Significance
The existence of progenitors within pancreatic ducts has been studied for decades, but the hypothesis that they may help regenerate the adult endocrine compartment (chiefly insulin-producing β-cells) remains contentious. Here, we examine the single-cell transcriptome of the human ductal tree. Our data confirm the paradigm-shifting notion that specific lineages, long thought to be cast in stone, are in fact in a state of flux between differentiation stages. In addition to pro-ductal and pro-acinar transcriptomic gradients, our analysis suggests the existence of a third (ducto-endocrine) differentiation axis. Such prediction was experimentally validated by transplanting sorted progenitor-like cells, which revealed their tri-lineage differentiation potential. Our findings further indicate that progenitors might be activated in situ for therapeutic purposes.
Keywords: human pancreatic progenitors, type 1 diabetes, islet regeneration, transplantation, single-cell RNA sequencing
Abstract
We have described multipotent progenitor-like cells within the major pancreatic ducts (MPDs) of the human pancreas. They express PDX1, its surrogate surface marker P2RY1, and the bone morphogenetic protein (BMP) receptor 1A (BMPR1A)/activin-like kinase 3 (ALK3), but not carbonic anhydrase II (CAII). Here we report the single-cell RNA sequencing (scRNA-seq) of ALK3bright+-sorted ductal cells, a fraction that harbors BMP-responsive progenitor-like cells. Our analysis unveiled the existence of multiple subpopulations along two major axes, one that encompasses a gradient of ductal cell differentiation stages, and another featuring cells with transitional phenotypes toward acinar tissue. A third potential ducto-endocrine axis is revealed upon integration of the ALK3bright+ dataset with a single-cell whole-pancreas transcriptome. When transplanted into immunodeficient mice, P2RY1+/ALK3bright+ populations (enriched in PDX1+/ALK3+/CAII− cells) differentiate into all pancreatic lineages, including functional β-cells. This process is accelerated when hosts are treated systemically with an ALK3 agonist. We found PDX1+/ALK3+/CAII− progenitor-like cells in the MPDs of types 1 and 2 diabetes donors, regardless of the duration of the disease. Our findings open the door to the pharmacological activation of progenitor cells in situ.
The existence of progenitor-like cells in the human pancreatic duct tree, despite conflicting findings in mice (1, 2), is supported by experimental evidence (reviewed in ref. 3). We have recently characterized one such cell population in human major pancreatic ducts (MPDs) and pancreatic duct glands (PDGs) (4, 5). These cells express the ductal/β-cell marker pancreatic and duodenal homeobox-1 gene (PDX1) and the bone morphogenetic protein (BMP) receptor 1A (BMPR1A), also known as activin-like kinase-3 (ALK3). Antibodies against ALK3 and the purinergic receptor P2Y1 (P2RY1), which we have established as a surrogate surface marker for PDX1 (4), can be used to sort live progenitor-like cells that expand in vitro in the presence of BMP-7 and differentiate along all three adult pancreatic lineages (endocrine, acinar, and ductal) upon removal of this factor (4).
This marker combination (P2RY1+/ALK3bright+) specifically excludes islets, which are ALK3− (4). Another salient feature of these cells is that, notwithstanding their location in the MPD epithelium, they lack protein expression of carbonic anhydrase II (CAII), a marker previously thought to be pan-ductal (6). Thus, PDX1+/ALK3+/CAII− cells, which we hypothesized to be progenitors, are readily detected in human MPDs and PDGs, often intercalated with regular ductal PDX1+/ALK3+/CAII+ cells (4).
In this report, we conducted single-cell RNA-sequencing (scRNA-seq) analysis of sorted human pancreatic ALK3bright+ cells, which offers a “high-magnification” view of all of the cell types within the ductal tree. This analysis shows the existence of multiple ductal subpopulations that run the gamut from mature functional ductal cells to undifferentiated/transitional cells, including progenitor-like cells with a transcriptional profile consistent with stress-induced de-differentiation. P2RY1+/ALK3bright+ sorting enriches in CAII− cells from such progenitor-like clusters. We demonstrate that sorted P2RY1+/ALK3bright+ cells engraft in immunocompromised mice, where they spontaneously mature into multiple pancreatic endocrine and exocrine cell types. This process was accelerated by the systemic administration of THR-123, a cyclic peptide with ALK3 agonism activity. Next, we sought to explore whether PDX1+/ALK3+/CAII− existed in samples from donors with type 1 and 2 diabetes (T1D/T2D). We confirm that progenitor-like cells exist in the MPD/PDG epithelium of all patients examined, regardless of the duration of the disease. Our findings that human P2RY1+/ALK3bright+ cells can be pharmacologically stimulated in vivo, combined with the observation that similar cells may be intact in T1D/T2D patients, have potential therapeutic implications.
Results
scRNA-Seq Analysis of ALK3+ Cells Reveals Ductal Heterogeneity and Suggests Transitional Phenotypes.
We have previously determined that the P2RY1+/ALK3bright+ fraction of the human exocrine pancreas is enriched in PDX1+/ALK3+/CAII− progenitor-like cells that can be cultured in defined conditions, expand in the presence of BMP-7, and exhibit multipotency when BMP-7 is removed. To further analyze in depth the composition of the human ALK3+ ductal compartment, we conducted scRNA-seq of sorted ALK3bright+ cells from human nonendocrine pancreatic tissue obtained after islet isolations at the Diabetes Research Institute (two donors) and the University of Pittsburgh (one donor) (SI Appendix, Fig. S1 A–E and Table S1). Our rationale was that, by analyzing the ALK3bright+ population at large, we would be able to study the PDX1(P2RY1)+/ALK3+ cells (a subpopulation thereof) in their broader context within the ductal tree. Fig. 1A presents a scheme of the experimental plan. The expected outcome of this sorting strategy is the selection of epithelial cells from the MPDs and PDGs with the exclusion of a significant percentage of ALK3dim+ cells (likely from small ducts), as determined in Qadir et al. (4). We also depleted contaminating mesenchymal cells by CD90− selection, further enriching in epithelial cells.
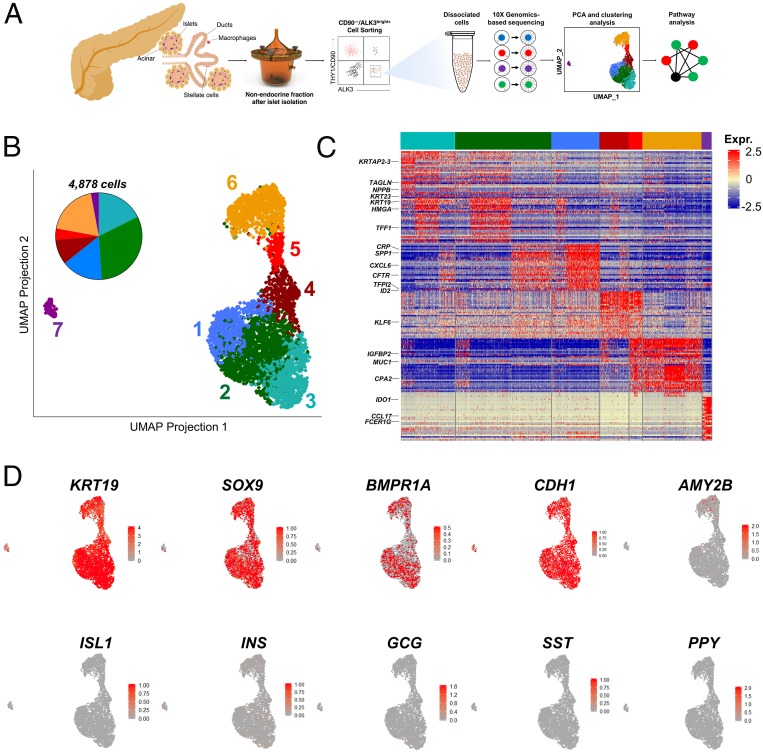
Fig. 1.
scRNA-seq identifies cellular heterogeneity across human ALK3bright+ pancreatic ductal cells. (A) Scheme of the experimental approach employed to isolate ALK3bright+ cells and derive single-cell libraries using the 10X Genomics microfluidics system. Data generated from sequenced single-cell libraries are analyzed to dissect cellular substructure. (B) UMAP plots of ductal populations from individual donor pancreas (n = 3, 4,878 cells). Each dot represents the transcriptome of a single ALK3bright+ cell, with color coding defining clusters of cells having similar transcriptional identities. (C) Gene-expression heatmap of the top 20 most DE genes in each cluster compared to all other clusters. Genes are represented in rows and cell clusters in columns. Select genes are indicated to the left. (D) Single-cell gene expression of known markers of ductal (KRT19 and SOX9), epithelial (CDH1), acinar (AMY2B), and endocrine (ISL1, INS, GCG, SST, PPY) cells. Scale bars represent z-test–normalized gene expression in C and gene counts in D.
Approximately 2,000 cells per preparation were sequenced at 250,000 reads per cell (Genewiz). Only single cells with mitochondrial RNA less than 20% of total RNA were included (SI Appendix, Fig. S1H). Using principal component analysis and clustering with the R v.3.5.3 package Seurat 3.1.1 (SI Appendix), an uniform manifold approximation and projection (UMAP) plot of the combined three preparations (4,878 cells) was generated (Fig. 1B and SI Appendix, Fig. S1 F–L). UMAP is a novel nonlinear dimensionality reduction algorithm for the analysis of high-dimensional data (7), which has proven superior to t-distributed stochastic neighborhood embedding for resolving subtle differences in cell populations (8–11). A scatterplot of differentially expressed (DE) genes and a heatmap with the top 20 DE genes/cluster (vs. all of the other clusters) are presented in SI Appendix, Fig. S1I and Fig. 1C, respectively. Clustree v0.4.1 (12) was used to choose the highest clustering resolution without cluster destabilization or mixing (SI Appendix, Fig. S1J). SI Appendix, Fig. S1 K and L present the individual contribution of each donor to the combined dataset. The overall epithelial/ductal identity of the sorted cells was confirmed by the widespread expression of the ductal markers KRT19 and SOX9, the epithelial marker CDH1, and the absence of any meaningful expression of the archetypal acinar marker AMY2B and the endocrine markers ISL1, insulin (INS), glucagon (GCG), somatostatin (SST), and PPY (Fig. 1D).
The only exception was the smaller cluster 7, which was largely KRT19− and SOX9−. The analysis of DE genes and gene ontology (GO)-term pathways (Fig. 2 A–D and Datasets S1 and S2) suggests an immune cell identity. BMP signaling regulates proliferation/differentiation in the immune system (13), and ALK2/ALK3 are functional BMP receptors in macrophages. Therefore, it is likely that ALK3+ sorting resulted in the carryover of this small immune-related subpopulation.
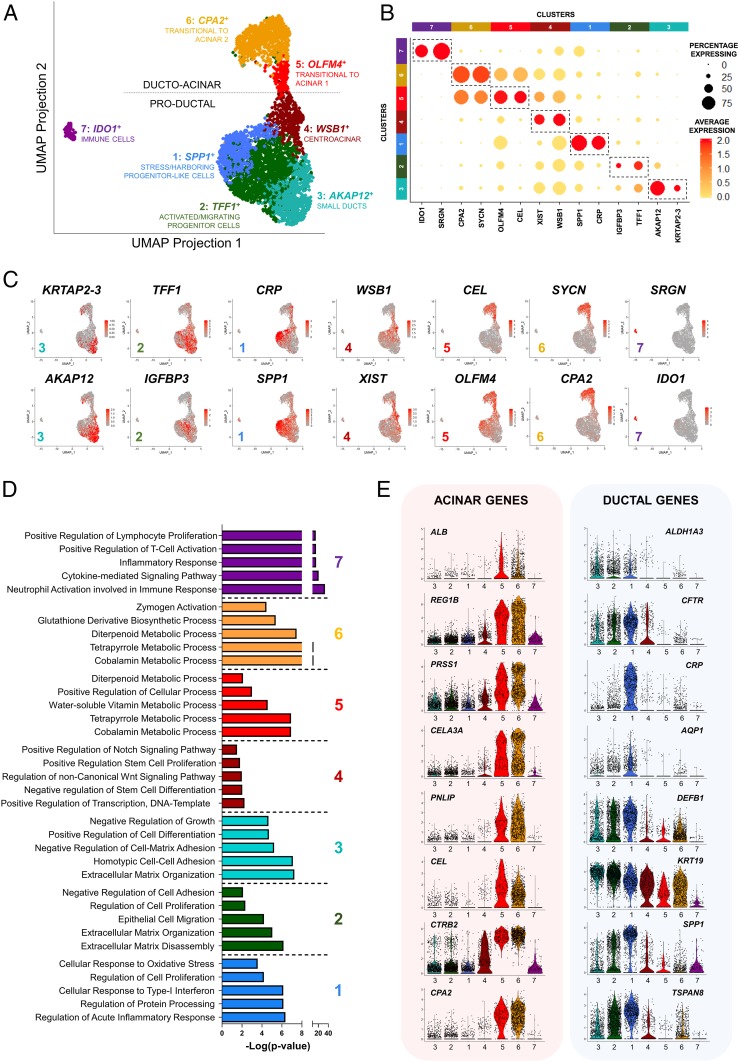
Fig. 2.
Identification of multiple subpopulations of ALK3bright+ ductal cells in the human pancreas. (A) UMAP plot mapping multiple subclusters across the single-cell dataset (n = 3). Each cluster is defined by a specific color and a representative DE gene. The dotted line divides ducto-acinar or “transitional to acinar” clusters (Upper) from “productal” clusters (Lower). (B) Gene-expression dot plot of select DE genes in each cluster. Rows depict clusters, while columns depict genes. The intensity of any given point indicates average expression, while its size represents the proportion of cells expressing a particular gene. Dotted regions identify select genes for a particular cluster. The scale bar represents average expression in gene counts. (C) UMAP expression plots of representative DE genes for clusters 3 (small ducts; KRTAP2-3, AKAP12); 2 (activated/migrating progenitor cells; TFF1, IGFBP3); 1 (stress/harboring progenitor-like cells; CRP, SPP1); 4 (centroacinar; WSB1, XIST); 5 (transitional to acinar 1; CEL, OLFM4); 6 (transitional to acinar 2; SYCN, CPA2); and 7 (immune cells; SRGN, IDO1). (D) GO-driven pathway analysis of DE genes with a ≥twofold change across clusters 1 to 7. (E) Violin plots for the expression of acinar (Left) and ductal genes (Right). Expression in each cell is shown along with the probability density of gene expression, denoted by the shape of the plot.
Fig. 2A shows the predicted identity of each cluster according to the above analysis. Representative top DE genes and GO pathways are indicated in Fig. 2D and SI Appendix, Fig. S2B, as well as Datasets S1 and S2. For identification purposes, we selected those whose expression is most specific to any given cluster vs. the rest. We detected two major groups of cells: Productal (clusters 1 to 4 in Fig. 2 D, Lower) and ducto-acinar (clusters 5 to 6 in Fig. 2 D, Upper). The distinction between ducto-acinar and productal is based on the DE of numerous acinar genes in clusters 5 to 6 (Fig. 2E and SI Appendix, Fig. S2A)
Cluster 1 is represented by osteopontin/secreted phosphoprotein 1 (SPP1). This gene was previously identified as a marker of undifferentiated pancreatic progenitors in mice (14), and also by scRNA-seq in the developing mouse pancreas as a marker of “proliferative ducts” (15). Cluster 1 also has the highest PDX1 and inhibition of differentiation (ID)-1, -2, and -4 expression in the entire dataset (SI Appendix, Fig. S2 C and D). High expression of ID1-4 is typical of progenitors, where they sequester basic helix–loop–helix transcription factors, leading to the inhibition of lineage-specific and cell cycle-inhibitory genes (16). ID2, in particular, has been shown to regulate BMP-dependent pancreatic progenitor expansion in mice (17). Elevated ID expression was also a hallmark of the P2RY1+/ALK3bright+ cells previously described by our team as progenitor-like, especially after the addition of BMP-7 (4). In line with the hypothesis that cluster 1 may harbor the multipotent progenitors therein described, among the most DE genes we also found HES1, FOS, and JUN (Datasets S1 and S2). HES1 is an inhibitor of differentiation up-regulated by Notch signaling, which is associated with progenitor proliferation during pancreatic development (18). FOS and JUN are the two components of the activator protein 1 early response transcription factor, which is essential for the proliferation of ductal epithelial cells in the context of pancreatic cancer. While there is no evidence that the donors used in this study had any malignancy, we conducted a search for DE pancreatic cancer pathways (19) to rule out any such conditions, which may have confounded the interpretation of our data. This search was negative, further reinforcing the notion that this cluster may harbor progenitor cells instead. In fact, also among the top DE genes in this cluster is TFPI2, a pancreatic tumor suppressor (20).
Inflammation/stress responses were also recurrent themes from the DE and GO pathway analyses conducted for cluster 1. In fact, C-reactive protein (CRP) was the top DE gene. The most likely source of stress in the ducts is protease secretion. Several serpins [a superfamily of proteins that protect exocrine cells against their own secreted proteases (21)] are indeed among the top DE genes in this cluster (Datasets S1 and S2). Therefore, the above progenitor-like signature is also consistent with potential de-differentiation processes associated with stress. Although our data do not offer direct confirmation of this hypothesis, stress-mediated de-differentiation has been extensively reported in the ductal tree (reviewed in ref. 3).
To validate gene-expression findings, we examined the immunostaining of select DE expressed markers for each cluster in the Human Protein Atlas, a comprehensive resource of more than 24,000 antibodies and 13 million immunohistochemistry images (22, 23). Since differential expression is not exclusive expression, we often found that markers with a positive signal were not restricted to specific pancreatic compartments. Due to this, we focused on the regions that exhibited the most defined staining for any given DE gene. For some of the top DE genes in cluster 1 (FOS, TSPAN8, SPP1) (Datasets S1 and S2), a strong signal was commonly observed within larger ductal structures containing a fibromuscular wall and in connection with PDGs (SI Appendix, Fig. S3). The presence of Mucin 20 among the DE genes further confirms the potential link of these cells to PDGs, which have long been proposed to be niches of pancreatic progenitors (24–27).
DE GO pathways in cluster 2 (trefoil factor 1, TFF1+) reflect active extracellular matrix remodeling, as evidenced by the presence of multiple metalloproteinases (MMP1, MMP7, MMP10) among the top DE genes. This list is headed by TFF1, with TFF2 close behind. The concerted action of these two genes mediates the migration of cells out of PDGs following inflammatory injury (26). Epithelial cell migration, alongside extracellular matrix remodeling are, in fact, the top GO pathways differentially up-regulated for this cluster. Another interesting DE gene is c19orf33, which encodes the hepatocyte growth factor activator inhibitor type 2-related small peptide (H2RSP). This protein is expressed in the epithelium of the gastrointestinal tract, where it is up-regulated in response to injury. The nuclear translocation of H2RSP has been suggested to have a role in the transition from proliferation to differentiation (28). Progenitor markers PDX1, and ID1-4 are also relatively elevated (SI Appendix, Fig. S2C). HMGA1 [a transcription factor that mediates motility and has been associated with pancreatic adenocarcinoma invasiveness (29, 30)] and S100A6 [calcium-binding protein also associated with pancreatic cancer cell motility (31)] are two other DE genes in this cluster that further suggest a migratory phenotype. As before, a search for DE pancreatic cancer pathways (19) was negative. Immunostaining for these markers shows that the cells with the strongest signal are either in major ductal/PDG structures or seemingly delaminating from them (SI Appendix, Fig. S3). Single cells positive for these markers can be detected throughout the parenchyma, including acini and islets.
Cluster 3 (AKAP12+) exhibits the DE of multiple cytokeratins (KRT17, KRT81, KRT23, KRT7, KRTAP2-3), which are markers of epithelial differentiation (32). GO analysis shows the differential elevation of homotypic cell–cell adhesion, positive regulation of cell differentiation, and negative regulation of growth pathways. Taken together, these features suggest terminally differentiated ducts. Immunostaining of some of the top DE genes (KRT17, KRT23, ANXA1, ANXA3, LAMA3) (SI Appendix, Fig. S4) confirms widespread staining in small ducts, with larger ones sometimes displaying intermittent signal patterns.
GO pathway analysis of cluster 4 (WSB1+) reveals cells with an active transcriptional profile and the up-regulation of pathways, such as negative regulation of differentiation, Notch, and Wnt signaling. Interestingly, the immunostaining of some of the top DE genes for this cluster suggests a centroacinar location (SI Appendix, Fig. S4B). Centroacinar cells are difficult to study because of the lack of markers to distinguish them from the adjacent intercalated small ductal cells (33). Studies in mice and zebrafish suggest that these cells express high levels of aldehyde dehydrogenases (ALDH), which have been proposed as markers for populations enriched in progenitors (34). However, this has not been confirmed in humans, where centroacinar ALDH activity has only been detected recently in children and during pathological processes (35). We did not observe DE of ALDHs in this cluster, and the Human Protein Atlas does not show differential centroacinar expression of these markers. However, ALK3bright+ sorting may have excluded many centroacinar cells, as ALK3 expression is brightest in major ducts but tends to be dimmer in smaller ducts (4). Therefore, it is possible that our dataset comprises only a fraction of the overall centroacinar population. Still, we base the hypothetical assignment of centroacinar identity to this cluster on several observations. First, its position at the crossroads between productal and ducto-acinar axes is consistent with cell types known to be at the interface between acinar and ductal tissues. Second, as indicated above, the immunostaining profile of some DE markers (SI Appendix, Fig. S4B), including epithelium-specific transcription factor-3 (ELF3), vacuole membrane protein-1 (VPMP1), or WD repeat and SOCS box containing-1 (WSB1) suggests a centroacinar location. Many studies suggest a direct involvement of centroacinar cells in neoplastic transformation trough centroacinar-to-ductal transdifferentiation (36). In this context, mutated forms of the above DE markers ELF3 (37), VMP1 (38), and WSB1 (39) have all been associated to neoplastic transformation of the pancreas. Finally, Notch signaling, which has been characteristically associated to centroacinar cells (40), is one of the top DE pathways in GO analysis. Taken together, the above evidence is consistent with the hypothesis that cells from cluster 4 are centroacinar.
The ducto-acinar clusters (5, 6), while predominantly ductal in nature (they are mostly negative for the archetypal acinar marker AMY2B and positive for the ductal marker KRT19), also feature the differential expression of acinar-specific genes, such as CEL, REG3A, REG1A, or CPA1 for cluster 5 (OLFM4+) and CLPS, CELA2A/3A/3B, or PGA5 for cluster 6 (CPA2+) (Fig. 2 B and E and SI Appendix, Fig. S2A). The latter already shows up-regulation of typical acinar cellular processes, such as zymogen activation (41) and the metabolization of glutathione (42). Immunostaining of most of these DE genes for clusters 5 and 6 is widespread throughout the pancreas and difficult to pinpoint to specific locations, since proteases are secreted by the acinar tissue into the ducts. However, some of these makers exhibit a distinct centroacinar position (SI Appendix, Fig. S4).
Taken together, our data suggest that the ALK3bright+-sorted population harbors an assortment of ductal cell types, ranging from cells with progenitor-like features to mature ducts, as well as subpopulations that exhibit some acinar-like expression patterns.
Pseudotemporal Analysis Suggests Two Transitional Axes in the ALK3bright+ Ductal Compartment.
Given the indications of potential differentiation gradients between ductal and acinar cells in clusters 4-5-6, as well as between different ductal cell types in clusters 1-2-3, we used Monocle v2.10.1 to arrange the cells in an unbiased pseudotemporal manner; that is, following a “developmental/transitional” path according to their transcriptomic similarity. This strategy has been used to determine lineage dynamics during pancreatic development (43). The results of this analysis after the bioinformatics removal of the immune cluster 7 are shown in Fig. 3 A and B. The top “ducto-acinar” arm of the graphic in Fig. 3A indicates that there is a pseudotemporal continuum, with a progressive acquisition of acinarity following the sequence from clusters 4 (centroacinar) to 6 (transitional to acinar 2). Such inference, while solely based on bioinformatics tools, strongly suggests the existence of cells transitioning from ductal to acinar phenotypes (4-5-6) or vice versa (6-5-4). Similarly, the bottom “productal” arms are also arranged in a manner that could be interpreted as progenitor cells (cluster 1) progressively becoming, upon activation and migration, cells of cluster 2. These cells lie at the crossroads between two differentiation paths: One toward centroacinar-acinar phenotypes (2-4-5-6) and another toward differentiated ducts (2-3). The widespread distribution of the cells of cluster 1 across the entirety of the productal arms suggests that these cells may give rise to (or result from the de-differentiation of) all mature ductal cell types.
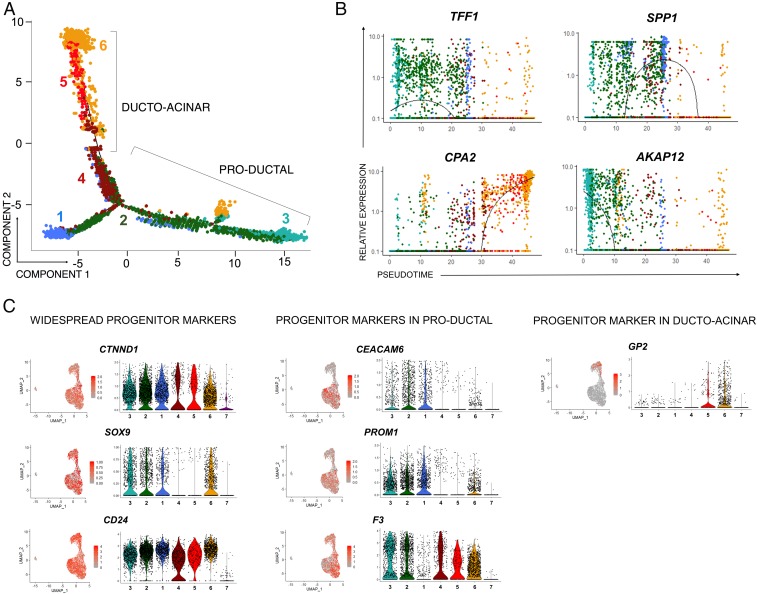
Fig. 3.
Ductal cells show transitional dynamism in pseudotime. (A) Monocle-generated plots presenting pseudotime ordering and differentiation trajectory of cell clusters. Color coding is as shown in Fig. 2A. Clusters 4-5-6 are arranged in a manner that suggests ductal-acinar transition (bottom left arm), while activated/migrating progenitor-like cells (cluster 2) are saddled between the productal and ducto-acinar clusters. (B) Monocle-generated plots showing pseudotime-ordered expression of selected marker genes: TFF1 (cluster 2), SPP1 (cluster 1), CPA2 (cluster 6), and AKAP12 (cluster 3). The color of a cell shows its cluster identity (as in Fig. 2A). Lines denote relative expression of each marker in pseudotime. (C) UMAP plots and corresponding violin plots showing relative gene expression for markers previously associated with ductal progenitors. Each dot in the violin plots represents the expression of a single cell, while the shape of the plot denotes probability density. Scale bars denote average expression in gene counts.
The notion that adult ducts harbor progenitor cells has been extensively studied, and the literature describes numerous markers for such cells, both in mice and humans. Our analysis of the ductal progenitor cell niche allowed us to map some of these against the UMAP plot. As shown in Fig. 3C, several have a widespread distribution (i.e., are found in both productal and ducto-acinar axes). That is the case of CTNND1 (44) and CD24 (45). While the expression of SOX9 (27, 46) is weaker in clusters 4 and 5, it is robustly expressed in clusters that are in both the productal axis (1–3) as well as in the proacinar cluster 6. Other markers, such as CEACAM6 (47), Prominin-1/CD133 (48), and F3/CD142 (49, 50), are detected preferentially in the productal axis. Interestingly, GP2 (51, 52) is expressed almost exclusively in the ducto-acinar axis (clusters 5 and 6), which is consistent with the recent discovery that this gene marks progenitor-like de-differentiated acinar cells (49). Taken together, our data confirm the existence of two major lineage differentiation axes in the adult ALK3bright+ ductal compartment.
Alignment of the ALK3bright+ Dataset Against a Single-Cell Resolution Transcriptome of the Whole Pancreas Confirms the Ducto-Acinar Transitional Axis and Suggests an Association between Productal and Endocrine Compartments.
To ascertain the position of sorted ALK3bright+ populations in the larger context of the pancreas, we plotted our dataset against two other datasets previously reported for human islets (47, 53), which are known to contain residual amounts of ductal and acinar cells and therefore offer a rudimentary atlas of most cell types in the organ (53–55). Fig. 4A and SI Appendix, Fig. S5 present the combined UMAP plot, cluster resolution, heatmap, and dot-plot of select DE genes of the ALK3bright+ clusters vs. the whole-pancreas integrated dataset, respectively. The cluster color coding of our original dataset has been preserved in the integrated graphic. The fact that the original clusters are largely maintained following the integration with a whole-pancreas dataset further confirms their distinctive identity. The only difference is that, upon integration, former cluster 2 (activated/migrating progenitors) is split into two subclusters, now identified by two different shades of green, and cluster 4 (centroacinar cells) disappears. Representative genes for each cluster are indicated in Fig. 4A (heatmap in SI Appendix, Fig. S5B). The contribution of the ALK3bright+ dataset (teal) is shown against the scRNAseq “map” of the whole pancreas (coral) in the circular inset of Fig. 4A. This analysis confirms that ALK3bright+ cells fall under any of the two categories described above: Productal (bottom area in Fig. 4A) or ducto-acinar (top right area in Fig. 4A). The latter appears to seamlessly merge with the population defined as “acinar” in the whole-pancreas dataset. SI Appendix, Fig. S5C shows that the whole-pancreas–derived acinar cells are more acinar than the ALK3bright+-sorted “transitional to acinar” clusters (higher CPA1/AMY2A and expression of AMY2B, which is undetectable in the ALK3bright+ dataset). This, coupled with the expression of KRT19 in those clusters, further confirms their transitional nature. Importantly, no cells from the ALK3bright+ dataset correspond to INS-expressing cells (i.e., all INS+ cells derive from the whole-pancreas dataset) (SI Appendix, Fig. S5C).
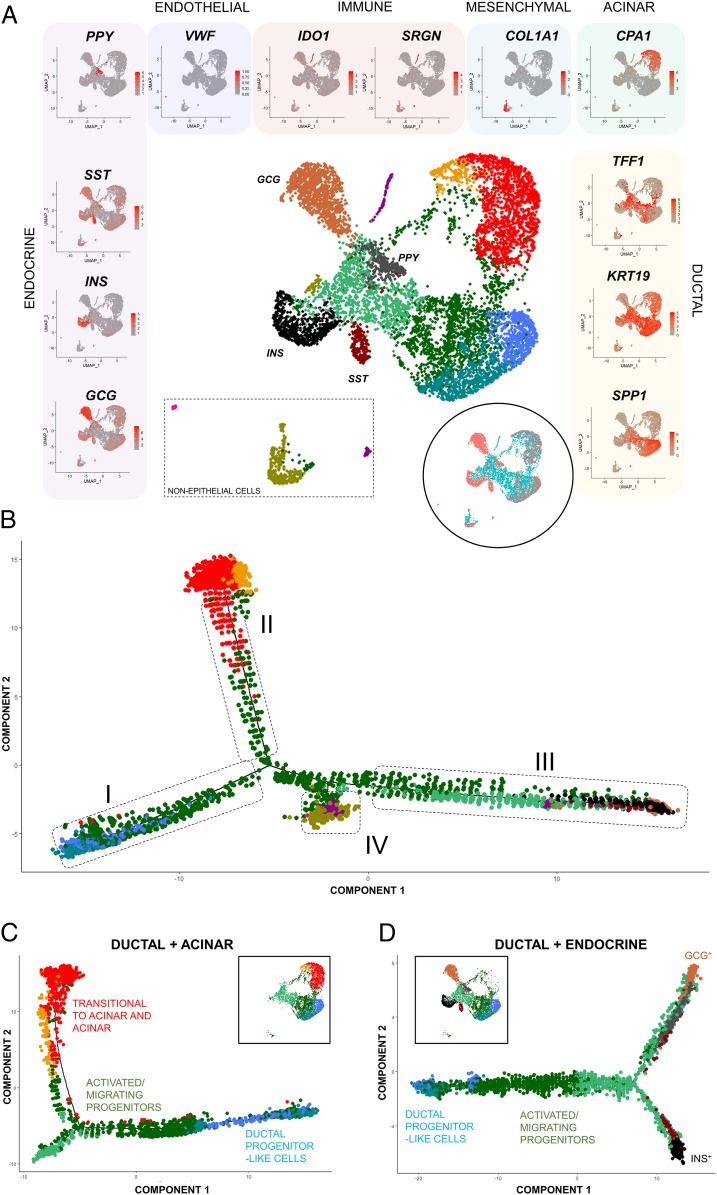
Fig. 4.
Alignment of the ALK3bright+ clusters against a scRNA-seq dataset of the whole pancreas confirms the ducto-acinar transitional axis and suggests potential association of the productal and endocrine compartments. (A) UMAP plot showing cellular distribution when the ALK3bright+ dataset (n = 3, 4,878 cells) is mapped against two combined islet datasets claimed to represent the whole pancreas (3,277 cells). Clustering of pancreatic subtypes reflects differential gene expression and was validated by visualizing gene expression in UMAP plots (left side: endocrine; top: endothelial, immune, mesenchymal and acinar; right side: ductal). Scale bars for each marker denote average expression in gene counts. (Inset) UMAP plot showing distribution of ALK3bright+ cells (teal color) vs. the whole-pancreas dataset (coral color). (B) Single-cell arrangement based on pseudotime calculations of the integrated whole-pancreas and ALK3bright+ datasets. Cell ordering analysis results in four main arms: I (productal), II (ducto-acinar), III (ducto-endocrine), and IV (immune/mesenchymal). (C) Pseudotime-ordered ductal and acinar components of the integrated whole-pancreas and ALK3bright+ cell datasets. (Inset) UMAP plot showing the distribution of clusters based on cellular identity. (D) Pseudotime-ordered ductal and endocrine components of the integrated whole-pancreas and ALK3bright+ cell datasets. (Inset) UMAP plot showing the distribution of clusters based on cellular identity.
The Monocle arrangement of the integrated dataset is depicted in Fig. 4B. Comparing this to the ALK3bright+ dataset alone, cells of cluster 2 (activated/migrating ductal progenitors) remain at the crossroads between the productal axis (Fig. 4B, box I), the ducto-acinar arm (Fig. 4B, box II) and what appears to be a ducto-endocrine branch (Fig. 4B, box III). Box IV in Fig. 4B indicates nonepithelial cells (mesenchymal, endothelial, immune). This pseudotemporal arrangement suggests that the differentiation axes previously identified within ALK3-sorted ductal cells are not only conserved upon integration with the whole-pancreas dataset, but may also span the endocrine compartment as well. This would be consistent with the observation of a spatial continuity between ductal and β-cell clusters (56), as well as with the historical evidence of ductal-to-endocrine β-cell neogenesis (reviewed in ref. 3).
Fig. 4 C and D show additional pseudotemporal analyses focusing specifically on ductal+acinar cells (i.e., excluding endocrine and immune/mesenchymal clusters) and ductal+endocrine cells (i.e., excluding acinar and immune/mesenchymal clusters). The first (Fig. 4C) is similar to that previously shown in Fig. 3A for the sorted ALK3bright+ cells, which is not surprising, since most cells derive from that dataset. The analysis of the ductal+endocrine clusters (Fig. 4D) further supports the existence of a differentiation gradient from progenitors (cluster 1, blue) to activated/migrating progenitors (cluster 2, green), which then bifurcates into GCG+ (top arm in Fig. 4D) and INS+ (bottom arm in Fig. 4D) arms.
Regular UMAP plots are bidimensional reductions of multidimensional data. While 2D renderings sometimes distort the relative position of clusters, this was not the case in the integrated dataset, whose 3D analysis showed maintenance of the previously determined spatial cluster distribution (Movie S1).
Sorted Human P2RY1+/ALK3bright+ Cells Exhibit Multilineage Differentiation Potential In Vivo, and This Outcome Is Enhanced upon Systemic Administration of a BMPR Agonist.
We have previously shown that sorted ALK3bright+/P2RY1+ cells exhibit a progenitor-like behavior following exposure to BMP receptor agonists in vitro (4). The replication of such outcomes in vivo may set the stage for therapies aimed at pharmacologically restoring islet mass. In an attempt to correlate our ALK3bright+ scRNA-seq findings and the previous analyses on a subset of such cells (ALK3bright+/P2RY1+), we observed that PDX1 (the marker for which P2RY1 is a surrogate) is almost exclusively expressed in cluster 1 (Fig. 5 A, Left). We hypothesized earlier that this cluster harbors progenitor-like cells, characterized by high PDX1 and ID1-2 and -4 expression. Sorted ALK3bright+/P2RY1+ cells are highly depleted of BMP-7–unresponsive CAII+ cells (4) (SI Appendix, Fig. S6 A–F). Intriguingly, cluster 1 had a significant number of CAII+ cells (Fig. 5 A, Center). However, we found that a vast majority of the cells with high PDX1 expression did not coexpress CAII (Fig. 5 A, Right). Taken together, these observations suggest that 1) only a subset of cells from cluster 1 is progenitor-like and 2) ALK3bright+/P2RY1+ sorting enriches for the progenitor-like cells (PDX1+/ALK3+/CAII—), which are preferentially found within cluster 1.
Fig. 5.
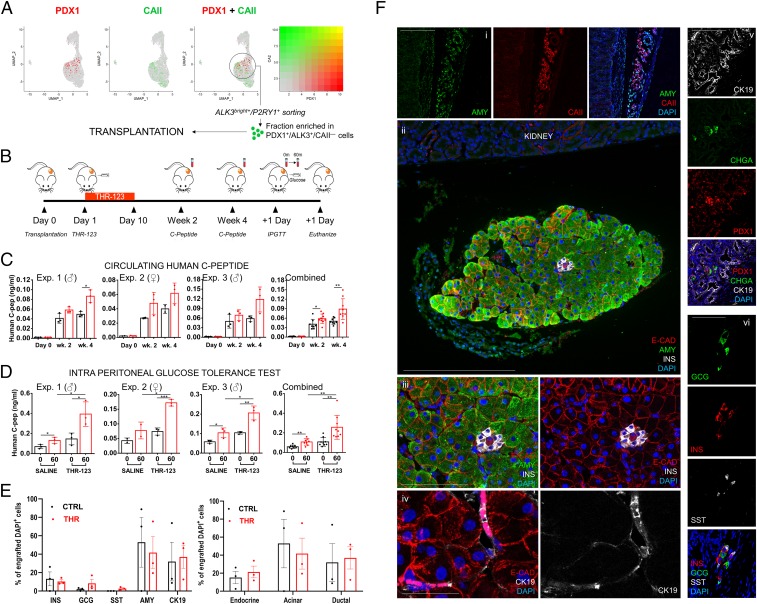
Sorted P2RY1+/ALK3bright+ cells display multilineage differentiation potential upon transplantation in immunodeficient mice. (A) The ALK3bright+/P2RY1+ fraction is enriched in PDX1+/ALK3+/CAII− cells, which map chiefly in cluster 1, as the expression of PDX1 (the marker for which P2RY1 is a surface surrogate; green) and that of CAII (red) are mutually exclusive. (B) Experimental outline for the transplantation of FACS-sorted P2RY1+/ALK3bright+ cells under the kidney capsule of immunodeficient nu/nu mice. (C) Circulating peripheral hC-pep measured at day 0 and weeks 2 and 4. (D) Circulating peripheral hC-pep measured during an IPGTT at 0 min and 60 min after glucose challenge. Each of the first three charts in C and D represents an individual experiment, with the sex of the recipient mice indicated on top. The fourth chart, in a larger size, is the combination of all of the experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars: SD. (E, Left) Quantification of the percentage of endocrine (INS, GCG, SST), acinar (AMY), and ductal (CK19) DAPI+ cells in the grafts of n = 3 transplanted, THR-123–treated animals. Only transplanted pancreatic cells (defined as expressing any of the above markers plus DAPI) were counted in the graft region, and values represent percentages vs. the total counted cells. (Right) Relative estimated percentages of engrafted cells according to their general lineage (endocrine, acinar, ductal). Error bars: SD. (F) Representative confocal immunofluorescence imaging of z-stack maximal projections showing expression of select markers of the grafted cells in the kidney subcapsular space of recipient mice. (i) Example of disorganized acinar (AMY, green) and ductal (CAII, red) tissue. (Scale bar, 100 µm.) (ii) Example of well-organized engrafted tissue showing fully formed acini (AMY, green), typical pancreatic epithelial architecture (E-cadherin, ECAD, red), and a small cluster of INS-producing cells (INS, gray). Details are shown in (iii). [Scale bars, 100 µm in (ii) and 50 µm in (iii).] (iv) Primitive ductal structures are shown in magenta (E-CAD, red + cytokeratin 19, CK19, white). (Scale bar, 25 µm.) (v) Endocrine clusters surrounded by ductal structures: Chromogranin A (CHGA, green); PDX1 (red); CK19, gray; and DAPI (blue). (Scale bar, 50 µm.) (vi) Example of endocrine cluster showing multiple single-hormone expressing cells: GCG (green); INS (red), SST (gray); and DAPI (blue). (Scale bar, 25 µm.) Merged images are depicted following those of single channels (n = 3 biological replicates with 6 mice per group).
Based on the above, we transplanted ALK3bright+/P2RY1+ cells to test the prediction that this subset of cells that maps in the region of cluster 1 would act as progenitors in vivo. We reasoned that this approach would be preferable to the alternative of transplanting cells sorted using yet-undefined surface markers specific for this cluster (which may or may not yield separation between CAII+ and CAII− cells). These experiments were conceived as proof-of-principle that human progenitor-like cells can differentiate into adult phenotypes in vivo, and not to model any potential therapeutic intervention. Therefore, we only transplanted subtherapeutic cell dosages in nondiabetic animals.
Transplantation of sorted P2RY1+/ALK3bright+ cells was done under the kidney capsule of immunodeficient (nu/nu) mice. These cells are hormone-negative (4) (Figs. 1D and 4A and SI Appendix, Fig. S5C). The strategy is schematized in Fig. 5B. Five- to 6-wk-old nu/nu mice received 5 × 105 P2RY1+/ALK3bright+ cells sorted from n = 3 donors in three independent experiments (demographics in SI Appendix, Table S1). Six mice were transplanted in each of the three experiments. Males were used in Exps. 1 and 3, whereas in Exp. 2 only females were used.
Although the primary endpoint was to test the hypothesis that sorted progenitor-like cells spontaneously differentiate upon transplantation, we also wanted to test whether the process could be enhanced by treating the recipient animals with a BMP receptor agonist. Thus, half the mice in each experiment (three) also received daily intraperitoneal injections (10 mg/kg of body weight) of THR-123, a small cyclic peptide with ALK3 agonism activity (5, 57). The treatment was for 10 d, starting 24 h after the transplantation. The other three animals per experiment received a similar regimen of saline injections.
Circulating human C-peptide (hC-pep) values were measured in both groups at 2 and 4 wk after transplantation. As shown in Fig. 5C, there was measurable hC-pep in all three experiments at both time points, indirectly establishing the formation of INS-secreting cells from the engrafted P2RY1+/ALK3bright+ cells. Average basal hC-pep in plasma at 4 wk was ∼0.1 ng/mL. This figure is in line with hC-pep values reported at any time point prior to week 8 postprocedure when human embryonic stem cell-derived stage 4 undifferentiated progenitors are transplanted (58), despite a 10× lower cell dosage per mouse. Of note, the concentration of hC-pep in blood was higher in the THR-123 group than in saline-injected mice (P < 0.05 at week 2 and P < 0.001 at week 4). An intraperitoneal glucose tolerance test (IPGTT) conducted at day 15 revealed that engrafted cells in both groups release INS in response to glucose stimulation (Fig. 5D), further supporting that INS-expressing cells are functional neogenic β-like cells. Again, while this was observed in both groups, responses were significantly stronger in those animals treated with THR-123. No statistically significant differences were observed between males and females. All animals were killed at day 16, and immunofluorescence of the grafts was performed. The quantification of the percentage of endocrine (INS+, GCG+, SST+), acinar (AMY+), and ductal (CK19+) cells was done by sampling transplanted kidney sections of n = 3 THR-123–treated (Fig. 5E, red columns) and n = 3 saline-treated (Fig. 5E, black columns) mice and counting pancreatic cells within the graft by FIJI/ImageJ (Fig. 5E). We observed a 2.7-fold reduction in the overall number of engrafted cells in sampled sections of controls vs. THR-123–treated mice (P = 0.025). In particular, there was a 2.5-fold (P = 0.08, not significant, n.s.) increase in the number of INS+ cells detected in the THR-123 group vs. controls. These observations are aligned with our previous determination that BMP-7 increased the mitotic rate of sorted ALK3bright+/P2RY1+ cells in vitro by ∼2.5-fold (4). However, there were no statistical differences between both groups in terms of relative percentages of specific cell types (excluding SST-expressing cells, undetected in controls). Therefore, differences in circulating C-peptide are not due to differentiation biases, but probably only reflect a differential expansion of the transplanted undifferentiated cells when animals were treated with THR-123 or saline. Of note, these values represent only estimations based on the sampling of a small number of animals.
Fig. 5F shows representative images of pancreatic cell types in the grafts of THR-123–treated mice. Acinar tissue was detected in discrete regions either in a disorganized fashion, intermingled with ductal cells (Fig. 5 F, i), or in well-developed acinar structures (shown in Fig. 5 F, ii with an endocrine cluster) with typical epithelial morphology (Fig. 5 F, iii) and small ductules (Fig. 5 F, iv). Although CK19 is a marker already found in the sorted fraction (KRT19) (Fig. 1D), the fact that we observed CAII+ cells when P2RY1+/ALK3bright+ cells are nearly 100% CAII− (4) (Fig. 5A and SI Appendix, Fig. S6 A–F) suggests that they have arisen by differentiation. AMY+ cells may have also differentiated along the ducto-acinar axis. Our data do not offer evidence of draining into ducts, and we cannot discard some autodigestion. However, pancreatic amylase is allosterically activated by chloride in the duodenum (59, 60). Other pancreatic enzymes also require the duodenum/intestinal mucosa microenvironment (absent in grafts) for activation. Therefore, we speculate that zymogen activation levels in the graft may be insufficient for extensive autodigestion.
Endocrine clusters were small (8 to 15 cells) and contained monohormonal cells expressing either INS, GCG, or SST (Fig. 5 F, v and vi). Considering that the transplanted populations are devoid of endocrine cells (4) (Figs. 1D and 4A and SI Appendix, Fig. S5C) and that the circulating hC-pep levels increase with time (Fig. 5C), we hypothesize that these clusters also arise by differentiation. Of note, while we did not transplant the negative fraction as a negative control, our previous experiments strongly suggest that it is strongly depleted in mature pancreatic markers, and completely devoid of progenitor-like cells (4). In summary, sorted progenitor-like cells transplanted into immunodeficient animals differentiate along all pancreatic lineages.
PDX1+/ALK3+/CAII− Progenitor-like Cells Exist in Pancreata of T1D/T2D Donors Regardless of the Duration of the Disease.
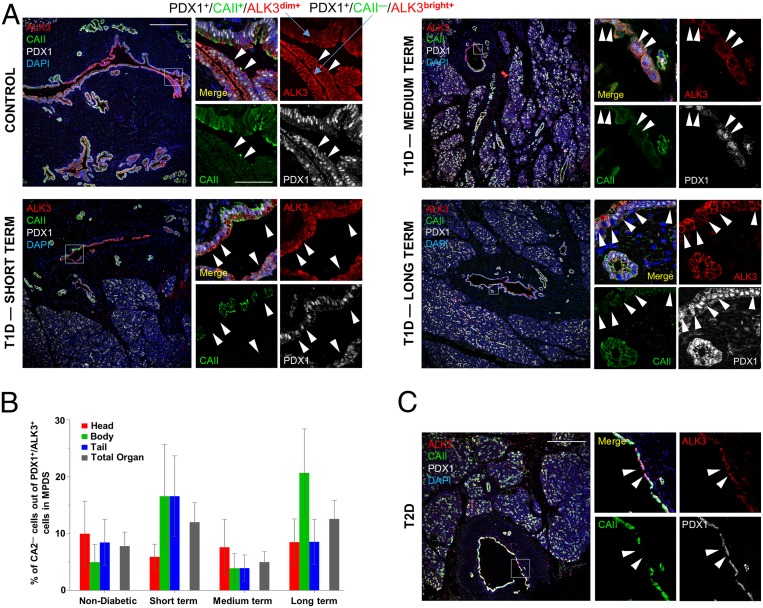
If progenitor-like cells were unaffected by T1D autoimmunity, such discovery may open the door to therapies aimed at their in situ activation for regenerative purposes. We have determined that the P2RY1+/ALK3bright+ fraction is highly enriched in cells with a PDX1+/ALK3+/CAII− phenotype (4) (SI Appendix, Fig. S6 A–F), which is quantifiable by immunofluorescence in human samples. To determine if such cells are present in ducts after disease onset, we examined the expression of PDX1, ALK3, and CAII in short- (0 to 5 y), medium- (6 to 10 y), and long- (>10 y) term T1D donor samples. Nondiabetic controls were used for comparison; n = 3 donors per group, plus one T2D donor, were selected for analysis (demographics in SI Appendix, Table S1). For each donor, three histological sections containing MPDs (characterized by a fibromuscular layer) were used for each of the three anatomical regions of the pancreas (head, body, and tail).
Immunofluorescence of PDX1 in paraffin-embedded nPOD (Network for Pancreatic Organ Donors with Diabetes) pancreatic samples is challenging and has not been reported thus far. Therefore, we optimized the protocol using control and T1D sections that encompassed both islets and ducts in the same field. We found that PDX1 signal was very abundant not only in islets (as expected), but also throughout the entirety of the ductal tree (Fig. 6 A and C and SI Appendix, Fig. S6G). SI Appendix, Fig. S6G shows gradual loss of INS, PDX1, and P2RY1 in islets as T1D progresses. However, PDX1 signal remains strong in ductal structures regardless of the stage of the disease. This protocol is described in SI Appendix.
Fig. 6.
PDX1+/ALK3+/CAII− cells are present in the large ducts of T1D/T2D donors. (A) Representative confocal immunofluorescence imaging of a z-stack maximal projection showing expression of ALK3 (red), CAII (green), PDX1 (gray), and DAPI (blue) from human donors. (Left) Nondiabetic (Upper) and short-term T1D (Lower). (Right) Medium-term (Upper) and long-term (Lower) T1D. White arrowheads indicate cells with a PDX1+/ALK3+/CAII− phenotype. (B) Quantification of PDX1+/ALK3+/CAII− cells calculated as a percentage of overall PDX1+/ALK3+ ductal cells in large human pancreatic ducts across the head, body, and tail of the pancreas, for control, short-term, medium-term, and long-term diabetic donors. Error bars: SE. No statistically significant differences were detected between groups. (C) Representative confocal immunofluorescence imaging of a z-stack maximal projection showing the presence of PDX1+/ALK3+/CAII− cells in a T2D donor. (Scale bars, 200 µm for low-magnification and 50 µm for high-magnification images.)
We also detected nuclear PDX1 in mature ductal cells that also expressed CAII (Fig. 6 A and C). While scRNA-seq indicated that PDX1 expression is restricted to a small proportion of ALK3+/CAII− cells, predominantly within cluster 1, this analysis was done on sorted ALK3bright+ cells, which represent only a fraction of all ALK3+ cells in the ductal tree. In fact, this analysis confirmed our previous observation (4) that PDX1+/CAII+ are generally ALK3dim+, whereas PDX1+/CAII− cells are invariably ALK3bright+ (Fig. 6A). This also explains why the ALK3bright+/P2RY1+ sorting conducted for transplantation results in the depletion of most CAII+ cells (4) (Fig. 5A and SI Appendix, Fig. S6 A–F).
PDX1+/ALK3+/CAII− ductal cells, often in an intercalated pattern with PDX1+/ALK3+/CAII+ cells, were detected in head, body, and tail of all donors, regardless of the duration of the disease (Fig. 6A). The percentage of PDX1+/ALK3+ cells vs. DAPI, and that of CAII− and CAII+ cells within PDX1+/ALK3+ cells, was calculated by FIJI/ImageJ for the epithelial lining of all of the major ducts found in each section (Fig. 6B). The overall percentage of PDX1+/ALK3+/CAII− cells ranged between 5% and 12% of all PDX1+/ALK3+ ductal cells examined (the remaining being CAII+). There were no statistically significant differences between control, short-, medium-, or long-term samples. No meaningful differences were observed either when comparing the different regions of the pancreas within each donor. These cells were also detected in a sample from a T2D donor (Fig. 6C), but no quantification was attempted. Taken together, these results indicate that cells with a progenitor-like immunofluorescence signature (PDX1+/ALK3+/CAII−) exist in the ductal tree of T1D patients regardless of the duration of the disease. It remains to be established whether these cells exhibit the same multipotency previously established for nondiabetic donors.
Discussion
We present the high-resolution characterization of a human pancreatic ductal progenitor cell niche. All previous analyses of the human pancreas by scRNA-seq have been performed on isolated islets (53–56, 61), which contain remnants of the acini and ducts. However, the number of sequenced ductal cells was invariably low, and probably depleted of the major ductal cells that are the subject of our study. Furthermore, the ductal identity of any given cell was determined indirectly by bioinformatics a posteriori. Our study design circumvents these limitations by focusing on a specific lineage of the pancreas and prevalidating its identity by cell sorting.
scRNA-seq of a sorted ductal population previously shown to harbor cells with progenitor-like characteristics (4) revealed an unexpected degree of heterogeneity, with cells spanning a broad range of differentiation stages across two major axes, one encompassing mature ductal cell types and the other showing progressive acinarity. The existence of a gradient of expression of acinar genes in clusters 5 and 6 rules out the possibility of ambient contamination of acinar RNA during the procedure (which would be equally detected in all clusters), and suggests an active transition between the cells of these clusters. Acinar-to-ductal metaplasia is a phenomenon by which, in response to inflammation or stress, acinar cells de-differentiate into progenitor-like (metaplastic) ductal cells. Such cells are then endowed with the capacity to regenerate the damaged organ (62–64). This process only becomes irreversible when cells acquire oncogenic Kras mutations or are subjected to persistent/aberrant signaling, which prevent redifferentiation and initiate further progression into pancreatic adenocarcinoma (65). We believe that the ducto-acinar ALK3bright+ axis described herein represents cells in the process of transitioning between these two lineages, which suggests that this reversible transdifferentiation may be more common than previously thought. Consistent with this hypothesis is the observation that GP2, an acinar-to-ductal transitional marker (49), is almost exclusively detected in the ducto-acinar clusters 5 and 6 (Fig. 3D).
Cluster 1 cells displayed strong inflammation/stress responses. While dissociation and sorting may induce stress, it is doubtful that this is the reason behind our observation. If that were the case, we would also probably observe it in the other clusters, which have also undergone the same process. Stress in the pancreatic microenvironment is a well-studied phenomenon. The exocrine pancreas is the organ with the highest protein synthesis, producing liters of pancreatic juices daily. Leaving aside the potential risk of intracellular activation of digestive enzymes (the main cause of pancreatitis), protein synthesis itself is a key stressor of the exocrine pancreas at all times (66). Stress has been associated with de-differentiation and acquisition of progenitor-like characteristics in ducts (67–69), but whether the stress signature of cells in cluster 1 is linked to de-differentiation is merely presented as a hypothesis.
This cluster also had the highest levels of PDX1 and ID1-2, and -4, known housekeepers of progenitor-like phenotypes (4, 17). Of note, while P2RY1+ cells were also found throughout the dataset, their scarcity made it difficult to analyze them. This is because P2RY1 expression is very low. As shown in SI Appendix, Fig. S6F, the relative level of expression (2−ΔCt) of P2RY1 was 5-fold lower than that of PDX1 and 60-fold lower than that of KRT19. Our findings are confirmed by the entry for pancreas in the Human Protein Atlas, which shows an average of transcripts per million across 248 samples to be 0.8 (i.e., there is a probability of finding 0.8 transcripts of P2RY1 per one million reads). As reference, PDX1 has 6.2 and INS has 3535.7. As recently reviewed in ref. 70, scRNA-seq cannot be used to draw meaningful conclusions from low-expressing genes.
Interestingly, albeit relatively widespread throughout all ALK3bright+ clusters, the expression of the progenitor marker CTNND1 (p120ctn) was highest in cluster 1. During development, cells with high p120ctn expression define “trunk” (major) ductal populations that give rise to both ducts and endocrine cells, whereas p120ctnlow cells, typically located in the tip of the ducts, only differentiate into acini (44). Similarly, cluster 1 had the highest expression of CEACAM6, which was independently predicted to be a progenitor marker in the pancreas (47); as well as PROM1, a marker of tripotent colony-forming progenitor cells in the adult murine pancreas (48). Enrichment in progenitor-like cells from cluster 1 was predicted to occur by P2RY1+/ALK3bright+ sorting, since P2RY1 (PDX1) selection mostly excludes the nonprogenitor CAII+ fraction therefrom.
We did not expect to detect ductal-to-endocrine transitional cells in our dataset due to the very design of the experiment, where endocrine cells (ALK3−) are specifically excluded by gating on the ALK3bright+ fraction. Our study is based on a static snapshot of the pancreas, and therefore conclusions about potential transitions are necessarily speculative and based on bioinformatics predictions. Even so, the integration with the whole-pancreas datasets, and especially the pseudotemporal arrangement analysis, strongly suggests the existence of a ducto-endocrine axis. In this particular setting, where none of the donors had diabetes and sources of stress appear to be largely physiological, it is plausible that there may be a basal level of endocrine cell formation from de-differentiated ductal cells, even if the full activation of ductal-mediated regenerative mechanisms may require more extensive damage (3, 71).
Transplantation is stressful for dissociated pancreatic epithelial cells (4). In this context, we further hypothesized that xenotransplanting sorted P2RY1+/ALK3bright+cells may offer us a glimpse of their potency under stress in vivo. We show here that these cells, which are hormone-negative at the time of transplantation, can differentiate into monohormonal GCG-, SST-, and INS-expressing cells, as well as cells of the acinar and ductal lineages. While some regions of the graft did not exhibit mature histological patterns, the organization of other regions in acini, small endocrine clusters, and ductules was remarkable, considering that only single cells sorted for ALK3 and P2RY1 (none of which is expressed in human acinar tissue) were transplanted. Our ultimate goal is not transplantation, but rather the activation of progenitor cells in situ. As indicated earlier, these experiments were only designed to establish proof-of-concept of in vivo multilineage differentiation, and not to revert diabetes in a disease model. Therefore, we did not conduct any comparison with islets or extensive functional studies. Still, we show that engrafted INS+ cells secreted INS in response to glucose stimulation in vivo, which is the hallmark of functional β-like cells. This process was enhanced when the animals were treated systemically with THR-123, an ALK3 receptor agonist (57). This finding, coupled with our discovery that PDX1+/ALK3+/CAII− cells (a marker combination signature equivalent to P2RY1+/ALK3bright+) (4) are present in T1D/T2D donors, suggests potential therapeutic avenues to induce β-cell regeneration in situ. To determine if that is a possibility, future studies will explore whether cells sorted from T1D/T2D donors also exhibit multipotency.
The existence of progenitor cells with the capacity to regenerate the endocrine and exocrine compartments in the adult human pancreas remains a contentious issue. However, the recent emergence of high-resolution analytical tools is bringing much needed nuance to the debate. Lineage barriers are now blurrier than ever, a discovery that calls into question the validity of many earlier lineage-tracing conclusions. The high degree of developmental heterogeneity supports the dynamic fate plasticity and the constant state of flux of the ductal compartment. If harnessed, these findings could be the basis for the development of pharmacological therapies for endocrine regeneration.
Materials and Methods
Human pancreata were processed as detailed in the SI Appendix. Methods for FACS of cells for scRNA-seq analysis and transplantation are also expanded therein. Animal experiments were conducted under the oversight of the University of Miami Institutional Animal Care and Use Committee. Procedures for transplantation and metabolic analyses of in vivo differentiation as well as immunofluorescence analyses are detailed in the SI Appendix. Single-cell capture, library preparation, and sequencing were performed at Genewiz (South Plainfield, NJ). Details on library preparation, computational analyses, and statistics are provided in SI Appendix.
Data Availability.
Sequencing data have been deposited in the Gene Expression Omnibus (GEO), accession no. GSE131886. Previously published scRNA-seq data reanalyzed here are available at under GEO accession nos. GSE81076/GSE85241. Source data for figures are provided in the SI Appendix reagent table. Code/Rscript files for the analyses reported are available in the GitHub repository https://github.com/JDBLab/Pancreas_ductal_scRNAseq (72).
Supplementary Material
Acknowledgments
We thank Andrew Butler, Paul Hoffman, Tim Stuart, and others at the Satija laboratory for help with Seurat’s GitHub, as well as the curators of the Human Protein Atlas (http://www.proteinatlas.org); Michael Bellio, Maria Boulina, Joel Szust, Armando Méndez, and the staff of the Imaging Core, Preclinical Cell Processing and Translational Models Core, and the Clinical Chemistry, Biomarkers, and Immunoassay Laboratory (all at the Diabetes Research Institute) for their technical contributions; Rita Bottino (University of Pittsburgh) and Alberto Pugliese (Diabetes Research Institute) for helpful discussions; our colleagues at the Human Islet Research Network for input and discussion; and the donors and their families for their contribution to science. This research was performed with support from the Network for Pancreatic Organ Donors with Diabetes (nPOD; RRID:SCR_014641), a collaborative type 1 diabetes research project sponsored by the Juvenile Diabetes Research Foundation (nPOD:5-SRA-2018-557-Q-R) and The Leona M. & Harry B. Helmsley Charitable Trust (Grant 2018PG-T1D053). The content and views expressed are the responsibility of the authors and do not necessarily reflect the official view of nPOD. Organ Procurement Organizations partnering with nPOD to provide research resources are listed at https://www.jdrfnpod.org/for-partners/npod-partners/. This work was funded by the Diabetes Research Institute Foundation, Inserra family, Fred and Mabel R. Parks Foundation, Tonkinson Foundation, American Diabetes Association Grant 1-19-ICTS-078, and NIH Grants 1R43DK105655-01, 2R44 DK105655-02, and U01DK120393. M.M.F.Q. was funded by the Foreign Fulbright Scholarship Board and the International Institute of Education. J.D.-B. and R.L.P. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for their integrity and the accuracy of the analysis.
Footnotes
Competing interest statement: The University of Miami and J.D.-B. and C.R. hold, but do not receive royalties for intellectual property used in this study. They are also equity owners in Ophysio, Inc., licensee of the intellectual property.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession code GSE131886). Source data for figures are provided in SI Appendix and reagent table therein. The code/Rscript files for the analyses reported in this paper are available in a GitHub coding repository maintained by laboratories of the corresponding authors (https://github.com/JDBLab/Pancreas_ductal_scRNAseq).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918314117/-/DCSupplemental.
References
- 1.Dor Y., Brown J., Martinez O. I., Melton D. A., Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Xiao X., et al. , No evidence for β cell neogenesis in murine adult pancreas. J. Clin. Invest. 123, 2207–2217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominguez-Bendala J., Qadir M. M. F., Pastori R. L., Pancreatic progenitors: There and back again. Trends Endocrinol. Metab. 30, 4–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qadir M. M. F., et al. , P2RY1/ALK3-Expressing cells within the adult human exocrine pancreas are BMP-7 expandable and exhibit progenitor-like characteristics. Cell Rep. 22, 2408–2420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein D., et al. , BMP-7 induces adult human pancreatic exocrine-to-endocrine conversion. Diabetes 64, 4123–4134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inada A., Nienaber C., Fonseca S., Bonner-Weir S., Timing and expression pattern of carbonic anhydrase II in pancreas. Dev. Dyn. 235, 1571–1577 (2006). [DOI] [PubMed] [Google Scholar]
- 7.McInnes L., Healy J., Melville J., UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 (6 December 2018).
- 8.Becht E., et al. , Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol., 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zappia L., Oshlack A., Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience 7, giy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuczma M., Kraj P., Bone morphogenetic protein signaling regulates development and activation of CD4(+) T cells. Vitam. Horm. 99, 171–193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilic G., Wang J., Sosa-Pineda B., Osteopontin is a novel marker of pancreatic ductal tissues and of undifferentiated pancreatic precursors in mice. Dev. Dyn. 235, 1659–1667 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sneddon J. B., et al. , Stem cell therapies for treating diabetes: Progress and remaining challenges. Cell Stem Cell 22, 810–823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasorella A., Benezra R., Iavarone A., The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 14, 77–91 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Hua H., et al. , BMP4 regulates pancreatic progenitor cell expansion through Id2. J. Biol. Chem. 281, 13574–13580 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Jensen J., et al. , Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Yin H., Wang S., Zhang Y. H., Cai Y. D., Liu H., Analysis of important gene ontology terms and biological pathways related to pancreatic cancer. BioMed Res. Int. 2016, 7861274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato N., et al. , Epigenetic inactivation of TFPI-2 as a common mechanism associated with growth and invasion of pancreatic ductal adenocarcinoma. Oncogene 24, 850–858 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Loftus S. K., et al. , Acinar cell apoptosis in Serpini2-deficient mice models pancreatic insufficiency. PLoS Genet. 1, e38 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Uhlen M., et al. , Towards a knowledge-based human protein atlas. Nat. Biotechnol. 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Butler A. E., et al. , In the setting of β-cell stress, the pancreatic duct gland transcriptome shows characteristics of an activated regenerative response. Am. J. Physiol. Gastrointest. Liver Physiol. 315, G848–G854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moin A. S., Butler P. C., Butler A. E., Increased proliferation of the pancreatic duct gland compartment in type 1 diabetes. J. Clin. Endocrinol. Metab. 102, 200–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi J., et al. , Pancreatic duct glands (PDGs) are a progenitor compartment responsible for pancreatic ductal epithelial repair. Stem Cell Res. 15, 190–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpino G., et al. , Progenitor cell niches in the human pancreatic duct system and associated pancreatic duct glands: An anatomical and immunophenotyping study. J. Anat. 228, 474–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naganuma S., et al. , Nuclear translocation of H2RSP is impaired in regenerating intestinal epithelial cells of murine colitis model. Virchows Arch. 448, 354–360 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Veite-Schmahl M. J., Joesten W. C., Kennedy M. A., HMGA1 expression levels are elevated in pancreatic intraepithelial neoplasia cells in the Ptf1a-Cre; LSL-KrasG12D transgenic mouse model of pancreatic cancer. Br. J. Cancer 117, 639–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liau S. S., Jazag A., Whang E. E., HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 66, 11613–11622 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Vimalachandran D., et al. , High nuclear S100A6 (Calcyclin) is significantly associated with poor survival in pancreatic cancer patients. Cancer Res. 65, 3218–3225 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Fuchs E., Keratins as biochemical markers of epithelial differentiation. Trends Genet. 4, 277–281 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Qu X., Nyeng P., Xiao F., Dorantes J., Jensen J., Growth factor independence-1 (Gfi1) is required for pancreatic acinar unit formation and centroacinar cell differentiation. Cell. Mol. Gastroenterol. Hepatol. 1, 233–247.e1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassalli G., Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cell. Int. 2019, 3904645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Socorro M., et al. , Identification of newly committed pancreatic cells in the adult mouse pancreas. Sci. Rep. 7, 17539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanger B. Z., et al. , Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 8, 185–195 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Yachida S., et al. , Genomic sequencing identifies ELF3 as a driver of ampullary carcinoma. Cancer Cell 29, 229–240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loncle C., et al. , The pancreatitis-associated protein VMP1, a key regulator of inducible autophagy, promotes Kras(G12D)-mediated pancreatic cancer initiation. Cell Death Dis. 7, e2295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archange C., et al. , The WSB1 gene is involved in pancreatic cancer progression. PLoS One 3, e2475 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beer R. L., Parsons M. J., Rovira M., Centroacinar cells: At the center of pancreas regeneration. Dev. Biol. 413, 8–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sphyris N., Logsdon C. D., Harrison D. J., Improved retention of zymogen granules in cultured murine pancreatic acinar cells and induction of acinar-ductal transdifferentiation in vitro. Pancreas 30, 148–157 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Neuschwander-Tetri B. A., Presti M. E., Wells L. D., Glutathione synthesis in the exocrine pancreas. Pancreas 14, 342–349 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Byrnes L. E., et al. , Lineage dynamics of murine pancreatic development at single-cell resolution. Nat. Commun. 9, 3922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyeng P., et al. , p120ctn-Mediated organ patterning precedes and determines pancreatic progenitor fate. Dev. Cell 49, 31–47.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Jiang W., et al. , CD24: A novel surface marker for PDX1-positive pancreatic progenitors derived from human embryonic stem cells. Stem Cells 29, 609–617 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Farley A. M., et al. , Antibodies to a CA 19-9 related antigen complex identify SOX9 expressing progenitor cells in human foetal pancreas and pancreatic adenocarcinoma. Sci. Rep. 9, 2876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grün D., et al. , De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin L., et al. , Cells with surface expression of CD133highCD71low are enriched for tripotent colony-forming progenitor cells in the adult murine pancreas. Stem Cell Res. 16, 40–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldan J., Houbracken I., Rooman I., Bouwens L., Adult human pancreatic acinar cells dedifferentiate into an embryonic progenitor-like state in 3D suspension culture. Sci. Rep. 9, 4040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelly O. G., et al. , Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat. Biotechnol. 29, 750–756 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Ameri J., et al. , Efficient generation of glucose-responsive beta cells from isolated GP2+ human pancreatic progenitors. Cell Rep. 19, 36–49 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Cogger K. F., et al. , Glycoprotein 2 is a specific cell surface marker of human pancreatic progenitors. Nat. Commun. 8, 331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muraro M. J., et al. , A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baron M., et al. , A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360.e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segerstolpe Å., et al. , Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabol. 24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enge M., et al. , Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugimoto H., et al. , Activin-like kinase 3 is important for kidney regeneration and reversal of fibrosis. Nat. Med. 18, 396–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rezania A., et al. , Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014). [DOI] [PubMed] [Google Scholar]
- 59.D’Amico S., Gerday C., Feller G., Structural similarities and evolutionary relationships in chloride-dependent alpha-amylases. Gene 253, 95–105 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Maurus R., et al. , Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase. Protein Sci. 14, 743–755 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y. J., et al. , Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan F. C., et al. , Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140, 751–764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murtaugh L. C., Keefe M. D., Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 77, 229–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen J. N., et al. , Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology 128, 728–741 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Storz P., Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 14, 296–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Logsdon C. D., Ji B., The role of protein synthesis and digestive enzymes in acinar cell injury. Nat. Rev. Gastroenterol. Hepatol. 10, 362–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X., et al. , Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 132, 197–207 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Valdez I. A., et al. , Proinflammatory cytokines induce endocrine differentiation in pancreatic ductal cells via STAT3-dependent NGN3 activation. Cell Rep. 15, 460–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Téllez N., Montanya E., Gastrin induces ductal cell dedifferentiation and β-cell neogenesis after 90% pancreatectomy. J. Endocrinol. 223, 67–78 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Mawla A. M., Huising M. O., Navigating the depths and avoiding the shallows of pancreatic islet cell transcriptomes. Diabetes 68, 1380–1393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin-Pagola A., et al. , Insulin protein and proliferation in ductal cells in the transplanted pancreas of patients with type 1 diabetes and recurrence of autoimmunity. Diabetologia 51, 1803–1813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qadir M. M. F., Dominguez-Bendala J, Pastori R., High-resolution single cell RNAseq of AlK3-expressing human pancreatic ductal cells. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131886. Deposited 28 April 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the Gene Expression Omnibus (GEO), accession no. GSE131886. Previously published scRNA-seq data reanalyzed here are available at under GEO accession nos. GSE81076/GSE85241. Source data for figures are provided in the SI Appendix reagent table. Code/Rscript files for the analyses reported are available in the GitHub repository https://github.com/JDBLab/Pancreas_ductal_scRNAseq (72).