Abstract
Fresh produce vegetables are colonized by different bacterial species, some of which are antagonistic to microbes that cause postharvest losses. However, no comprehensive assessment of the diversity and composition of bacteria inhabiting surfaces of fresh pepper plants grown under different conditions has been conducted. In this study, 16S RNA amplicon sequencing was used to reveal bacterial communities inhabiting the surfaces of red and green pepper (fungicides-treated and non-fungicides-treated) grown under hydroponic and open field conditions. Results revealed that pepper fruit surfaces were dominated by bacterial phylum Proteobacteria, Firmicutes, Actinobacteria, and, Bacteroidetes. The majority of the bacterial operation taxonomic units (97% similarity cut-off) were shared between the two habitats, two treatments, and the two pepper types. Phenotypic predictions (at phylum level) detected a high abundance of potentially pathogenic, biofilm-forming, and stress-tolerant bacteria on samples grown on open soils than those from hydroponic systems. Furthermore, bacterial species of genera mostly classified as fungal antagonists including; Acinetobacter, Agrobacterium, and Burkholderia were the most abundant on the surfaces. These results suggest that peppers accommodate substantially different bacterial communities with antagonistic activities on their surfaces, independent of employed agronomic strategies and that the beneficial bacterial strains maybe more important for peppers established on open fields, which seems to be more vulnerable to abiotic and biotic stresses.
Subject terms: Biotechnology, Computational biology and bioinformatics, Microbiology, Molecular biology
Introduction
Fresh products such as apples, grapes, peaches, and tomatoes are known to harbour diverse bacterial populations1–3. Plant species, geographic location, climatic conditions, ripening stage and application of agrochemicals, are some of the factors that determine distribution of microorganisms on the surface of these products4. Bacterial species that colonise fruit surfaces (epiphytes) are introduced from the soil to the host plants by insects, air currents and other animal species5–7. Among these microorganisms, some are beneficial to plants, for example, several Sphingomonas strains induce resistance to Fusarium head blight caused by Fusarium culmorum in the host plant8; while others are phytopathogens (e.g., Phoma and Pantoea) known to cause economic loses1,9. Therefore, understanding the diversity and ecology of epiphytic bacteria may be important to develop new biocontrol agents10.
Previously, the identities of the members of microbial communities were established using culture-dependent methods11. However, these methods are known to underestimate microbial diversity, as only 0.1–8.4% of environmental bacteria are considered cultivable12,13. Data gathered using these methods only provide limited information on the vast majority of microbes present in a given sample. Nowadays, the diversity of bacterial communities is usually assessed by culture-independent techniques that include the analysis of the 16S rRNA gene fragments14. Such methods have allowed for instance the investigation of the microbial diversity of tomato, grape, peach and apple fruits15,16. However, information on bacterial communities associated with the surface of fresh sweet pepper fruits is still limited, despite this being vital in identifying microbes that can antagonize the effects of pathogenic strains which may contribute to postharvest loses.
The primary goal of this study was to investigate, using 16S rRNA gene Illumina amplicon sequencing, how the effect of growing conditions (hydroponic system versus direct sowing), inorganic pesticides treatment (i.e., application of a fungicide) and maturity status (green versus red), could influence the structure and composition of bacterial communities on the surfaces of fresh pepper fruits. Additionally, we aimed to predict the phenotypic changes in the microbiota of pepper samples and also, to identify bacterial taxa with potential to minimize postharvest losses of peppers.
We hypothesized that, regardless of agronomic management approaches, pepper fruits can accommodate antagonistic bacteria on its surfaces that can potentially minimize damage that maybe induced by its potential pathogens, and that some of these antagonists, may contribute in reduction of post-harvest loses.
Results and discussion
Analysing the bacterial communities associated with the surface of Capsicum annuum fruits, we obtained 1,586,400 bacterial high-quality reads, which resulted in 1,137 OTUs (97% cut-off). The majority of bacterial OTUs were shared between the habitats, treatments and pepper sample types (56.4%, 58.9% and 59.4%, respectively) (Supplementary Fig. S1). Microbial diversity (Supplementary Fig. S2) tended to be higher in the fungicide-treated compared to fungicide-untreated samples, in open field compared to the hydroponic system samples, and in the green compared to the red samples, although they did not differ significantly (P > 0.05). This implies that microbial diversity on the surfaces of peppers is not affected by growth stage, growing system and treatment with fungicides. For habitats, diversity results were as expected, as it is well known that the organic matter in the soil is an important source of nutrients for microorganisms and contains higher levels of fungal and bacterial propagules than hydroponic systems17. The diversity in treatments is in agreement with a study by Schaeffer, et al.18, which showed fungicides application on nectar have no observable effect on bacterial OTU richness or community compositions. Furthermore, higher bacterial populations observed on the immature (green) fruit surfaces compared to the mature samples corroborated with findings by Palumbo, et al.19, who found greatest bacterial diversity on early summer mature almond fruits than on the late summer mature almonds. The possible explanation for this observation is that the intact hulls during the immature growing stage of fruits will still be metabolically active and therefore, could be ideal sources of carbon and water for microbial survival.
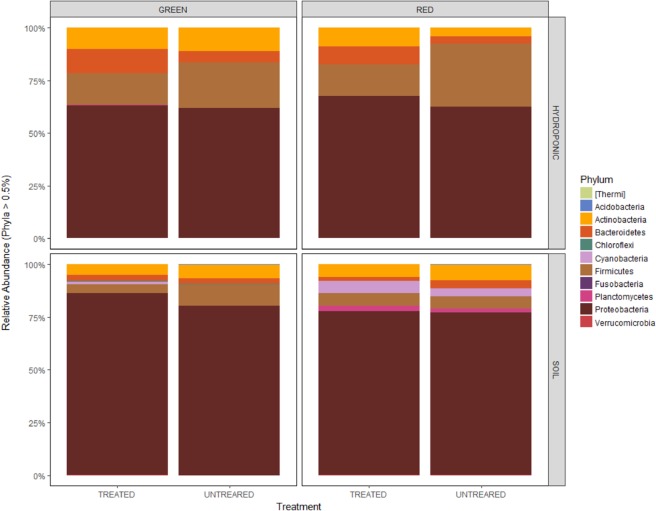
A total of 17 distinct bacterial phyla were detected across all 80 samples. The most abundant sequences in all the 80 samples were affiliated with the phylum Proteobacteria (71%), followed by Firmicutes (13%), Actinobacteria (7%) and Bacteroidetes (5%) (Fig. 1). Other phyla were also represented, although in lower proportions. There were significant differences in Proteobacteria abundance between the two habitats, with the phylum being more abundant in open soil as compared to the hydroponic habitat (Kruskal-Wallis: P < 0.001), but non-significant differences were observed between the two treatment groups (Kruskal-Wallis: P = 0.55) and the two pepper sample types (Kruskal-Wallis: P = 0.53). For Firmicutes, significant differences in abundance were shown between the habitats (Kruskal-Wallis: P < 0.001) and treatments (Kruskal-Wallis: P = 0.03), but non-significant differences in abundance were noted between the pepper sample types (Kruskal-Wallis: P = 0.71). Moreover, abundance of Actinobacteria did not differ between habitats (Kruskal-Wallis: P = 0.34), treatments (Kruskal-Wallis: P = 0.68) and pepper sample types (Kruskal-Wallis: P = 0.34). Additionally, significant differences in abundance were shown between habitats (Kruskal-Wallis: P < 0.001) and pepper sample types (Kruskal-Wallis: P = 0.03) for the Bacteroidetes, while abundances between treatment groups were not significant (Kruskal-Wallis: P = 0.06). The trend in abundance of Proteobacteria in habitats could be explained by the fact that this phylum is commonly identified as being copiotrophic (i.e., they thrive in conditions of elevated carbon availability and exhibit relatively rapid growth rates and compete successfully when organic resources are abundant), possibly because they associate with nematodes soil layers where organic matter, plant roots, and other resources are more abundant20,21. Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes have been shown to be widely represented on the surfaces of fruits of other plants such as grape22. They represent various taxonomic groups and different ecological statuses, such as antagonist, symbionts (especially, endophytes) and saprophytes23. Their dominance on fruit surfaces could be attributed to the fruit’s ability to use a wide variety of carbon sources such as carbohydrates, amino acids, and lipids, which could help resist different environmental changes that occur during fruit development24,25.
Figure 1.
Mean relative abundances of taxa (phylum); (a) between hydroponic and soil habitats samples, (b) green and red samples, (c) treated and untreated samples. The abundance of each taxon calculated as the percentage of sequences per location for a given microbial group.
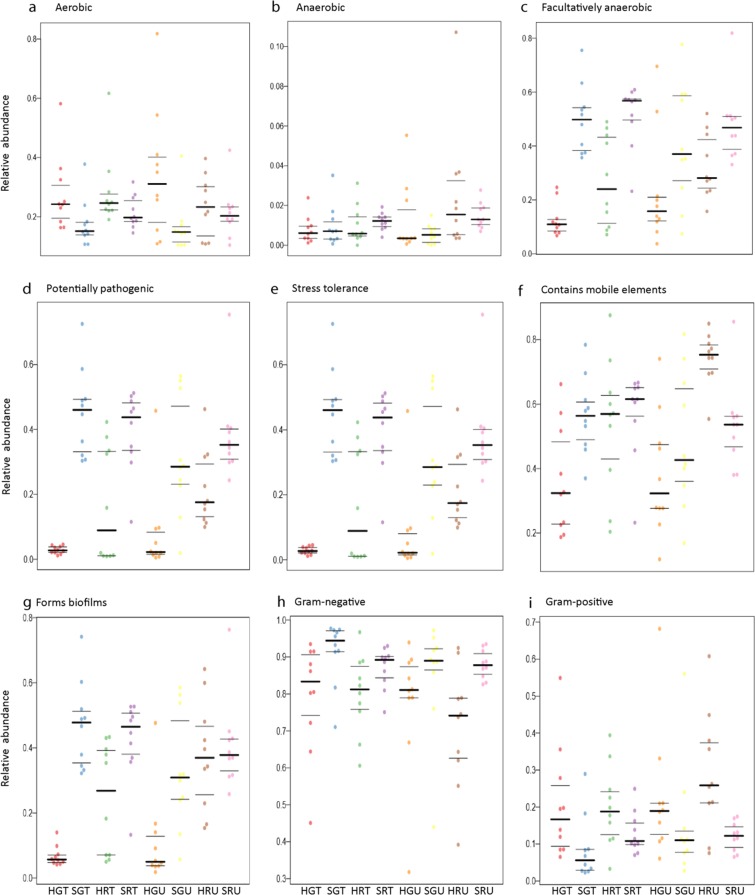
Potential prediction of phenotypic functions of bacterial communities (at phylum level) on the surfaces of the different pepper samples detected nine potential microbial phenotypes including; aerobic, anaerobic, facultative anaerobic, mobile elements carriers, biofilm forming, Gram-negative, Gram-positive and pathogens (Fig. 2; Supplementary Table S1). In general, aerobic bacteria were more abundant on fungicide-treated compared to untreated samples and this was opposite for anaerobic bacterial populations. This could suggest that the rise in abundance of aerobic bacteria is associated with the capability of degrading fungicides by these bacteria as described by Megadi et al.26. On another note, potentially pathogenic bacteria showed to be more overrepresented on surfaces of both, immature (green) and mature (red) peppers grown on open field (both fungicide-treated and untreated). This was not the case with peppers grown under the hydroponic system, and this clearly demonstrates that, growing peppers using the hydroponic system maybe an effective agronomic management strategy in comparting yield constraining effects of microbial pathogens of peppers. Therefore, using the hydroponics technology, crops can be grown with minimal negative effects on ecosystems and biodiversity, which are usually profound in cropping systems dependent on synthetic pesticides for control of pests and diseases27–29. Although the initial investment of hydroponic systems in huge30, it may tend to be a cheaper method of growing high-value, horticultural crops such as peppers, since production costs will be minimized by reduction in pesticide requirements, which are generally very expensive31,32. Hydroponic systems have been adopted in production of some high value crops such as tomato and lettuce33 and in seedling production in nurseries33,34.
Figure 2.
Phenotypic prediction based on BugBase analysis. Prediction of phenotypic differences from 16S rRNA sequence data associated with aerobic, potentially pathogenic, stress tolerance, mobile element, biofilms formation, Gram-negative bacteria and Gram- positive bacteria from sample between hydroponic and soil treated and untreated pepper samples.
Additionally, it is not surprising that stress tolerance functions were predicted to be more abundant on surfaces of peppers grown on open field than those grown on the hydroponic system. It is widely known that hydroponic systems raise plants free from most abiotic stresses (e.g., drought and nutrient stress) as well as biotic stresses (diseases and weeds). In addition, it is highly likely that the biofilm forming function, predicted to be present on surfaces of peppers grown on open field than hydroponic-produced peppers, is necessary to compart the various crop growth constraining factors known to be more common on the open fields. It is also highly probable that bacterial antagonists to deter potential pathogens could be exhibited by biofilm function. Likewise, pathogenicity can also be promoted by biofilm formation. Biofilms are defined as a collective of one or more types of microorganisms that can grow on many different surfaces35.
Differences in abundance of all the predicted nine phenotypic functions were significant (Supplementary Table S1), implying that the bacterial communities with these functions were affected by how pepper plants were managed, (e.g., fungicide treatment versus non-fungicide treatment or hydroponic versus open soil planting). It is also worth mentioning that most of these predicted functions are present in the bacterial phylum, Proteobacteria and Firmicutes (Fig. 1, Supplementary Fig. 2). According to our knowledge, no phenotypic functions on surface bacterial communities for fresh produce grown in hydroponic and open field soil or in other farming practices such as in organic and conventional practices have been reported before.
At the genus level, significant differences in abundance of Microbispora, Sphingobium, Paenibacillus and Lactococcus were noted between the treated and untreated red and green peppers produced in hydroponics. A similar observation was recorded for peppers grown in soil (Table 1). Microbispora species have the ability to produce phenazine-1-carboxylic acid, which is capable of controlling southern blight disease caused by the phytopathogenic fungus, Sclerotium rolfsii, which causes large economic losses in many crops such as Zea mays36. Sphingobium species were reported to produce a volatile inhibitory compound 2-methyl-1-propanol against fungus Pseudogymnoascus destructans37, while Paenibacillus is known to be capable of producing various plant hormones, antibiotics and hydrolytic enzymes with ability to suppress Fusarium wilt of cucumber (Cucumis sativus), which is caused by Fusarium oxysporum f. sp. cucumerinum in non-sterile, soil-less potting medium38. The bacterial genus Lactococcus, a bacteriocin producing Lactic acid bacteria (LAB), isolated from fresh fruits Chryso-phyllum cainito (star apple) and Solanum stramofolium (pea eggplant), was reported to show inhibitory activities against both Gram-positive pathogens such as Bacillus cereus and Staphylococcus aureus and the Gram-negative pathogens (e.g., Salmonella typhimurium)39,40.
Table 1.
Comparison of bacterial genera showing significance differences; between hydroponic treated and untreated green samples, soil treated and untreated green pepper samples, hydroponic treated and hydroponic untreated red pepper samples, and between soil treated and untreated red pepper samples.
| Genus | HGT | HGU | P-value | ||
|---|---|---|---|---|---|
| Mean | St. error | Mean | St. error | ||
| Brevundimonas | 0.310 | 0.039 | 0.000 | 0.000 | <0.001 |
| Chitinophaga | 0.317 | 0.009 | 0.174 | 0.020 | <0.001 |
| Chryseobacterium | 0.377 | 1.123 | 0.170 | 0.015 | <0.001 |
| Clostridium | 0.414 | 0.025 | 0.214 | 0.038 | <0.001 |
| Microbispora | 6.929 | 0.042 | 1.577 | 0.014 | <0.001 |
| Myroides | 0.660 | 0.083 | 0.000 | 0.000 | <0.001 |
| Ochrobactrum | 0.088 | 0.001 | 0.000 | 0.000 | <0.001 |
| Paenibacillus | 1.178 | 0.008 | 0.359 | 0.105 | <0.001 |
| Pedobacter | 0.383 | 0.041 | 0.000 | 0.000 | <0.001 |
| Phenylobacterium | 0.417 | 0.028 | 0.217 | 0.051 | 0.002 |
| Sphingobacterium | 0.721 | 0.010 | 0.213 | 0.017 | 0.026 |
| SGT | SGU | ||||
| Sphingobium | 0.363 | 0.004 | 0.118 | 0.001 | 0.001 |
| HRT | HRU | ||||
| Agromyces | 0.373 | 0.001 | 0.014 | 0.025 | <0.001 |
| Azospirillum | 0.340 | 0.010 | 0.141 | 0.017 | <0.001 |
| Bacteroides | 0.523 | 0.000 | 0.000 | 0.091 | <0.001 |
| Cellvibrio | 0.384 | 0.000 | 0.000 | 0.045 | <0.001 |
| Chitinophaga | 0.474 | 0.015 | 0.000 | 0.000 | <0.001 |
| Clostridium | 0.355 | 0.000 | 0.000 | 0.016 | <0.001 |
| Corynebacterium | 0.477 | 0.006 | 0.126 | 0.037 | <0.001 |
| Exiguobacterium | 0.400 | 0.004 | 0.104 | 0.044 | <0.001 |
| Geobacillus | 1.158 | 0.052 | 0.230 | 0.120 | <0.001 |
| Paenibacillus | 1.645 | 0.008 | 0.642 | 0.097 | <0.001 |
| Phenylobacterium | 0.370 | 0.012 | 0.149 | 0.039 | <0.001 |
| Serratia | 0.358 | 0.000 | 0.000 | 0.603 | <0.001 |
| SRT | SRU | ||||
| Agromyces | 0.688 | 0.000 | 0.000 | 0.008 | <0.001 |
| Clostridium | 0.488 | 0.004 | 0.015 | 0.056 | <0.001 |
| Comamonas | 0.701 | 0.004 | 0.115 | 0.037 | <0.001 |
| Corynebacterium | 0.950 | 0.008 | 0.133 | 0.242 | 0.002 |
| Geobacillus | 0.406 | 0.014 | 0.200 | 0.027 | <0.001 |
| Klebsiella | 0.364 | 0.001 | 0.098 | 0.054 | <0.001 |
| Lactococcus | 2.312 | 0.116 | 0.657 | 0.007 | 0.001 |
Average relative abundance of sequences assigned to genus (Mean) constituting 0.3% or more sequences in either of the sample, standard error of the corresponding average (St. error) and p-value (p < 0.05 significant) describing the significance of the differential abundance observed between the two sample sources.
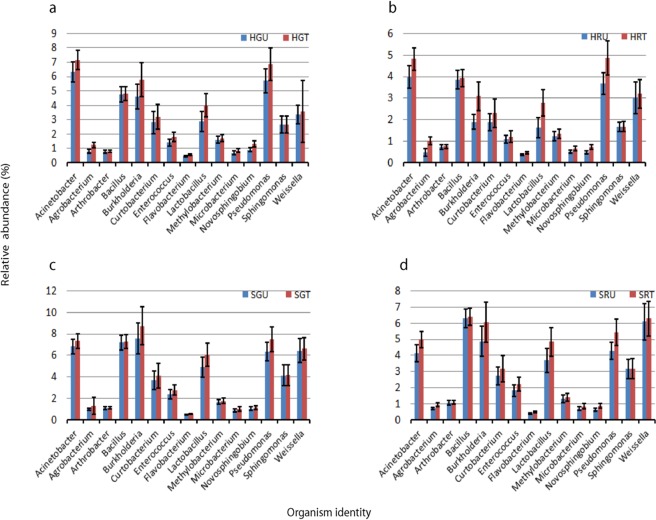
Other well-known bacterial genera such as: Acinetobacter, Agrobacterium, Arthrobacter, Bacillus, Burkholderia, Curtobacterium, Enterococcus, Flavobacterium, Lactobacillus, Methylobacterium, Microbacterium, Novosphingobium, Pseudomonas, Sphingomonas and Weissella, were represented by the majority of sequences, but no significant differences in abundance for these genera were observed between the hydroponic-green-treated (HGT) and the hydroponic-green-untreated (HGU) samples, the soil-green-treated (SGT) and the soil-green-untreated (SGU) pepper samples, the hydroponic-red-treated (HRT) and the hydroponic-red-untreated (HRU) samples as well as between the soil-red-treated (SRT) and the soil-red-untreated (SRU) pepper samples (Fig. 3, Supplementary Table S2a,b). These genera are known to have an antagonistic action against fungal pathogens, reducing cucumber Fusarium wilt, Fusarium oxysporum, and other fungal pathogens while stimulating growth of other vegetable and fruit crops such as cucumbers and chickpeas41–46. These findings suggest that the abundance of these bacterial genera on surfaces of peppers are not affected by changes in growing conditions, maturity stage and pesticide treatment. Hence, these antagonists may be recommended for use in integrated pest management (IPM) programs, were both biological and chemical methods of pest control are recommended47. Similar results were obtained when the fruit surface bacterial communities living on apple fruits under conventional and organic management were compared, where only low abundance groups differed between the two environments48. A study by Telias, et al.15 also showed that these bacterial genera were highly abundant and variable on the surfaces of tomato fruits, but with no significant differences detected between the tomato fruit samples sprayed with surface water and groundwater. Acinetobacter, Pseudomonas and Sphingomonas were also identified in high abundance in the phyllosphere of some Atlantic rainforest tree species and cottonwood15,49, as well as on the leaves of field-grown tomatoes50.
Figure 3.
Relative proportion of bacterial antagonists (mean ≥0,3); (a) between hydroponic green untreated and hydroponic treated green samples, (b) hydroponic red untreated and hydroponic red treated samples, (c) soil green untreated and soil green treated samples, (d) soil red untreated and soil red treated samples. Error bars indicate mean ± SE.
In general, the abundance of all genera was consistently higher in pesticides-treated compared to pesticides-untreated pepper samples grown under both hydroponic and open field conditions. The same scenario was observed in the case of fruit maturity, where the relative abundances of all genera were higher in green compared to red sample types. For treatments, similar trends were observed in studies conducted by Johnsen, et al.51, which showed that some microbial groups are capable of using the applied pesticides as a source of energy and nutrients to multiply. For instance, benomyl insecticides have been found to stimulate Pseudomonas sp, which use the insecticide as a carbon source for growth52. Some pesticides inhibit certain groups of microorganisms and outnumber other groups by releasing them from competition. For example, a study by Hussain, et al.53 demonstrated that fungicide applications inhibited fungal activity of Fusarium and Colletotrichum, which led to a rapid flush in bacterial activity of Bacillus, Acinetobacter and Rhodobacter. Trends observed for fruit maturity could be explained by the fact that, (i) a cyclic changes are observed in temperature and water availability during fruit development in early summer and (ii) progressive desiccation of fruits during maturation in late summer causing pepper to become less susceptible to many bacterial species. These conditions are selective for few species including Bacillus as described by Nicholson, et al.54.
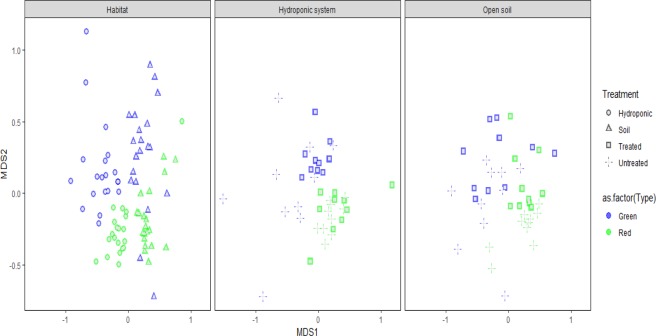
Ordinating bacterial communities data using NMDS plots grouped the bacterial communities separately according to their habitats, treatements and sample type, observing distinct microbial assemblages (Fig. 4). Permutation tests revealed significant effects of habitats, pepper types and treatments on bacterial community structure and composition (i.e., PERMANOVAHabitat, F1 = 23.99, P < 0.001; PERMANOVATreatment, F1 = 2.89, P < 0.001; and, PERMANOVAType, F1 = 8.80, P < 0.001). Although differences in pepper surface bacterial community structure have been reported between organic and conventional farming practices15, no differences have been reported for hydroponic and open field soil. Results from the present study indicate that the bacterial community in hydroponic surface pepper is distinct from those in open field soil surface pepper, regardless of pesticides application and pepper types.
Figure 4.
An NMDS plot showing differences in bacterial structure; (a) between hydroponic and soil habitat, (b) green and red samples under hydroponic habitat, (c) green and red samples under soil habitat.
In conclusion, we have demonstrated that pepper (Capsicum annum) harbors diverse bacterial communities on its surfaces, independent of growing conditions, sample treatment, and sample type, which influenced their composition and abundances. Some of these bacteria are potential antagonists, which may interact with and inhibit postharvest pathogens. The likely biocontrol mechanisms by these genera involve multifaceted interactions between the host, pathogen and the antagonists which include production of extracellular cell wall degrading enzymes, competition for space, nutrients and space, production of various plant hormones, mycoparasitsm, and production of volatile organic compounds36–38,41–44,46,55. A large group of taxa were common across habitats, treatments and sample type. These taxa represented more than 50% bacterial phylotypes. Phenotypic predictions (at phylum level) seemed to suggest that the agronomic decision of whether to grow peppers on hydroponics or on open fields can be key as a disease control measure, as potentially pathogenic bacteria were predicted to be more abundant on samples grown on open fields than those from hydroponic systems. This finding demonstrated that hydroponic systems can be key in reducing production costs, in the long-run, as well as in preserving the integrity of ecosystems, which have for long, been under threat from high-input crop production systems that rely much of heavy inorganic pesticide and fertilizer applications. Additionally, many of the bacterial genera observed in high abundance in samples collected on plants grown under hydroponic and open field conditions are known to contain bacterial strains with plant growth promoting abilities for example Acinetobacter, Arthrobacter, Bacillus, Burkholderia, Curtobactarium and Microbacterium56–59 and those that act as antagonists against fungal plant pathogens42–46. However, a further investigation of these beneficial bacteria using culture-based approaches will help in isolating and characterizing the effects of the antagonists against bacterial pathogens of pepper. Overall, peppers can accommodate different bacterial taxa on its surfaces, some of which with beneficial functional attributes such as pathogenic microbe antagonism, but these beneficial functions will be more important for plants grown under open soils, since they will be more exposed to both, biotic and abiotic stress factors.
Materials and methods
Study sites and crop management
Sweet peppers were grown in the summer and autumn seasons from October 2014 to March 2015, at the Agricultural Research Council-Vegetable and Ornamental Plants (ARC-VOP), Roodeplaat, Pretoria, South Africa (25°59′S; 28°35′E and at an altitude of 1200 m above sea level). Plants were grown under both hydroponic (40% black and white shade net structure) and field conditions. The mean temperature for hydroponic growing conditions were 33 °C day/15 °C night. In the open field, temperatures of 34.5 °C day/15 °C night were recorded. The experimental design was a 2 (treatments) × 2 (growing conditions) × 2 (maturity stages) factorial, with ten replicates (n = 80). The treatments were fungicide-treated (T) and fungicide-untreated (U); the growing conditions hydroponic (H) and field (S); and the two maturity stages red (R) and green (G).
For the field experiment, seven-week-old sweet-pepper seedlings of cultivar ‘King Arthur’ of indeterminate growth habit were transplanted onto 20 cm-high ridges, with an intra-row spacing of 0.3 m and an inter-row spacing of 1.5 m. Plants were pruned to three stems and supported by horizontal twines to box the plants between horizontal twine until the height of 1.5 m. The soil was composed of a mixture of sandy, clay and loam (68%, 8% and 24%, respectively). The chemical composition of the soil (pH 7.3) was as follows: 73.1 mg.kg−1 phosphorus (P), 182 mg.kg−1 potassium (K), 978 mg.kg−1 calcium (Ca), 189 mg.kg−1 magnesium (Mg), and 51.1 mg.kg−1 sodium (Na). Nitrogen was applied at the rate of 180 kg.ha−1 and was incorporated into the soil by banding with three split applications. The first application of nitrogen was at transplanting (50%), the second four weeks after transplanting (WAT) at WAT (25%) and the last at eight WAT (25%). Superphosphate (Ca (H2PO4)2) and potassium sulphate (K2SO4) were applied at planting at the rate of 20 kg.ha−1 (10.5% P) and 40 kg.ha−1 (42% K), respectively. Drip irrigation supplied 550 mm water. The total rainfall received during the growing season was 40 mm.
For the hydroponic experiment, sweet-pepper seedlings as above were transplanted into 10 L plastic bags filled with sawdust as a growing medium. The drip irrigation system, with one dripper per plant, delivering 2.1 litres of nutrient solution per hour was used to fertilize the plants as described by Maboko and Du Plooy60. The plants were pruned to three stems at four WAT. Each stem was trellised by twisting twine around the main stem and fixing it to a stay wire 2 m above the ground surface to support the plant. Side branches were removed weekly to maintain the three-stem system.
For both hydroponic and field conditions, after two WAT plants were sprayed with the following fungicides to control powdery mildew, blight and leaf spot: COPPER-COUNT N (5 mL/L), SPOREKILL (1 mL/L), BINOMYL (50 g mL/L), BRAVO (210 mL/L) and RIDMOL (360 mL/L). Insecticides ACTARA (50 mL/L), HUNTER (40 mL/L), DIOZINON (160 mL/L), BIOMECTINE (60 mL/L), and SAVAGE (40 mL/L)) were also applied to control white flies, red spider mites and aphids.
Sample collection and processing
Fresh, intact and healthy green and red (10 and 14 weeks after planting, respectively) sweet pepper fruit samples were aseptically collected, stored in sterile Ziploc bags and kept at 4 °C in the lab. A total of 80 samples were harvested: 10 Hydroponic-Green-Treated (HGT), 10 Hydroponic-Red-Treated (HRT), 10 Hydroponic-Green-Untreated (HGU), 10 Hydroponic-Red-Untreated (HRU), 10 Soil-Green-Treated (SGT), 10 Soil-Red-Treated (SRT), 10 Soil-Green-Untreated (SGU) and 10 Soil-Red-Untreated (SRU). Microbial biofilms on the surfaces of the pepper fruits were retrieved using sterile cotton swabs soaked in a solution containing 0.15 M NaCl and 0.1% Tween 20, as described by Paulino, et al.61. The swabs were then transferred to micro centrifuge tubes and stored at −80 °C until DNA extraction was performed.
DNA extraction and fragment amplification and high- throughput sequencing
Genomic DNA was isolated from the 80 samples using the ZR Fungal/Bacterial DNA extraction kit (ZYMO Research, Irvine, CA, USA) according to the manufacturer’s instructions. Bacterial 16S rRNA gene amplicons were amplified using primers, 515F (5′-GTGYCAGCMGCCGCGGRA-3′) and 909 R (5′-CCCCGYCAATTCMTTTRAG-3′), targeting the V4 hypervariable region62. PCR was conducted in a single step using a barcoded forward primer and HotStarTaq Plus Master Mix Kit (QIAGEN, Valencia, CA). The thermocycling conditions were initial denaturation at 94 °C for 3 minutes, followed by 28 cycles of 94 °C for 30 seconds, 53 °C for 40 seconds and 72 °C for 1 minute, then a final elongation step at 72 °C for 5 minutes. PCR products were separated by electrophoresis on 2% agarose gel to observe the expected band sizes. All samples were pooled in equal proportions and purified using calibrated Ampure XP beads (Agencourt Bioscience Corporation, MA, USA). Sequencing was performed on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) at the Molecular Research LP next generation sequencing service (http://www.mrdnalab.com, Shallowater, TX, USA) according to the manufacture’s guidelines.
Bioinformatics analysis
The generated 16S rRNA gene sequence data was analyzed using QIIME v1.9.163. Joined sequences <200 bp long, with more than two ambiguous bases, had a quality score of <25 or more than one mismatch to the sample-specific barcode or to the primer sequences, were discarded. Chimeric sequences were discarded using USEARCH V6.164. Good quality reads were clustered into operational taxonomic units (OTUs) at 97% similarity level based on the Greengenes reference sequence database (version 13.8) and the de novo OTU picking algorithm. The taxonomic affiliations of the OTUs were determined using the naive Bayesian rRNA classifier65 at the 80% confidence level. Singletons, chloroplast and archaea species were filtered out from the OTU table and each sample was randomly subsampled (rarefied) to 28,646 reads, which was the lowest number of sequences obtained in a given sample.
Statistical analyses
Alpha diversity was assessed by computing richness and Shannon index using the ‘diversity’ function in the Vegan66 R package. Statistical differences were evaluated using Kruskal-Wallis tests67. The number of shared OTUs between communities/samples was visualized using the ‘Venn’ function in Gplots (cran.r-project.org/package = gplots). The OTU table was Hellinger-Transformed and the Bray-Curtis distances was used to generate a dissimilarity matrix. The structure of the microbial communities was visualized using non-metric multidimensional scaling (nMDS) plots. Permutational analysis of variance (PERMANOVA)68 using the ‘Adonis’ function in the Vegan R package was used to test for differences in bacterial composition and structure. Bugbase (http://github.com/danknights/bugbase) was used to calculate differences between both groups in terms of microbial phenotypes.
Supplementary information
Acknowledgements
The authors would like to thank Mr Phathutshedzo Ramudingana and Mr Silence Chiloane for assistance in sampling. We thank Prof Angel Valverde and Dr Nyaradzai Kamutando for their constructive criticism on the manuscript. This work was supported by funding from the Agriculture Research Council and National Research Foundation, South Africa. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the experiment: O.A.A., M.M.M. and O.O.B. Performed the experiments and analysed the data: T.P.M. Supervised the Research and contributed research material: O.A.A., M.M.M. and O.O.B. Wrote the paper: T.P.M., O.A.A., M.M.M. and O.O.B.
Data availability
The raw Illumina sequencing reads for this project have been submitted to the National Centre for Biotechnology Information Short Read Archive (SRA) database with accession no. PRJNA529905. This Targeted Locus Study (TLS) project have been deposited at DDBJ/EMBL/GenBank under the accession KDDL00000000. The version described in this paper is the first version, KDDL01000000.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-65587-9.
References
- 1.Abdelfattah A, Wisniewski M, Droby S, Schema L. Spatial and compositional variation in the fungal communities of organic and conventionally grown apple fruit at the consumer point-of-purchase. Horticulture Research. 2016;3:16047. doi: 10.1038/HORTRES.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira M, Usall J, Vin˜as I, Anguera M, Gatius F. Microbiological quality of fresh lettuce from organic and conventional production. Food Microbiology. 2010;27:679–684. doi: 10.1016/j.fm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. The ISME Journal. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto C, et al. Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Frontiers in Microbiology. 2015;6:905. doi: 10.3389/fmicb.2015.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valero E, Cambon B, Schuller D, Casal M, Dequin S. Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Research. 2007;7:317–329. doi: 10.1111/j.1567-1364.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- 6.Burrows SM, Elbert W, Lawrence MG, Pöschl U. Bacteria in the global atmosphere—Part 1: Review and synthesis of literature data for different ecosystems. Atmospheric Chemistry and Physics. 2009;9:9263–9280. [Google Scholar]
- 7.Stefanini I, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proceedings of the National Academy of Sciences of the United States of America. 2015;109:13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Sui Y, Wisniewski M, Droby S, Liu Y. Review: utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. International Journal of Food Microbiology. 2013;167:153–160. doi: 10.1016/j.ijfoodmicro.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Applied Environmental Microbiology. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller. T, Silke. R. Progress in Cultivation-Independent Phyllosphere Microbiology. FEMS Microbiology Ecology. 2014;87:2–17. doi: 10.1111/1574-6941.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner M, Amann R, Lemmer H, Schleifer KH. Probing activated-sludge with oligonucleotides specific for Proteobacteria - inadequacy of culture-dependent methods for describing microbial community structure. Applied Environmental Microbiology. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acosta-Martínez V, Dowd SE, Sun Y, Allen VG. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biology and Biochemistry. 2008;40:2762–2770. [Google Scholar]
- 13.Aremu BR, Babalola OO. Construction of specific primers for rapid detection of South African exportable vegetable macergens. International Journal of Environmental Research and Public Health. 2015;12:12356–12370. doi: 10.3390/ijerph121012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamutando CN, et al. Soil nutritional status and biogeography influence rhizosphere microbial communities associated with the invasive tree Acacia dealbata. Scientific reports. 2017;7:6472. doi: 10.1038/s41598-017-07018-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telias A, White JR, Pahl DM, Ottesen AR, Walsh CS. Bacterial community diversity and variation in spray water sources and the tomato fruit surface. BMC Microbiology. 2011;11:81–93. doi: 10.1186/1471-2180-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leff JW, Fierer N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One. 2013;8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postma, J., van OS, E. & Bonanitas, P. J. M. Pathogen detection and management strategies in soilless plant growing systems. In: Soiless culture: Theory and practice (eds. Raviv, M. & Lieth, H. J) 425–457 (Elsevier, 2008).
- 18.Schaeffer RN, Vannette RL, Brittain C, Williams NM, Fukami T. Non-target effects of fungicides on nectar-inhabiting fungi of almond flowers. Environmental Microbiology Reports. 2017;9:79–84. doi: 10.1111/1758-2229.12501. [DOI] [PubMed] [Google Scholar]
- 19.Palumbo JD, Baker JL, Mahoney NE. Isolation of Bacterial Antagonists of Aspergillus flavus from almonds. Microbial Ecology. 2006;52:45–52. doi: 10.1007/s00248-006-9096-y. [DOI] [PubMed] [Google Scholar]
- 20.Fierer N, Bradford M, Jackson. R. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 21.Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biology and Biochemistry. 2010;42:896–903. [Google Scholar]
- 22.Anderson MJ. A new method for non‐parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 23.Leveau JHJ, Tech JJ. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by High-throughput sequence analysis of 16S rRNA amplicons. Acta Horticulture. 2011;905:31–42. [Google Scholar]
- 24.Peighamy-Ashnaei S, Sharifi-Tehrani A, Ahmadzadeh M, Behboudi K. Effect of carbon and nitrogen sources on growth and biological efficacy of Pseudomonas fluorescens and Bacillus subtilis against Rhizoctonia solani, the causal agent of bean damping-off. Communications in Agricultural and Applied Biological Sciences. 2006;72:951–956. [PubMed] [Google Scholar]
- 25.Kazakov AE, et al. Comparative genomics of regulation of fatty acid and branched chain amino acid utilization in Proteobacteria. Journal of Bacteriology. 2009;191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Megadi VB, Tallur PN, Hoskeri RS, Mulla SI, Ninnekar HZ. Biodegradation of pendimethalin by Bacillus circulans. Indian Journal of Biotechnology. 2010;9:173–177. [Google Scholar]
- 27.Römbke J, Schmelz RM, Pélosi C. Effects of Organic Pesticides on Enchytraeids (Oligochaeta) in Agroecosystems: Laboratory and Higher-Tier Tests. Frontiers in Environmental Science. 2017;5:20. doi: 10.3389/fenvs.2017.00020. [DOI] [Google Scholar]
- 28.Tunstall-Pedoe H, et al. Pesticide pollution remains severe after clean up of a stock pile of obsolete pesticides at Vikuge, Tanzania. AMBIO A Journal of the Human Environment. 2004;33:503–508. doi: 10.1579/0044-7447-33.8.503. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Yan C, Guo Q, Zhang J, Ruiz-Menjivar J. The impact of agricultural chemical inputs on environment: global evidence from informetrics analysis and visualization. International Journal of Low-Carbon Technologies. 2018;13:338–352. [Google Scholar]
- 30.Sulma V, Régio G, Binotto E. Economic viability for deploying hydroponic system in emerging countries: A differentiated risk adjustment proposal. Land Use Policy. 2019;1:357–369. [Google Scholar]
- 31.Aktar W, Sengupta D, Chowdhury A. Impact of pesticides use in agriculture: their benefits and hazards. Interdisciplinary Toxicology. 2009;2:1–12. doi: 10.2478/v10102-009-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alavanja MCR. Pesticides use and exposure extensive worldwide. Reviews on Environmental Health. 2009;24:303–309. doi: 10.1515/reveh.2009.24.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen NT, McInturf SA, Mendoza-Cózatl DG. Hydroponics: A Versatile System to Study Nutrient Allocation and Plant Responses to Nutrient Availability and Exposure to Toxic Elements. Journal of Visualized Experiments. 2016;113:54317. doi: 10.3791/54317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang NN, Kitaya Y, Shibuya T, Endo R. Development of an in vitro hydroponic culture system for wasabi nursery plant production—Effects of nutrient concentration and supporting material on plantlet growth. Scientia Horticulturae. 2019;245:237–243. [Google Scholar]
- 35.Davey ME, O’toole GA. Microbial Biofilms: from Ecology to Molecular Genetics. Microbiology and Molecular Biology Reviews. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patil NN, et al. Potential of Microbispora sp. V2 as biocontrol agent against Sclerotium rolfsii, the causative agent of southern blight of Zea mays L (Baby corn)- in vitro studies. Indian Journal of Experimental Biology. 2014;52:1147–1150. [PubMed] [Google Scholar]
- 37.Micalizzi EW, Mack JN, White GP, Avis TJ, Smith. ML. Microbial inhibitors of the fungus Pseudogymnoascus destructans, the causal agent of white-nose syndrome in bats. Plos One. 2017;12:e0179770. doi: 10.1371/journal.pone.0179770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garzón K, Ortega C, Tenea GN. Characterization of Bacteriocin-Producing Lactic Acid Bacteria Isolated from Native Fruits of Ecuadorian Amazon. Polish Journal of Microbiology. 2017;66:473–481. doi: 10.5604/01.3001.0010.7037. [DOI] [PubMed] [Google Scholar]
- 39.Azhar NS, Zin NHM, Hamid THTA. Lactococcus Lactis Strain A5 Producing Nisin-like Bacteriocin Active against Gram Positive and Negative Bacteria. Tropical Life Sciences Research. 2017;28:107–118. doi: 10.21315/tlsr2017.28.2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larran S, Simon MR, Moreno MV, Siurana MPS, Perell A. Endophytes from wheat as biocontrol agents against tan spot disease. Biological Control. 2016;92:17–23. [Google Scholar]
- 41.Madhaiyan M, et al. Plant Growth–Promoting Methylobacterium Induces Defence Responses in Groundnut (Arachis hypogaea L.) Compared with Rot Pathogens. Current Microbiology. 2006;53:270–276. doi: 10.1007/s00284-005-0452-9. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Liang Y, Shen T, Yang H, Shen B. Biological characteristics of Streptomyces albospinus CT205 and its biocontrol potential against cucumber Fusarium wilt. Biocontrol Science and Technology. 2016;26:1–23. [Google Scholar]
- 43.Palmieri D, Vitullo D, De Curtis F, Lima G. A microbial consortium in the rhizosphere as a new biocontrol approach against fusarium decline of chickpea. Plant and Soil. 2016;412:425–439. [Google Scholar]
- 44.Garbeva P, Veen JA, Elsas JDV. Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiology Ecology. 2004;47:51–64. doi: 10.1016/S0168-6496(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 45.Mata L, Chaves C, Rodríguez-Herrera R, Hernández-Castillo D, Aguilar C. Growth inhibition of some phytopathogenic bacteria by cell-free extracts from Enterococcus sp. British Biotechnology Journal. 2013;3:359–366. [Google Scholar]
- 46.Wachowska U, Kucharska K, Jedryczka M, Łobik N. Microorganisms as biological control agents against fusarium pathogens in winter wheat. Polish Journal of Environmental Studies. 2013;22:591–597. [Google Scholar]
- 47.Gangwar RK. Role of biological control agents in integrated pest management approaches. Acta Scientific Agriculture. 2017;1:9–11. [Google Scholar]
- 48.Ottesen AR, White JR, Skaltsas DN, Newell MJ, Walsh CS. Impact of organic and conventional management on the phyllosphere microbial ecology of an apple crop. Journal of Food Protection. 2009;72:2321–2325. doi: 10.4315/0362-028x-72.11.2321. [DOI] [PubMed] [Google Scholar]
- 49.Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR. Bacterial diversity in tree canopies of the Atlantic forest. Science. 2006;312:1917–1917. doi: 10.1126/science.1124696. [DOI] [PubMed] [Google Scholar]
- 50.Enya J, et al. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microbial Ecology. 2007;53:524–536. doi: 10.1007/s00248-006-9085-1. [DOI] [PubMed] [Google Scholar]
- 51.Johnsen K, Jacobsen CS, Torsvik V. Pesticides effects on bacterial diversity in agricultural soils—A review. Biology and Fertility of Soils. 2001;33:443–453. [Google Scholar]
- 52.Pandey CB, Singh GB, Singh K, Singh RK. Soil nitrogen and microbial biomass carbon dynamics in native forests and derived agricultural land uses in a humid tropical climate of India. Plant and Soil. 2010;333:453–467. [Google Scholar]
- 53.Hussain S, Siddique T, Saleem M, Arshad M, Khalid A. Impact of pesticides on soil microbial diversity, enzymes, and biochemical reactions. Advances in Agronomy. 2009;102:159–200. [Google Scholar]
- 54.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extra terrestrial environments. Microbiology and Molecular Biology Reviews. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamphogoro, T. P., Babalola, O. O. & Aiyegoro, O. A. Sustainable management strategies for bacterial wilt of sweet peppers (Capsicum annuum) and other Solanaceous crops. Preprint at, 10.1111/jam.14653 (2020). [DOI] [PubMed]
- 56.Huddedar SB, et al. Isolation, characterization and plasmid pUPI126 mediated indole 3 acetic acid (IAA) production in Acinetobacter strains from rhizosphere of wheat. Applied Biochemistry and Biotechnology. 2002;102:21–29. doi: 10.1385/abab:102-103:1-6:021. [DOI] [PubMed] [Google Scholar]
- 57.Raj S, Vikas V, PatelbJay K, Singh S. Plant growth promoting Curtobacterium albidum strain SRV4: An agriculturally important microbe to alleviate salinity stress in paddy plants. Ecological Indicators. 2019;105:553–562. [Google Scholar]
- 58.Singh T, Singh DK. Rhizospheric Microbacterium sp. P27 Showing Potential of Lindane Degradation and Plant Growth Promoting Traits. Current Microbioogyl. 2019;76:888–895. doi: 10.1007/s00284-019-01703-x. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharyya PN, Jha DK. “Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture,”. World Journal of Microbiology and Biotechnology. 2012;28:1327–1350. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 60.Maboko MM, Du Plooy CP. Effect of Plant Growth Regulators on Growth, Yield, and Quality of Sweet Pepper Plants Grown Hydroponically. HortScience. 2015;50:383–386. [Google Scholar]
- 61.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions. Journal of Clinical Microbiology. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Qian PY. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. Plos One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Garrity GM, Tiedj JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences in to the new bacterial taxonomy. Applied and Environmental Microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oksanen, J. et al. Vegan: Community Ecology Package, vR package version 2.0–2, http://cran.r-project.org/package=vegan (2007).
- 67.R Development Core Team. R: A language and environment for statistical computing. R foundation for statistical computing, http://www.r-project.org/ (2014).
- 68.Hollander, M. & Wolfe, D. A. Nonparametric Statistical Methods (ed. John, W.) 115–120 (Wiley & Sons, 1973).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Illumina sequencing reads for this project have been submitted to the National Centre for Biotechnology Information Short Read Archive (SRA) database with accession no. PRJNA529905. This Targeted Locus Study (TLS) project have been deposited at DDBJ/EMBL/GenBank under the accession KDDL00000000. The version described in this paper is the first version, KDDL01000000.