Abstract
Cell pluripotency, spatial restriction, and development are spatially and temporally controlled by epigenetic regulatory mechanisms that occur without any permanent loss or alteration of genetic material, but rather through modifications “on top of it.” These changes modulate the accessibility to transcription factors, either allowing or repressing their activity, thus shaping cell phenotype. Several studies have demonstrated the possibility to interact with these processes, reactivating silenced genes and inducing a high plasticity state, via an active demethylating effect, driven by ten-eleven translocation (TET) enzymes and an overall decrease of global methylation. In agreement with this, TET activities have been shown to be indispensable for mesenchymal to epithelial transition of somatic cells into iPSCs and for small molecule-driven epigenetic erasure. Beside the epigenetic mechanisms, growing evidences highlight the importance of mechanical forces in supporting cell pluripotency, which is strongly influenced by 3D rearrangement and mechanical properties of the surrounding microenvironment, through the activation of specific mechanosensing-related pathways. In this review, we discuss and provide an overview of small molecule ability to modulate cell plasticity and define cell fate through the activation of direct demethylating effects. In addition, we describe the contribution of the Hippo signaling mechanotransduction pathway as one of the mechanisms involved in the maintenance of pluripotency during embryo development and its induction in somatic cells.
Keywords: Cell plasticity, Epigenetics, Mechanosensing, Reprogramming, TET activities
Pluripotency, cell commitment, and epigenetic restriction
Adult somatic cells are highly specialized and have a specific molecular pattern that regulates their functions and physiology. Although genetically identical, since derived from one-single cell with half-genome from each parent, they can display any of the over the 230 different cell types that are present in a complete multicellular organism. During the early phases of mammalian embryonic development, three germ layers, namely endoderm, mesoderm, and ectoderm, are formed. Subsequently, each one of that responds to specific developmental cues giving rise to different set of tissue types and contributing to organ formation. These processes are driven by several factors both extrinsic and intrinsic to the cell [1] that induce tissue-specific gene expression, timely regulated by epigenetic restrictions. Cell commitment and differentiation are indeed fortified by the cell’s own machinery that chemically modify the DNA, without any permanent loss or alteration of genetic material [2, 3], also referred to as “epigenetic modifications.” These latter gradually limit cell potency [4] to a more specific phenotype-related transcription pattern, resulting in a progressive restriction in cell options [5].
More than 60 years ago, Conrad Waddington used for the first time the term “epigenetics” in his very famous landscape to describe the idea that a phenotype arises by a program defined by the genome, under the influence of the organism’s environment [6]. In his metaphor, Waddington represents the cell of an embryo as a ball rolling from a non-committed, pluripotent condition down the hill, to a specific cell fate. The hill possesses slopes and valleys that represent the many different and complex processes characterizing the events leading to cell differentiation. The ball is addressed towards a favored position, along a progressively more restricted potency pathway, down to the bottom of the hill, where the cell is unipotent and characterized by a tissue-specific differentiated state.
To date, it is widely known that embryo development, pluripotent cell differentiation, and the acquisition of tissue-specific marks are driven by epigenetic modifications that regulate the accessibility to transcription factors, in either a positive or negative manner. The two major mechanisms involved are DNA methylation and histone modifications [7]. The first is a biochemical process characterized by a covalent addition of a methyl (CH3) group at the 5-carbon of the cytosine. The added CH3 group physically impedes the binding of transcription factors to the gene [8] or recruits the methyl-CpG-binding domain proteins (MBDs), forming compact, inactive chromatin, known as heterochromatin. Histone modifications that involve acetylation, phosphorylation, methylation, SUMOylation, citrullination, ADP-ribosylation, or ubiquitination [9] encourage or prevent transcription factor and other protein access to DNA. All these processes are responsible for the so called epigenetic memory and underly the stable maintenance of cell differentiated phenotype that has been considered irreversible until not long ago [7, 10–13].
In this review, we focus on the recent knowledge of epigenetic mechanisms that modulate cell plasticity and define cell fate. We also discuss and provide an overview of the well- established methods to interact with the epigenetic signature of a cell, inducing and maintaining pluripotency. Interestingly, a surprising overlap among the molecular mechanisms that control cell reprogramming and the regulatory pathways acting in the early embryonic development phases was found. The understanding of these processes is therefore not only an intriguing topic of study but also sheds light on the genetic and environmental factors that affect the oocyte epigenetic landscape and determine the phenotypes of individuals and their offspring [14, 15].
Erasing cell “epigenetic memory”
During the last years, many studies demonstrated that, although generally stable in vivo, a terminally differentiated cell can be reversed and forced in an upstream, counter-current direction up the Waddington’s hill, along different states of increased potency [16].
The first attempt to modify the somatic cell identity dates back to the 1960s when a somatic nucleus was “reprogrammed” by exposure to the oocyte’s environment [17, 18]. This early result paved the way for studying the mechanisms involved in the erasure of “epigenetic memory” and the re-establishment of pluri- or totipotency. It was therefore demonstrated that an adult somatic cell can be brought back a to a high plasticity state, using cell fusion techniques [19] or overexpressing master regulator transcription factors (TF), such as in the creation of induced pluripotent cells (iPSCs) [20]. However, one of the fundamental aspects for iPSC generation is the need for high levels of gene expression to reach an increased potency state for the requirement of elevated “activation energy” that allows initiation of cell reprogramming in an upstream, counter-current direction [4]. This demands the use of TF, such as OCT4, kruppel-like factor 4 (KLF4), SOX2, and MYC that are carried into the cell by retroviral vectors, which in turn, integrate in the host genome, resulting in the reactivation of endogenous genes [21–24]. Currently, various methodologies avoiding the viral stable integration have been established for iPSC derivation, from virus-free [22, 25] to removable PiggyBac transposons [26], minicircle systems [27], episomal systems [28], synthetic mRNAs [29–31], and microRNAs [32, 33]. Although these advances have reduced safety concerns, iPSC induced pluripotent state remains stable and unphysiological, making them prone to acquire genomic alterations that increase the risk of mutagenesis [34].
Adult mature cells can also be pushed into a “less committed state” through the use of small molecules and epigenetic modifiers. Following the pioneering work of Taylor and Jones [35], many groups have indeed reported the possibility to directly convert an adult cell type into another [36–45]. These methods are based on the use of chemical compounds able to interact and modify the cell epigenetic signature, avoiding the use of transgenes stably integrated into the genome, but increasing plasticity for a short transient time-window, which is however sufficient to redirect cells towards a different lineage. The first paper reporting the possibility to use the small molecule “reversine” to induce myoblasts into multipotent mesenchymal progenitor cells was published in 2004 [46]. Since then, several approaches that involve the use of epigenetic modifiers have been described. The general concept at the base of these new protocols is that, among the several mechanisms that drive cell differentiation, DNA methylation plays a fundamental role during both early embryonic development and cell lineage specification. For this reason, the well-characterized DNA methyltransferase (DNMT) inhibitor 5-azacytidine (5-aza-CR) has often been used to remove the epigenetic “blocks” that are responsible for tissue differentiation [47]. Thanks to its powerful effects, 5-aza-CR is able to induce global DNA hypo-methylation [48, 49] and silenced gene reactivation [50], promoting a high plasticity state that facilitates somatic cell switch from one phenotype to another [35, 37, 51]. In agreement with these findings, we demonstrated that an adult somatic cell can be reverted into a fully permissive state after an 18 h exposure to 5-aza-CR [40, 41, 43, 45, 52]. Erased cells underwent considerable changes in their phenotype and gene expression patterns, which were accompanied by a significant decrease in global DNA methylation. More in detail, following exposure to the demethylating agent, cells exhibited the most common features of ESCs, iPSCs, and more in general, of pluripotent cells [53], namely reduced dimensions, large nuclei, global chromatin decondensation, and the expression of pluripotency-related genes, such as OCT4, NANOG, REX1, and SOX2. It is interesting to highlight that this condition was transient and reversible, and when cells were returned to their standard culture medium, they reverted to their original phenotype, gradually turning down the expression of pluripotency related genes within a few days [40, 41, 45, 54]. On the other hand, this short high plasticity window was sufficient to allow cell transition towards a different phenotype (such as functional pancreatic beta [40–42, 44, 45, 55], muscle [43], neural progenitor-like [38, 39, 56], or mature Schwann cells [57]), in response to specific differentiation stimuli, demonstrating the acquisition of fully functional high plasticity state.
TET demethylation waves and cell plasticity
At the beginning of development, DNA demethylation plays a key role in shaping the identity of mammalian embryos. In particular, demethylation waves allow the acquisition of the distinctive totipotent state in the zygote as well as the confinement of pluripotency to the inner cell mass (ICM). The specific epigenetic profiles of paternal and maternal gametes are erased shortly after fertilization and syngamy through active ten-eleven translocation (TET)-mediated and passive DNMT-related demethylation processes. These allow embryos to activate transcriptional functionality and, together with polyadenylation regulatory mechanisms, to modulate the expression of specific genes [58], re-establishing pluripotency (Fig. 1). In agreement with this, it has been demonstrated that decreased global 5-mC methylation in ESCs is crucial for maintaining their naive state and for antagonizing differentiation signals [59]. Conversely, cell fate definition and differentiation are accompanied by a progressive increase in DNA methylation that silences pluripotency genes and establishes a phenotype-specific epigenetic pattern [60, 61]. This hypermethylated state is then stably maintained in terminally differentiated somatic cells by copying the specific pattern onto daughter DNA strands during cell replication and division [62–64]. However, as described above, it has been recently demonstrated that cell phenotype can be reverted, either through the use of specific reprogramming factors or by exposure to chemical compounds, generating iPSCs or “epigenetically erased” cells, respectively. Since cell differentiated state is strictly controlled and stably maintained by specific methylation profiles, it is necessary to remove the epigenetic “blocks” and restrictions to allow the transition to a permissive state. Recent studies have shown that reprogramming events require active demethylation processes driven by the TET family of enzymes (Fig.1), which catalyze the oxidation of cytosine-5 methylation into 5hydroxymethyl-cytosine [65–67], leading to the transcription of previously silenced pluripotency genes. Similarly, oocyte TET enzymes showed reprogramming ability by reactivating pluripotency genes during early embryonic development, after both nuclear transfer and natural fertilization [68]. All these findings point to the possibility that TET enzymes play a key role in somatic cell reprogramming to iPSCs, specifically in mesenchymal to epithelial transition (MET). This hypothesis was confirmed in experiments carried out using TET-deficient murine fibroblasts that failed to undergo MET, blocking their reprogramming potential [69]. In addition, recent studies demonstrated that the epigenetic eraser 5-aza-CR interferes with DNA methylation through a direct TET2-mediated mechanism that accompanies the well-known indirect DNMT-related effect (Fig.1). This indicates the possibility that 5-aza-CR action on cell plasticity may occur through alternative mechanisms that require the involvement of novel cellular targets [54]. Even more intriguing, these data suggest that TET proteins play a pivotal role in epigenetic tuning cell potency, and that from the acquisition of the totipotent state of the zygote to somatic cell reprogramming and chemical erasing, they are the common actors controlling methylation levels that facilitate the acquisition of a high plasticity state.
Fig. 1.
TET demethylation waves and cell plasticity. Transitions where TET proteins play a key role in loss or gain of methylation are indicated as waves
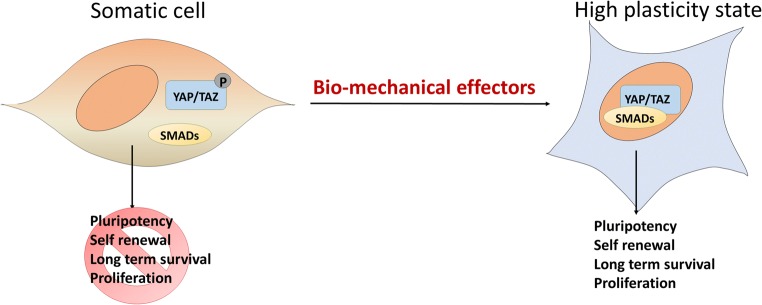
Biomechanical effectors and cell plasticity
During the last years, increasing evidences have highlighted the importance of signal transduction pathways correlated to mechanosensing, which play a fundamental role in cell behavior, both in physiological conditions and in many pathological situations. All cells respond to stimuli exerted by the surrounding microenvironment, such as traction or compression forces from neighboring cells or elasticity and stiffness of the extracellular matrix (ECM). These signals are constantly communicated across cell–ECM and cell–cell adhesion sites, influencing cell own stiffness and cytoskeleton organization [70–73]. However, to date, in vitro and in vivo mechanical cues that control cell fate and the molecular pathways that perceive and transduce this information still remain poorly understood.
Currently, the term “mechanotransduction” is used to describe cell ability to recognize the surrounding microenvironment and appropriately respond to external physical stimuli. The main actors that regulate these processes belong to the Hippo signaling pathway and are the Yes-associated protein (YAP) and the WW domain-containing transcription regulator protein 1 (WWTR1 or TAZ) [22]. Recent studies have demonstrated that these two molecules respond to cytoskeletal-mediated mechanosensing cues that control survival, proliferation, and differentiation in somatic cells (Fig. 2), while no specific role has been fully identified neither in oogenesis nor in spermatogenesis [34]. However, they are present in both human and mouse gametes as well as in early stages of embryonic development [27, 28, 35, 36]. In particular, it was shown that YAP/TAZ are maternally accumulated and highly expressed in early zygotic stage, when maternal RNAs and proteins are exhausted [34], suggesting a developmental stage-specific function for these molecules. In agreement with this, it was demonstrated that distinct changes in TAZ/YAP localization are essential for determining early differentiation, where their nuclear/cytoplasmic compartmentalization defines the first cell fate choice. Indeed, at the blastocyst stage, YAP/TAZ distribution is strictly compartmentalized to the nucleus in the inner cell mass (ICM), while it appears more diffused in the outer trophoblast cells. This localization allows the two molecules to elicit their transcriptional co-activator functions. More in detail, once sorted to the nucleus, YAP and TAZ directly interact with SMAD2/3 [39]. This newly formed YAP/TAZ-SMAD2/3 complex binds to TEAD1/3/4 transcription factors as well as to OCT4, sustaining pluripotency-related gene transcription, buffering pluripotency, and repressing differentiation processes [37, 38] (Fig. 3). Conversely, nuclear exclusion of YAP/TAZ is directly related to the specification of the trophectoderm (TE) with the induction of CDX2 expression [40, 41]. These observations are in line with recent study, demonstrating YAP/TAZ upregulation and specific nuclear compartmentalization in parthenogenetic ESCs [42]. This distinctive behavior of the two proteins results in higher ability to form outgrowths [43–45], generate 3D spheroid colonies, and increase high plasticity [46–48] in mono-parental cells, when compared with their bi-parental counterparts, suggesting that the exclusive maternal origin of these cells may be the main possible cause.
Fig. 2.
Mechanosensing cues and cell plasticity. YAP/TAZ respond to cytoskeletal-mediated mechanosensing cues, controlling cell survival, proliferation, and pluripotency vs. differentiation
Fig. 3.
Biomechanical effectors and cell plasticity. YAP/TAZ nuclear compartmentalization leads to direct interaction with SMAD2/3, sustaining pluripotency-related gene transcription, buffering pluripotency, and repressing differentiation processes
Biomechanical effectors and epigenetic erasing
The data currently present in the literature point to the possibility to combine cell reprogramming techniques with mechanosensing in order to achieve a stable high plasticity state. A relationship between cell fate commitment and 3D rearrangement was recently reported by Harrison et al. [74], who co-cultured murine ESCs and extra-embryonic trophoblast stem cells (TSCs) onto 3D scaffolds, generating aggregates whose morphogenesis was remarkably comparable with in vivo embryos. Similarly, other studies described the possibility to obtain structures referred to as “organoids” [75] or “blastoids” [76, 77] through the use of 3D in vitro cultures that mimic the natural conditions and the biomechanical effectors at work during in vivo embryogenic processes and differentiation [78].
Presently, organoids have been generated from different cell types using encapsulation systems that boost the formation of functional cell aggregation and provide optimal gas exchange between the interior liquid and the surrounding environment [52, 79–83]. A successful strategy, in particular, involved the use of the super hydrophobic synthetic compound polytetrafluoroethylene (PTFE) to produce microbioreactors that allow scale down experiments and work in smaller volumes, amenable for higher throughput applications [52, 83]. Previous studies reported that PTFE efficiently encouraged cell aggregation, facilitating the formation of embryoid bodies (EBs) from murine ESCs [84] and the establishment of olfactory ensheathing cell (OEC) spheroid structures [85]. Consistent with these observations, microbioreactor culture systems were demonstrated to induce epigenetically erased cells to self-assemble and form multicellular spheroids, displaying a uniform size geometry. This was paralleled by a global DNA demethylation and elevated transcription of pluripotency markers, suggesting that the use of PTFE microbioreactors may encourage cell aggregation and may boost the acquisition and maintenance of a long-term high plasticity state. Interestingly, these molecular and morphological modifications were accompanied by the activation of the Hippo signaling pathway, with distinctive changes in YAP/TAZ localization. In particular, 3D cell confinement in PTFE encouraged their nuclear retention that was mirrored by a parallel nuclear accumulation of SMAD2/3 and the activation of the OCT4 dependent pluripotency pathways [52].
Altogether, this evidence suggests that biomechanical effectors can be used to support cell plasticity, eliciting YAP/TAZ nuclear compartmentalization, as it happens in the blastocyst, and encouraging YAP/TAZ interaction with SMAD2/3, buffering pluripotency, and repressing differentiation processes.
Conclusions
Thanks to the abundant data accumulated in the literature, our understanding of pluripotency has significantly expanded in the past decade. Pluripotency, as we presently see it, is no longer a singular property but rather a flexible and dynamic state along the cell developmental program.
In vivo, it is a unique feature of the inner cell mass/epiblast cells that can give rise to all cell lineages of the developing and adult organism. Embryonic pluripotency is short lived and lacks self-renewal, since it is limited to cells transiting through early stages of development.
However, it can be captured and stabilized indefinitely in vitro, under artificial culture conditions, as it happens in ESC. Similarly, in vitro pluripotency can also be reactivated in differentiated somatic cells, using several alternative approaches (Fig.4), some of which were discussed in the present review. All these strategies that may involve direct reprogramming with exogenous factors or the induction of an hypomethylated state with an epigenetic modifier or the key contribution of biomechanical effectors have the common ability to activate the network of transcription factors that prevents cell differentiation and maintains self-renewal, encouraging cells to return to a pluripotent state.
Fig. 4.
All roads lead to Rome: the many ways to pluripotency
All this suggests that the finely tuned spatio-temporal signaling system, driving cell differentiation, can be covered in opposite directions, either specifying terminal cell differentiation or restoring its pluripotency-related functions. Similarly to what happened in ancient Rome, pluripotency is the Golden Milestone (the Milliarium Aureum) of the network; it is the point of origin but also the specific point towards which all roads lead back to, after covering distances and territories within the Empire.
Acknowledgments
Generously supported by the Carraresi Foundation. The authors are members of the COST Action CA16199 “In vitro 3-D total cell guidance and futness” (CellFit)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci U S A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell, 2007;128:635–8. [DOI] [PubMed]
- 3.Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 7.Pennarossa G, Zenobi A, Gandolfi CEE, Manzoni EFMF, Gandolfi F, Brevini TALA. Erase and rewind: epigenetic conversion of cell fate. Stem Cell Rev. 2015;12:163–170. doi: 10.1007/s12015-015-9637-1. [DOI] [PubMed] [Google Scholar]
- 8.Choy MK, Movassagh M, Goh HG, Bennett MR, Down TA, Foo RS. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics. 2010;11:519. doi: 10.1186/1471-2164-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jost D. Bifurcation in epigenetics: implications in development, proliferation, and diseases. Phys Rev E Stat Nonlinear Soft Matter Phys. 2014;89:10701. doi: 10.1103/PhysRevE.89.010701. [DOI] [PubMed] [Google Scholar]
- 12.Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–119. doi: 10.1038/nature13458. [DOI] [PubMed] [Google Scholar]
- 13.Brevini TAL, Pennarossa G, Maffei S, Gandolfi F. Phenotype switching through epigenetic conversion. Reprod Fertil Dev. 2015;27. [DOI] [PubMed]
- 14.Sendžikaitė G, Kelsey G. The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 2019;63:691–705. doi: 10.1042/EBC20190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xavier MJ, Roman SD, Aitken RJ, Nixon B. Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum Reprod Update. 2019;25:519–541. doi: 10.1093/humupd/dmz017. [DOI] [PubMed] [Google Scholar]
- 16.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 18.Gurdon JB. The developmental capacity of nuclei taken from differentiating endoderm cells of Xenopus laevis. J Embryol Exp Morpholog. 1960;8:505–526. [PubMed] [Google Scholar]
- 19.Miller RA, Ruddle FH. Pluripotent teratocarcinoma-thymus somatic cell hybrids. Cell. 1976;9:45–55. doi: 10.1016/0092-8674(76)90051-9. [DOI] [PubMed] [Google Scholar]
- 20.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 21.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 22.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science (80- ) 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–76. [DOI] [PubMed]
- 24.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 25.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Chau KF, Vodyanik MA, Jiang J, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. [DOI] [PMC free article] [PubMed]
- 32.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature MicroRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Hong SG, Dunbar CE, Winkler T. Assessing the risks of genotoxicity in the therapeutic development of induced pluripotent stem cells. Mol Ther. 2013;21:272–281. doi: 10.1038/mt.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 36.Chandrakanthan V, Yeola A, Kwan JC, Oliver RA, Qiao Q, Kang YC, et al. PDGF-AB and 5-Azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells. Proc Natl Acad Sci U S A. 2016;113:E2306–E2315. doi: 10.1073/pnas.1518244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris DM, Hazan-Haley I, Coombes K, Bueso-Ramos C, Liu J, Liu Z, et al. Transformation of human mesenchymal cells and skin fibroblasts into hematopoietic cells. PLoS One. 2011;6:e21250. doi: 10.1371/journal.pone.0021250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirakhori F, Zeynali B, Rassouli H, Salekdeh GH, Baharvand H. Direct conversion of human fibroblasts into dopaminergic neural progenitor-like cells using TAT-mediated protein transduction of recombinant factors. Biochem Biophys Res Commun. 2015;459:655–661. doi: 10.1016/j.bbrc.2015.02.166. [DOI] [PubMed] [Google Scholar]
- 39.Mirakhori F, Zeynali B, Kiani S, Baharvand H. Brief azacytidine step allows the conversion of suspension human fibroblasts into neural progenitor-like cells. Cell J. 2015;17:153–158. doi: 10.22074/cellj.2015.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pennarossa G, Maffei S, Campagnol M, Rahman MM, Brevini TAL, Gandolfi F. Reprogramming of pig dermal fibroblast into insulin secreting cells by a brief exposure to 5-aza-cytidine. Stem Cell Rev Rep. 2014;10:31–43. doi: 10.1007/s12015-013-9477-9. [DOI] [PubMed] [Google Scholar]
- 41.Brevini TAL, Pennarossa G, Acocella F, Brizzola S, Zenobi A, Gandolfi F. Epigenetic conversion of adult dog skin fibroblasts into insulin-secreting cells. Vet J. 2016;211:52–56. doi: 10.1016/j.tvjl.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 42.Brevini TAL, Pennarossa G, Maffei S, Zenobi A, Gandolfi F. Epigenetic conversion as a safe and simple method to obtain insulin-secreting cells from adult skin fibroblasts. J Vis Exp. 2016; (109). 10.3791/53880. [DOI] [PMC free article] [PubMed]
- 43.Brevini TAL, Pennarossa G, Rahman MM, Paffoni A, Antonini S, Ragni G, et al. Morphological and molecular changes of human granulosa cells exposed to 5-azacytidine and addressed toward muscular differentiation. Stem Cell Rev Rep. 2014;10:633–642. doi: 10.1007/s12015-014-9521-4. [DOI] [PubMed] [Google Scholar]
- 44.Pennarossa G, Santoro R, Manzoni EFM, Pesce M, Gandolfi F, Brevini TAL. Epigenetic erasing and pancreatic differentiation of dermal fibroblasts into insulin-producing cells are boosted by the use of low-stiffness substrate. Stem Cell Rev Reports. 2018;14:398–411. [DOI] [PubMed]
- 45.Pennarossa G, Maffei S, Campagnol M, Tarantini L, Gandolfi F, Brevini TAL. Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proc Natl Acad Sci U S A. 2013;110:8948–8953. doi: 10.1073/pnas.1220637110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Zhang Q, Wu X, Schultz PG, Ding S. Dedifferentiation of lineage-committed cells by a small molecule. J Am Chem Soc. 2004;126:410–411. doi: 10.1021/ja037390k. [DOI] [PubMed] [Google Scholar]
- 47.Brevini TAL, Pennarossa G, Manzoni EFM, Zenobi A, Gandolfi F. Mountain high and valley deep: epigenetic controls of pluripotency and cell fate. Anim Reprod. 2017;14:61–68.
- 48.Christman JK. 5-Azacytidine and 5-aza-2[prime]-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. [DOI] [PubMed]
- 49.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 50.Jones PA. Effects of 5-azacytidine and its 2′-deoxyderivative on cell differentiation and DNA methylation. Pharmacol Ther. 1985;28:17–27. doi: 10.1016/0163-7258(85)90080-4. [DOI] [PubMed] [Google Scholar]
- 51.Glover TW, Coyle-Morris J, Pearce-Birge L, Berger C, Gemmill RM. DNA demethylation induced by 5-azacytidine does not affect fragile X expression. Am J Hum Genet. 1986;38:309–318. [PMC free article] [PubMed] [Google Scholar]
- 52.Pennarossa G, Manzoni EFM, Ledda S, de Eguileor M, Gandolfi F, Brevini TAL. Use of a PTFE micro-bioreactor to promote 3D cell rearrangement and maintain high plasticity in epigenetically erased fibroblasts. Stem Cell Rev Rep. 2019;15:82–92. doi: 10.1007/s12015-018-9862-5. [DOI] [PubMed] [Google Scholar]
- 53.Tamada H, Van Thuan N, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, et al. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manzoni EFM, Pennarossa G, Deeguileor M, Tettamanti G, Gandolfi F, Brevini TAL. 5-azacytidine affects TET2 and histone transcription and reshapes morphology of human skin fibroblasts. Sci Rep. 2016;6:37017. doi: 10.1038/srep37017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brevini TAL, Pennarossa G, Manzoni EFM, Gandolfi F. Safety and efficacy of epigenetically converted human fibroblasts into insulin-secreting cells: a preclinical study. Adv Exp Med Biol. 2018;1079 p. 151–62. [DOI] [PubMed]
- 56.Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, et al. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;25:645–646. doi: 10.1038/cr.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thoma EC, Merkl C, Heckel T, Haab R, Knoflach F, Nowaczyk C, et al. Chemical conversion of human fibroblasts into functional Schwann cells. Stem Cell Reports. 2014;3:539–547. doi: 10.1016/j.stemcr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brevini TAL, Cillo F, Colleoni S, Lazzari G, Galli C, Gandolfi F. Expression pattern of the maternal factor zygote arrest 1 (Zar1) in bovine tissues, oocytes, and embryos. Mol Reprod Dev. 2004;69:375–380. doi: 10.1002/mrd.20140. [DOI] [PubMed] [Google Scholar]
- 59.Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, et al. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berdasco M, Esteller M. DNA methylation in stem cell renewal and multipotency. Stem Cell Res Ther. 2011;2:42. doi: 10.1186/scrt83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oda M, Kumaki Y, Shigeta M, Jakt LM, Matsuoka C, Yamagiwa A, et al. DNA methylation restricts lineage-specific functions of transcription factor gata4 during embryonic stem cell differentiation. Greally JM, editor. PLoS Genet. 2013;9:e1003574. doi: 10.1371/journal.pgen.1003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oda M, Kito S, Tanaka T, Nishida I, Awano S, Fujita Y, et al. Prevalence and imaging characteristics of detectable tonsilloliths on 482 pairs of consecutive CT and panoramic radiographs. BMC Oral Health. 2013;13:54. doi: 10.1186/1472-6831-13-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J-Y, Pu M-T, Hirasawa R, Li B-Z, Huang Y-N, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science (80- ) 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (80- ) 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabó PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 69.Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 70.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science (80- ) 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mammoto A, Ingber DE. Cytoskeletal control of growth and cell fate switching. Curr Opin Cell Biol. 2009;21:864–870. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 73.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. [DOI] [PubMed]
- 75.Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216:31–40. doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sozen B, Cox AL, De Jonghe J, Bao M, Hollfelder F, Glover DM, et al. Self-organization of mouse stem cells into an extended potential blastoid. Dev Cell. 2019;51:698–712.e8. doi: 10.1016/j.devcel.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset J-C, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk C, Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- 78.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 79.Arbatan T, Al-Abboodi A, Sarvi F, Chan PP, Shen W. Tumor inside a pearl drop. Adv Heal Mater. 2012;1:467–469. doi: 10.1002/adhm.201200050. [DOI] [PubMed] [Google Scholar]
- 80.Sarvi F, Jain K, Arbatan T, Verma PJ, Hourigan K, Thompson MC, et al. Cardiogenesis of embryonic stem cells with liquid marble micro-bioreactor. Adv Heal Mater. 2015;4:77–86. doi: 10.1002/adhm.201400138. [DOI] [PubMed] [Google Scholar]
- 81.Tian J, Fu N, Chen XD, Shen W. Respirable liquid marble for the cultivation of microorganisms. Colloids Surf B Biointerfaces. 2013;106:187–90. [DOI] [PubMed]
- 82.Serrano MC, Nardecchia S, Gutierrez MC, Ferrer ML, del Monte F. Mammalian cell cryopreservation by using liquid marbles. ACS Appl Mater Interfaces. 2015;7:3854–3860. doi: 10.1021/acsami.5b00072. [DOI] [PubMed] [Google Scholar]
- 83.Brevini TALL, Manzoni EFMM, Ledda S, Gandolfi F. Use of a super-hydrophobic microbioreactor to generate and boost pancreatic mini-organoids. Methods Mol Biol. 2017:291–9. [DOI] [PubMed]
- 84.Sarvi F, Arbatan T, Chan PPY, Shen WA. A novel technique for the formation of embryoid bodies inside liquid marbles. RSC Adv. 2013;3:14501–14508. [Google Scholar]
- 85.Vadivelu RK, Ooi CH, Yao RQ, Tello Velasquez J, Pastrana E, Diaz-Nido J, et al. Generation of three-dimensional multiple spheroid model of olfactory ensheathing cells using floating liquid marbles. Sci Rep. 2015;5:15083. doi: 10.1038/srep15083. [DOI] [PMC free article] [PubMed] [Google Scholar]