexed for 30 s and incubated at 55 °C for 10 min. The mixture was transferred into a Zymo-spin™ IC column in a collection tube and then centrifuged at 10,000×g for 1 min. The collection tube with the fluid was then discarded. The Zymo-spin™ Column was put into a new collection tube, 200 µl of DNA pre-wash buffer added and centrifuged at 10,000×g for 1 min. The genomic DNA was further washed by adding 500 µl of g-DNA wash buffer to the spin column and centrifuged at 10,000×g for 1 min. The DNA was eluted in 1.5 ml Eppendorf tube by adding 50 µl of DNA Elution Buffer, incubated at room temperature for 2–5 min and then centrifuged at 16,500×g for 30 s. The eluted DNA was then stored at − 20 °C until use.
Amplification of PCS20 fragment
A nested PCR, targeting partial region of pSC20 gene, specific for E. ruminantium was performed to amplify E. ruminantium DNA from cattle and Amblyomma ticks found on each animal. Two pairs of primer sets, U24, L24 and AB128, AB129 (Table 1) obtained from Bioneer Inc, USA were used for the primary and nested PCR, respectively. The reactions were performed in 20 µl final volume containing 10 µl of 2x PCR premix, with dye (SydLabs, Inc. USA), 8 µl of nuclease free water (Qiagen, USA) and 0.5 µl (40 ng) each of the forward and reverse primers and 1 µl of the genomic DNA.
Table 1.
Primer sets used for nested PCR to detect E. ruminantium from the blood and Amblyomma ticks obtained from extensively grazed cattle from Nigeria
The reaction conditions were as follows for the primary reactions: initial denaturation of DNA was 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min, this was followed by final extension of 72 °C for 10 min. For the nested reaction, 1 µl of the product from the primary reaction was added to 10 µl of the 2x PCR premix with dye, 8 µl of nuclease free water and 0.5 µl (40 ng) each of the AB128 and AB129 primer set, with the same conditions as the primary reaction. It should be noted the amplifications were carried out blindly as we did not optimize the synthesized primers with a known positive sample but in every reaction, nuclease free water was used as negative control.
Ten microliter of the PCR products was electrophoresed through 1% agarose gel stained with Gel Red (Phoenix Research Product, Candler, NC, USA) in 1 x TAE buffer solution at 90 V for 60 min. 10 µl of a 100 bp DNA ladder (BiolabInc, USA) was also electrophoresed alongside.
Sequencing and phylogenetic analysis
To validate our results, four PCR products (consisting of two each for cattle and Amblyomma) from those showing the expected band size were randomly selected and unidirectionally sequenced using the forward (AB129) of the secondary primer in a commercial molecular laboratory (Sequetech, Mountain View, California, USA). The nucleotide sequences obtained were viewed on FinchTV (Geospiza, Inc. Seattle, WA, USA) and manually cleaned. The sequences were subjected to BLAST search for homology in the GenBank of NCBI data base and highly similar nucleotide sequences of E. ruminantium from the GenBank and those from this study were aligned in BioEdit software. The phylogenetic tree was constructed using Maximum Likelihood (ML) algorithm in the Molecular Evolutionary Genetic Analysis (MEGA) software version 5.
Nucleotide sequence data reported in this paper are available in the DDBJ databases under the accession numbers: LC385765, LC385766 and LC385767.
Data analysis
Estimate of the sample size required to generate a true prevalence was based on an equation described by Thrusfield (1995). Where n = [Z2 × P × (1 − P)] ÷ d2. At a confidence level of 95% (Z = 1.96), with an expected prevalence of 19% in cattle (P = 0.19) and precision of 5% (d = 0.05) the calculated minimum sample size (n) to obtain a statistically significant estimate of true prevalence was 250. The expected prevalence of 19% adopted in this study was that of Faburay et al. (2007) who employed similar detection method used in this study. Descriptive statistic was used to summarize the data obtained from this study. The prevalence was compared with Chi square in SPSS version 19.
Results
Animals and tick identification
Two hundred and fifty-nine cattle consisting of 90 (34.7%) males and 169 (65.3%) females were sampled. The Bos indicus sampled consisted of 179 White Fulani, 21 Sokoto Gudali, 12 Red Bororo, 4 Keteku and 2 Jali while the Bos taurus included 24 N′ Dama and 17 Muturu. Their ages were grouped into two, < 24 months referred to as young (109) and ≥ 24 months referred to as adult (150). 252 engorged ticks were all identified Amblyomma species.
Detection of Ehrlichia DNA by PCR
The electrophoresed PCR products obtained after amplification with primary and secondary primers sets revealed amplicon sizes of about 350 bp and 280 bp, respectively. These corresponded with the expected band sizes of the amplified partial region of pSC20 gene using the listed primers. The sequences obtained from this study had between 95 and 99% homology with those sequences derived from the NCBI data base (Table 2). Among the 259 cattle and 252 A. variegatum ticks screened for E. ruminantium, 23.55% (61/259) and 50% (126/252) respectively showed amplicon size 280 bp following the nested PCR. The prevalence of E. ruminantium DNA in ticks was significantly (p < 0.05) higher than in the corresponding cattle blood.
Table 2.
Autochthonous sequences compared to reference isolates with their accession number, percentage homology, source and country of origin
| Isolate | Accession number | Identity (%) | Source | Strain/country of origin |
|---|---|---|---|---|
| ErB338Nig | AB218277 | 96 | A. lepidum | Gedaref/Sudan |
| AY236061 | 95 | NS | Gardel/Caribbean | |
| DQ631922 | 96 | R. evertsi | Ree5_2/S. Africa | |
| JQ039931 | 96 | A. variegatum | Dumbo/Cameroun | |
| GU797236 | 96 | A. gemma | 20/Ethiopia | |
| ErT389Nig | AB218277 | 99 | A. lepidum | Gedaref/Sudan |
| DQ631925 | 99 | Cattle | MB9_02/S. Africa | |
| AY236060 | 99 | MO | Vosloo/S. Africa | |
| JQ039923 | 99 | A. variegatum | Buea55/Cameroun | |
| ErT390Nig | JQ039939 | 97 | A. variegatum | Dumbo48/Cameroun |
| GU644448 | 97 | A. gemma | 31/Ethiopia | |
| DQ655712 | 97 | CS | Kiswani/S. Africa | |
| JQ039914.1 | 98 | A. variegatum | Buea20/Cameroun | |
| GU797236.1 | 98 | A. gemma | 20/Ethiopia |
Key: ErB338Nig; Ehrlichia ruminantium from blood of cattle, ErT389Nig; Ehrlichia ruminantium from haemolymph of Amblyomma variegatum, ErT390Nig; Ehrlichia ruminantium from haemolymph of Amblyomma variegatum, NS not stated, MO multiple origin, CS Cultured strains
Effect of age, sex and breed on the prevalence of E. ruminantium
The prevalence of E. ruminantium in different age and sex are shown in Table 3. There was no significant (p > 0.05) difference in the prevalence of E. ruminantium either amongst the sexes or age groups, but there was significant difference (p < 0.05) in the prevalence of E. ruminantium among the breeds of cattle sampled with the highest prevalence in White Fulani cattle (25.70%; 46/179). Also, there was significant (p ≤ 0.05) difference between the E. ruminantium prevalence in A. variegatum collected on male and female and on young and adult groups.
Table 3.
Effect of age and sexon the prevalence of Ehrlichia ruminantium in cattle
| Age Group | Blood | Tick | ||
|---|---|---|---|---|
| Number of cattle | Number positive (%) | Number of cattle | Number positive (%) | |
| Young | 109 | 24 (22.01)a | 107 | 48 (44.86)a |
| Adult | 150 | 36 (24.10)a | 145 | 78 (53.79)b |
| Total | 259 | 60 (23.17) | 251 | 126 (50.0) |
| Sex | ||||
| Male | 90 | 24 (26.67)a | 87 | 36 (41.38)b |
| Female | 169 | 37 (21.89)a | 165 | 90 (54.55)a |
| Total | 259 | 61 (23.55) | 252 | 126 (50.0) |
Variables with different superscript are significantly (p < 0.05) different
Sequencing and phylogenetic analysis
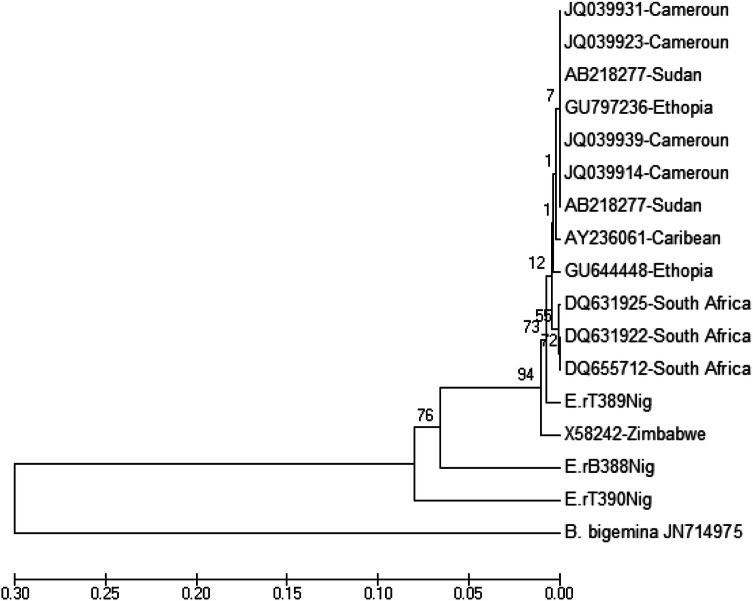
Four PCR products were sent for sequencing but only three of the obtained sequences were readable and were used for the construction of phylogenetic tree. The sequences were deposited in GenBank with accession number LC385765 (ErB388Nig), LC385766 (ErT389Nig) and LC385767 (ErT390Nig). The lengths of the three sequences were 248 bp, 248 bp and 399 bp, respectively with G-C content that range from 34.3 to 50.4%. The aligned autochthonous sequences (Fig. 1) revealed that the three sequences were polymorphic. The phylogenetic trees inferred by ML show topology with two autochthonous sequences, one each from cattle and tick, clustering together in one clade and the other clustering between those sequences from South Africa and Zimbabwe (Fig. 2).
Fig. 1.
Aligned sequences of pCS20 gene of Ehrlichia ruminantium detected in Amblyomma variegatum ticks and blood of cattle with natural infection in Abeokuta, Nigeria
Fig. 2.
Phylogenetic relationship based on cytochrome Oxidase III (pCS20) gene sequences of Ehrlichia ruminantium detected in Amblyomma variegatum and cattle in Abeokuta. The phylogenetic analysis was carried using Maximum Likelihood (ML), involving a bootstrap procedure with 1000 replicate and evolutionary distance adjusted using the Kimurra-2 parameter
Discussion and conclusion
A better understanding of the epidemiological distribution and genetic variation of E. ruminantium in cattle and tick vector infesting them is important for effective control of the disease cause by the parasite. To contribute to this, we assessed the presence of E. ruminantium DNA in the blood of cattle and the engorged A. variegatum infesting them by nested pCS20-PCR and sequence analysis.
Majority of the cattle sampled were White Fulani. This observation is consistent with the reports of RIM (1993) and Ahamefule et al. (2007) in Nigeria. The preference of farmers for this breed has been associated with their high calving and feed conversion rate (Synge 1980; Otchere 1983) and tolerance to trypanosomosis in tsetse infested areas.
The E. ruminantium prevalence of 23.55% reported in the blood of cattle sampled in this study is much higher than 1.1% reported by Lorusso et al. (2016) who utilized Reverse Line Blotting (RLB) for the parasite detection.
The variation in the results may not be unconnected with PCR probes utilized in the study, as many reports have adjudged the pCS20-PCR detection of E. ruminantium as the most specific and sensitive (Peter et al. 2000; Steyn et al. 2003; Mahan et al. 2004; Steyn et al. 2008); whereas the PCR detection method in the report of Lorusso et al. (2016) utilized a catch it all PCR (generic for Anaplasma and Ehrlichia species), the sensitivity and specificity of which has not been determined. The higher prevalence recorded in cattle in this study may underscore the importance of cattle as reservoir of E. ruminantium in the study area, especially to small ruminants. But the high prevalence of 50% recorded in the engorged Amblyomma spp. in this study could not be compared due to paucity of information on the prevalence of the parasite in A. variegatum ticks in Nigeria. However, a study in two states (Plateau and Nassarawa) of Nigeria by Reye et al. (2012) indicated detection of E. ruminantium in two unidentified ticks whereas, in a similar study conducted by Ogo et al. (2012) in the same states, no E. ruminantium DNA was detectable in all the ticks sampled including A. variegatum. Though both studies utilized generic primers, there is need for further studies that employs species specific primers to throw more light on the possible role play by other genera of ticks in the epidemiology of heartwater. This suggestion becomes imperative as reports on the detection of E. ruminantium in tick genera, other than Amblyomma, is on the increase (Allsopp et al. 2007; Reye et al. 2012; Biguezoton et al. 2016). The prevalence of heartwater in cattle and A. variegatum has been studied extensively in Africa and the Caribbean Islands (Bell-Sakyi et al. 1996; Awa 1997; Knopf et al. 2002 Faburay et al. 2007 and Molia et al. 2008) but all their reports are at variant with the prevalence reported in this study. For instance, studies of heartwater carried out by Molia et al. (2008) in the Carribean, Faburay et al. (2007) in the Gambia and Esemu et al. (2012) in Cameroun reported prevalences of 19.1%, 1.6–15.1% and 28.4%, respectively in A. variegatum ticks using pCS20-PCR.
The polymorphism shown by the aligned autochthonous pCS20 sequences may suggest that more than one strain of E. ruminantium exists in the study area which has been reported in other part of West Africa (Adakal et al. 2010). The pathogenicity of these strains may need further investigation as our study did not assess the clinical/health status of the sampled cattle. Though the cattle looked apparently healthy, which may suggest that the local breeds seem not to suffer from E. ruminantium infection but act as sentinel of the infection to other susceptible ruminants especially exotic breeds.
The phylogenetic tree of the E. ruminantium pCS20 partial sequences from this study separated to two different clades, the autochthonous sequences. This may support the suggestion vide supra that there may be more than one strain of the parasite in circulation in the study area. One of the sequences has affinity for those sequences from the South Africa and Zimbabwe, and the remaining two sequences were well separated into a separate clade which suggests that those autochthonous sequences represent two distinct genotypes. It must be mentioned that the analysis of these three sequences may not be good enough to draw genetic diversity inference in Nigeria therefore, there is the need to carry out extensive molecular detection and sequences analysis in other regions in order to understand the epidemiological distribution of different strains and genotypes among E. ruminantium in ruminants and ticks in Nigeria. The clustering together of those sequences from South Africa may be an indication that the pCS20 gene is highly conserved as against those sequences obtained from this study that exhibited some level of variation. This variation is not in agreement with the report of Heerden et al. (2004) and Allsopp and Allsopp, (2007) who suggested that pCS20 gene is more conserved in West the African strain of E. ruminantium.
In conclusion, this study revealed higher prevalence of E. ruminantium in engorged Amblyomma ticks than in the blood of infected cattle from the study area. Hence, it is suggested that the amplification that targets the pCS20 gene in engorged ticks may be more suitable to determine the E. ruminantium carrier status of cattle.
Acknowledgements
We thank Mr Olugbogi, E. I. of the Department of Veterinary Microbiology and Parasitology for technical support rendered during the course of this study.
Compliance with ethical standards
Ethical consideration
Ethical approval, Number FUNAAB/COLVET/CREC/001/18 was obtained from the College of Veterinary Medicine Ethical Committee, College of Veterinary Medicine, Federal University of Agriculture Abeokuta, Nigeria, before commencing the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adakal H, Stachurski F, Konkobo M, Zoungrana S, Meyer DF, Pinarello V, Aprelon R, Marcelino I, Alves PM, Martinez D, Lefrancois T, Vachiéry N. Efficiency of inactivated vaccines against heartwater in Burkina Faso: impact of Ehrlichia ruminantium genetic diversity. Vaccine. 2010;28(29):4573–4580. doi: 10.1016/j.vaccine.2010.04.087. [DOI] [PubMed] [Google Scholar]
- Ahamefule FO, Ibeawuche JA, Okereke SN, Anyanwu AC. Reproductive performance of White Fulani, N’dama and their crossbred in a hot humid environment. J Anim Vet Adv. 2007;6(8):955–958. [Google Scholar]
- Allsopp BA. Natural history of Ehrlichia ruminantium. Vet Parastiol. 2010;167:123–135. doi: 10.1016/j.vetpar.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Allsopp MTEP, Allsopp BA. Extensive genetic recombination occurs in the field between genotypes of Ehrlichia ruminantium. Vet Microbiol. 2007;124(1–2):58–65. doi: 10.1016/j.vetmic.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Allsopp MTEP, Van Strijp MF, Faber E, Josemans AI, Allsopp BA. Ehrlichia ruminantium variants which do not cause heartwater found in South Africa. Vet Microbiol. 2007;120(1–2):158–166. doi: 10.1016/j.vetmic.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Awa DN. Seroligical survey of heartwater relative to the distribution of the vector Amblyomma variegatum and other ticks species in north Cameroun. Vet Parasitol. 1997;68(1–2):165–173. doi: 10.1016/S0304-4017(96)01058-8. [DOI] [PubMed] [Google Scholar]
- Bell-Sakyi L, Koney EB, Dogbey O, Sumption KJ. Heartwater in Ghana: implications for control ticks. Trop Anim Health Prod. 1996;28:59–64. doi: 10.1007/BF02310701. [DOI] [PubMed] [Google Scholar]
- Biguezoton A, Noel V, Adehan S, Adakal H, Dayo G, Zoungrana S, Farougou S, Chevillon C. Ehrlichia ruminantium infects Rhipicephalus microplus in West Africa. Parasit Vector. 2016;9:354. doi: 10.1186/s13071-016-1651-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitanga S, Gaff H, Mukaratirwa S. Tick-borne pathogens of potential zoonotic importance in the southern African region’. J S Afr Vet Assoc. 2014;85(1):1–3. doi: 10.4102/jsava.v85i1.1084. [DOI] [PubMed] [Google Scholar]
- Deem SL, Noval RAI, Yonow T, Peter TF, Mahan SM, Burridge MJ. The epidemiology of heartwater: establishment and maintenance of endemic stability. Trend Parasitol. 1996;12(10):402–405. doi: 10.1016/0169-4758(96)10057-0. [DOI] [PubMed] [Google Scholar]
- Esemu SN, Ndip LM, Ndip RN. Ehrlichia Species, Probable Emerging Human Pathogens in Sub-Saharan Africa: environmental Exacerbation. Rev Environ Health. 2011;26(4):269–279. doi: 10.1515/REVEH.2011.034. [DOI] [PubMed] [Google Scholar]
- Esemu SN, Besong WO, Ndip R, Ndip L. Prevalence of Ehrlichia ruminantium in adult Amblyomma variegatum collected from cattle in Cameroun. Exp Appl Acarol. 2012;59(3):377–387. doi: 10.1007/s10493-012-9599-9. [DOI] [PubMed] [Google Scholar]
- Faburay B, Geysen D, Munstermann S, Faoufik A, Postigo M, Jongejan F. Molecular detection of Ehrlichia ruminantium infection in Amblyomma variegatum ticks in The Gambia. Exp Appl Acarol. 2007;42(1):61–74. doi: 10.1007/s10493-007-9073-2. [DOI] [PubMed] [Google Scholar]
- Hart A, Kocan KM, Bezuidenhou JD, Prozeskyc L. Ultrastructural morphology of Cowdria ruminantium in midgut Epithelial cells of adult Amblyomma hebraeum female ticks. Onderstepoort J Vet Res. 1991;58:187–193. [PubMed] [Google Scholar]
- Heerden HV, Steyn HC, Allsopp MTEP, Allsopp BA. Characterization of pCS20 region of different of Ehrlichia ruminantium isolates. Vet Microbiol. 2004;101(4):279–291. doi: 10.1016/j.vetmic.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Kasari TR, Miller RS, James AM, Freier JE. Recognition of the threat of Ehrlichia ruminantum infection in domestic and wild ruminants in the continental United State. J Am Vet Med Assoc. 2010;237(5):520–530. doi: 10.2460/javma.237.5.520. [DOI] [PubMed] [Google Scholar]
- Knopf L, Komoin-Oka C, Betschart B, Jongejan F, Gottstein B, Zinsstag J. Seasonal epidemiology of ticks and aspect of cowdriosis in N’dama Village cattle in the Central Guinean savannah of Cote D’lvoirs. Prevent Vet Med. 2002;53(1–2):21–30. doi: 10.1016/S0167-5877(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Leeflang P, Ilemobade AA. Tick-borne diseases of domestic animals in northern Nigeria. Trop Anim Health Prod. 1977;9(4):211–218. doi: 10.1007/BF02240342. [DOI] [PubMed] [Google Scholar]
- Lorusso V, Wijnveld M, Majekodunmi AO, Dongkum C, Fajinmi A, Dogo AG, Thrusfield M, Mugenyi A, Vaumourin E, Igweh AC, Jongejan F, Welburn SC, Picozzi K. Tick-borne pathogens of zoonotic and veterinary importance in Nigerian cattle. Parasit Vector. 2016;9:217. doi: 10.1186/s13071-016-1504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw M, Allsopp MTEP, Meyer EC. Ehrlichia ruminantium, an emerging human pathogen—a further report. S Afr Med J. 2006;95(12):948–950. [PubMed] [Google Scholar]
- Mahan SM, Waghela SD, McGuire TC, Rurangirwa FR, Wassink LA, Barbet AF. A cloned DNA probe for Cowdria ruminantium hybridizes with eight heartwater strains and detects infected sheep. J Clin Microbiol. 1992;30:981–986. doi: 10.1128/JCM.30.4.981-986.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan SM, Simbi BH, Burridge MJ. The pCS20 PCR assay for Ehrlichia ruminantium does not cross-react with the novel deer ehrlichial agent found in white-tailed deer in the United States of America. Onderstepoort J Vet Res. 2004;71:99–105. doi: 10.4102/ojvr.v71i2.271. [DOI] [PubMed] [Google Scholar]
- Martinez D, Vachiery N, Starchursky F, Kandassamy Y, Raliniaina M, Aprelon R, Gueye A. Nested PCR for detection and genotyping of Ehrlichia ruminantium: use in genetic diversity analysis. Ann N Y Acad Sci. 2004;1026:106–113. doi: 10.1196/annals.1307.014. [DOI] [PubMed] [Google Scholar]
- Molia S, Frebling M, Vachiery N, Pinareno V, Petitclerc M, Rousteau A, Martinez D, Lefrancois T. Amblyomma variegatum in cattle in Marie Galante, French Antilles; prevalence, control measure and infection by Ehrlichia ruminantium. Vet Parasitol. 2008;153(3–4):338–346. doi: 10.1016/j.vetpar.2008.01.046. [DOI] [PubMed] [Google Scholar]
- Ogo N, Fernandez de Mera II, Scimeca RC, Okubanjo OO, Inuwa HM, Agbede RIS, Torina A, Alongi A, Vicente J, Gortazar C, Dela Fuente J. Molecular identification of ticks borne pathogen in Nigerian ticks. Vet Parasitol. 2012;187(3–4):572–577. doi: 10.1016/j.vetpar.2012.01.029. [DOI] [PubMed] [Google Scholar]
- Okello-Onen J, Hassan SM, Essuman S. Taxonomy of African ticks. Nairobi: ICIPE Science Press; 2006. [Google Scholar]
- Otchere EO (1983) The productivity of Bunaji (White Fulani) cattle in pastoralist herds on the Kaduna plains of Nigeria. International Livestock Centre for Africa (ILCA) Sub-humid Programme Kaduna. Internal Comm. No. 46. pp27
- Peter TF, Barbet AF, Alleman AR, Simbi BH, Burridge MJ, Mahan SM. Detection of the agent of heartwater, Cowdria ruminantium, in Amblyomma Ticks by PCR: validation and application of the assay to field ticks. J Clin Microbiol. 2000;38(4):1539–1544. doi: 10.1128/JCM.38.4.1539-1544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reye AL, Arinola OG, Hübschen JM, Mullera CP. Pathogen prevalence in ticks collected from the vegetation and livestock in Nigeria. Appl Environ Microbiol. 2012;78(8):2562–2568. doi: 10.1128/AEM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIM (Resource Inventory and Management) (1993) Nigerian Livestock Reserves RIM report: Vol. I-IV. Federal Livestock Department and Pest Control Services, Federal Ministry of Agriculture, Abuja
- Sayler KA, Loftis AD, Mahan SM, Barbet AF. Development of a quantitative PCR Assay for differentiating the agent of heartwater disease, Ehrlichia ruminantium, from Panola Montain Ehrlichia. Transbound Emerg Dis. 2016;63(6):e260–e269. doi: 10.1111/tbed.12339. [DOI] [PubMed] [Google Scholar]
- Steyn HC, Van Heerden H, Allsopp MTEP, Allsopp BA. Variability of pCS20 gene sequences among different Ehrlichia ruminantium isolates. Ann N Y Acad Sci. 2003;990:723–725. doi: 10.1111/j.1749-6632.2003.tb07450.x. [DOI] [PubMed] [Google Scholar]
- Steyn HC, Pretorius A, McCrindle CME, Steinmann CML, Van Kleef M. A quantitative real-time PCR assay for Ehrlichia ruminantium using pCS20. Vet Microbiol. 2008;131(3–4):258–265. doi: 10.1016/j.vetmic.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Synge BA (1980) Factors limiting cattle productivity in highland areas of Nigeria II. Centre for Tropical Veterinary Medicine, Easter Bush, Roslin, Midlothian, Scotland, p 50
- Takeet MI, Fagbemi BO, De Donatos M, Yakubu A, Rodulfo HE, Peters SO, Wheto M, Imumorin GI. Molecular survey of pathogenic trypanosomes in naturally infected Nigerian cattle. Res Vet Sci. 2013;94(3):555–561. doi: 10.1016/j.rvsc.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Thrusfield M. Veterinary epidemiology. 2. London: Black Well; 1995. Sampling; pp. 179–284. [Google Scholar]
- Van de Pypekamp HE, Prozesky L. Heartwater: an overview of the clinical signs, susceptibility and differential diagnoses of the disease in domestic ruminants. Onderstepoort J Vet Res. 1987;54(3):263–266. [PubMed] [Google Scholar]
- Yunker CE. Heartwater in sheep and Goats: a review. Onderstepoort J Vet Res. 1996;63:159–170. [PubMed] [Google Scholar]