Significance
This study investigates how proteins with new biochemical and biological activities arise. Venom toxins are good examples of biochemical novelties because many different kinds of toxins have evolved in different kinds of venomous animals to subdue prey. We uncover a series of genetic events involved in the genesis of an important family of toxins in rattlesnakes, the metalloproteinases. We trace the origin of this family to a single locus in reptiles that was duplicated, modified, and massively expanded by 30 genes in the course of rattlesnake evolution. We suggest that one potential evolutionary force driving this expansion was selection for increased production of toxin.
Keywords: evolution, gene duplication, novelty, venom
Abstract
The genetic origins of novelty are a central interest of evolutionary biology. Most new proteins evolve from preexisting proteins but the evolutionary path from ancestral gene to novel protein is challenging to trace, and therefore the requirements for and order of coding sequence changes, expression changes, or gene duplication are not clear. Snake venoms are important novel traits that are comprised of toxins derived from several distinct protein families, but the genomic and evolutionary origins of most venom components are not understood. Here, we have traced the origin and diversification of one prominent family, the snake venom metalloproteinases (SVMPs) that play key roles in subduing prey in many vipers. Genomic analyses of several rattlesnake (Crotalus) species revealed the SVMP family massively expanded from a single, deeply conserved adam28 disintegrin and metalloproteinase gene, to as many as 31 tandem genes in the Western Diamondback rattlesnake (Crotalus atrox) through a number of single gene and multigene duplication events. Furthermore, we identified a series of stepwise intragenic deletions that occurred at different times in the course of gene family expansion and gave rise to the three major classes of secreted SVMP toxins by sequential removal of a membrane-tethering domain, the cysteine-rich domain, and a disintegrin domain, respectively. Finally, we show that gene deletion has further shaped the SVMP complex within rattlesnakes, creating both fusion genes and substantially reduced gene complexes. These results indicate that gene duplication and intragenic deletion played essential roles in the origin and diversification of these novel biochemical weapons.
Evolutionary novelties enable species to adopt new lifestyles, exploit new niches, and can spur adaptive radiations. Such traits may entail the formation of new morphological features or new biochemical activities, or both. Because of their importance in shaping the course of evolution across the tree of life, the genetic mechanisms underlying the origins of novelties have long been of special interest in evolutionary biology.
Most new protein functions appear to evolve from preexisting proteins via varying degrees of modification (1, 2). But the relative contributions of structural changes, expression changes, and gene duplication to the origin of novel functions are not well understood in general, and the order of such events in the origin of any particular novel protein are challenging to untangle.
Since Susumu Ohno’s landmark treatise 50 y ago (3), gene duplication has played a predominant role in models explaining the origins of biochemical novelties. Ohno proposed that gene duplication was a necessary prerequisite to the origin of new functions because the duplication of a gene would free one copy to incorporate new functional mutations (neofunctionalization), while the other copy preserved the ancestral function. The widespread distribution of tandemly duplicated and diversified gene complexes appeared to support a strong link between gene duplication and biochemical novelty.
However, one serious challenge for the neofunctionalization model is that the newly duplicated gene is assumed to be neutral and must acquire new, innovative, selectable, and presumably rare mutations (4), before acquiring inactivating mutations (deletions, frameshifts, nonsense mutations) which would be more likely (5, 6). It is now appreciated that gene duplications may occur and be preserved without the generation of any novelty. In the duplication-degeneration-complementation (DDC) model, Force et al. (5) posit that one outcome of gene duplication is for the two copies to each accumulate mutations that inactivate any separable functional or regulatory elements (degeneration) and thus subdivide the function of the ancestral gene among its daughter genes (complementation). The DDC model thus accounts for the maintenance of gene duplicates but does not explain how a gene with a new function might evolve.
To surmount the theoretical issues surrounding novel mutations arising after duplication, alternative models have been proposed and empirical examples have been demonstrated in which innovative mutations occur prior to duplication, or without duplication occurring at all. For example, in a circumstance dubbed “gene sharing” vertebrate crystallin proteins evolved from various proteins by gaining very high expression levels in the lens while maintaining their expression in nonlens tissues, that is, without gene duplication (7).
In the escape from adaptive conflict (EAC) model, a protein with dual activities is constrained from acquiring mutations that optimize either function due to negative effects on the second function. This “adaptive conflict” can be relieved by gene duplication which frees the separate copies to improve each function by acquiring previously forbidden mutations (8), as has been shown, for example, in the case of the yeast Gal1/Gal3 galactokinase/coinducer proteins (9). Similarly, the innovation-amplification-divergence model (IAD) begins with innovative mutations, which provide a weak, secondary function. Selection then drives the amplification of gene number via the retention of gene duplicates, which boost the activity of the weak secondary function, and duplicates in turn may acquire additional mutations that optimize the novel function (10). The operational difference between EAC and IAD appears to be mostly a matter of the degree to which a second function has evolved. The origin of one novelty, of antifreeze proteins in the Antarctic eelpout from an ancestral sialic acid synthase (SAS) protein, has even been cited as an example of both models (1, 11). Because the genomic origins of biochemical novelties have been traced for only a handful of prominent examples, the relative prevalence of any particular evolutionary path remains unknown.
Snake venoms are biochemical novelties comprised of mixtures of proteins with a diverse array of molecular functions and physiological effects. Snake toxins belong to about two dozen well-established protein families containing many nonvenom proteins, and it appears that representatives of different protein families became incorporated into venoms at different times during the evolution of major venomous snake lineages (12). With respect to the evolution of novelty, there are several major mechanistic questions to address including: 1) What is the genomic origin of individual toxins?; 2) what changes have occurred to protein structure and activity?; and 3) how did expression in the venom gland arise?
Like other biochemical novelties, tracing the origins of venom genes and proteins (from nonvenom genes or proteins) requires high-quality genomic data, which has been sparse for snakes until recently. It has been shown, for example, that the array of crotalid phospholipase A2 toxin genes evolved from a single, snake-restricted ancestral gene (13), and that certain subsequent mutations were key to the genesis of a novel heterodimeric neurotoxin (14). But the genetic origins of most other snake toxins have not been elucidated.
The snake venom metalloproteinases (SVMPs) are a family of multidomain proteins that are highly abundant in viperid venoms (∼50% of toxin proteins in Crotalus atrox) (15) and to a much lesser degree in elapid venom (16, 17). There are three main classes of SVMPs (P-I, P-II, and P-III) that differ by the presence/absence of three distinct domains: all SVMPs contain a metalloproteinase domain; class P-III SVMPs additionally possess both disintegrin-like and C-terminal cysteine-rich domains; class P-II proteins lack the cysteine-rich domain, but possess a disintegrin domain; and P-I SVMPs possess only the metalloproteinase domain (18, 19).
The SVMPs are in turn part of a large and diverse family of vertebrate metalloproteinase proteins known as the reprolysin or ADAM (a disintegrin and metalloproteinase) protein family. In addition to the metalloproteinase, disintegrin, and cysteine-rich (MDC) domains present in the SVMPs, most ADAMs also have an EGF-repeat, a transmembrane domain, and a cytoplasmic tail (20). Previous work has shown that the SVMPs form a monophyletic group that is most closely related to the vertebrate ADAM7, ADAM28, and decysin-1 proteins (12, 21). Casewell (21) utilized phylogenetic analysis of viperid venom-expressed SVMPs to reveal the monophyletic origin of P-II SVMPs from a P-III SVMP ancestor, but P-I SVMPs appear to have arisen independently at least eight times from class P-II ancestors. The large number of SVMP proteins and their diverse activities have prompted many authors to suggest a role for gene duplication and sequence divergence during the expansion of advanced snakes (16, 19, 22–24). In the king cobra (an elapid), Vonk et al. (16) found two venom-expressed SVMPs adjacent to the ADAM28 locus and suggested that the two cobra SVMPs evolved by duplication from the ADAM28 ancestor. However, the genomic origins of the diverse array of viperid SVMPs are not well understood. This is primarily because of limited genomic data which is essential for a detailed understanding of SVMPs and related adam28 loci, and not attainable solely through examination of expressed transcripts or protein products.
Here, we traced the origin and diversification of the SVMP gene family in the Crotalus genus. We show that a large tandem array of SVMP genes expanded from a single nonvenom ancestral adam28 gene and detail the subsequent, stepwise duplication and intragenic deletion events that generated the three classes of SVMP genes. We further show how additional whole gene deletions modified the SVMP complex (and venom composition) within rattlesnakes, creating both fusion genes and substantially reduced gene complexes.
Results
The C. atrox SVMP Complex Contains 31 Tandemly Arrayed ADAM-like Metalloproteinases.
About 50% of the toxin proteins in C. atrox venom are metalloproteinases (MPs) (15). Several proteomic and transcriptomic studies identified at least eight distinct variants, including members from each of the three classes (P-I, P-II, and P-III) (25–28). Therefore, at the outset, we anticipated finding at least eight SVMP loci in the C. atrox genome and hoped to identify closely related gene family members that are not expressed in venom.
In order to minimize potential gaps in sequence coverage or ambiguities about gene number, we elected to screen a bacterial artificial chromosome (BAC) library made from C. atrox to physically isolate large genomic DNA fragments containing C. atrox SVMP genes and then sequence those clones with PacBio long-read technology (13). To our surprise, we discovered 30 SVMP genes with intact open reading frames (ORFs) tandemly arrayed in a single complex spanning 1.3 Mb of genomic sequence (Fig. 1A and SI Appendix, Fig. S1). Each of the 30 SVMP gene loci spans between 17 and 30 kb. Interspersed between the 31 genes of the complex are 12 sets of orphaned SVMP exons (SI Appendix, Fig. S1). These exons exist singly or in groups of 2 to 3, possibly as remnants of past duplications and/or deletions of SVMPs, as many exons contain degenerate splice junctions and/or frame shift mutations. The complex also contains a considerable number of transposable elements, with over 10% of the 1.3-Mb complex deriving from retroelements, with long interspersed nuclear element (LINE)/CR1 elements being the most abundant.
Fig. 1.
The C. atrox SVMP complex contains 30 tandemly arrayed genes. (A) Schematic of the C. atrox SVMP complex. Gene loci are represented by arrows with each color representing a different SVMP class (ADAM: dark blue; P-III: green; P-II: yellow/orange; P-I: red). The small rectangles and triangles interspersed among the genes represent orphaned SVMP exons. (B) The venom protein composition of the animal used for genome sequencing was characterized by mass spectrometry (Dataset S3). A colored box below a gene arrow indicates the genes for which unique peptides were identified. Shared peptide hits unique to a pair of highly similar genes (mad-5a/b) are represented by a connection between two colored boxes. (C) Colored boxes denote matches to protein database entries from prior studies on C. atrox venom (Dataset S1). The white box represents a second mpo paralog, atrolysin C (Atr-C), found by Hite et al. (25), which we did not find in our snake. (D) Transcript abundance in the venom gland is calculated by RNAseq by expectation-maximization (RSEM) with FPKM values plotted on an inverted graph. Expression levels differ greatly across the gene complex but adam28 and mdc-1 transcripts are inactive. Overlapping the FPKM graph is the number of peptides that map uniquely, with 100% identity, to a given gene plotted with blue bars. Colored lines above the schematic represent segmental duplications.
We sought to identify which among this unexpectedly large set of genes encoded previously identified C. atrox SVMPs, and which were previously unknown. This task was challenging because disparate naming strategies over decades of research have led to a confusing, often species-specific nomenclature for snake SVMP genes without incorporating orthology/paralogy relationships. Therefore, we integrated historical precedent with our genomic and phylogenetic data to develop a unified nomenclature that both adheres to the critical SVMP class designation and enables evolutionarily informed comparisons across species (see Fig. 1 and below).
We constructed protein phylogenies using hypothetical translations of the C. atrox SVMP complex genes and known venom metalloproteases from other viperid and elapid species. Most database entries for known SVMP proteins derive largely from sequenced venom peptides or venom gland cDNAs; we also utilized the hypothetical translations of genomic sequences from three additional Crotalus species, a European viper (Vipera berus) and the king cobra (Ophiophagus hannah) to capture a more expansive set of snake SVMP genes (Dataset S1).
At the base of our phylogenetic tree is a clade that contains a set of 25-exon metalloproteinase genes structurally similar to the mammalian adam28 gene, (ADAM; dark blue, Fig. 2). The P-III SVMPs then populate a series of well-supported clades of 17-exon genes, which we designate the mdc genes (metalloproteinase, disintegrin, and cysteine-rich). The most basal of these, mdc-1 (Fig. 2, light blue), share a gene structure with venom SVMPs but mdc-1 transcripts or peptides have not been detected in any published viperid venom gland transcriptomes or venom proteomes. The orthologs of adam28 and mdc-1 are physically adjacent to each other at the 5′ end of the C. atrox complex (Fig. 1A).
Fig. 2.
Protein phylogeny of snake SVMP proteins. Protein phylogeny of full-length SVMP proteins using sequences from available protein databases and hypothetical translations from our gene models. SVMP classes (P-I, P-II, and P-III) are color coded (ADAM: blue; P-III/ MDC: green shades: P-II/ MAD: orange shades; MPO/ P-I: red). Gene paralogs are numbered (e.g., atx_MDC-4, atx_MAD-6), and are part of distinct, well-supported clades (bootstrap value >85) containing entries from more than one species. Two or more genes that are found to have high sequence identity (>90%), fall into a shared paralog group, or that appear to be part of a segmental duplication are considered duplicates and denoted with a letter at the end of the gene name (i.e., atx_MDC-5a, atx_MDC-5b). atx, C. atrox; dur, C. durissus; horA, C. horridus (neurotoxic [A type]); horB, C. horridus (hemorrhagic [B type]); scuA, C. scutulatus (neurotoxic [A type]); scuB, C. scutulatus (hemorrhagic [B type]); Vber, V. berus; Ohan, O. hannah; mus, Mus musculus; Lcha, Latimeria chalumnae. Green star indicates the well-supported clade of all viperid SVMPs excluding ADAM28 and MDC-1. For a full list of protein sequences and references see Dataset S1.
The remaining viperid SVMPs, including all known venom genes, form a large, well-supported monophyletic clade (Fig. 2, green star) that includes all of the remaining genes in the C. atrox SVMP complex. The majority of P-II genes fall into a weakly supported monophyletic group of genes distinct from the P-III mdc genes (Fig. 2, green wedge) that we designate as the mad genes (metalloproteinase and disintegrin) (Fig. 2, orange wedge). The two notable exceptions, which we call the mpf genes (mp fusion), result from the fusion of genomic regions from a mdc and mad gene that could make a hypothetical P-II product (see below, SI Appendix, Fig. S2). The Crotalus P-I MPs, which we designate mpo (metalloproteinase only) (Fig. 2, red wedge), form a monophyletic clade nested within the mad genes. Altogether, the C. atrox SVMP complex contains 16 P-III mdc MPs, 11 P-II mad genes, two P-II mpf genes, a single P-I mpo gene, and a single adam28 gene (Figs. 1 and 2). In C. atrox there are multiple paralogs that fall into the same clade, are highly similar, and appear to be recent duplicates. Most of these have arisen as products of multigene duplications involving three separate clusters of three genes (Fig. 1, mdc-3a:mad-1a:mad-2a/mdc-3b:mad-1b:mad-2b, mad-3a:mad-4a:mdc-5a/mad-3b:mad-4b:mdc-5b, and mad-5a:mpf-1a:mdc-8a/mad-5b:mpf-1b:mdc-8b).
A Large Subset of SVMP Genes Is Expressed in the Venom Gland.
The larger than expected number of SVMP genes raised the question of whether any previously unknown genes are expressed in the venom gland. To identify which genes encoded proteins expressed in venom, peptides generated by mass spectrometry of venom from the same individual snake used for genomic analysis were mapped to hypothetical translations from the 31 gene loci. Despite the high level of conservation between SVMP paralogs, we were able to assign unique peptides to 14 of the 31 genes (Fig. 1B, boxes; Datasets S3 and S4) plus an additional set of peptides that map uniquely to a pair of nearly identical gene duplicates (mad-5a/b) (Fig. 1B, linked boxes; Dataset S3). This family of 15 to 16 expressed SVMPs is larger than the 8 C. atrox SVMPs detected across previous proteomic studies (Fig. 1C) (15, 26, 29, 30), 7 of which have a high identity match to a gene in this SVMP complex (Fig. 1C; we did not detect unique peptides corresponding to atrolysin-C/D, see below).
The absence of protein expression of any SVMP could be due to the lack of transcription or due to posttranscriptional mechanisms. To examine which loci were transcriptionally active, we sequenced venom gland cDNA fragments (RNA-sequencing [RNA-seq]) isolated from this specific individual snake and mapped the reads to the gene complex. We found that the majority of venom RNA expression derives from mpo-1 (P-I) and mdc-4 (P-III) but there is also expression from other loci such as mad-3a/b (P-II) and mdc-2 (P-III) (Fig. 1D). Expression levels (fragments per kilobase of transcript per million mapped reads [FPKM]) above 2,000 are found for 8 genes, and this expression level correlates with detection in venom of the corresponding protein products. If we consider genes with expression levels of FPKM greater than 200, then over half of the 31-gene complex is transcriptionally active in this single animal.
The one previously reported SVMP that we did not detect is the P-I class SVMP atrolysin-C/D (25). Our SVMP complex has a single class P-I SVMP (mpo-1) that is nearly identical to atrolysin-B and mass spectrometry analysis recovered unique peptide hits to atrolysin-B (mpo-1), but not to atrolysin-C/D. Furthermore, we found no unique RNA reads that corresponded to atrolysin-C/D. We also performed genomic PCR but found no evidence of a second, class P-I gene in this individual.
The Secreted C. atrox SVMPs Are Descended from an Ancestral, Transmembrane ADAM Metalloproteinase.
The number and diversity of SVMP genes and proteins expressed in C. atrox venom raises the fundamental question of their evolutionary origin(s). The tandem arrangement of SVMP genes is evidence of gene duplication, but which gene(s) is the ancestor(s) and which is the descendant(s)?
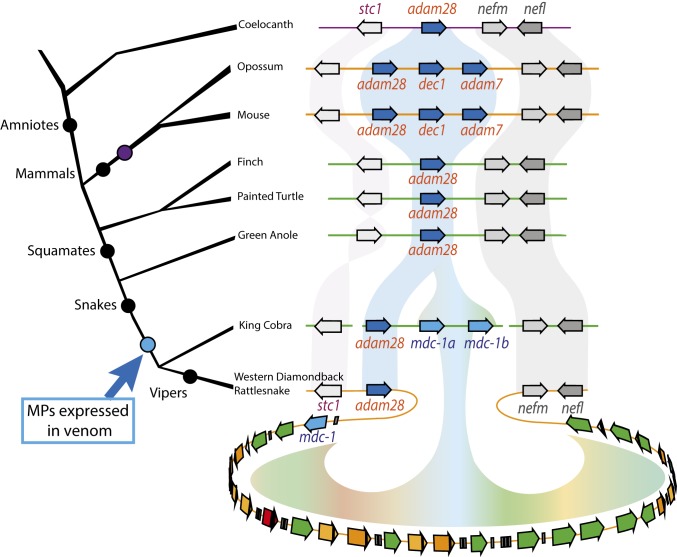
A key observation is the presence of two, nonvenom-expressed SVMP genes adam28 and mdc-1 at the 5′ end of the complex, immediately adjacent to the highly expressed mdc-2 (vap1) gene (Fig. 1) (28). We find this same arrangement of adam28 and mdc-1 in other crotalids (e.g., Crotalus scutulatus) (31) and in other snake species we have examined including the king cobra (an elapid) (Fig. 3). In addition, the SVMP genes are flanked on the 5′ end of the complex by stanniocalcin 1 (stc1) and on the 3′ end of the complex by the gene pair neurofilament light/medium (nefl/nefm). This synteny is conserved in the genomes of other reptiles (painted turtle, anole), birds (ground finch), and the coelacanth, where a single adam28 homolog is flanked by stc1 and nefl/nefm, as well as in mammals (mouse, opossum) where an independently derived cluster of adam28 paralogs is situated between stc1 and nefl/nefm (Fig. 3) (32). Taken together, these syntenic relationships and the SVMP phylogenetic tree robustly confirm the prior suggestion that adam28 is the closest ancestor of SVMPs (16), and show that the locus is the direct ancestor of the massive expansion of SVMPs found in C. atrox.
Fig. 3.
The SVMP complex expanded from a single adam28 gene. Schematized vertebrate phylogeny showing representative genomic regions for the adam28 locus and surrounding genes. The adam28 gene (dark blue arrow) and its homologs remain in a conserved syntenic arrangement with flanking genes stc1 (white) and nefm/nefl (gray). A single adam28 gene is found in the genomes of other reptiles and birds. An independent expansion (purple dot) in the mammalian lineage resulted in three ADAM genes (adam28, dec1, and adam7) (32). mdc genes (blue) are found in king cobra (O. hannah) (16). The SVMP complex of vipers has undergone massive expansion as exemplified by the 30 SVMP genes of the C. atrox (Western diamondback rattlesnake) complex.
A Series of Stepwise Deletions Have Shaped SVMP Diversity.
It is important to note, however, that snake venom SVMPs are secreted molecules, whereas adam28, and the majority of other mammalian adam genes encode membrane-bound metalloproteinases (33, 34). In addition, while ADAMs and P-III class snake venom SVMPs possess three domains in common (a metalloproteinase domain, a disintegrin domain, and a cysteine-rich domain), P-II and P-I class MPs lack one or two of these domains, respectively (19). Therefore, it is possible that structural changes may have occurred to the adam28 gene or its descendants in order to: 1) evolve secreted SVMPs that could diffuse within envenomated prey and 2) generate SVMPs with fewer domains. We sought to trace the possible genomic changes that generated the various forms of venom MPs.
Detailed inspection of the 25-exon adam28 gene reveals that it encodes a transmembrane domain that is absent from all adjacent SVMP genes (Fig. 4A). Evolving a soluble SVMP from a membrane-bound ancestor would appear then to have been a critical step in venom SVMP evolution. Comparison of the C. atrox adam28 and mdc-1 loci reveals homologous stretches of sequence and a similar exon/intron structure up to the 17th exon. However, that homology ends midway through the 17th exon, coincident with a novel early stop codon in mdc-1. Furthermore, immediately 3′ of the 17th exon of mdc-1 is a cluster of three long terminal repeat of an endogenous retrovirus (LTR/ERV) transposable elements and a Line/CR1 element. A Line/CR1 element is also located 3′ of the adam28 locus, making it a candidate boundary of a genomic deletion that eliminated the transmembrane domain coding exons and gave rise to a secretable SVMP (Fig. 4B). Based on these observations, we infer that the P-III class mdc-1 gene arose via: 1) duplication of part of the adam28 locus or 2) duplication of the entire adam28 locus followed by the subsequent intragenic deletion of part of exon 17 through exon 25.
Fig. 4.
Stepwise intragenic deletions gave rise to the three classes of MP. Schematics of the genomic regions from exon 12 to the 3′ ends of multiple snake SVMP genes are shown. At top and bottom is a generic SVMP protein with colored blocks showing which exons contribute to the distinct protein domains (red, metalloproteinase domain; orange, disintegrin domain; green, cysteine-rich domain; blue, transmembrane domain). (A) The C. atrox adam28 gene has 25 coding exons, including several that encode a putative transmembrane domain (exons 18 to 24). (B) P-III SVMP (mdc) genes from the king cobra (O. hannah), European viper (V. berus), and C. atrox share a shortened (compared to the adam28 gene) 17th exon with a stop codon. A LTR/ERV transposable element (brown circle) adjacent to a Line/CR1 element (pink circle) exists 3′ to the 17th exon in all snake SVMP genes analyzed, except adam28 homologs. (C) P-II SVMP (mad) genes from vipers and C. atrox share an identical 4-kb deletion (∆Cys-4k) that interrupts exon 14 and extends to exon 16. The deletion results in a stop codon that immediately follows the disintegrin domain coding sequence effectively deleting the cysteine-rich domain (see also SI Appendix, Fig. S3). (D) The P-I SVMP (mpo-1) genes from C. atrox and C. durissus have, in addition to the ∆Cys-4k deletion of mad genes (above), a stop codon in the 12th exon (red octagon) and an ∼2-kb deletion (∆Dis-2k) that extends from just after exon 12 into exon 14, which effectively removes all of the exons that encode the disintegrin domain.
We note, however, that while mdc-1 is present in the genomes of at least five Crotalids, it is not expressed in the C. atrox venom gland (Fig. 1D) nor has it been reported in any Crotalid venom gland transcriptomes or proteomes (35–39). Therefore, gene duplication and partial deletion alone are not sufficient to account for the genesis of a P-III class venom MP. Rather, since the adjacent mdc-2 gene is highly expressed in venom, we infer that both a duplication event and a gain of gene expression in the venom gland were required.
The 11 P-II (mad) SVMPs genes found in the C. atrox SVMP complex encode proteins that lack the C-terminal cysteine-rich domain present in class P-III proteins. Casewell et al. (18) and our phylogenetic analyses suggest that all class P-II (mad) SVMPs are derived from a single P-III (mdc) ancestor, but the support is not robust [Fig. 2, bootstrap <70 (18), posterior probability <0.95]. Therefore, we also traced the origin of these genes by detailed comparisons of C. atrox P-II and P-III SVMP gene sequence and structure. We discovered a shared ∼4.6-kb genomic deletion extending from the middle of the 14th exon to the end of the 16th exon in all P-II genes (∆Cys-4k, Fig. 4C). This deletion generates a premature stop codon in the 14th exon in the region between the encoded disintegrin and cysteine-rich domains (SI Appendix, Fig. S3). This deletion is also shared across all Crotalus class P-II genes as well as those from two distantly related vipers, V. berus and Echis ocellatus (40). We conclude that these conserved genomic features support a single evolutionary origin of the P-II class of MPs.
The P-I class SVMPs encode proteins that lack both cysteine-rich and disintegrin domains due to a stop codon in the 12th exon at the end of the metalloproteinase domain. Detailed examination of the C. atrox mpo-1 locus reveals that it also bears the ∆Cys-4k deletion that is characteristic of the P-II clade of MPs, as well as a second deletion which spans from the 12th exon to the beginning of the 14th exon (∆Dis-2k, Fig. 4D). This deletion leaves a small genomic remnant of the 14th exon, effectively removing the entire disintegrin domain. This pair of deletions is shared by the P-I SVMPs from at least two other Crotalus species, Crotalus durissus (Fig. 4D) and Crotalus helleri (SI Appendix, Fig. S3). These rare genomic events serve as useful landmarks in tracing the evolutionary history of the complex; ∆Cys-4k marks all known viperid P-II mad genes as descendants from a single P-III mdc ancestor while the ∆Dis-2k + stop codon is characteristic of the Crotalus P-I mpo genes. The ∆Dis-2k-derived P-I mpo molecules from Crotalus have a distinct origin from the P-I gene from E. ocellatus (40), which formed via a splice-site mutation that bypasses a disintegrin-coding exon resulting in a frame shift that terminates the protein early. Together, these findings are consistent with the inference of multiple origins of P-I SVMPs from P-II SVMPs within vipers (18).
Genomic Deletions Have Resulted in the Fusion of Paralogous Loci and Shape SVMP Complex Gene Number Among Crotalus Species.
The P-I, P-II, and P-III class genes are not the only forms of SVMP loci we find in C. atrox. We also identified two gene fusions (mpf genes) of a mdc gene with a mad gene. C. atrox mpf-1a/b both possess the ∆Cys-4k deletion, which is an identifying character of the P-II mad genes; however, they cluster with the P-III mdc-8 genes in our protein phylogenies (Fig. 2, multicolored outer arc). To better understand this association, we closely compared the genomic regions of these loci. Blast alignment of the mpf and mdc-8 genomic regions 5′ of exon 14 match with an average identity of >96% compared with 90% to the mad genes. In addition, the mpf and mdc-8 loci share a number of distinct genomic features. There is a shared 1.8-kb insertion in the 13th intron, which is marked by a Line/CR1 element (Fig. 5, small light blue oval), and the mpf loci also share several small deletions (3 to 12 bp) in the 12th intron with the mdc-8 genes and they lack a 9-bp deletion that is shared among the other P-II mad genes (SI Appendix, Fig. S4). In total, this evidence suggests that the mpf locus is most likely the product of a fusion of genomic sequences from mdc-8 and mad genes between the 13th intron and 14th exon. Although the mpf locus appears otherwise intact and capable of producing a P-II metalloproteinase, we have no evidence from either the transcriptome or mass spectrometry data that this gene is expressed in venom.
Fig. 5.
mpf-1 is a fusion of mdc and mad genomic loci. Schematics are shown for the genomic regions from exon 12 to the end of the loci for multiple C. atrox SVMP genes. At top is a generic SVMP protein with colored blocks showing which exons contribute to the distinct protein domains (red, metalloproteinase domain; orange, disintegrin domain; green, cysteine-rich domain). In a full protein phylogeny, C. atrox MPF-1a/b cluster with P-III (MDC) SVMPs (Fig. 2). The mpf loci also share a number of genomic features with the P-III mdc-8 gene; they share several small deletions in the 12th intron (green dots; see SI Appendix, Fig. S4) but not a P-II, mad-specific, 9-bp deletion (orange dot). The mpf genes also possess a Line/L1 insertion (light blue oval) into the 13th intron, similar to mdc-8b/c. The mpf-1a locus has an additional, large, Line/CR1 element (long pink oval) inserted into the Line/L1 insertion. However, mpf-1a/b do share the ∆Cys-4K deletion common to all P-II (mad) genes (see also SI Appendix, Fig. S3). Thus, mpf-1a/b is the result of a fusion between a mdc and mad genes.
Such gene fusions appear to be the products of deletion events that result in the contraction of SVMP gene number in other species. For example, Dowell et al. (31) described a similar deletion event that created the 5-gene SVMP complex of A-type C. scutulatus from the larger 16-gene complex of B-type animals. In that case a deletion spanning ∼335 kb occurred between the sixth introns of two mad genes in the B haplotype, to form a fusion gene (Fig. 6) (31).
Fig. 6.
Fusions of SVMP loci reduce the C. scutulatus SVMP complex. Schematics are shown of the SVMP complexes in C. atrox and of hemorrhagic (type B) and neurotoxic (type A) individuals from two subpopulations of C. scutulatus. Regions of high nucleotide sequence identity as determined by Blastn are shown. Gray bands connect homologous regions with shading, representing the extent of sequence identity (dark gray >95%, gray >90%, light gray >85%). Known C. scutulatus venom-expressed SVMPs are indicated (mdc-4: scutiarin; mad-3: mojastin). Mapping blocks of high nucleotide (nt) sequence identity reveals the hybrid origin of some SVMP genes, as they may derive from genomic regions that are distinct and distant in another species. We designate the C. scutulatus mad-2 a mad gene because the entirety of the mature protein (x7 to x14) is homologous to C. atrox mad-2a. The exon closest to the approximate fusion boundary is denoted above the C. scutulatus complex (e.g., x4, x7).
Given the different sizes of SVMP complexes between C. atrox and C. scutulatus type B, we wondered whether this type of gene deletion/fusion process has shaped the sizes of SVMP gene complexes. For instance, the C. atrox complex (31 genes) is much larger than the C. scutulatus B complex (16 genes). To search for potential deletions, we aligned the two SVMP complexes using BLAST and mapped the regions of high sequence identity (Fig. 6). Overall synteny across the complexes is maintained except for several regions of the C. atrox complex that are absent from the corresponding C. scutulatus complex. Although the C. scutulatus complex is large, it appears that it is derived from an even larger complex and that there have been multiple deletions including multiple instances of gene fusion (Fig. 6). For instance, the C. scutulatus mad-2 gene appears to be the result of a deletion that joined mdc-3a and mad-2a at the seventh intron, and mdc-6b and mdc-8 also appear to be fusion genes created by genomic deletion. The C. scutulatus mdc-9 gene also appears to be a fusion of the first four exons of a mad-1b gene and an mdc paralog that is seemingly absent from C. atrox but found in other Crotalus species such as Crotalus horridus (SI Appendix, Fig. S2). Note that the deletion that created mdc-9 may have also removed three genes (mad-1b, mad-2b, and mpo-1) that correspond to highly expressed venom proteins in C. atrox, and thus such deletions may contribute significantly to differences in venom content.
Discussion
We have shown that the C. atrox genome contains many more venom metalloproteinase genes than were previously known from proteomic studies, and that this 30-gene complex has been massively expanded in the rattlesnake lineage from a single, deeply conserved ancestral disintegrin and metalloproteinase (adam28) gene. In addition, we have identified a series of stepwise intragenic deletions that gave rise to three major classes of secreted SVMP toxins by the successive removal of a membrane-tethering domain, a cysteine-rich domain, and a disintegrin domain, respectively. These findings allow us to reconstruct the genetic path of snake venom SVMP innovation—the relative order of key gene duplications, expression changes, and structural changes in the origin and diversification of this family of biochemical novelties and to consider which evolutionary forces may best explain their genesis.
A Surprisingly Large C. atrox SVMP Gene Complex.
We did not expect to discover so many SVMP genes with intact ORFs in the C. atrox genome. Previous proteomic studies had detected up to nine distinct SVMP proteins in C. atrox venom (15). We detected 30 tandemly arrayed SVMP loci in a contiguous span from the highly expressed mdc-2 gene to the mdc-8c gene. The majority of these genes express RNA, and several express proteins that were not previously detected. However, some genes, while intact, do not make significant amounts of RNA or protein in adult venom glands.
The first question these observations raise is: Why did we detect more genes and proteins than prior studies? We suggest that the key to uncovering previously unknown genes was our methodology of physically isolating BAC clones spanning the complex and sequencing with long-read technology, as opposed to assembling with short reads from whole genome shotgun sequencing (41). By sequencing BAC clones, we were better able to identify several pairs or trios of genes that are very similar in sequence but were located on different physical pieces of genomic DNA (different BAC clones). In obtaining high quality genomic sequence across the complex, we were then able to accurately annotate the total set of SVMP genes which was necessary to precisely map unique peptides isolated from venom to the exact hypothetical gene translations. In the absence of high-quality genomic data, proteomic, transcriptomic, and short-read genomic approaches all struggle to distinguish close paralogs from, for example, allelic variants or alternative splice forms.
But our genomic analysis also raises a paradox: Why are genes such as mdc-3a/b and mad-6, that are not expressed at significant levels in adult venom, maintained intact? One might expect selection to be relaxed on such genes and inactivating mutations to accumulate within them. Potential explanations for the preservation of these genes are that they might be expressed at higher levels in other individuals, or at a different life stage, or under different environmental conditions, or in a different tissue, but we have no evidence yet for any of these possibilities.
The Genetic Path of Innovation.
Previous protein phylogenetic studies identified snake venom SVMPs as members of the ADAM family (12, 22) and indicated that vertebrate adam28, adam7, or dec-1 was the most likely ancestral gene (18). The genomic and comparative synteny data presented here are unambiguous that the large snake SVMP complex arose via expansion of the adam28 locus. Importantly, we identified the nonvenom-expressed mdc-1 gene as the most likely immediate ancestor of snake venom SVMP genes. The generation of the entire C. atrox complex and of SVMP diversity appears to have involved the following steps, which are schematized in Fig. 7:
Fig. 7.
The origin of snake venom SVMPs via gene duplication and intragenic deletion. Schematic of the proposed steps involved in generating the diverse array of snake metalloproteinase genes. Protein domains are indicated by color (metalloproteinase domain, red; disintegrin domain, yellow; cysteine-rich domain, green; transmembrane domain, blue). Stars indicate duplication events and ∆ represent deletions. At Right are the protein products that would be produced at each step. Purple line indicates the venom recruitment event with venom-expressed products to the Right and nonvenom-expressed genes (adam28, mdc-1) to the Left. See text for detailed explanation of the steps.
Invention of a mdc gene.
The 17-exon p-III class mdc1 gene is most closely related to and adjacent to the 25-exon adam28 gene, but lacks sequences spanning from part of exon 17 through exon 25, which encode the transmembrane domain. We infer that mdc1 was generated by a duplication of the 25-exon adam28 gene, which occurred via a partial duplication of the first 17 exons of adam28, or a full gene duplication with a subsequent intragenic deletion spanning from exon 17 through 25, and created a novel 17-exon gene encoding just the metalloproteinase, disintegrin and cysteine-rich domains (Fig. 7, step i).
Recruitment of a mdc gene into venom.
mdc1 and adam28 are not expressed in C. atrox venom but many of the adjacent SVMP genes such as mdc2 are highly expressed in venom (Fig. 1). This observation indicates that in the course of the evolution and expansion of the SVMP complex, certain SVMP genes attained high level expression in the venom gland. There are two general ways in which divergent expression of paralogs could have occurred: 1) an ancestral gene was not expressed in the venom gland (or its precursor tissue), but after duplication, its paralog gained venom gland expression (12); or 2) an ancestral gene was expressed in the venom gland (or precursor tissue) and after duplication the paralog refined (likely elevated) venom gland expression (24). We favor the former scenario because neither adam28 nor mdc-1 are expressed in extant rattlesnake venom glands. We also note that two mdc1-related genes are found in the king cobra genome, one of which is expressed in venom (Fig. 2) (16). It appears reasonable then to infer that a closely related paralog of mdc1 was recruited into snake venom early in the evolution of advanced snakes. This recruitment may have occurred after a duplication of mdc1 (Fig. 7, step ii).
Expansion of the P-III class SVMP genes.
The C. atrox genome contains 15 P-III mdc genes. Subsequent steps in the evolution of the gene family involved additional gene duplication events (Fig. 7, step iii). Gene trees indicate that most mdc genes arose from single gene duplications, although mdc-5a/b and mdc-3a/b occur within a larger duplication of three SVMP genes. The interspersal of other SVMP type genes among mdc genes, and the greater number of mdc genes in C. atrox relative to Vipera indicates that mdc gene duplications have occurred at various times during the evolution of the C. atrox lineage.
Invention of the P-II class SVMP gene.
The P-II class mad genes encode SVMPs that lack the C-terminal cysteine-rich domain present in P-III MPs. All mad genes, including those in V. berus and E. ocellatus, form a monophyletic group that share the ∆Cys-4k genomic deletion extending from the middle of the 14th exon to the end of the 16th exon (Fig. 4). Therefore, a key step in the invention of the P-II class mad genes was the occurrence of this deletion in a mdc gene in a common ancestor of Vipera and Crotalus (Fig. 7, step iv).
Expansion of the P-II class SVMP genes.
The C. atrox SVMP complex contains 11 P-II class mad genes so there have been numerous duplication events since the origin of this class of MPs. Gene trees indicate that several mad genes arose from single gene duplications, but many (mad-1/2, mad3/4, and mad-5) were subsequently part of larger, multigene segmental duplications. The presence of more mad genes in C. atrox than Vipera and their interspersal among mdc genes indicates that mad gene duplications have also occurred numerous times in the course of the evolution of the C. atrox lineage (Fig. 7, step v).
Invention of the P-I class SVMP gene.
The P-I class mpo genes encode SVMPs that lack both cysteine-rich and disintegrin domains. The lone C. atrox mpo-1 locus bears the deletion that is characteristic of the P-II clade of MPs, a stop codon in the 12th exon, as well as a second unique deletion which spans from the 12th intron to the beginning of the 14th exon (∆Dis-2k, Fig. 3 and Fig. 6, step vi). This pair of deletions is shared by the P-I SVMPs from at least two other Crotalus species, C. durissus and C. helleri (Fig. 4 and SI Appendix, Fig. S3). Genomic data for other P-I SVMPs is only available from E. ocellatus, where one P-I gene (Eoc00028) contains the ∆Cys-4k deletion but not the 12th exon stop codon or ∆Dis-2k deletion (40). These data indicate that the Crotalus and Echis P-I loci have independent origins, which is consistent with the prior inference of multiple origins of P-I genes (18).
It is interesting to note that while there appear to be multiple evolutionary paths toward making what is essentially a truncated metalloproteinase-only gene, none of them involve the genetic truncation of a P-III gene.
Evolutionary Forces That Shaped the Genesis of Snake Venom SVMPs.
The most challenging issue concerning the role of gene duplication in the evolution of any novelty is the determination of the evolutionary forces responsible for the retention of gene duplicates. The principal differences among evolutionary models of gene duplication and innovation concern whether: 1) the initial duplicate is neutral or positive; 2) the duplicate is fixed by drift or selection; and 3) whether innovative mutations take place before and/or after gene duplication (for reviews see refs. 1, 42, 43).
We endorse the view of Hahn (42) that, in the absence of functional data on individual duplicates (as demonstrated for example by refs. 9–11, 44), inferences about evolutionary forces acting on the retention of specific duplicates are perilous. However, there is no doubt that the massive expansion and diversification of SVMP genes that encode major components of prey-killing venom has involved episodes of positive selection. Indeed, we suggest that the lineage specificity of this expansion, which occurred in Viperids and Crotalids but not other snakes or reptiles, is itself a tacit signature of selection, just as the lineage-specific expansion of antifreeze genes in Antarctic fish (11) or salivary amylase genes in starch-eating modern humans (45) are also signs of positive selection for adaptations in lifestyle. Moreover, Casewell et al. have found evidence for accelerated evolution of and positive selection on surface-exposed residues of SVMP domains (46).
Therefore, despite the absence of paralog-specific functional data, we think it is constructive to consider when and how positive selection might act in the making of such a large and diverse gene complex, particularly around the major steps highlighted above (Fig. 7) that generated the three distinct types of secreted SVMPs produced by the venom gland. As Casewell et al. (18) underscored previously, the SVMPs present a case of neofunctionalization via domain loss. It is worthwhile to consider the evolutionary forces that may have enabled the generation of paralogs that lack particular domains. In general, we can envision either IAD (amplification of a minor activity) or EAC (resolution of adaptive conflict) scenarios leading to the retention of duplicates and the generation of SVMP types.
A crucial first step was the evolution of a mdc gene and secreted P-III SVMP from adam28 (Fig. 7, step i). It has been shown in some vertebrates that alternative splicing of adam28 transcripts produces a soluble isoform without a transmembrane domain or cytoplasmic tail (reviewed in ref. 47). If this soluble isoform had a distinct and selectable activity, it may have been amplified in an IAD scenario by a partial duplication of adam28 resulting in a 17-exon mdc gene. Alternatively, if the activities of the soluble and membrane-bound form of ADAM28 could not each be optimized (i.e., were in adaptive conflict) the duplicate could have been retained in an EAC scenario.
Similarly, amplification of a minor activity or escape from adaptive conflict between the cysteine-rich, disintegrin, or metalloproteinase domains could explain the evolution of P-II and P-I genes. It has recently been reported that alternative splicing of the SVMP genes in the habu snake (an Asian viper) can generate transcripts which encode different combinations of domains (48). Gene deletions that reduce the proteins from three to two domains (mdc to mad) or two to one domain (mad to mpo) would relieve potential conflict among isoforms and allow for optimization of minor activities.
But these scenarios would account for perhaps a handful of different SVMP genes, not the 29 loci that evolved subsequent to mdc-1. What other forces might explain the amplification of SVMP gene number? We would like to draw attention to recent work from this laboratory which reported two pertinent observations from the analysis of the evolution of increased alcohol dehydrogenase (Adh) gene activity in certain Drosophila species. Specifically, it was found that the expression of tandem gene duplicates is often greater than twofold that of single genes (49) and that a tandem gene duplication produced the single largest quantitative effect on Adh protein activity of a variety of individual mutations among several Drosophila species (49). The relevant implication of these studies is that selection for greater gene expression is sufficient to favor the retention of a gene duplicate without any prior or subsequent innovative mutations. We note that Ohno (3) raised this general idea long ago, which he described as “duplication for the sake of producing more of the same” and numerous examples have been highlighted (50, 51). Therefore, we suggest that SVMPs (and other venom proteins), which are often expressed at very high concentrations, may initially experience positive selection simply for increased production via increased gene dosage. Once retained, duplicates may then acquire innovative mutations.
Methods
Specimen Collection and Biological Sample Preparation.
Animal and tissue sample collections were described previously (13, 31). The BAC library was generated from a female C. atrox individual that originated in southern Texas and was housed at the serpentarium at the Texas A&M Kingsville National Natural Toxins Research Center (NNTRC). A venom sample from this individual was collected and lyophilized before tissue collection at NNTRC (Institutional Animal Care and Use Committee approval 2010-09-01A).
BAC Library Construction and Screening.
Frozen liver tissue (C. atrox) was sent to Amplicon Express (Pullman, WA) for high molecular weight genomic DNA extraction and library construction. The resulting genomic libraries consisted of ∼73,000 clones with an insert length of 80 to 150 kb (5 to 7× genome coverage for an estimated 1.4-Gb genome for C. atrox) arrayed in 190 384-well plates. A combinatorial pooling strategy was carried out that facilitated PCR-based screening (a list of PCR primer sequences used for screening are at the following link: https://figshare.com/s/d00e4d7fb95085d945b2 and in Dataset S3) (52). PCR-positive clones were picked from the library, streaked on plates, and single colonies grown overnight at 37 °C in 500 mL Luria Broth (LB) containing chloramphenicol and processed using the standard Qiagen midi-prep protocol. We identified >24 BAC clones positive for SVMP; see SI Appendix, Fig. S1 and Dataset S5 for a summary of BAC clones presented in this study.
BAC Clone Library Sequencing and Assembly.
The University of Michigan DNA sequencing core prepared the Pacific Biosciences sequencing libraries using 10 μg of BAC DNA according to the standard protocol with a size selection of large (>10,000 bp) DNA fragments. Single molecule real-time (SMRT) sequencing of each individual BAC clone library was carried out on a PacBio RSII Sequencer (Pacific Biosciences) in a single SMRT cell except for three clones (77K10, 180P3, and 192F22) that were pooled prior to library generation and sequenced in a single SMRT cell. The raw reads for each clone were assembled using the accuracy optimized HGAP2 (hierarchical genome assembly protocol) algorithm (53) or Canu (v1.9) (54). BAC vector sequence was removed from the assembled sequences before manual assembly of the whole SVMP complex.
Complex Assembly.
We identified >24 C. atrox BAC clones positive for SVMP. BAC sequences were stitched together if the overlap was >10 kb with an identity match >99.9%. Nearly all of the errors are due to homopolymer indels. There is a single break in the complex; see SI Appendix, Fig. S1 and Dataset S5 for greater detail on complex assembly.
Annotation of Venom Loci.
Computational annotation of venom genes in our long read-assembled complexes is complicated by two factors: 1) Small indels in homopolymer stretches, which are the most prevalent error in long read assemblies can create artificial frame shifts in coding regions (55) and 2) the presence of orphaned exons can lead to aberrant transcript calls. We have found manual annotation to be far more reliable; potential exons are identified through a reiterative process of BLAST (tblastn) using individual exon protein sequences as query sequences, which are then refined by identifying exon/intron boundaries AG_exon_GT and alignment to sequenced transcripts. We annotated publicly available genomes for king cobra (O. hannah) (16) and V. berus (European Adder Genome Project; https://www.hgsc.bcm.edu/reptiles/european-adder-genome-project) and renamed annotations for Crotalus SVMP complexes described by Dowell et al. (31). A complete list of genes, corresponding translations, and accession numbers can be found in Dataset S1.
Additional Methods.
Methods pertaining to the analysis of RNA, proteins, and phylogenies can be found in SI Appendix.
Data Availability.
The assembled sequences have been deposited in GenBank; accession numbers are presented in Dataset S1. Raw reads for BAC clones have been deposited in the NCBI database under BioProject ID PRJNA613473.
Supplementary Material
Acknowledgments
We thank Elda Sanchez and Mark Hockmuller of the National Natural Toxins Research Center, Texas A&M University-Kingsville for their help in obtaining snake tissues and venom. We thank Emily Hubbard for technical assistance and Alejandra Panzera for comments on the manuscript. This work was supported by the Howard Hughes Medical Institute (S.B.C.) and the Andrew and Mary Balo and Nicholas and Susan Simon Endowed Professorship in Biology at the University of Maryland (S.B.C.).
Footnotes
The authors declare no competing interest.
Data deposition: The assembled sequences have been deposited in GenBank; accession numbers are presented in Dataset S1, column E. Raw reads for BAC clones have been deposited in the National Center for Biotechnology Information (NCBI) database under BioProject ID PRJNA613473.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920011117/-/DCSupplemental.
References
- 1.Andersson D. I., Jerlström-Hultqvist J., Näsvall J., Evolution of new functions de novo and from preexisting genes. Cold Spring Harb. Perspect. Biol. 7, a017996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soskine M., Tawfik D. S., Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Ohno S., Evolution by gene duplication, (Springer Verlag, New York, NY, 1970). [Google Scholar]
- 4.Nadeau J. H., Sankoff D., Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 147, 1259–1266 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Force A. et al., Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch M., Conery J. S., The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Piatigorsky J., Wistow G., The recruitment of crystallins: New functions precede gene duplication. Science 252, 1078–1079 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Hughes A. L., The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256, 119–124 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Hittinger C. T., Carroll S. B., Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Näsvall J., Sun L., Roth J. R., Andersson D. I., Real-time evolution of new genes by innovation, amplification, and divergence. Science 338, 384–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng C., Cheng C. H. C., Ye H., He X., Chen L., Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict. Proc. Natl. Acad. Sci. U.S.A. 107, 21593–21598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry B. G., From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 15, 403–420 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowell N. L. et al., The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 26, 2434–2445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittington A. C., Mason A. J., Rokyta D. R., A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol. Biol. Evol. 35, 887–898 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Calvete J. J., Fasoli E., Sanz L., Boschetti E., Righetti P. G., Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 8, 3055–3067 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Vonk F. J. et al., The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 110, 20651–20656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu N. et al., Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteomics 159, 19–31 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Casewell N. R., Wagstaff S. C., Harrison R. A., Renjifo C., Wüster W., Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evol. 28, 2637–2649 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Fox J. W., Serrano S. M. T., Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 45, 969–985 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Takeda S., ADAM and ADAMTS family proteins and snake venom metalloproteinases: A structural overview. Toxins (Basel) 8, E155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casewell N. R., On the ancestral recruitment of metalloproteinases into the venom of snakes. Toxicon 60, 449–454 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Moura-da-Silva A. M., Theakston R. D. G., Crampton J. M., Evolution of disintegrin cysteine-rich and mammalian matrix-degrading metalloproteinases: Gene duplication and divergence of a common ancestor rather than convergent evolution. J. Mol. Evol. 43, 263–269 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Calvete J. J. et al., Snake venom disintegrins: Evolution of structure and function. Toxicon 45, 1063–1074 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Hargreaves A. D., Swain M. T., Hegarty M. J., Logan D. W., Mulley J. F., Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol. Evol. 6, 2088–2095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hite L. A., Jia L. G., Bjarnason J. B., Fox J. W., cDNA sequences for four snake venom metalloproteinases: structure, classification, and their relationship to mammalian reproductive proteins. Arch. Biochem. Biophys. 308, 182–191 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Jia Y., Pérez J. C., Molecular cloning and characterization of cDNAs encoding metalloproteinases from snake venom glands. Toxicon 55, 462–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarborough R. M. et al., Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J. Biol. Chem. 268, 1058–1065 (1993). [PubMed] [Google Scholar]
- 28.Masuda S., Hayashi H., Araki S., Two vascular apoptosis-inducing proteins from snake venom are members of the metalloprotease/disintegrin family. Eur. J. Biochem. 253, 36–41 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Bjarnason J. B., Tu A. T., Hemorrhagic toxins from Western diamondback rattlesnake (Crotalus atrox) venom: Isolation and characterization of five toxins and the role of zinc in hemorrhagic toxin e. Biochemistry 17, 3395–3404 (1978). [DOI] [PubMed] [Google Scholar]
- 30.Masuda S., Araki S., Yamamoto T., Kaji K., Hayashi H., Purification of a vascular apoptosis-inducing factor from hemorrhagic snake venom. Biochem. Biophys. Res. Commun. 235, 59–63 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Dowell N. L. et al., Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol. 28, 1016–1026.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bates E. E., Fridman W. H., Mueller C. G., The ADAMDEC1 (decysin) gene structure: Evolution by duplication in a metalloprotease gene cluster on chromosome 8p12. Immunogenetics 54, 96–105 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Giebeler N., Zigrino P., A disintegrin and metalloprotease (ADAM): Historical overview of their functions. Toxins (Basel) 8, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seals D. F., Courtneidge S. A., The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 17, 7–30 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Strickland J. L., Mason A. J., Rokyta D. R., Parkinson C. L., Phenotypic variation in mojave rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins (Basel) 10, E135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durban J. et al., Integrated venomics and venom gland transcriptome analysis of Juvenile and adult Mexican rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus revealed miRNA-modulated ontogenetic shifts. J. Proteome Res. 16, 3370–3390 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Rokyta D. R., Margres M. J., Ward M. J., Sanchez E. E., The genetics of venom ontogeny in the eastern diamondback rattlesnake (Crotalus adamanteus). PeerJ 5, e3249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rokyta D. R., Wray K. P., McGivern J. J., Margres M. J., The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98, 34–48 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Hofmann E. P. et al., Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 8, 15534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanz L., Calvete J. J., Insights into the evolution of a snake venom multi-gene family from the genomic organization of Echis ocellatus SVMP genes. Toxins (Basel) 8, E216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huddleston J. et al., Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 24, 688–696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn M. W., Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100, 605–617 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Innan H., Kondrashov F., The evolution of gene duplications: Classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Zhang Y. P., Rosenberg H. F., Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Perry G. H. et al., Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casewell N. R., Wüster W., Vonk F. J., Harrison R. A., Fry B. G., Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Wei S. et al., Conservation and divergence of ADAM family proteins in the Xenopus genome. BMC Evol. Biol. 10, 211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogawa T. et al., Alternative mRNA splicing in three venom families underlying a possible production of divergent venom proteins of the Habu Snake, Protobothrops flavoviridis. Toxins (Basel) 11, E581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loehlin D. W., Ames J. R., Vaccaro K., Carroll S. B., A major role for noncoding regulatory mutations in the evolution of enzyme activity. Proc. Natl. Acad. Sci. U.S.A. 116, 12383–12389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondrashov F. A., Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279, 5048–5057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kondrashov F. A., Rogozin I. B., Wolf Y. I., Koonin E. V., Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vu G. T. H., Caligari P. D. S., Wilkinson M. J., A simple, high throughput method to locate single copy sequences from Bacterial Artificial Chromosome (BAC) libraries using high resolution melt analysis. BMC Genom. 11, 301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin C. S. et al., Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Koren S. et al., Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ely B. et al., Genome comparisons of wild isolates of caulobacter crescentus reveal rates of inversion and horizontal gene transfer. Curr. Microbiol. 76, 159–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled sequences have been deposited in GenBank; accession numbers are presented in Dataset S1. Raw reads for BAC clones have been deposited in the NCBI database under BioProject ID PRJNA613473.