Significance
Cyclic nucleotide-gated (CNG) and hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channels generate the primary electrical signals underlying mammalian phototransduction, olfaction, and cardiac pacemaking. These channels activate upon binding of cyclic nucleotide ligands (cAMP and/or cGMP), but the allosteric mechanism that couples ligand binding to gating is not well understood. Using double electron-electron resonance (DEER) spectroscopy, we reveal an allosteric conformational transition in the bacterial CNG channel SthK upon cAMP binding that involves outward movement of the C-linker domain in relation to the pore axis. Computational models constrained by DEER data also predict an upward movement of the cytoplasmic domain toward the membrane and suggest key interactions that may stabilize the agonist-induced conformational change.
Keywords: DEER, EPR, ion channel, allostery, cyclic nucleotide
Abstract
Cyclic nucleotide-gated (CNG) ion channels are essential components of mammalian visual and olfactory signal transduction. CNG channels open upon direct binding of cyclic nucleotides (cAMP and/or cGMP), but the allosteric mechanism by which this occurs is incompletely understood. Here, we employed double electron-electron resonance (DEER) spectroscopy to measure intersubunit distance distributions in SthK, a bacterial CNG channel from Spirochaeta thermophila. Spin labels were introduced into the SthK C-linker, a domain that is essential for coupling cyclic nucleotide binding to channel opening. DEER revealed an agonist-dependent conformational change in which residues of the B′-helix displayed outward movement with respect to the symmetry axis of the channel in the presence of the full agonist cAMP, but not with the partial agonist cGMP. This conformational rearrangement was observed both in detergent-solubilized SthK and in channels reconstituted into lipid nanodiscs. In addition to outward movement of the B′-helix, DEER-constrained Rosetta structural models suggest that channel activation involves upward translation of the cytoplasmic domain and formation of state-dependent interactions between the C-linker and the transmembrane domain. Our results demonstrate a previously unrecognized structural transition in a CNG channel and suggest key interactions that may be responsible for allosteric gating in these channels.
Cyclic nucleotide-gated (CNG) ion channels generate the primary electrical signals in response to visual and olfactory stimuli in vertebrates (1). Their activity is regulated by second messenger cyclic nucleotides (cAMP and/or cGMP), which bind directly to a cytoplasmic domain of the channel and promote opening of the ion conduction pathway (2). In this manner, CNG channels couple transient fluctuations in cellular cyclic nucleotide concentrations to the polarization state of the plasma membrane, ultimately regulating neurotransmitter release. Genetic mutations in rod (CNGA1/B1), cone (CNGA3/B3), and olfactory (CNGA2/A4/B1) channels give rise to autosomal recessive retinitis pigmentosa, achromatopsia—also known as total color blindness—and impaired olfaction, respectively, underscoring their importance in mammalian sensory processing (3, 4). CNG channels are related—both structurally and functionally—to the hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channels that control rhythmic activity in the sinoatrial node of the heart and in thalamocortical relay neurons (1, 5), and these two classes of channels share similar structures and mechanisms of cyclic nucleotide-dependent regulation (1).
CNG and HCN channels are members of the larger superfamily of voltage-gated ion channels (VGIC) and are composed of four protein subunits assembled around a central ion-conductive pore (Fig. 1A). Each subunit possesses six transmembrane-spanning helices (S1-S6), with helices S1-S4 comprising the voltage-sensing domain (VSD) and helices S5, S6, and the intervening pore helix making up the pore domain. In contrast to many other members of the VGIC family, CNG and HCN channels adopt a nondomain-swapped transmembrane architecture in which a short S4–S5 linker allows interaction between the VSD and pore domain of the same protein subunit (6–9). In addition, while most VGICs are activated by membrane depolarization, CNG channels are voltage independent and HCN channels are activated by membrane hyperpolarization—despite having similarly charged voltage-sensing S4 helices. It is possible that the nondomain-swapped transmembrane arrangement may play a role in the unique voltage-sensing properties of these channels (10–14).
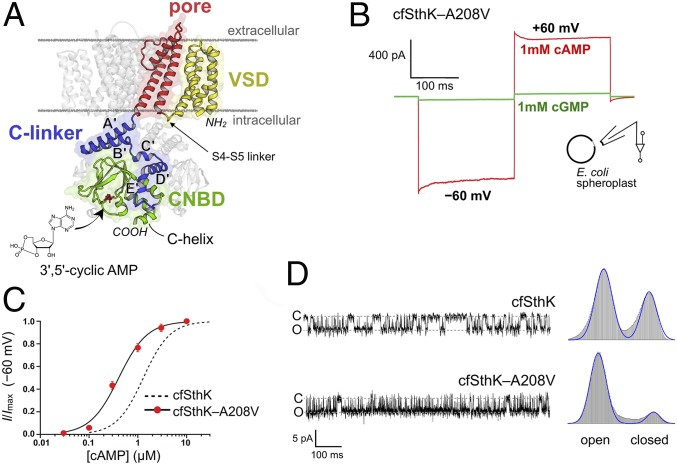
Fig. 1.
A cysteine-free bacterial CNG channel with high open probability in the presence of cAMP. (A) Cartoon representation of SthK (PDB ID code 6CJU) with a single subunit colored. The voltage sensing domain (VSD, yellow), pore domain (red), C-linker domain (blue), and cyclic nucleotide-binding domain (CNBD, green) are highlighted. C-linker helices A′-E′ are labeled, and the cAMP-binding site is indicated. (B) Representative current response of an inside-out patch from an E. coli spheroplast expressing cfSthK–A208V with voltage steps from 0 mV to −60 mV and +60 mV. Perfusion of 1 mM cAMP (red) elicits large currents at both negative and positive voltages, whereas 1 mM cGMP (green) displays only weak current response. (C) cAMP dose–response of cfSthK–A208V (red circles, mean ± SEM). The solid line is the fit of the Hill equation to the data (K1/2 = 0.4 ± 0.2, h = 1.5 ± 0.2, n = 3). The cAMP dose–response curve for cfSthK (dotted line) from previous work is shown for comparison (28). (D) Single-channel time traces of cfSthK (Upper) and cfSthK–A208V (Lower) recorded at −80 mV and perfused with saturating concentrations of cAMP. All-points histograms fit with two Gaussians are shown on the right and demonstrate increased open probability resulting from the A208V mutation.
The main feature distinguishing CNG and HCN channels from other members of the VGIC family is activation by cyclic nucleotide ligands, which is conferred by a large intracellular carboxyl-terminal domain consisting of four cyclic nucleotide-binding domains (CNBDs) that are connected to the pore by way of a C-linker domain (Fig. 1A). Previous work has uncovered the conformational changes that occur in the CNBD of CNG/HCN channels upon cyclic nucleotide binding, but precisely how this conformational change is coupled to pore opening is still an open question (15–18). The C-linker domain has previously been shown to allosterically couple the agonist-induced conformational change in the CNBD to the opening of the channel pore (19). Structurally, the C-linker is composed of six helices (A′–F′), with A′–D′ arranged as two antiparallel helix-turn-helix motifs (20). This region makes extensive intrasubunit and intersubunit interactions within the channel, with the A′-B′ helices of one subunit interacting with the C′-D′ helices of the neighboring subunit in an “elbow-on-shoulder” configuration that gives rise to a fourfold symmetric gating ring beneath the channel pore (20, 21). Despite several recent cryogenic electron microscopy (cryo-EM) structures of members of the CNBD channel family (6–9, 14, 22), the molecular mechanism underlying allostery in the C-linker domain remains poorly understood.
In this study, we employed electron paramagnetic resonance (EPR) spectroscopy to probe the cyclic nucleotide-dependent conformations of the C-linker region of the bacterial CNG channel from Spirochaeta thermophila, SthK. Bacterial CNG channels have emerged as promising models for studying CNG channel structure and function, and SthK in particular has recently been characterized both structurally and functionally (9, 23–28). We used site-directed spin-labeling (SDSL) and double electron-electron resonance (DEER) spectroscopy to measure select intersubunit distance distributions between residues of the B′-helices in full-length, tetrameric SthK and found an agonist-dependent conformational rearrangement in this region. We then used our DEER results as distance constraints in the construction of a Rosetta-based structural model of the resting-to-active conformational change in the C-linker.
Results and Discussion
A Cysteine-Free SthK Construct with Increased Open Favorability.
In order to study ligand-induced structural transitions by DEER, it is important to have a channel in which cyclic nucleotides produce a large change in the open probability (PO). Previous studies have indicated that SthK has a fairly low PO even in saturating concentrations of cAMP (23, 26), although reported values vary significantly depending on the amino acid sequence of the protein construct used as well as on the membrane composition (28). The largest PO value reported thus far for WT SthK in the presence of cAMP is 0.90 ± 0.02 at +60 mV, recorded on the full-length channel expressed in Escherichia coli spheroplasts (28). The native lipid environment is maintained in the E. coli spheroplasts and may be responsible, in part, for the higher PO. Since SthK is known to display weak depolarization-dependent activation, this PO at +60 mV corresponds to a PO of about 0.7 at 0 mV, the conditions experienced in our EPR experiments. The reported PO of SthK was essentially zero in the absence of cyclic nucleotide or in the presence of cGMP (28). Additionally, a cysteine-free construct of SthK—termed “cfSthK” (SthK–C153V,C387S)—was reported to have functional properties in E. coli spheroplasts virtually identical to those of the WT channel (28).
Previously we found that an alanine-to-valine mutation in transmembrane helix S6 (A208V), in combination with other mutations, drastically increased the open probability of cfSthK in E. coli spheroplasts (28). Here, we investigated if the A208V mutation alone was sufficient to achieve high PO values in the presence of cAMP. As seen in Fig. 1B, cfSthK–A208V in inside-out patches from E. coli spheroplasts produced large currents at both negative and positive voltages in the presence of cAMP. Similar to the WT channel, the A208V construct passed no current in the absence of cyclic nucleotides and displayed only weak activation in the presence of cGMP, consistent with the classification of cGMP as a partial agonist. The cAMP dose–response curve of cfSthK–A208V was shifted to lower ligand concentrations relative to cfSthK, with an apparent affinity of 0.4 ± 0.2 μM (n = 3) compared to K1/2 = 1.1 ± 0.1 μM for cfSthK (n = 3) (Fig. 1C) (28). The Hill coefficients (h) extracted from the dose–response curves were indistinguishable between cfSthK (h = 1.5 ± 0.1) and cfSthK–A208V (h = 1.5 ± 0.2), suggesting that the A208V mutation did not substantially affect the gating mechanism.
To more accurately determine the effect of the A208V mutation on the PO of SthK, we performed single-channel recordings on cfSthK and cfSthK–A208V expressed in E. coli spheroplasts. Fig. 1D shows representative single-channel traces from cfSthK or cfSthK–A208V channels recorded at −80 mV in inside-out patches and perfused with saturating concentrations of cAMP. The single-channel recordings displayed bursting behavior with brief flickers from the open state to the closed state, as well as longer-duration, closed-state events. It is apparent from Fig. 1D that the A208V mutation resulted in significantly reduced closed-state dwell times. All-points histograms constructed from the single-channel recordings were fit with the sum of two Gaussians, giving PO = 0.61 for cfSthK and PO = 0.88 for cfSthK–A208V at −80 mV. Assuming that cfSthK–A208V retained the same voltage dependence as WT SthK in spheroplasts (28), we estimate the PO of cfSthK–A208V in saturating cAMP to be ∼0.92 at 0 mV, which should allow for near-complete transition to the open state of the channel for EPR experiments. We therefore chose cfSthK–A208V as the base construct for all spin labeling EPR experiments.
Site-Directed Spin Labeling of Residues in the B′-Helix.
In order to probe cyclic nucleotide-dependent conformational changes occurring in the C-linker upon channel activation, we introduced single-cysteine residues in the C-linker of cfSthK–A208V. Six cysteine residues were individually introduced on the exposed surface of the B′-helix for SDSL with the thiol-reactive nitroxide spin label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSL) (Fig. 2A). As previously demonstrated, SthK confers marked toxicity when expressed in E. coli, presumably due to activation by endogenous cAMP, which is particularly problematic with SthK constructs displaying high PO such as cfSthK–A208V (28). We therefore expressed our SthK constructs in an engineered strain of E. coli deficient in adenylate cyclase, the enzyme responsible for cAMP synthesis in bacteria (SI Appendix, Fig. S1) (28). All six single-cysteine mutants displayed robust expression and were monodispersed after purification from the bacterial membrane (SI Appendix, Fig. S2).
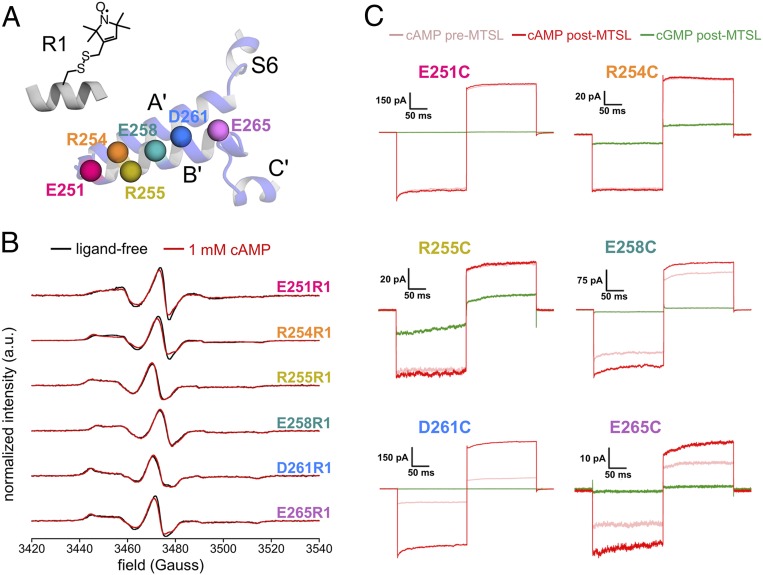
Fig. 2.
Spin labeling of the SthK C-linker. (A) Sites of cysteine mutations along the B′-helix and chemical structure of the spin-labeled side chain R1. (B) Continuous-wave EPR spectra of spin-labeled SthK mutants recorded at room temperature. Ligand-free spectra (black) are overlaid with spectra recorded in the presence of 1 mM cAMP (red). Spectra are normalized to the number of spins in the sample as determined by double integration of the baseline-corrected EPR spectrum. (C) Representative macroscopic currents of spheroplasts expressing single-cysteine B′ mutants of cfSthK–A208V. Inside-out patches were subjected to voltage steps to −60 and +60 mV after perfusion with 1 mM cAMP (red), 1 mM cGMP (green), and in the presence of 1 mM cAMP after perfusion with MTSL (pink).
Fig. 2B shows the room-temperature continuous-wave (CW) EPR spectra of the six SthK constructs after reaction with MTSL to give the spin-labeled side chain R1. All constructs showed robust labeling, with labeling efficiencies of >90%, indicating that all mutation sites were accessible to solvent in the absence of ligand. These EPR spectra are typical of nitroxide radicals with rotational correlation times on the order of tens of nanoseconds, as would be expected for helical sites on a large protein. Addition of 1 mM cAMP produced only minor changes to the spectra, suggesting that ligand binding does not result in large-scale redistributions of the R1 rotamer populations and/or mobilities, nor does it cause local unfolding of secondary structures. Small but detectable changes in the EPR line shape are apparent for three of the spin-labeled sites (E251R1, R254R1, and E265R1). In all three cases, we observed an increase in the intensity of the low-field component of the spectrum relative to the central line upon addition of cAMP, indicating that the side chains at these locations are somewhat less mobile in the ligand-bound state.
To determine if the cysteine mutations or SDSL had an effect on SthK function, we expressed each single-cysteine mutant in E. coli and prepared spheroplasts for characterization with patch-clamp recording. Inside-out patches containing each SthK variant were excised from the spheroplasts, and macroscopic currents were recorded in the absence and presence of 1 mM cAMP, with both negative (−60 mV) and positive (+60 mV) voltage steps (Fig. 2C). Cysteine residues on the B′-helix were then modified by perfusion of MTSL to the intracellular side, and currents were again recorded in saturating concentrations of both cAMP and cGMP. In general, our spin-labeled cfSthK–A208V constructs retain the overall characteristics of the WT channel. Namely, they maintain strong activation by cAMP, whereas cGMP elicits little or no measurable current under the conditions tested and, therefore, likely represents a very weak partial agonist. A notable exception is the R255C mutation where there appears to be significant activation in the presence of cGMP, indicating that the overall favorability of channel opening has been increased. This is not surprising given that the analogous residues in the mammalian channels CNGA1 and HCN2 are known to participate in intersubunit and intrasubunit interactions important for maintaining the C-linker in the resting conformation (19, 29). In CNGA1 and HCN2, a lysine residue at the analogous position participates in a “salt bridge triad” with a glutamate on the D′-helix of the neighboring subunit and an aspartate on the CNBD β-roll of the same subunit. In the closed-state structures of SthK, the intersubunit salt bridge to the D′-helix is not conserved; however, the intrasubunit interaction with the β-roll is maintained (9, 24). It is likely that mutation of R255 in SthK abolishes the salt bridge interaction with β-roll residue E326 and results in a channel with substantially increased propensity for the open state.
Another interesting feature observed in our functional studies is the apparent increase in cAMP-induced current upon MTSL modification of D261C (and, to a lesser extent, R254C and E258C). These data suggest that the open probabilities of these cysteine-mutant channels are lower than cfSthK–A208V and are increased upon modification with MTSL. Based on the available structures of SthK (9, 24), these residues appear to reside on the top surface of the B′-helix, with side chains facing upward toward the transmembrane domain. The functional data presented here (Fig. 2C) suggest that these residues may participate in open state-dependent interactions with the transmembrane domain or the membrane itself that are disrupted upon substitution with the small hydrophilic side chain of cysteine but can be partially restored upon conjugation with the longer R1 side chain.
DEER Reveals cAMP-Induced Structural Changes in the C-Linker.
To investigate cyclic nucleotide-dependent conformational changes of the C-linker region, we employed DEER spectroscopy on our SthK constructs spin-labeled throughout the B′-helix. A primary advantage of the DEER experiment is that the DEER signal encodes information about the full interspin distance distribution and not simply a weighted average of distances in the sample (30). The homotetrameric nature of SthK dictates that for a singly spin-labeled protein construct, we will see a minimum of two intersubunit distance populations, provided they are within the distance sensitivity of the DEER experiment. As illustrated in Fig. 3A, a fourfold symmetric channel will give one population of distances arising from spin labels on adjacent subunits (r) and a second population centered at a distance of √2 × r resulting from spin label pairs that are diagonally opposed in the tetramer. With uniform excitation of all spins in the sample, these two populations have an expected 2:1 ratio of integrated intensities in the DEER distance distribution.
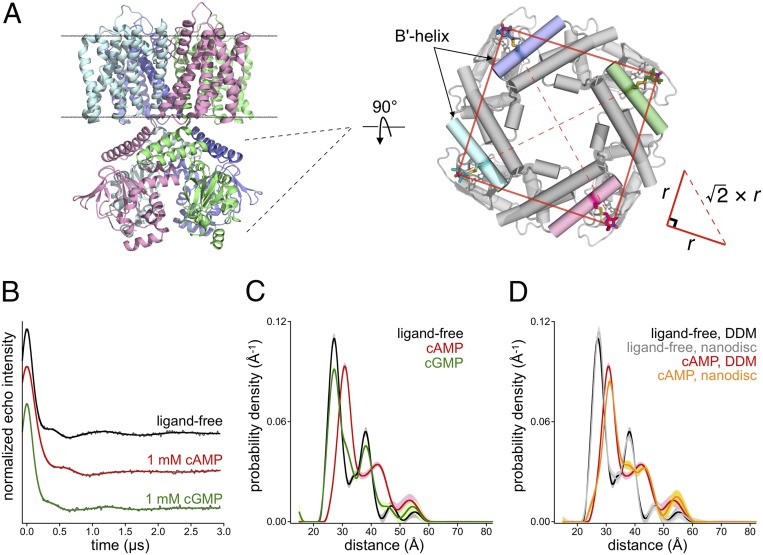
Fig. 3.
DEER reveals agonist-dependent conformational rearrangement in the C-linker. (A) Ribbon diagram of SthK (PDB ID code 6CJQ) and top-down view of the cytoplasmic domain with R1 labels at position 261 in the B′-helix. The relationship of adjacent and diagonal interspin distances is shown. (B and C) Background-corrected time-domain DEER traces (B) and distance distributions for cfSthK–A208V,D261R1 recorded in DDM detergent (C). The presence of cAMP (red) produces an increase in interspin distance relative to the ligand-free (black) and cGMP (green) conditions. (D) Comparison of the ligand-free and cAMP-bound distance distributions from cfSthK–A208V,D261R1 recorded in detergent (black and red, respectively) versus channels reconstituted into lipid nanodiscs (gray and orange, respectively).
The background-corrected four-pulse DEER time-domain data and distance distributions, respectively, of cfSthK–A208V,D261R1 in n-dodecyl-β-d-maltopyranoside (DDM) detergent micelles is shown in Fig. 3 B and C. In the absence of cyclic nucleotide, the distance distribution shows two prominent peaks at 27.2 and 38.1 Å, with an intensity ratio of roughly 2:1, representing the adjacent and diagonal distances within the tetramer, respectively. Interestingly, an addition of 1 mM cAMP shifts the distance distribution to longer interspin distances with the adjacent and diagonal distances shifted to 30.8 and 42.3 Å, respectively. An addition of 1 mM cGMP, a weak partial agonist, results in adjacent and diagonal interspin distances identical to those of the ligand-free channel but with a slightly broadened distribution as evidenced by the lower amplitude of the main peaks relative to the ligand-free distribution. In combination with our functional data from spheroplast patches (Fig. 2C), these results suggest that the conformational change observed by DEER with cAMP is associated with channel activation. It is interesting to note the presence of a small shoulder in the distribution at ∼31 Å in 1 mM cGMP (Fig. 3C), indicating that a minor population of channels possessing an active C-linker may exist when bound with partial agonist.
The presence and composition of biological lipid membranes is known to affect the structures, energetics, and conformational dynamics of membrane proteins (31, 32). Indeed, the activity of SthK in a synthetic bilayer was shown to depend on the lipid environment, with channel opening being potentiated in the presence of negatively charged lipids, particularly cardiolipin (26). To test whether the conformational change we observed by DEER is affected by the nonnative conditions of the detergent micelle, we reconstituted purified cfSthK–A208V,D261R1 into lipid nanodiscs composed of E. coli polar lipids and the membrane scaffolding protein MSP2N2 (33). The E. coli polar lipids should closely mimic the lipid environment of our spheroplast electrophysiological recordings. Fig. 3D shows the DEER distance distributions from cfSthK–A208V,D261R1 in lipid nanodiscs, with and without 1 mM cAMP, along with the corresponding distributions recorded in DDM. Although there appears to be a minor population of the resting C-linker conformation in the nanodisc distribution with cAMP as evidenced by small shoulders at ∼25 Å and ∼38 Å, it is clear that the same conformational rearrangement observed in DDM is also present in the context of a native-like membrane environment. Like the detergent-solubilized channels, cfSthK–A208V,D261R1 in nanodiscs shows no distance increase in the presence of the partial agonist cGMP and gives a distance distribution nearly identical to that in the absence of ligand (SI Appendix, Fig. S4). Given that the transition from resting to active C-linker appears to be similar in both detergent and in a lipid membrane, we performed our DEER experiments on the remaining B′-helix mutants in the presence of DDM.
DEER distance distributions were obtained for all six constructs with spin-labels spanning the B′-helix in the absence of cyclic nucleotide as well as in the presence of 1 mM cAMP or 1 mM cGMP (Fig. 4A and SI Appendix, Fig. S5). Sufficiently long dipolar evolution times were recorded to reliably resolve not only the interspin distances arising from adjacent subunits, but also from the diagonally facing subunits, an important prerequisite for analysis of channel symmetry. These diagonal distances typically exceeded 50 Å, and in the case of E251R1 were about 70 Å, requiring dipolar evolution times approaching 7 μs. Uncertainties arising from noise and imperfect background separation in the DEER signal are quantified as error bands on the distance distributions shown in Fig. 4A.
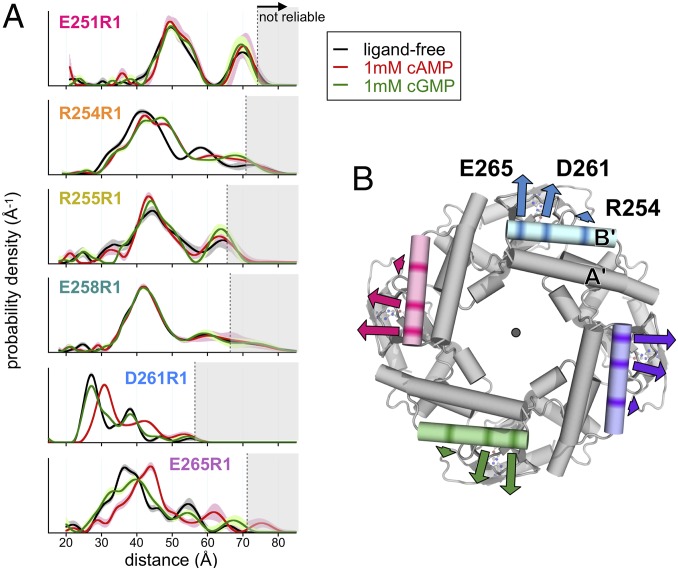
Fig. 4.
DEER indicates outward movement of the B′-helix upon channel activation. (A) Distance distributions for spin-labeled SthK constructs in the absence of ligand (black), and in the presence of 1 mM cAMP (red) or 1 mM cGMP (green). Gray shaded areas represent distance regions where the analysis is unreliable due to the limited length of the time-domain DEER data trace. Error bars on the distributions represent mean ± SD based on stochastic variation of the signal noise and background function (Methods). (B) Cartoon depiction of the SthK cytoplasmic domain (PDB ID code 6CJQ) viewed from the extracellular side. Arrows indicate the outward movement relative to the symmetry axis at residues 254, 261, and 265 in the presence of cAMP, as observed by DEER.
In addition to the cAMP-induced distance increase at D261R1 just described, we observed a large agonist-dependent increase in interspin distance between labels attached at residue 265, positioned approximately one helical turn from D261 at the C-terminal end of the B′-helix (Fig. 4A). Here, the most probable nearest-neighbor distance shifts by more than 7 Å, from 36.5 Å in the absence of ligand to 43.9 Å in the presence of 1 mM cAMP. In the presence of the partial agonist cGMP, the distance distribution for E265R1 displays noticeable broadening, suggesting a greater mixture of conformational or rotameric populations; however, the distribution as a whole does not shift to larger distances as it does with cAMP. The DEER results from D261R1 and E265R1 together indicate a sizable agonist-induced conformational change occurring at the C-terminal portion of the B′-helix in which intersubunit distances are increased in the active conformation of the channel.
In contrast to residues at the C-terminal end of the B′-helix, spin labels introduced at residues at the N-terminal end of the B′-helix undergo only minor distance shifts and/or changes in the shape of the distribution in the presence of cyclic nucleotide. For the SthK construct spin-labeled at E251—positioned closest to the N terminus of the B′-helix—there is essentially no change in the most populated distance between ligand-free and cAMP-bound conditions. The R255R1 construct also displays only slight changes in the position of the distance distribution; however, this site shows a detectable narrowing of the distance distribution in the presence of cAMP, indicating a reduction of the conformational and/or rotameric heterogeneity of this region in the active C-linker conformation.
An interesting exception to the general trend of smaller ligand-induced changes at N-terminal sites relative to C-terminal sites in the B′-helix is seen in the DEER distributions for SthK spin-labeled at residue R254. Here, we observed a clear shift to longer interspin distances in the presence of cyclic nucleotide, as well as a splitting of the main distance peak into a bimodal distribution. Interestingly, we observed this change in both cAMP- and cGMP-bound conditions, despite the fact that this construct produces only small fractional activation by cGMP in E. coli spheroplasts (Fig. 2C). A possible explanation for this result is that both cAMP and cGMP stabilize a population of side-chain conformations that produce an overall shift in the interspin distance but are not associated with channel activation, whereas the resting-to-active transition in the channel does not result in a significant change in interspin distance at this site. The CW EPR spectra of R254R1 does indeed show a noticeable change in the low-field region upon addition of cAMP, consistent with a ligand-induced redistribution of side-chain rotamers and/or mobilities (Fig. 2B).
Interestingly, the distance distributions obtained for the E258R1 construct, which is located one helical turn N-terminal to D261 and near the middle of the B′-helix, are virtually identical between ligand-free, cAMP-bound, and cGMP-bound states. Since the E258R1 mutant is still clearly activated by cAMP (and not cGMP) in E. coli spheroplasts (Fig. 2C), this residue may represent either a static element or pivot point around which the C-linker structural transition occurs. Taken together, our DEER results point to an allosteric mechanism whereby agonist binding at the CNBDs produces an outward movement of the B′-helix relative to the symmetry axis (Fig. 4B). This dilation appears to be more pronounced for residues near the C-terminal portion of the B′-helix than for those at the N-terminal end or in the middle of the helix.
Previous studies have suggested that the structural mechanism of ligand-induced gating in CNG and HCN channels involves a change in the quaternary architecture of the C-terminal cytoplasmic domains—with a hypothesized transition from a dimer-of-dimers arrangement to a fourfold symmetric complex upon cyclic nucleotide activation (34, 35). Our ability to simultaneously measure adjacent and diagonal distance distributions provides a way to test this hypothesis. For cfSthK–A208V,D261R1 in the absence of ligand, the ratio of diagonal to neighboring distances as judged by the respective maxima of the DEER distributions is 1.38 and 1.40 for the nanodisc and DDM distributions, respectively. These ratios are within 2% of the value expected for a perfectly symmetric complex (√2 ≈ 1.41). The cAMP-bound DEER distributions indicate a similar level of symmetry in the activated conformation, with a distance ratio between 1.37 and 1.38 (within 3% of ideal fourfold symmetry). This indicates that SthK is fourfold symmetric in the absence of ligand and retains this symmetry upon cyclic nucleotide activation—at least at the level of the C-linker domain.
A Structural Model of C-Linker Movement.
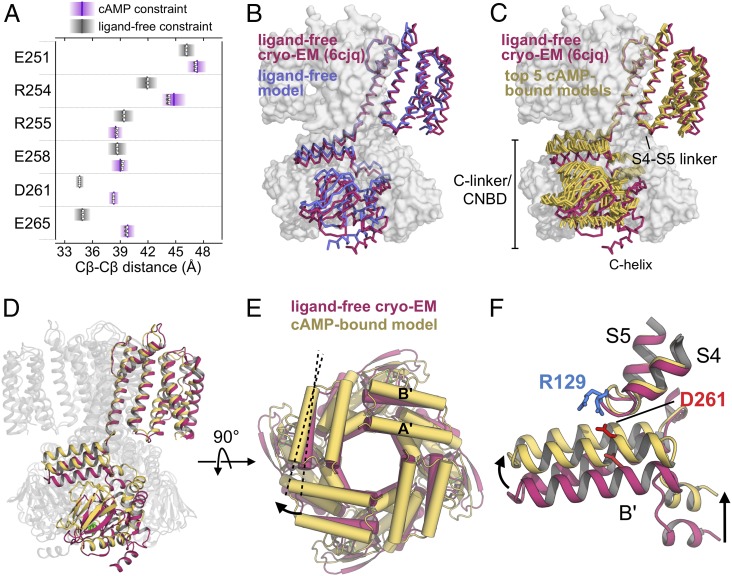
To better understand the resting-to-active allosteric transition of the C-linker, we used Rosetta-based structural modeling to generate SthK models capable of producing interspin distances consistent with our DEER results. Rosetta is a software package for macromolecular structure prediction that is capable of integrating experimental data as constraints (36, 37). We first built starting models of SthK in both ligand-free and cAMP-bound states using previously published structures of SthK variants as templates (9, 24, 38). These template cryo-EM structures of SthK are virtually identical in the ligand-free and cAMP-bound states and are thought to represent closed conformations of the C-linker and pore (9). As such, the starting Rosetta models are also representative of the resting C-linker domain (SI Appendix, Fig. S6). Next, we used Rosetta to impose Cβ‒Cβ distance constraints derived from our DEER distance distributions (SI Appendix, Fig. S7) and generated SthK models that best satisfied these constraints. The resulting models all satisfied the imposed DEER distance constraints at the six B′-helix residues and converged to similar structures regardless of the starting model used (Fig. 5A and SI Appendix, Fig. S8). The best-scoring Rosetta model for ligand-free SthK was comparable in the C-linker domain (Cα rmsd = 1.0 Å) to the ligand-free cryo-EM structure (PDB ID code 6CJQ), indicating that the addition of atom-pair constraints derived from ligand-free DEER distributions did not significantly change the C-linker conformation from that observed in the cryo-EM structure (Fig. 5B).
Fig. 5.
Modeling the cyclic nucleotide-dependent transition. (A) Constraint satisfaction for 100 Rosetta models (open circles) with DEER-derived Cβ-Cβ distance constraints for ligand-free (gray) and cAMP-bound (purple) states. The center of the constraint is shown as a line. (B) Superposition of the best-scoring ligand-free Rosetta model (blue) with the ligand-free cryo-EM structure (PDB ID code 6CJQ, red) aligned by the pore domain (S5-S6) (9). (C) Superposition of the five best-scoring Rosetta models of the cAMP-bound state (yellow) with the cryo-EM structure in the absence of ligand (red) (9). (D) Ribbon diagram showing the final cAMP-bound Rosetta model (yellow) aligned by the pore domain with the ligand-free cryo-EM structure (red). (E) Top-down view of the aligned Rosetta and cryo-EM structure cytoplasmic domains showing outward movement of the A′- and B′-helices in the ligand-bound model. (F) Interaction of the A′/B′-helices with the S4-S5 linker resulting from upward movement of the C-linker/CNBD in the cAMP-bound model. A salt bridge interaction is predicted in the ligand-bound state between R129 of the S4-S5 linker and D261 of the B′-helix.
In contrast to the ligand-free models, the DEER-constrained Rosetta models for the cAMP-bound state of SthK showed pronounced differences in the C-linker conformation compared to that of the ligand-free structure. The five best-scoring models for cAMP-bound SthK are shown in Fig. 5C superimposed on the ligand-free cryo-EM structure. These models also readily satisfy the Cβ‒Cβ distance constraints we derived from DEER distributions recorded in cAMP (Fig. 5A). The models are highly similar to each other in the C-linker domain (Cα rmsd = 0.7 ± 0.2 Å), indicating convergence of the structural models. Moreover, the C-linker domains are substantially different from that of the best-scoring Rosetta model in the ligand-free state (Cα rmsd = 2.4 ± 0.2 Å) and from the ligand-free cryo-EM structure (Cα rmsd = 2.6 ± 0.1 Å). The best-scoring cAMP-bound models all display a pronounced outward movement of the C-terminal end of the B′-helix of ∼5‒6 Å across the channel diagonal relative to the ligand-free structure (Fig. 5E and Movie S1), as predicted from the increased distances seen for D261R1 and E265R1 by DEER. Computational modeling of the R1 side chain onto each labeling site in our final Rosetta models produced similar predicted distance changes as those seen by DEER, further supporting the consistency of our models with the experimental distributions (SI Appendix, Fig. S9).
The cyclic nucleotide-induced conformational change known to occur in the CNBD of CNG and HCN channels is nicely recapitulated in the Rosetta models (16, 18, 20, 24, 39–42). Cyclic nucleotide binding at the β-roll is accompanied by closure of the distal B- and C-helices, with C-helix residue E421 forming a hydrogen-bond interaction with N6 of cAMP. Surprisingly, however, all of the cAMP-bound models also showed a large (∼6 Å) upward translation of the entire cytoplasmic C-terminal domain, including the C-linker and CNBDs (Fig. 5 D and F and Movie S2). The turn motif between the A′- and B′-helices—which constitutes the “elbow” of the C-linker domain—moved ∼7 Å toward the transmembrane domain in comparison to the ligand-free cryo-EM structure, resulting in A′- and B′-helices that were more parallel to the plane of the plasma membrane. This nearly parallel configuration of the A′- and B′-helices with respect to the membrane is reminiscent of the conformations seen in recent cryo-EM structures of the cAMP-bound bacterial CNG channel LliK and the cGMP-bound eukaryotic CNG channel TAX-4 (7, 8). Our models suggest that cyclic nucleotide-induced closure of the C-helix results in upward movement of the B- and A-helices of the CNBD. The A-helix of the CNBD resides immediately below and interacts tightly with the D′-helix of the C-linker “shoulder.” The cAMP-induced translation of the CNBD results in movement of the C-linker toward the membrane, which would, in turn, exert force on the A′/B′-helices (“elbow”) of the neighboring subunit, producing upward translation and a more parallel orientation of the A′/B′ helices with respect to the membrane.
The upward movement of the C-linker/CNBD upon channel activation predicted by our Rosetta models is consistent with previous experimental data on both SthK and the mammalian channel CNGA1 (27, 43, 44). Upon addition of cAMP, SthK channels were observed by atomic force microscopy (AFM) to undergo a vertical displacement of the C-linker/CNBD toward the membrane of at least 6 Å, similar to what is predicted in our Rosetta models (27). In CNGA1, cyclic nucleotide-dependent activation is potentiated by the transition metal ions Co2+ and Ni2+ (43, 45), an effect that was localized to a single histidine residue at the C-terminal end of the A′-helix (H420 in bovine CNGA1) (43). More recently, Co2+ was found to be coordinated by H420 and a high-affinity metal binding site located at the plasma membrane, suggesting that interaction between the A′-helix and the lipid membrane stabilizes the open state of the channel (44). In combination with these data, our DEER-constrained Rosetta models lend further support to a mechanism for cyclic nucleotide-dependent activation whereby the C-linker moves toward the membrane during channel activation.
Given the consistent prediction of upward translated C-linker/CNBDs in our cAMP-bound Rosetta models, it is possible that interactions between the C-linker and transmembrane domains might stabilize this conformation. Indeed, a salt bridge interaction between R129 in the transmembrane S4-S5 linker and D261 in the C-linker B′-helix is present in four of the five best-scoring cAMP-bound models (Fig. 5F). By contrast, none of the five best-scoring ligand-free models contain an intact salt bridge between these residues. Interestingly, the analogous B′-helix residue of TAX-4 (D453) is observed to interact with an arginine residue at the bottom of the S4-helix (R296) in the cGMP-bound open-state cryo-EM structure (7), suggesting that polar interactions between the lower S4 or S4-S5 linker and the C-terminal end of the B′-helix may be a hallmark of the active CNG conformation. The cAMP-bound Rosetta models also show interactions between the cytoplasmic S2-S3 loop and the A′/B′ helices of the C-linker that are not present in the ligand-free models. For example, K68 of the S2-S3 loop is predicted to form an interaction with both E265 and T266 of the B′-helix, whereas S70 is predicted to interact with K247 of the neighboring A′-B′ loop. However, residues of the S2-S3 loop are unresolved in the resting-state cryo-EM structures and were rebuilt de novo by Rosetta in our models, so the existence of these interactions in the active C-linker conformation remains uncertain.
Recent evidence from cryo-EM and AFM experiments has given rise to a model of cyclic nucleotide-dependent activation in CNG/HCN channels involving a clockwise rotation of the cytoplasmic domains relative to the transmembrane domains when viewed from the intracellular side of the membrane (6, 8, 11, 27). This model—referred to as the “twist-open model” or “iris diaphragm mechanism”—posits that the C-linker domain in the resting conformation exerts an autoinhibitory force on the right-handed four-helix bundle formed by the C-terminal segments of the S6 helices, holding the channel closed. Upon binding of the cyclic nucleotide agonist, clockwise rotation of the C-linker/CNBD relieves the tonic inhibition on the S6 bundle-crossing, allowing the unwinding and subsequent dilation of the helical bundle, and permitting ion permeation through the pore. Our models do not show relative rotation between the cytoplasmic and transmembrane domains, and the pore remains closed. This may be due to the insensitivity of our DEER distributions to rigid-body rotations around the symmetry axis. Experimental distance constraints between the transmembrane domain and the C-linker, as well as between the four transmembrane domains, may be needed for Rosetta to sufficiently sample the open conformation of the channel.
The experiments presented and analyzed here show a cyclic nucleotide-dependent conformational change in the tetrameric C-linker of a full-length CNG channel and provide insight into the structural transitions that occur during channel activation. Structural models built using our data suggest specific interactions that may be critical for coupling between the C-linker and the pore domain. Nevertheless, questions remain about precisely how this C-linker conformational change is translated into opening of the channel pore. Future experiments examining C-linker-to-pore coupling are likely to provide further mechanistic understanding of gating processes in CNG, HCN, and related members of the CNBD channel family.
Methods
SthK Expression and Purification.
Full-length cysteine-free SthK constructs (C153V, C387S) (UniProt accession no. E0RR11) were expressed from the pETM11 vector and contained an N-terminal 6xHis tag followed by a TEV protease cleavage site. Single cysteine substitutions for SDSL were introduced using a modified QuikChange protocol (46) with PhusionHF polymerase (New England BioLabs) and verified by DNA sequencing. Expression was performed in C43 (DE3) cells (Lucigen) engineered to be deficient in adenylate cyclase (cyaA−) (28). Transformed cells were grown in terrific broth medium containing 50 μg/mL kanamycin to an OD600 ∼1.0 before induction with 0.5 mM isopropyl β-ᴅ-1-thiogalactopyranoside (IPTG). Induction was carried out at 19 °C for 17–19 h, after which cells were harvested by centrifugation and resuspended in lysis buffer (150 mM KCl, 2 mM β-mercaptoethanol [β-ME], 50 mM Tris, pH 8.0) containing EDTA-free protease inhibitors (Pierce) and ∼100 μg/mL DNase I. All subsequent lysis and purification steps were carried out on ice or at 4 °C unless otherwise noted. Cells were lysed by three passes through an EmulsiFlex-C3 cell disruptor at 15,000–20,000 psi, and the lysate was clarified by centrifugation at 25,000 × g for 30 min. Membranes were harvested from the supernatant by ultracentrifugation at 140,000 × g for 2 h and resuspended 1:1 (wt/vol) in resuspension buffer (150 mM KCl, 2 mM β-ME, 20% [vol/vol] glycerol, 25 mM Tris, pH 8.0). Resuspended membrane aliquots were frozen in liquid nitrogen and stored at −80 °C until purification.
Thawed aliquots of resuspended membranes were extracted with an equal volume of extraction buffer (150 mM KCl, 50 mM DDM, 5 mM cholesteryl hemisuccinate [CHS], 2 mM β-ME, 25 mM Tris, pH 8.0) with EDTA-free protease inhibitors (Pierce) and nutated for 1 h. Insoluble material was pelleted by centrifugation at 40,000 × g for 45 min, and the supernatant was applied to 1/10 bed-volume of TALON resin (Clontech) preequilibrated with 150 mM KCl, 50 mM imidazole, 25 mM Tris, pH 8.0 in a 20-mL Poly-Prep column (Bio-Rad). Batch binding proceeded with gentle nutation for ∼1 h, after which the resin was washed with 10 bed-volumes wash buffer (150 mM KCl, 0.5 mM DDM, 0.05 mM CHS, 20 mM imidazole, 2 mM β-ME, 25 mM Tris, pH 7.7), followed by 10 bed-volumes wash buffer containing 300 mM KCl, and finally an additional 10 bed-volumes of wash buffer. Protein was eluted with elution buffer (wash buffer containing 200 mM imidazole) and immediately desalted (PD-10, GE Healthcare) into low-salt anion exchange chromatography (AEX) buffer (50 mM KCl, 0.4 mM DDM, 0.04 mM CHS, 2 mM β-ME, 20 mM Tris, pH 7.6). Desalted protein was then applied to a 5 mL bed-volume ANX (high-sub) anion exchange column (GE Healthcare) equilibrated with low-salt AEX buffer at a flow rate of 1 mL/min. The column was washed with low-salt AEX buffer for several column volumes until a baseline absorbance at 280 nm was reached, at which point the protein was eluted with a linear gradient to 1 M KCl over 30 mL. Protein fractions were supplemented with 10 mM tris(2-carboxyethyl)phosphine and concentrated with a 100-kDa molecular-weight cutoff (MWCO) spin concentrator (GE Healthcare). Concentrated protein was heated in a 50 °C water bath for 10 min, filtered (0.2 μm), and loaded onto a Superose 6 10/300 GL size-exclusion column (GE Healthcare) equilibrated with SEC buffer (150 mM KCl, 0.4 mM DDM, 0.04 mM CHS, 20 mM Hepes, pH 7.2).
Spin-Labeling and EPR Sample Preparation.
Single-cysteine SthK constructs were reacted immediately following SEC with 100 μM MTSL (Toronto Research Chemicals) at room temperature for ∼4 h, protected from light. Unreacted spin-label was then removed by desalting (PD-10) into SEC buffer followed by concentration (100-kDa MWCO). Removal of residual free spin-label and exchange into deuterated buffer (20 mM Hepes, 150 mM KCl, 0.4 mM DDM, 0.04 mM CHS, pH 7.2 in D2O) for DEER spectroscopy was achieved by microdialysis (Pierce). d8-glycerol (Cambridge Isotopes) was supplemented to achieve a final glycerol concentration of 26% (vol/vol). For samples containing cyclic nucleotide ligand, 3′,5′-cAMP or 3′,5′-cGMP (Sigma) in deuterated buffer was included at 1 mM. Samples for CW EPR were loaded into 1.0 mm outer diameter (OD), 0.7 mm inner diameter (ID) quartz capillaries (Sutter), sealed with wax, and maintained at room temperature until measurement. Samples for DEER spectroscopy were loaded into 1.5 mm OD/1.1 mm ID quartz tubes (Sutter) with flame-sealed bottoms and flash frozen in liquid nitrogen. Samples were stored at −80 °C until measurement. CW EPR and DEER samples of nanodisc-reconstituted cfSthK–A208V,D261R1 were prepared as described above, with the exception that detergent was omitted in all buffers.
Nanodisc Reconstitution.
Membrane scaffolding protein MSP2N2 was expressed in E. coli and purified as previously described (47). Purified MSP2N2 was digested with TEV protease to remove the N-terminal 6xHis tag and repurified by reverse immobilized-metal affinity chromatography. E. coli polar lipids (Avanti) from a 25 mg/mL solution in chloroform were dried under a stream of argon and further dried overnight in a vacuum desiccator at room temperature. The lipid film was resuspended to 10 mM (assuming a molecular weight of 770 g/mol) in buffer containing 14 mM DDM, 1.4 mM CHS, 150 mM KCl, 20 mM Hepes, at pH 7.4. This lipid stock solution was subjected to five freeze/thaw cycles in liquid nitrogen and bath sonicated until the solution was clear. Spin-labeled cfSthK–A208V, D261R1 in SEC buffer was concentrated to ≈100 μM and mixed with the lipid stock (10 mM) and MSP2N2 stock (∼82 μM) to achieve a final molar ratio of 1:1.5:200 (SthK monomer:MSP2N2:lipid). This mixture was incubated on ice for 1 h before detergent removal was initiated with 20 mg (wet weight) of BioBeads SM-2 (Bio-Rad). The reaction was nutated at room temperature in the dark for 4 h before another 20-mg addition of BioBeads was added and the reaction was nutated at 4 °C overnight. The reconstitution reaction was separated from the BioBeads by centrifugation, filtered (0.2 μm), and purified by Superose 6 SEC in detergent-free buffer (150 mM KCl, 20 mM Hepes, pH 7.2).
Electrophysiology.
Electrophysiological recordings on SthK constructs in E. coli spheroplasts was performed as described previously (28). Briefly, spheroplasts for macroscopic patch-clamp recordings were produced from C43(DE3) E. coli (Lucigen) expressing SthK constructs cloned into to the pETM11 vector, with the exception of cfSthK–A208V and cfSthK–A208V,R255C, which were expressed in a modified pET vector containing a C-terminal GFP fusion (pCGFP) (48). Transformed bacteria were grown at 37 °C in 2× yeast tryptone medium containing 50 μg/mL kanamycin (pETM11) or 100 μg/mL carbenicillin (pCGFP) to OD600 ∼ 0.3 and then diluted 1:10 into fresh medium prewarmed to 42 °C and supplemented with 60 μg/mL cephalexin. After 1.5-h incubation at 42 °C and 180 rpm, the culture was cooled to 19 °C and expression was induced with 0.4 mM IPTG and incubated overnight at 19 °C and 150 rpm. For single-channel recordings, cfSthK and cfSthK–A208V constructs in the pCGFP vector were treated identically except that IPTG induction was carried out at 37 °C for ∼20 min. Spheroplast production was carried out exactly as described previously (28), and spheroplasts were stored as aliquots at −80 °C. Spheroplasts were thawed immediately prior to electrophysiological recording and were patched within 1 h of thawing.
Patch-clamp recording was performed in the inside-out configuration with symmetric solutions of 10 mM Hepes, 150 mM KCl, 500 mM sucrose, 20 mM MgCl2, pH 7.4 in both the pipette and in the bath. Patch pipettes were pulled from borosilicate glass to an open resistance of 2‒4 MΩ for macroscopic currents and 5‒8 MΩ for single-channel recordings. Data were acquired using an Axopatch 200A amplifier with Patchmaster (HEKA Elektronik), and perfusion was achieved with an RSC-100 rapid solution changer (BioLogic). For macroscopic recordings, the data were acquired at a sampling rate of 10 kHz and low-pass filtered at 2 kHz. Macroscopic current traces were leak-subtracted using an identical voltage protocol in the absence of ligand. Single-channel recordings were acquired at 20 kHz, low-pass filtered at 2 kHz, and were recorded at a holding potential of −80 mV unless otherwise noted.
Electrophysiology data were analyzed with Igor (Wavemetric) and QuB Express (49). Dose–response curves from macroscopic currents were fit with the Hill equation,
| [1] |
where Imax is the maximal current at saturating concentrations of cAMP, K1/2 is the concentration of cAMP producing half-maximal current, and h is the Hill coefficient. Single-channel PO values at −80 mV for cfSthK and cfSthK–A208V were determined from fits to the all-points histograms of single-channel time traces with the sum of two Gaussian distributions. The PO of cfSthK–A208V at 0 mV was estimated from the conductance–voltage relationship of WT SthK in E. coli spheroplasts reported previously (28).
EPR Experiments.
Continuous-wave EPR spectra were recorded at room temperature on a Bruker EMX spectrometer operating at X-band frequency (∼9.8 GHz) and equipped with a Bruker ER 4123D dielectric resonator. Spectra were recorded with 100-kHz field modulation with a sweep rate of 1.8 G/s and a modulation amplitude of 2 G. CW EPR spectra were background subtracted and baseline corrected in LabVIEW with a routine written by Eric D. Walter. Spin concentrations were calculated by double integration of the field-modulated spectrum and comparison to a standard curve of 4-hydroxy TEMPO free radical (Sigma). Labeling efficiencies were calculated as the spin concentration obtained by double integration divided by the total protein concentration obtained from optical absorbance at 280 nm, using an extinction coefficient for 6xHis-SthK of 49,640 M−1·cm−1.
Pulsed EPR experiments were performed at Q-band frequency (∼34 GHz) using a Bruker EleXsys E580 spectrometer equipped with an overcoupled Bruker EN 5107D2 resonator. Pulses were generated with a Bruker SpinJet AWG and amplified with a 300 W TWT amplifier. DEER experiments were performed at 50 K using a variable-temperature cryogen-free system (Bruker). The deadtime-free, four-pulse DEER sequence [(𝜋/2)probe ― 𝜏1 ― (𝜋)probe ― 𝜏1 + t ― (𝜋)pump ― 𝜏2 – t ― (𝜋)probe ― 𝜏2 ― (echo)] was employed with a 400-ns 𝜏1 delay and 𝜏2 delays ranging from 3,200 ns to 7,000 ns depending on the sample (SI Appendix, Fig. S3) (50). Probe pulses were 30 ns (𝜋/2) and 60 ns (𝜋) Gaussian-shaped pulses at a frequency corresponding to the maximum of the resonator response function and a magnetic field value corresponding to the high-field shoulder of the echo-detected field-swept spectrum (SI Appendix, Fig. S3) (51). The pump pulse was implemented as a 200-ns sech/tanh pulse centered at a frequency 80 MHz higher than the probe frequency and corresponding to the maximum of the nitroxide field-swept spectrum (SI Appendix, Fig. S3) (52). The pump pulse had a bandwidth of 80 MHz, a truncation parameter (β) of 4, and was not compensated for the resonator bandwidth. Raw time-domain DEER traces were background-corrected using LongDistances (by Christian Altenbach, available at www.biochemistry.ucla.edu/Faculty/Hubbell), and the resulting signals were power-scaled in MATLAB to suppress sum and difference peaks arising from multispin effects (SI Appendix, Fig. S5) (53). Distance distributions were then calculated from the scaled and background-corrected DEER traces by Tikhonov regularization in LongDistances. Error bands in the distance distribution were calculated by stochastic addition of random noise to the DEER signal and variation of the background parameters followed by recalculation of the distance distribution using the error analysis feature of LongDistances with default values. Error bands are plotted as the mean ±1 SD from 100 independent calculations with parameters varied as described above. The raw time-domain DEER traces (phase-corrected) for all experiments are provided as Dataset S1. All plots were generated in KaleidaGraph (Synergy Software).
Rosetta Modeling.
For all modeling, C4 symmetry was assumed and implemented into Rosetta as previously described (54). Starting models for DEER-constrained Rosetta modeling were generated using the RosettaCM routine (37). For the ligand-free model, the cryo-EM structure of SthK in the absence of ligand (PDB ID code 6CJQ) (9) was used as the sole template structure. Residues of the S2-S3 loop that were unresolved in the cryo-EM structure were modeled de novo, as were unresolved or missing residues of the C-helix, in order to include all amino acids known to interact with the cyclic nucleotide ligand. For the starting cAMP-bound model, we used both the cryo-EM structure of SthK with cAMP bound (PDB ID code 6CJU) (9) and the crystal structure of the SthK cytoplasmic domain with cAMP bound (PDB ID code 3D7T) (24) as template structures. One hundred twenty models were calculated with RosettaCM for ligand-free and cAMP-bound states, and the 10 best-scoring models for each condition were manually examined. Best-scoring models that retained core-packing interactions known to exist based on the cryo-EM and crystal structures of SthK were selected for further refinement. Final models for the ligand-free and cAMP-bound conformations of SthK were generated with the RosettaRelax routine with experimental DEER constraints imposed as Cβ-Cβ distance constraints (SI Appendix, Fig. S7 and Datasets S2 and S3). Models were run with atom-pair constraints on all Cα atoms within 8 Å and separated in primary sequence by at least four residues. These were “top-out”constraints, which are roughly harmonic up to 1 Å from the minimum, and flat beyond. The weights of these constraints were increased until Cα rmsd ≤2 Å was achieved over the full tetramer in the absence of DEER-derived constraints. Cβ-Cβ distance constraints derived from DEER were implemented as harmonic constraints with a variance (𝜎) corresponding to the half-width at half-maximal probability of the DEER distributions. Center values of the harmonic constraints in the ligand-free harmonic models were set to the Cβ-Cβ distances from the ligand-free cryo-EM structure (9), and constraints for the cAMP-bound models were set at a value ΔrDEER from the cryo-EM Cβ-Cβ distances (SI Appendix, Fig. S7). One hundred models were generated for each condition and ranked by their Rosetta score.
Data Availability.
The raw time-domain DEER traces (phase-corrected) for all experiments are provided as Dataset S1. The constraint files used in the structural modeling are provided as Datasets S2 and S3.
Supplementary Material
Acknowledgments
We thank Dr. Zach James for invaluable discussions and materials, and all the members of the S.S. and W.N.Z. laboratories for helpful conversations and support. This work was supported by the Raymond and Beverly Sackler Scholars Program in Integrative Biophysics (fellowship to E.G.B.E.); NIH Training Grants T32-EY007031 (to E.G.B.E.) and T32-HL007312 (to J.L.W.M.); and NIH Grants R01-EY010329 (to W.N.Z.) and R01-GM127325 (to W.N.Z. and S.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916375117/-/DCSupplemental.
References
- 1.Craven K. B., Zagotta W. N., CNG and HCN channels: Two peas, one pod. Annu. Rev. Physiol. 68, 375–401 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Fesenko E. E., Kolesnikov S. S., Lyubarsky A. L., Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature 313, 310–313 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Michalakis S., Becirovic E., Biel M., Retinal cyclic nucleotide-gated channels: From pathophysiology to therapy. Int. J. Mol. Sci. 19, E749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biel M., Michalakis S., Function and dysfunction of CNG channels: Insights from channelopathies and mouse models. Mol. Neurobiol. 35, 266–277 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Biel M., Wahl-Schott C., Michalakis S., Zong X., Hyperpolarization-activated cation channels: From genes to function. Physiol. Rev. 89, 847–885 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Lee C.-H., MacKinnon R., Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M. et al., Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James Z. M. et al., CryoEM structure of a prokaryotic cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 114, 4430–4435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rheinberger J., Gao X., Schmidpeter P. A., Nimigean C. M., Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. eLife 7, e39775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn G. E., Zagotta W. N., Insights into the molecular mechanism for hyperpolarization-dependent activation of HCN channels. Proc. Natl. Acad. Sci. U.S.A. 115, E8086–E8095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James Z. M., Zagotta W. N., Structural insights into the mechanisms of CNBD channel function. J. Gen. Physiol. 150, 225–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai G., Aman T. K., DiMaio F., Zagotta W. N., The HCN channel voltage sensor undergoes a large downward motion during hyperpolarization. Nat. Struct. Mol. Biol. 26, 686–694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lörinczi É. et al., Voltage-dependent gating of KCNH potassium channels lacking a covalent link between voltage-sensing and pore domains. Nat. Commun. 6, 6672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whicher J. R., MacKinnon R., Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matulef K., Flynn G. E., Zagotta W. N., Molecular rearrangements in the ligand-binding domain of cyclic nucleotide-gated channels. Neuron 24, 443–452 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Taraska J. W., Puljung M. C., Olivier N. B., Flynn G. E., Zagotta W. N., Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat. Methods 6, 532–537 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puljung M. C., Zagotta W. N., A secondary structural transition in the C-helix promotes gating of cyclic nucleotide-regulated ion channels. J. Biol. Chem. 288, 12944–12956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puljung M. C., DeBerg H. A., Zagotta W. N., Stoll S., Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. U.S.A. 111, 9816–9821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craven K. B., Zagotta W. N., Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J. Gen. Physiol. 124, 663–677 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zagotta W. N. et al., Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 425, 200–205 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Johnson J. P. Jr., Zagotta W. N., The carboxyl-terminal region of cyclic nucleotide-modulated channels is a gating ring, not a permeation path. Proc. Natl. Acad. Sci. U.S.A. 102, 2742–2747 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., MacKinnon R., Cryo-EM structure of the open human ether-à-go-go-related K+ channel hERG. Cell 169, 422–430.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brams M., Kusch J., Spurny R., Benndorf K., Ulens C., Family of prokaryote cyclic nucleotide-modulated ion channels. Proc. Natl. Acad. Sci. U.S.A. 111, 7855–7860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kesters D. et al., Structure of the SthK carboxy-terminal region reveals a gating mechanism for cyclic nucleotide-modulated ion channels. PLoS One 10, e0116369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowal J. et al., Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1. Nat. Commun. 5, 3106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidpeter P. A. M., Gao X., Uphadyay V., Rheinberger J., Nimigean C. M., Ligand binding and activation properties of the purified bacterial cyclic nucleotide-gated channel SthK. J. Gen. Physiol. 150, 821–834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchesi A. et al., An iris diaphragm mechanism to gate a cyclic nucleotide-gated ion channel. Nat. Commun. 9, 3978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan J. L. W., Evans E. G. B., Zagotta W. N., Functional characterization and optimization of a bacterial cyclic nucleotide-gated channel. J. Biol. Chem. 294, 7503–7515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craven K. B., Olivier N. B., Zagotta W. N., C-terminal movement during gating in cyclic nucleotide-modulated channels. J. Biol. Chem. 283, 14728–14738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeschke G., DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Martens C. et al., Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 23, 744–751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Eps N. et al., Conformational equilibria of light-activated rhodopsin in nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 114, E3268–E3275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grinkova Y. V., Denisov I. G., Sligar S. G., Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng. Des. Sel. 23, 843–848 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D. T., Tibbs G. R., Paoletti P., Siegelbaum S. A., Constraining ligand-binding site stoichiometry suggests that a cyclic nucleotide-gated channel is composed of two functional dimers. Neuron 21, 235–248 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Ulens C., Siegelbaum S. A., Regulation of hyperpolarization-activated HCN channels by cAMP through a gating switch in binding domain symmetry. Neuron 40, 959–970 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Rohl C. A., Strauss C. E. M., Misura K. M. S., Baker D., Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Bender B. J. et al., Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry 55, 4748–4763 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Y. et al., High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varnum M. D., Black K. D., Zagotta W. N., Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron 15, 619–625 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Akimoto M. et al., A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP. J. Biol. Chem. 289, 22205–22220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldschen-Ohm M. P. et al., Structure and dynamics underlying elementary ligand binding events in human pacemaking channels. eLife 5, e20797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saponaro A. et al., Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. U.S.A. 111, 14577–14582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon S. E., Zagotta W. N., A histidine residue associated with the gate of the cyclic nucleotide-activated channels in rod photoreceptors. Neuron 14, 177–183 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Aman T. K., Gordon S. E., Zagotta W. N., Regulation of CNGA1 channel gating by interactions with the membrane. J. Biol. Chem. 291, 9939–9947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karpen J. W., Brown R. L., Stryer L., Baylor D. A., Interactions between divalent cations and the gating machinery of cyclic GMP-activated channels in salamander retinal rods. J. Gen. Physiol. 101, 1–25 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., Naismith J. H., An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie T. K. et al., Chapter 11–Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Nicolai C., Sachs F., Solving ion channel kinetics with the QuB software. Biophys. Rev. Lett. 8, 191–211 (2013). [Google Scholar]
- 50.Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W., Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Teucher M., Bordignon E., Improved signal fidelity in 4-pulse DEER with Gaussian pulses. J. Magn. Reson. 296, 103–111 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Spindler P. E., Glaser S. J., Skinner T. E., Prisner T. F., Broadband inversion PELDOR spectroscopy with partially adiabatic shaped pulses. Angew. Chem. Int. Ed. Engl. 52, 3425–3429 (2013). [DOI] [PubMed] [Google Scholar]
- 53.von Hagens T., Polyhach Y., Sajid M., Godt A., Jeschke G., Suppression of ghost distances in multiple-spin double electron-electron resonance. Phys. Chem. Chem. Phys. 15, 5854–5866 (2013). [DOI] [PubMed] [Google Scholar]
- 54.DiMaio F., Leaver-Fay A., Bradley P., Baker D., André I., Modeling symmetric macromolecular structures in Rosetta3. PLoS One 6, e20450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw time-domain DEER traces (phase-corrected) for all experiments are provided as Dataset S1. The constraint files used in the structural modeling are provided as Datasets S2 and S3.