Abstract
Obligate symbiotic associations are present in a wide variety of animals with a nutrient-restricted diet. Aphids (hemiptera: Aphididae) almost-universally host Buchnera aphidicola bacteria in specialised organs (called bacteriomes). These bacteria supply the aphid with essential nutrients lacking from their diet (i.e. essential amino acids and some B vitamins). Some aphid lineages, such as species from the Lacninae subfamily, have evolved co-obligate associations with secondary endosymbionts, deriving from a loss of biotin- and riboflavin-biosynthetic genes. In this study, I re-analyse previously published sequencing data from the banana aphid Pentalonia nigronervosa. I show that the metabolic inference results from De Clerck et al. (Microbiome 3:63, 2015) are incorrect and possibly arise from the use of inadequate methods. Additionally, I discuss how the seemingly biased interpretation of their antibiotic treatment analyses together with an incorrect genome-based metabolic inference resulted in the erroneous suggestion “that a co-obligatory symbiosis between B. aphidicola and Wolbachia occurs in the banana aphid”.
Keywords: Aphid, Buchnera, Wolbachia, Pentalonia nigronervosa, Co-obligate, Symbiont
Main text
In a previous study, De Clerck et al. [1] claimed to present evidence for a potential co-obligate association of Buchnera and Wolbachia in the aphid Pentalonia nigronervosa. They reach these conclusions mainly based on 4 lines of evidence: (1) the apparently fixed nature of Wolbachia in P. nigronervosa; (2) a genome-based metabolic inference coming from a pooled metagenomic assembly of extracted DNA from three P. nigronervosa populations sampled in Gabon, Madagascar, and Burundi; (3) an antibiotic treatment directed towards the elimination of the endosymbionts; and (4) the attempted detection of putatively missing genes through PCR. In this work, I have re-analysed the publicly available sequencing data and performed a genome-based metabolic inference of the biosynthetic capabilities of Buchnera. I find that this Buchnera has equivalent biosynthetic capabilities, concerning essential amino acids, B vitamins, and co-factors, to all published Buchnera strains from “mono-symbiotic” aphids (only harbouring Buchnera as the nutritional obligate symbiont), and thus should not need an additional partner to fulfil its nutritional role. I also critically discuss the interpretation of the experimental results presented by De Clerck et al. [1] and conclude that their suggestion of the nutritional-based co-obligate nature of Wolbachia in P. nigronervosa derives from inadequate analyses of their data as well as a seemingly biassed interpretation of their experimental results.
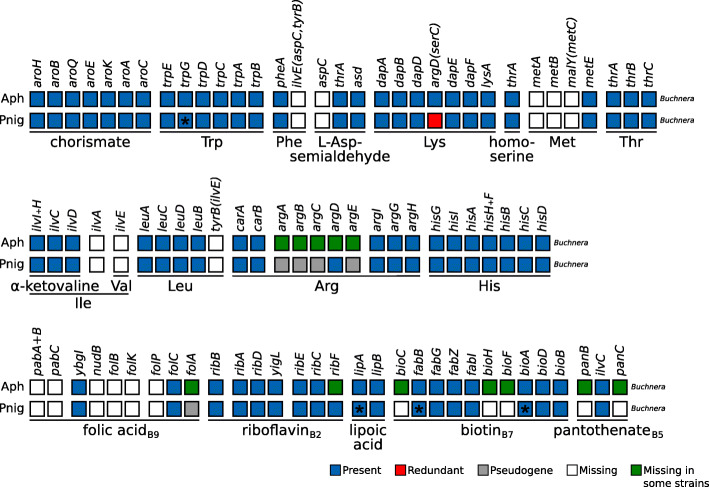
To produce a genome assembly, I downloaded the three datasets deposited in NCBI with project number PRJNA268300 and accession SRX766492. The pooled genome assembly of these reads resulted in 269,717 contigs that were then binned into 4 groups: Wolbachia, Buchnera, mitochondrion, and the aphid host. These resulted in 135 scaffolds for Buchnera with an average k-mer (77 bp) coverage of 110 and 1309 for Wolbachia with an average k-mer coverage of 396. The search for the genes involved in the biosynthesis of essential amino acids (EAAs), B vitamins, and other co-factors (hereafter referred to collectively as “nutritional genes”) revealed that Buchnera from P. nigronervosa retains all genes common to other Buchnera from aphids displaying a mono-symbiotic relationship with Buchnera (Fig. 1). Additionally, I found that all genes claimed by De Clerck et al. [1] to be missing from Buchnera in the biosynthetic pathways shown in Fig. 4 of the article are actually present, except for that of pgm (coding for a phosphoglucomutase which is absent in all currently sequenced Buchnera strains). The lipA, fabB, and bioA genes show frameshifts in low complexity regions (see GenBank file for details), which is not uncommon for Buchnera nor for other small A+T-biassed genomes. The expression of these genes is likely to be rescued by ribosomal frameshifting. Additionally, the trpG gene also displays a frameshift in a low complexity region and a stop codon in the consensus sequence. Closer inspection revealed that the “TAG” stop codon shows a variant (“CAG”) present at 13.40% in library SRR1662246. This could be explained by the collapsed assembly of the tandem trpEG units found in other Aphidinae, which tend to show pseudogenised variants [2, 3]. To test for the presence of the genes claimed as missing by De Clerck et al. [1] and “nutritional genes”, and in a similar fashion to De Clerck et al. [1], I performed read mapping of each library vs. the nucleotide sequence of each gene. This confirmed the presence of most of these genes in all three sequencing libraries (Supplementary Table S1 in Additional file 1, Supplementary Material online). Many Buchnera genes had very low coverages of ≤3 in sequencing library SRR1661114 and ≤15 in sequencing library SRR1662249. In fact, these two libraries had between 1 and 2% of the reads coming from Buchnera, contrasting library SRR1662249, where 25% of the reads mapped to the Buchnera scaffold bin (supplementary table S2 in Additional bluefile 1). The fact that the authors solely searched for intact protein-coding genes, using myRAST [4, 5], and their binning method based on a BLASTX search vs. the nr database of NCBI, rather than a narrower database consisting of expected associates, surely impacted both accurate binning and gene identification. Therefore, the genome-based metabolic inference results do not in fact support the nutritional need of a co-obligate symbiont in P. nigronervosa. The lack of amplification by PCR of these genes by De Clerck et al. [1] can be explained by the nucleotide sequence divergence between Buchnera harboured by not-so-distantly related aphids. Due to this divergence across Buchnera strains, using positive amplification of the target gene in a sample from Acyrthosiphon pisum was not an adequate control. Manual inspection of the alignments of the primers vs. the assembled reference sequences, produced here for Buchnera, reveals that designed primers did not matched the expected target sequences. Therefore, the lack of amplification is not surprising and should not have been taken as evidence for the lack of these genes in Buchnera.
Fig. 1.
Essential amino acid and selected B vitamin and co-factor biosynthetic metabolic capabilities of obligate symbiotic consortia of different aphid species. Diagram summarising the metabolic capabilities of Buchnera from “mono-symbiotic” Aphidinae aphids (Aph) and P. nigronervosa (Pnig). “Aph” rows are a collapsed representation of several Aphidinae species (see Supplementary Table S3 in Additional file 1). The names of genes coding for enzymes involved in the biosynthetic pathway are used as column names. Each row’s boxes represent the genes coded by the Buchnera genome. On the bottom, lines underlining the genes involved in the pathway leading to the compound specified by the name underneath the line. For amino acids, its three-letter abbreviation is used
Regarding the experimental analyses, I would argue that in spite of Wolbachia being seemingly fixed across different P. nigronervosa populations collected in different countries, the fact that this is a facultative endosymbiont cannot be discarded. While it is true that the authors convincingly show that the three populations sampled apparently harbour the same strain of Wolbachia, the widespread localisation of this symbiont does not suggest a change of tissue tropism for this strain, but rather what one would expect from a Wolbachia bacterium [6–8]. This greatly contrasts the localisation of the nutritional co-obligate Wolbachia strain of the bedbug Cimex lectularius, where “the infection is substantially restricted to the bacteriome” [9]. Finally, the reported “selective” elimination of the symbionts through antibiotic treatment does not in fact conclusively point towards the obligatory nature of Wolbachia. This could only be achieved either through the actual selective elimination of Wolbachia and the subsequent measure of biological parameters vs. controls (e.g. untreated and/or injected with distilled water), or through proving that the antibiotics solely decrease the quantity of Wolbachia and do not affect Buchnera. The authors base their conclusions on the treatments with two antibiotics: tetracycline and ampicillin. Regarding the former, the authors incorrectly conclude that since all the injected nymphs died before adulthood and dead nymphs were only positive for Buchnera in PCR assays, then the antibiotic treatment “eliminated Wolbachia, but only decreased Buchnera quantity, without eliminating it completely”. However, as the authors themselves mention, “tetracycline has been reported to have negative effect on Buchnera as well as on Wolbachia”. Therefore, it is reasonable to conclude that the impact of the decrease in Buchnera could also explain the observed mortality. However, without quantitative PCR and/or microscopy analyses, the impact of the antibiotic treatment on Buchnera cannot be correctly assessed. In regard to the ampicillin treatment, similar concerns arise. The fact that the authors could only detect Buchnera but failed to measure the impact on the abundance of this symbiont in the dead nymphs raises concerns regarding the decrease in Buchnera possibly being the responsible for the observed mortality.
Conclusion
In brief, De Clerck et al. [1] provide no evidence that the apparent fixation of Wolbachia in the analysed populations of P. nigronervosa is due to this symbiont being a nutritional co-obligate in this aphid. The re-analysis of the sequencing data revealed the genome-based conclusions for a nutritional co-obligate symbiosis in P. nigronervosa were erroneous. Additionally, inspection of the primer sequences revealed that most primer pairs do not match the targetted sequences in Buchnera from P. nigronervosa. Finally, the antibiotic treatment results have alternative explanations, which the authors failed to discuss, and that cannot be discarded without more sensitive analyses. Regardless, Wolbachia’s apparently consistent presence could nonetheless be explained by other processes, such as some sort of reproductive manipulation, protection against viral infections, or the aphid strains sampled actually belonging to one, or a few, “superclone(s)” originally infected with closely related Wolbachia endosymbionts.
Materials and methods
Sequencing data collection and genome assembly
The tree sequencing libraries (SRR1661114, SRR1662246, SRR1662249) from De Clerck et al. [1] were downloaded from the NCBI SRA database (accession SRX766492) from BioProject (https://www.ncbi.nlm.nih.gov/bioproject/268300, last accessed April 15, 2019). Inspection of the downloaded fastq files with fastQC v0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed April 15, 2019) revealed contamination of the reads with TruSeq adapter indexes 4, 5, and 6, as well as an unknown T+G-rich adapter (“TGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGT”). To trim these adapters from the read datasets and perform quality trimming, I used fastx_clipper followed by fastx_quality_trimmer which performed right-tail quality trimming (using a minimum quality threshold of 20) and reads shorter than 50 bp were dropped (options -t 20 -l 50). Additionally, PRINSEQ v0.20.4 [10] was used to remove reads containing undefined nucleotides as well as separate those left without a pair after the filtering and trimming process. Assembly of the three libraries was done using SPAdes v3.10.1 [11] with k-mers 33, 55, and 77.
Binning of assembled sequences to Buchnera and Wolbachia
First, all scaffolds shorter than 200 bp and with an average k-mer coverage lower than 3 were dropped. This resulted in 10,097 (3.74% of the total number of scaffolds) which were then used for a BLASTX-based binning. This was done by building a database of the proteomes of the Pea aphid and a set of Wolbachia and Buchnera bacteria. A separate database of the mitochondrion proteins encoded by the aphid Schizaphis graminun was built to detect mitochondria sequences. All these protein datasets were available before the time of publication of De Clerck et al. [1]. Accession numbers and information on strains can be found in supplementary table S4 (Additional bluefile 1). A BLASTX search against the aforementioned databases was done using the filtered scaffolds as query (-soft_masking true -seg yes -max_target_seqs 10000 -evalue 1e-3) followed by assigning the scaffold to the organism with the best BLASTX hit to each scaffold. These bins were then filtered by setting a minimum k-mer coverage threshold of 10 for both Buchnera and Wolbachia. This resulted in 135 and 1309 scaffolds being assigned to each bacterium, respectively.
Identification of “missing” and “nutritional genes” in Buchnera
For verifying the absence of the genes claimed as missing by De Clerck et al. [1] and the “nutritional genes”, I collected the amino acid sequences of these genes from Buchnera strain 5A, APS, and Bp. Then, a TBLASTN search of the aforementioned sequences vs. both the total filtered scaffolds and the Buchnera scaffold bin (-evalue 1e-03 -db_gencode 11) was performed. Afterwards, manual verification of each hit was performed using UGENE v1.29.0 and then BLASTX on the on-line BLAST server from NCBI vs. the nr database. If the gene was not found in the Buchnera scaffold bin, then the gene was verified from the TBLASTN vs. the filtered scaffolds. This revealed the misassignment of “NODE_9” to the Wolbachia bin. This manual curation also revealed remnants of the “TGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGTTGGGTGTGT” sequence inside the lipA and pgi genes. Further manual inspection of the reads revealed a consistent insertion of this sequence in a specific place of each gene, revealing the biassed insertion of the contaminant. Additionally, an alignment of each read library vs. the abovementioned Buchnera genes from P. nigronervosa was done with Bowtie2 v2.3.4.1 [12] for visualisation purposes. To avoid reads from Wolbachia mapping to sequences of orthologous genes from Buchnera, the Wolbachia scaffolds were also included in the Bowtie2 mapping.
The suitability of the reported primers in De Clerck et al. [1] (found in “Additional file 1: Table S1”) was judged by aligning them vs. the gene sequence identified and reported in the present work using AliView v1.26 [13].
Supplementary information
Additional file 1 Supplementary tables S1-S4. XLS-formatted supplementary tables S1-S4, including descriptions.
Acknowledgements
I am grateful to the genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing help and/or computing and/or storage resources.
Abbreviations
- EAAs
Essential amino acids
Authors’ contributions
AMM conceived and designed the experiments, contributed the analysis tools, and drafted the manuscript. The author read and approved the final manuscript.
Funding
This work was supported by the Marie-Curie AgreenSkills+ fellowship programme co-funded by the EU’s Seventh Framework Programme (FP7-609398). This publication has been written with the support of the AgreenSkills+ fellowship programme which has received funding from the EU’s Seventh Framework Programme under grant agreement no. FP7-609398 (AgreenSkills+ contract). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares that there are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40168-020-00865-2.
References
- 1.De Clerck C, Fujiwara A, Joncour P, Léonard S, Félix M-L, Francis F, Jijakli MH, Tsuchida T, Massart S. A metagenomic approach from aphid’s hemolymph sheds light on the potential roles of co-existing endosymbionts. Microbiome. 2015;3(1):63. doi: 10.1186/s40168-015-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai CY, Baumann P, Moran NA. The endosymbiont (Buchnera sp.) of the aphid Diuraphis noxia contains plasmids consisting of trpEG and tandem repeats of trpEG pseudogenes. Appl Environ Microbiol. 1996;62(2):332–9. doi: 10.1128/AEM.62.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann Linda, Clark Marta A., Rouhbakhsh Dadbeh, Baumann Paul, Moran Nancy A., Voegtlin David J. Endosymbionts ( Buchnera ) of the Aphid Uroleucon sonchi Contain Plasmids with trpEG and Remnants of trpE Pseudogenes. Current Microbiology. 1997;35(1):18–21. doi: 10.1007/s002849900204. [DOI] [Google Scholar]
- 4.Meyer F, Paarmann D, D’Souza M, Olson R, Glass E, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards R. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics; 1:386. 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed]
- 5.Aziz Ramy K, Bartels Daniela, Best Aaron A, DeJongh Matthew, Disz Terrence, Edwards Robert A, Formsma Kevin, Gerdes Svetlana, Glass Elizabeth M, Kubal Michael, Meyer Folker, Olsen Gary J, Olson Robert, Osterman Andrei L, Overbeek Ross A, McNeil Leslie K, Paarmann Daniel, Paczian Tobias, Parrello Bruce, Pusch Gordon D, Reich Claudia, Stevens Rick, Vassieva Olga, Vonstein Veronika, Wilke Andreas, Zagnitko Olga. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Valero Laura, Soriano-Navarro Mario, Pérez-Brocal Vicente, Heddi Abdelaziz, Moya Andrés, García-Verdugo José Manuel, Latorre Amparo. Coexistence of Wolbachia with Buchnera aphidicola and a Secondary Symbiont in the Aphid Cinara cedri. Journal of Bacteriology. 2004;186(19):6626–6633. doi: 10.1128/JB.186.19.6626-6633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed Muhammad Z., Li Shao-Jian, Xue Xia, Yin Xiang-Jie, Ren Shun-Xiang, Jiggins Francis M., Greeff Jaco M., Qiu Bao-Li. The Intracellular Bacterium Wolbachia Uses Parasitoid Wasps as Phoretic Vectors for Efficient Horizontal Transmission. PLOS Pathogens. 2015;11(2):e1004672. doi: 10.1371/journal.ppat.1004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider DI, Parker AG, Abd-alla AM, Miller WJ. High-sensitivity detection of cryptic Wolbachia in the African tsetse fly (Glossina spp.)BMC Microbiol; S1:140. 10.1186/s12866-018-1291-8. [DOI] [PMC free article] [PubMed]
- 9.Hosokawa T., Koga R., Kikuchi Y., Meng X.-Y., Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proceedings of the National Academy of Sciences. 2009;107(2):769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich Anton, Nurk Sergey, Antipov Dmitry, Gurevich Alexey A., Dvorkin Mikhail, Kulikov Alexander S., Lesin Valery M., Nikolenko Sergey I., Pham Son, Prjibelski Andrey D., Pyshkin Alexey V., Sirotkin Alexander V., Vyahhi Nikolay, Tesler Glenn, Alekseyev Max A., Pevzner Pavel A. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead Ben, Salzberg Steven L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson Anders. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30(22):3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplementary tables S1-S4. XLS-formatted supplementary tables S1-S4, including descriptions.