Abstract
Background and Objectives:
Endophytic actinomycetes have been known as a promising source for new antibiotics discovery against susceptible and resistant forms of pathogenic microorganisms. This study was aimed at determining antibacterial compound from Streptomyces sp. strain B-92 isolated from a medicinal plant Neesia altissima.
Materials and Methods:
Streptomyces sp. strain UICC B-92 was endophytic actinomycetes of N. altissima that obtained from Universitas Indonesia Culture Collection (UICC). Isolation and determination of bioactive compound were carried out using thin layer chromatography (TLC), nuclear magnetic resonance spectroscopy (NMR), and liquid chromatography mass spectrometry (LC-MS) analyses. An in vitro antibacterial assay of pure bioactive compound from the endophytic actinomycetes strain was performed against Bacillus cereus strain ATCC 10876, Escherichia coli strain ATCC 25922, Salmonella typhimurium strain ATCC 25241, Shigella flexneri strain ATCC 12022 and Staphylococcus aureus strain ATCC 25923.
Results:
The bioactive compound was identified as 4-((3S,4R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl) tetrahydro-2H-pyran-2-yloxy) phenazine-1-carboxylic acid. In vitro antimicrobial assay showed that bioactive compound of Streptomyces sp. strain UICC B-92 exhibited antagonistic activities against two Gram-positive bacteria, viz, B. cereus strain ATCC 10876 and S. aureus strain ATCC 25923.
Conclusion:
The findings of this research showed that, bioactive compound of Streptomyces sp. strain UICC B-92 is suggested a new compound based on glycoside structure and its position.
Keywords: Actinomycetes, Antibacterial, Endophyte, Gram-positive bacteria
INTRODUCTION
Accelerating number of bacterial and fungal infections worldwide has become problematic disease due to antibiotic resistance of those pathogenic microorganisms (1). The problem is worsened by the emergence of new pathogens with the potential for rapid global spread, such as Staphylococcus spp., Mycobacterium tuberculosis and Streptococcus spp. (2). There are more interests in discovery and development of new antimicrobial agents from various sources to combat microbial resistance, including microbial endophytes associated with various medicinal plants (3).
Each of plant species generally hosts to one or more microbial endophyte species. Majority of microbial endophytes produce bioactive compounds that inhibit the growth of other microbes, and even acquire the ability to synthesize the same bioactive compounds produced by their host plant (3). Therefore, microbial endophytes are potential source for discovery of novel bioactive compounds for use in medicine, agriculture, and other industries. Among various microbial groups, members of actinomycetes have been known for their pharmaceutically important compounds as they have contributed 52.73% of antibiotic production of which approximately 50% for human consumption (4). These include munumbicins produced by Streptomyces sp. strain NRRL 30562 (5), kakadumycins produced by Streptomyces sp. strain NRRL 30566 (6), bioactive compound of multicyclic indolosesquiterpenes, i.e. Xiamycin B [1b], Xiamycin A [1a], Indosespene [2], and Sespenine [3] isolated from Streptomyces sp. strain HKI 0595 (7), coronamycin produced by Streptomyces sp. strain MSU-2110 (8) and so on.
Neesia altissima (Blume) Blume (Indonesian: Bengang) is a tropical plant belong to family Malvaceae. This plant has different local names in several regions in Indonesia such as ki bengang (Sundanese), and Durian hantu or sibengang (Sumatera) (9). The plant is a large tree (grows up to + 40 m) and distributed primarily in the rainforest of Malaysia and Indonesia (Sumatera, Borneo, and Java islands). This plant is endemic to Indonesia and used as treatment of gonorrhea, diuretic, and diarrhea diseases (10). Our previous study on discovery of new compound from microbial endophytes associated with N. altissima reported that three endophytic bacterial strains, viz, Pseudomonas aeruginosa strain UICC B-40 and strain UICC B-93, and Pseudomonas azotoformans strain UICC B-91, exhibited antibacterial activities against Bacillus cereus ATCC 10876, Escherichia coli ATCC 25922, Salmonella Typhimurium ATCC 25241, Shigella flexneri ATCC 12022, and Staphylococcus aureus ATCC 25923 (11). The (2E,5E)-phenyltetradeca-2,5-dienoate (MW = 300 g/mol) bioactive compound was isolated and determined from fermentation extract of P. aeruginosa strain UICC B-40, a bacterial endophyte strain from N. altissima (12).
The naturally occurring phenazines and many of its derivatives are found in nature produced by Gram-positive bacteria, particularly members of Streptomyces spp. (13). These include S. griseoluteus (14), S. kebangsaanensis (13), S. lomondensis (15), S. anulatus (16), S. niveus (17) and several other species (14, 18, 19). Luo et al. (14) also noted that members of Streptomyces have commonly been used as a source of various phenazines complex such as griseolutein, phenacetin, lomofungin, and 5,10-dihydrophenazine. In this study, we determined and elucidated the phenazine derivative antibacterial compound that isolated from endophytic actinomycetes from N. altissima. A mechanism of the antibacterial compound is morphologically described using Scanning Electron Microscope (SEM).
MATERIALS AND METHODS
Microbial sources.
Streptomyces sp. strain UICC B-92 was obtained from Universitas Indonesia Culture Collection (UICC) (20). This endophytic actinomycetes strain was isolated from N. altissima using the modification method published by Cao et al. (21).
Isolation of bioactive compound.
Isolation of antibacterial compound from actinomycetes was conducted according to the method described by Pratiwi et al. (12) and Saha et al. (22) as follows: selected endophytic actinomycetes strain that showed highest antagonistic activity was sub-cultured on ISP2 medium and incubated for 7 days. Spores of the endophytic actinomycetes strain were inoculated into 500 mL of ISP2 broth medium (pH 6.75) in a 1 L Erlenmeyer flask and incubated in shaking incubator (100 rpm) at room temperature for 5 days. Fermented liquid was further extracted by ethyl acetate. The mixture was allowed to form two distinct layers and then filtrated. After filtration, 50 mL of fermented broth was taken in a 250 mL separating funnel. About 25 mL of different organic solvents (nonpolar to polar) were used to recover different products. The mixture was shaken for 15 min, and was kept in stationary condition for another 15 min to separate the solvent (ethyl acetate) from aqueous phase. The solvent was evaporated in a rotary evaporator. The obtained powder of crude extract was further dissolved in 2–5 mL methanol for thin layer chromatography (TLC) assay. The TLC plates were developed with CH 2 Cl 2 /10% MeOH solvent system. The plates were dried under hot air and were further stained with Anisaldehyde/ H 2 SO 4 and Ehrlich’s reagents, separately. Visualization was conducted under UV light (at λ254 nm and λ366 nm) for the appearance of different color bands. The components showing UV absorbance and fluorescence were marked and scanned.
Identification of bioactive compound.
After 5 days incubation of the culture, the fermentation was stopped. Cells of actinomycetes were removed from fermentation broth by filtration and the culture filtrate was extracted by ethyl acetate. Purification of crude bioactive compound was conducted according to the method described by Pratiwi et al. (23) in the silica gel chromatography column (22 × 5 cm, Silica gel 60, Merck) and eluted with gradient solvent system consisting of ethyl acetate + hexane. Elutions collected during column chromatography were concentrated and tested for their antimicrobial activity against B. cereus and S. aureus to screen bioactive fractions. Further purification of bioactive fractions was carried out in HPLC preparative column at 3 mL/min flow rate. Structural elucidation of pure bio-active compounds from the strain was carried out by LC-MS and 1 H-NMR spectral studies (24).
Antibacterial activity assay of pure bioactive compound.
An in vitro antibacterial assay of pure bioactive compound from the endophytic actinomycetes strain was performed against B. cereus strain ATCC 10876, E. coli strain ATCC 25922, S. Typhimurium strain ATCC 25241, S. flexneri strain ATCC 12022 and S. aureus strain ATCC 25923. All bacterial isolates used in this study are preserved at Universitas Indonesia Culture Collection (UICC).
The antibacterial activity assay was conducted using the disc diffusion method described by Pratiwi et al. (12) and Schwalbe et al. (25). Pathogenic bacterial isolates were inoculated onto 50 mL NB medium (Merck) in a 250 mL Erlenmeyer flask and were incubated at 37 ºC for 18 h. 200 μL of 10 5 CFU/mL of each pathogenic bacterial cultures was applied to the surface of the NA plate (15 cm diam.). The bioactive compounds were prepared at concentrations of 100; 1,000; 5,000; 10,000; 50,000; and 250,000 μg/mL in 1% DMSO. Tetracycline (100 μg/mL) was used as positive control, and DMSO without bioactive compound was used as negative control. Approximately 15 μL of each concentration was diffused onto a 6 mm diameter disc (Fuoroni), and then placed on NA plates. Antimicrobial activity was observed after 24–48 h incubation at 37 °C. Inhibition activity was determined by measuring the diameter of the inhibition zone.
Analysis of cellular morphology using scanning electron microscopy (SEM).
SEM was carried out according to the method described by Kai et al. (26). After treatment with antibacterial compound, 50 mL of bacterial cells with concentration of 10 8 CFU/mL were soaked in glutaraldehyde 2.5% for 2 h, then washed with cacodylate buffer, and soaked in osmium tetraoxide solution for 4 h. The cells were further washed with cacodylate buffer and soaked in tanin acid solution for 12 h. The cells were hydrated with 5–10 mL EtOH 50%, 70%, 80%, 90% and 99%. Centrifugation was conducted at 3,500 rpm for 10 min. Cells were dissolved in tert-BuOH and coated with gold using a vacuum (6–7 Pa) for 20 min. Appearance of each damaged cells was carried out with the SEM system (JOEL seri JSM-5310LV) at 5,000× magnification.
RESULTS
Isolation of bioactive compound.
Approximately 27 fractions were determined in TLC analysis of the extract from actinomycetes strain B-92. Among them, fractions 9–13 showed highest antibacterial activity based on bioautography analysis (data not shown). Retention factor (Rf) value of these fractions were 0.56–0.58 mm, indicating that the fractions belong to polar compound. These fractions were further analyzed using spectroscopy method.
Identification of bioactive compound.
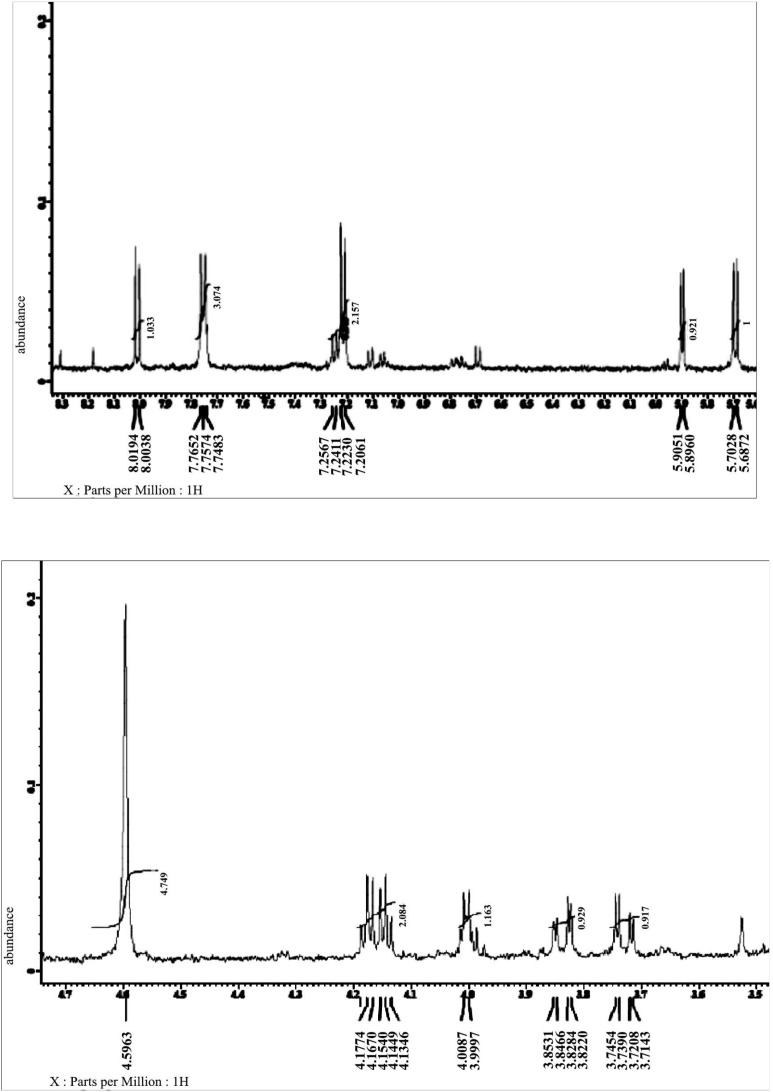
LC-MS analyses (Fig. 1) and 1 H-NMR spectra (Fig. 2) of the purified compound from actinomycetes strain B-92 showed that fractions 9–13 exhibited aromatic signal at δH 5.7 (H; d; 8.3 Hz) ppm (Fig. 3), phenazine structure at δH 7.75 (2H; d; 8.5 Hz); 7.22 (2H; d; 8.5 Hz) ppm (Fig. 4), and sugar at δH 5.9 (H; d; 4.6 Hz); 4.08 (H; d; 4.6 Hz); 3.74 (H; dd; 2.8; 12.3 Hz); 3.81 (H; dd; 2.8; 12.3 Hz); 4.0 (m); 4.14; 4.17 (H; t; 5.2 Hz) ppm (Fig. 5). Therefore, based on these analyses, the fractions from purified compound of actinomycetes strain B-92 was determined as 4-((3S,4R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl) tetrahydro-2H-pyran-2-yloxy) phena zine-1-carboxylic acid or 4-O-glucosyl, 1-carboxyl-phenazine with chemical formula C 19 H 18 N 2 O 8 (MW 402 g/mol) (Fig. 6 and Table 1).
Fig. 1.
Spectral LC-MS of bioactive compound from endophytic Streptomyces sp. strain UICC B-92
Fig. 2.
Spectral 1 H-NMR of bioactive compound from Streptomyces sp. strain UICC B-92
Fig. 3.
Aromatic signal of the purified compound from Streptomyces sp. strain B-92 at δH 5.7 (H; d; 8.3 Hz) ppm
Fig. 4.
Phenazine structure of the purified compound from Streptomyces sp. strain B-92 at δH 7.75 (2H; d; 8.5 Hz); 7.22 (2H; d; 8.5 Hz) ppm
Fig. 5.
Sugar of the purified compound from Streptomyces sp. strain B-92 at δH 5.9 (H; d; 4.6 Hz); 4.08 (H; d; 4.6 Hz); 3.74 (H; dd; 2.8; 12.3 Hz); 3.81 (H; dd; 2.8; 12.3 Hz); 4.0 (m); 4.14; 4.17 (H; t; 5.2 Hz) ppm
Fig. 6.
The structure of 4-O-glucosyl, 1-carboxyl-phenazine of the purified compound from Streptomyces sp. strain B-92: Prediction (a); Based on result (b)
Table 1.
Spectral 1H-NMR of the bioactive compound 4-((3S,4R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl) tetrahydro-2H-pyran-2-yloxy) phenazine-1-carboxylic acid (C 19 H 18 N 2 O 8 ) that have 402 of molecular weight
| No. | 1(CD3 OD, 500 MHz) | No. | 1 (CD3OD, 500 MHz) |
|---|---|---|---|
| δ1H(m, JHz) (ppm) | δ1(m, JHz) (ppm) | ||
| Bioactive compound of Streptomyces sp. strain UICC B-92 | Prediction of bioactive compound (using Chem Draw) | ||
| 1 | - | 1 | - |
| 2 | 8.01 (d; 8.3) | 2 | 7.40 (d) |
| 3 | 5.7 (d; 8.3) | 3 | 8.44 (d) |
| 4 | - | 4 | - |
| 5/8 | 7.75 (2H; d; 8.5) | 5/8 | 7.80 (d) |
| 6/7 | 7.22 (2H; d; 8.5) | 6/7 | 7.67 (d) |
| 1a | - | 1a | - |
| 4a | - | 4a | - |
| 5a | - | 5a | - |
| 8a | - | 8a | - |
| 1″ | 5.9 (d; 4.6) | 1″ | 5.88 (d) |
| 2″ | 4.08 (d; 4.6) | 2″ | 3.91 (d) |
| 3″ | 3.74 (dd; 2.8; 12.3) | 3″ | 3.49 (dd) |
| 4″ | 3.81 (dd; 2.8; 12.3) | 4″ | 3.40 (dd) |
| 5″ | 4.0 (m) | 5″ | 3.76 (m) |
| 6″ | 4.14; 4.17 (t; 5.2) | 6″ | 3.79; 3.54 (t) |
Antibacterial activity assay.
Disc diffusion method of the purified compound isolated from the actinomycetes strain UICC B-92 exhibited antibacterial activity against B. cereus ATCC 10876, S. aureus ATCC 25923, and S. flexneri ATCC 12022. However, negative results for the antibacterial test were found on E. coli strain ATCC 25922 and S. Typhimurium strain ATCC 25241. Based on the inhibition zone diameter, B. cereus ATCC 10876 were the most sensitive bacterium (Fig. 7), followed by S. aureus ATCC 25923 and S. flexneri ATCC 12022 (Table 2).
Fig. 7.
The bioautography visualization of the antibacterial bioactive compound inhibition zone from Streptomyces sp. strain UICC B-92 for B. cereus at 41st fraction with Methanol: Chloroform eluent = 2000 μL:250 μL (A); Methanol: Chloroform eluent = 2000 μL:400 μL (B); Methanol: Chloroform eluent = 2000 μL:500 μL (C); Methanol: Chloroform eluent = 2000 μL:600 μL (D); Methanol: Chloroform eluent = 2000 μL:750 μL (E)
Table 2.
Antibacterial activity of the purified compound from Streptomyces sp. strain UICC B-92
| Compound concentration (μg/mL) | Diameter of inhibitory zone (mm) | ||||
|---|---|---|---|---|---|
| B. cereus strain ATCC 10876 | E. coli strain ATCC 25922 | S. typhimurium strain ATCC 25241 | S. flexneri strain ATCC 12022 | S. aureus strain ATCC 25923 | |
| 250,000 | 10.00 | - | - | 8.00 | 6.70 |
| 50,000 | 7.10 | - | - | - | - |
| 10,000 | 7.00 | - | - | - | - |
| 5,000 | - | - | - | - | - |
| 1,000 | - | - | - | - | - |
| 100 | - | - | - | - | - |
| Tetracycline (1,000) | 9.00 | 9.00 | 9.00 | 9.00 | 9.00 |
| DMSO | - | - | - | - | - |
(−): without inhibition halo
The mechanism of action of the antibacterial compound.
The SEM analysis of pathogenic bacterial cells indicated a possible mechanism of action of the antibacterial compound produced by Streptomyces sp. strain UICC B-92 (Fig. 8). Selection of B. cereus ATCC 10876 in the SEM analysis due to lowest concentration (the minimal inhibitory concentration / MIC) of the bioactive compound from Streptomyces sp. strain UICC B-92 was found against this bacterium (10,000 μg/mL) (Table 2). The SEM analysis revealed that bacterial cells of B. cereus strain ATCC 10876 morphologically changed after treatment using the bioactive compound of Streptomyces sp. strain UICC B-92. A surface of the B. cereus strain ATCC 10876 cells in the control group were smooth and the bacteria cells were plump and cylindrical. However, the cell membranes became coarse, wrinkled, and distorted (Fig. 8B and C) after treatment with the antibacterial compound from Streptomyces sp. strain UICC B-92. Local rupture and pore formation were also observed in the cell membranes (Fig. 8B and C).
Fig. 8.
SEM analysis of B. cereus strain ATCC 10876 before and after antibacterial compound treatment from Streptomyces sp. strain UICC B-92. Negative control (1% DMSO), cells of B. cereus strain ATCC 10876 (A); cells of B. cereus strain ATCC 10876 bacterial cells after treatment with 250,000 μg/mL of the antibacterial compound from Streptomyces sp. strain UICC B-92 (B, C)
DISCUSSION
N. altissima has been known by the local people in Southeast Asian, in particular Indonesia, as a plant to treat infectious diseases such as gonorrhea, diuretic, and diarrhea diseases (10). The current study confirmed that endophytic bacterium Streptomyces sp. strain UICC B-92 isolated from N. altissima has the same ability as their host in producing active compounds. It is due to the active compounds produced by the endophytes actually serves to protect their hosts from the various environmental pressures such as plant diseases infection. Further challenges in this study include specific compound determination, mode of action, specificity, and the compound manipulation both physicochemically and genetically to increase yields of desired metabolites.
The phenazine compound synthesized by Streptomyces sp. UICC B-92, (4-[(3S,4R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy] phenazine-1-carboxylic acid or 4-O-glucosyl, 1-carboxyl-phenazine), is composed of aromatic compound, phenazine structure, and sugar group. The sugar group in this compound is considered rare due to not many phenazine derivatives synthesized by a member of the genus Streptomyces contains sugar structure (18, 19, 27). This phenazine compound (Figs. 3–6) has similarity with glycoside phenazine izuminoside A–C that synthesized by Streptomyces sp. IFM 11260 (19), however, a sugar group from Streptomyces sp. UICC B-92 belong to is different to that of Streptomyces sp. IFM 11260. The sugar group of Streptomyces sp. UICC B-92 belong to 4-O-glucosyl and located at the right side of the structure, while the latter belong to rhamnoside and located at the left side (19).
The current study showed that 4-((3S,4R,5S)-3,4,5- trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy) phenazine-1-carboxylic acid or 4-O-glucosyl, 1-carboxyl-phenazine compound exhibited antibacterial activity against two Gram-positive bacteria, B. cereus strain ATCC 10876 and S. aureus strain ATCC 25923 (Table 2). This compound was produced at the late stage of the Streptomyces sp. strain UICC B-92 growth phase. Most of the low molecular weight secondary metabolites produced by microorganisms generally occurs at the late growth phase (13), whereas phenazine production in liquid batch cultures typically occurs after the period of exponential phase (rapid growth), and phenazines are accumulated in the culture at the stationary phase. Phenazine [(C 6 H 4 ) 2 N 2 ] is an organic compound that possess important biological activities such as anti-tumor activity, and a broad-spectrum antimicrobial activity which is important for agricultural and medicine (28, 29). These include streptophenazine A (30), phenazine-1-carboxylic acid (31), antibiotic griseolutein (32), antibiotic lomofungin, endophenazines A–D (16), phenazinolins A–E (7), phenaziterpenes A–B (17). Currently, over 100 different phenazine derivatives have been identified from various sources in nature (33).
A detail antibacterial mechanism of the phenazine compound from Streptomyces sp. UICC B-92 is still unknown. However, the results from SEM analysis showed this compound damaged cell walls probably through a kind of lysis mechanism. A morphological change was apparent in the bacterial cells after treatment with the phenazine compound from Streptomyces sp. UICC B-92. Occurrence of local rupture or pore formation in the cell membranes is related to leakage of essential intracellular constituents from the cells. This condition causes failure for budding formation, nutrition absorption inability by the cells, and the reduction of the cell walls the metabolism rate and permeability (34), (35). In the end, these conditions cause bacterial cell death.
Many bacterial species produce one or more of the 50 known phenazine compounds, including Pseudomonas spp., Streptomyces spp., Nocardia spp., Sorangium spp., Brevibacterium spp., Burkholderia spp. and Pantoea agglomerans (36). Many phenazine-producing microorganisms belong to the bacterial phyla Actinobacteria and Proteobacteria, and the archaeal phylum Euryarcheota (37). The phenazine production among species, isolates, and even repeat cultivations of the same strain are often variable (38). It is probably due to subtle discrepancies in the regulatory networks and sensing mechanisms of the same strain (38). In natural systems, multiple derivatives or an individual phenazines play multiple roles in the bacterial interactions and behaviors (39), for example member of the genus Streptomyces (40). In addition, the differences between the gene clusters responsible for the phenazines biosynthesis among different Streptomyces species (13) that may lead to the diverse phenazine structural derivatives produced by the bacteria at different levels (39).
Although phenazines are generally isolated from Pseudomonas, Streptomyces and several other microbial genera from soil and marine habitats, however, many endophytic Streptomyces species associated with various plants are also capable in producing this secondary metabolite compounds (12, 13). This suggests that the endophytic microbial group is one source of potential natural sources in the discovery of new antimicrobial compounds. Strobel and Daisy (3) previously suggested that exploration and conservation of hidden microbial diversity such as microbial endophytes are urgently needed, because they might possess many unknown natural products that are useful for medicines and agrochemical agents. In addition, based on the complexity in a structure, the phenazines from Streptomyces are considered more promising than those of Pseudomonas (39, 41). Although an investigation on the biosynthesis of 4-((3S,4R,5S)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy) phenazine-1-carboxylic acid or 4-O-glucosyl, 1-carboxyl-phenazine compound and determination of genes that responsible for the phenazine biosynthesis in Streptomyces sp. strain UICC B-92 is still ongoing, however, the current antibacterial assay showed that this novel compound possesses a potential in the discovery of new agrichemical and medicinal compounds.
In conclusion, this research showed that, bioactive compound of Streptomyces sp. strain UICC B-92 is suggested a new compound based on glycoside structure and its position.
ACKNOWLEDGEMENTS
This research was supported by PPT Grant 2016 No. 003/SP2H/LT/DRPM/II/2016 and 214/SP2H/LT/ DRPM/III/2016 awarded to RHP and BSLN 2017 from KemenristekDikti. We thank Staff from Chemistry Laboratory at Research Center for Chemistry, Indonesian Institute of Sciences (LIPI), PUSPIPTEK Serpong for technical assistance.
REFERENCES
- 1.World Health Organization (2014). Antimicrobial resistance: global report on surveillance.
- 2.Nathan C, Cars O. Antibiotic resistance-problems, progress, and prospects. N Engl J Med 2014;371:1761–1763. [DOI] [PubMed] [Google Scholar]
- 3.Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 2003;67:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. [DOI] [PubMed] [Google Scholar]
- 5.Castillo U, Strobel GA, Ford EJ, Hess WM, Porter H, Jensen JB, et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology 2002;148:2675–2685. [DOI] [PubMed] [Google Scholar]
- 6.Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, et al. Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol Lett 2003;224:183–190. [DOI] [PubMed] [Google Scholar]
- 7.Ding L, Armin M, Heinz-Herbert F, Wen-Han L, Christian H. A family of multicyclic indolosesquiterpenes from a bacterial endophyte. Org Biomol Chem 2011;9:4029–4031. [DOI] [PubMed] [Google Scholar]
- 8.Ezra D, Castillo UF, Strobel GA, Hess WM, Porter H. Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology 2004;150:785–793. [DOI] [PubMed] [Google Scholar]
- 9.Chung RCK. (1998). Sarcotheca diversifolia. In: Plant Resources of South-East Asia No 5(3). Timber Trees: Lesser-Known Timbers. Eds, Sosef MSM, Hong LT, Prawirohatmodjo S. Backhuys Publishers, Leiden, the Netherlands, pp. 435–448. [Google Scholar]
- 10.Soepadmo S. (1960). A monograph of the genus Neesia Blume (Bombacaceae). In: Reinwardtia. Herbarium Bogoriense Publishing, 5th vol Botanical Garden, Bogor, Indonesia, pp. 481–508. [Google Scholar]
- 11.Pratiwi RH, Hidayat I, Hanafi M, Mangunwardoyo W. Identification and characterization of three endophytic bacteria from Neesia altissima (Malvaceae) antagonistic to diarrhea-causing bacteria. Malays J Microbiol 2016;12:300–307. [Google Scholar]
- 12.Pratiwi RH, Hidayat I, Hanafi M, Mangunwardoyo W. Antibacterial compound produced by Pseudomonas aeruginosa strain UICC B-40, an endophytic bacterium isolated from Neesia altissima. J Microbiol 2017;55:289–295. [DOI] [PubMed] [Google Scholar]
- 13.Remali J, Sarmin NM, Ng CL, Tiong JJL, Aizat WM, Keong LK, et al. Genomic characterization of a new endophytic Streptomyces kebangsaanensis identifies biosynthetic pathway gene clusters for novel phenazine antibiotic production. PeerJ 2017;5:e3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Q, Hu H, Peng H, Zhang X, Wang W. Isolation and structural identification two bioactive phenazines from Streptomyces griseoluteus P510. Chin J Chem Eng 2015;23:699–703. [Google Scholar]
- 15.Johnson LE, Dietz A. Lomofungin, a new antibiotic produced by Streptomyces lomondensis sp. Appl Microbiol 1969;17:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebhardt K, Schimana J, Krastel P, Dettner L, Rheinheimer J, Zeeck A, et al. Endophenazines, A.-D., new phenazine antibiotics from the arthropod associated endosymbiont Streptomyces anulatus. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot 2002;55:794–800. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Huang H, Chen Y, Ding J. Cytotoxic and antibacterial marfiraquinocins from the deep South China Sea-derived Streptomyces niveus SCSIO 3406. J Nat Prod 2013;76:2263–2268. [DOI] [PubMed] [Google Scholar]
- 18.Abdelfattah MS, Toume K, Ishibashi M. Izumiphenazines A–C: isolation and structure elucidation of phenazine derivatives from Streptomyces sp. IFM 11204. J Nat Prod 2010;73:1999–2002. [DOI] [PubMed] [Google Scholar]
- 19.Abdelfattah MS, Toume K, Ishibashi M. Isolation and structure elucidation of izuminosides A–C: a rare phenazine glycosides from Streptomyces sp. IFM 11260. J Antibiot 2011;64:271–275. [DOI] [PubMed] [Google Scholar]
- 20.Pratiwi RH, Hidayat I, Mangunwardoyo W. (2018). Molecular identification of endophytic bacteria from Neesia altissima Bl. (Malvaceae) antagonistic to diarrhea-causing bacteria. In: Proceedings of the 18th Science Council of Asia Conference: “Role of Science for Society: Strategies towards SDGs in Asia” Tokyo, Japan. pp. 6. [Google Scholar]
- 21.Cao L, Qiu Z, You J, Tan H, Zhou S. Isolation and characterization of endophytic Streptomycetes antagonists of fusarium wilt pathogen from surface sterilized banana roots. FEMS Microbiol Lett 2005;247:147–152. [DOI] [PubMed] [Google Scholar]
- 22.Saha BC, Cotta MA. Comparison of pretreatment strategies for enzymatic saccharification and fermentation of barley straw to ethanol. N Biotechnol 2010;27:10–16. [DOI] [PubMed] [Google Scholar]
- 23.Pratiwi RH, Hanafi M, Artanti N, Pratiwi RD. Bioactivity of antibacterial compounds produced by endophytic actinomycetes from Neesia altissima. J Trop Life Science 2018;8:37–42. [Google Scholar]
- 24.Narayana KJP, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y, Krishna PSJ. Study on bioactive compounds from Streptomyces sp. ANU 6277. Pol J Microbiol 2008;57:35–39. [PubMed] [Google Scholar]
- 25.Schwalbe R, Steele-Moore L, Goodwin AC. (2007). Antimicrobial Susceptibility Testing Protocols. 1st ed CRC Press; New York: pp. 75–79. [Google Scholar]
- 26.Kai J, Matoh M, Tsukidate K. A new method for preparing electron microscopic specimens of Helicobacter pylori. Med Electron Microsc 1999;32:62–65. [DOI] [PubMed] [Google Scholar]
- 27.Hui J, Wang W, Hu H, Peng H, Zhang X. Streptomyces griseoruber Y1B, a novel Streptomyces for 1-Hydroxyphenazine production. J Appl Biotechnol 2014;2:13–31. [Google Scholar]
- 28.Bunbamrung N, Dramae A, Srichomthong K, Supothina S, Pittayakhajonwut P. Streptophenazines I–L from Streptomyces sp. BCC21835. Phytochem Lett 2014;10:91–94. [Google Scholar]
- 29.Kennedy RK, Naik PR, Veena V, Lakshmi BS, Lakshmi P, et al. 5-methyl phenazine-1-carboxylic acid: a novel bioactive metabolite by a rhizosphere soil bacterium that exhibits potent antimicrobial and anticancer activities. Chem Biol Interact 2015;231:71–82. [DOI] [PubMed] [Google Scholar]
- 30.Guttenberger N, Blankenfeldt W, Breinbauer R. Recent developments in the isolation, biological function, biosynthesis, and synthesis of phenazine natural products. Bioorg Med Chem 2017;25:6149–6166. [DOI] [PubMed] [Google Scholar]
- 31.Sarmin NIM, Tan GYA, Franco CMM, Edrada-Ebel R, Latip J, Zin NM. Streptomyces kebangsaanensis sp. nov., an endophytic actinomycete isolated from an ethnomedicinal plant, which produces phenazine-1-carboxylic acid. Int J Syst Evol Microbiol 2013;63:3733–3738. [DOI] [PubMed] [Google Scholar]
- 32.Umezawa H, Hayano S, Maeda K, Ogata Y, Okami Y. On a new antibiotic, griseolutein, produced by Streptomyces. Jpn Med J (Natl Inst Health Jpn) 1950;3:111–117. [DOI] [PubMed] [Google Scholar]
- 33.Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 2006;44:417–445. [DOI] [PubMed] [Google Scholar]
- 34.Diver JM, Wise R. Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin. J Antimicrob Chemother 1986;18 Suppl D :31–41. [DOI] [PubMed] [Google Scholar]
- 35.Sasidharan S, Darah I, Jain K. Antimicrobial activity and wound healing potential on infected rat of Gracilaria changii methanolic extract. Pharmacologyonline 2008;2:661–670. [Google Scholar]
- 36.Delaney SM, Mavrodi DV, Bonsall RF, Thomashow LS. phzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30–84. J Bacteriol 2001;183:318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol 2010;76:866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakhtah H, Price-Whelan A, Dietrich LEP. (2013). Regulation of phenazine biosynthesis. In: Microbial Phenazines: Biosynthesis, Agriculture and Health. Eds, Chincholkar S, Thomashow L. Springer-Verlag, Heidelberg, Berlin, pp. 19–42. [Google Scholar]
- 39.Pierson LS, III, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 2010;86:1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haagen Y, Glück K, Fay K, Heide L. A gene cluster for prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042. Chembiochem 2006;7:2016–2027. [DOI] [PubMed] [Google Scholar]
- 41.Laursen J, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev 2004;104:1663–1686. [DOI] [PubMed] [Google Scholar]