Significance
Commercial strategies to deliver biomolecular cargo ex vivo (e.g., electroporation, lipofection) to clinically relevant cell lines are limited by toxicity, cost, and throughput. These technical limitations have inhibited development of these technologies into streamlined clinical platforms for manufacturing gene-modified stem cells and cancer immunotherapies. Here, we demonstrate an acoustofluidic platform capable of delivering plasmids with high throughput to human T lymphocytes, peripheral blood mononuclear cells, and CD34+ hematopoietic stem and progenitor cells. Acoustofluidic-treated cells showed evidence of cytosolic DNA delivery, endocytic DNA aggregation, and nuclear membrane rupture. Collectively, these observations demonstrate the utility of this method as a research tool for gene editing applications and mechanistic studies of plasma membrane and nuclear membrane repair.
Keywords: acoustofluidics, hematopoietic stem cells, intracellular delivery, gene therapy
Abstract
Advances in gene editing are leading to new medical interventions where patients’ own cells are used for stem cell therapies and immunotherapies. One of the key limitations to translating these treatments to the clinic is the need for scalable technologies for engineering cells efficiently and safely. Toward this goal, microfluidic strategies to induce membrane pores and permeability have emerged as promising techniques to deliver biomolecular cargo into cells. As these technologies continue to mature, there is a need to achieve efficient, safe, nontoxic, fast, and economical processing of clinically relevant cell types. We demonstrate an acoustofluidic sonoporation method to deliver plasmids to immortalized and primary human cell types, based on pore formation and permeabilization of cell membranes with acoustic waves. This acoustofluidic-mediated approach achieves fast and efficient intracellular delivery of an enhanced green fluorescent protein-expressing plasmid to cells at a scalable throughput of 200,000 cells/min in a single channel. Analyses of intracellular delivery and nuclear membrane rupture revealed mechanisms underlying acoustofluidic delivery and successful gene expression. Our studies show that acoustofluidic technologies are promising platforms for gene delivery and a useful tool for investigating membrane repair.
Intracellular delivery of plasmids to cells for gene modification is a critical step for clinical and research applications for treating genetic disorders. Of the variety of techniques that have been developed for inserting DNA or RNA into cells, viral-based delivery is the current standard for genetic engineering. Virus-based methods have been successful for establishing efficacious gene therapies for a range of diseases, including hemoglobinopathies and cancer (1, 2). However, viral carriers are expensive and are known to modify DNA semirandomly. Such indiscriminate chromosomal integration can lead to inefficient gene transfer and off-target effects, such as insertional mutations (3). The recent emergence of targeted endonuclease gene-editing strategies (e.g., clustered regularly interspaced palindromic repeats and Cas9 protein, CRISPR-Cas9) offer an exciting solution to investigate therapeutic approaches through coordinated gene disruption or insertion of new DNA sequences at preselected sites (4). Yet, these gene-editing systems require alternative intracellular delivery strategies to overcome the size limitations of viral vectors for simultaneously encapsulating editing enzymes and corrective DNA templates (5, 6). Engineered ribonucleoprotein complexes configured for base and prime editing could replace these templates but similarly require alternative delivery strategies for effective genome editing because of the large fusion protein constructs used in these systems (7, 8).
Nonviral ex vivo transfection strategies have been employed in both commercial and research settings to circumvent the limitations of viral delivery (9, 10). However, improving the cost, safety, speed, throughput, and efficiency of nonviral transfection remains a challenge for the broader application of gene therapies to patient care. Of note, nanoparticle delivery, microinjection, electroporation, and lipofection are efficient techniques but vary in efficacy and throughput, depending on the cell line or the platform (11–13). Several clinical trials have shown that a minimum of 2 million cells/kg of body weight is needed for the effective engraftment of CD34+-selected hematopoietic stem cell populations used for gene therapies (14). One such example is gene-modified treatments for inherited disorders such as adenosine deaminase-related severe combined immunodeficiency, which typically appears in the first years of life (15). Assuming a pediatric patient weighs 12 kg, this therapy would thus require efficient processing of ∼24 million cells, which is difficult to achieve quickly and efficiently with the aforementioned techniques. Cell-squeezing technologies offer a promising alternative to address these throughput limitations, using constricted microchannels to form pores in cell membranes that enable biomolecule delivery (16–18). Recent work has shown that these physical disruption strategies can preserve human T cell function after the delivery of CRISPR-Cas9 biomolecules, with minimal aberrancies in transcriptional responses (compared with electroporation) (19).
In parallel with the success of membrane-disruption techniques, emerging acoustic methods can address the technical limitations of electroporation, lipofection, and viral vectors. Fechhemeimer et al. demonstrated that ultrasonic waves to deliver exogenous DNA to cell populations via sonoporation (20). In the wake of this pioneering discovery, a variety of ultrasonic devices were developed, but they required ultrasound contrast agents (e.g., Albunex) to stimulate microbubble formation for intracellular delivery (21, 22). The rapid expansion of microbubbles from ultrasonic perturbation is known to lead to bubble collapse, also known as cavitation, resulting in high local temperatures and pressures that are detrimental to cell viability (23). Contrast agent-free acoustofluidic systems have also been explored, where cells are focused to nodal planes of pressure from ultrasonic waves (enabling physical and spatial manipulation) (24). Notably, Rodamporn et al. demonstrated gene delivery using a bulk acoustic resonator with microfluidics that relied on radiation forces from standing waves, establishing parameters for gene delivery to HeLa cells (25). Carugo et al. miniaturized these platforms for drug delivery to H9c2 cardiomyoblasts, optimizing frequency selection, device power, and flow conditions for increased cell viability (26). Note that these bulk resonators operate in frequency regimes that do not completely suppress cavitation, which can be a mechanism of intracellular delivery using sonoporation. Recently, Yeo and coworkers demonstrated device architectures that utilize surface acoustic waves at high frequencies (>10 MHz) to suppress cavitation for gene delivery to human embryonic kidney cells and porcine tissue (27, 28). Zhang et al. explored even higher frequencies (gigahertz) with bulk acoustic resonators and demonstrated intracellular delivery of doxorubicin and plasmids with high efficiency (29). As such, there is great promise in applying acoustic-based systems toward intracellular delivery to therapeutic and disease-relevant cell types (e.g., T cells, stem cells) with high throughput.
Herein, we report the design and operation of an acoustofluidic device that delivers plasmid DNA to immortalized and primary human cell types with a throughput of 200,000 cells/min. These devices induce pores and permeability in cell membranes, enabling intracellular delivery without contrast agents. The mechanism of delivery was also explored using Jurkat cells as a model system. AlexaFluor 546-labeled DNA (Cy3-DNA) was delivered and enabled device optimization of both plasmid delivery to Jurkat cells and nuclear membrane ruptures in mouse embryonic fibroblasts (MEFs). Optimized device parameters showed successful delivery of an enhanced green fluorescent protein (eGFP)-expressing plasmid as well as nuclear membrane ruptures in acoustofluidic-treated cells. These results prompted additional experiments with plasmid delivery to human primary cells, including peripheral blood mononuclear cells (PBMCs) and umbilical cord blood CD34+ hematopoietic stem and progenitor cells (CD34+ HSPCs). All cell types tested showed eGFP expression and >80% viability over 72 h, providing strong evidence for long-term protein expression. Altogether, these data indicate that this acoustofluidic-mediated gene-delivery approach could make it possible to manufacture gene-modified therapeutic cell products at doses appropriate for pediatric patients within 2 h, making it a viable approach for gene-editing applications. Further optimization with simultaneous cell processing in multiple channels is straightforward and is currently under development.
Results and Discussion
Cell Manipulation with Acoustofluidic Devices.
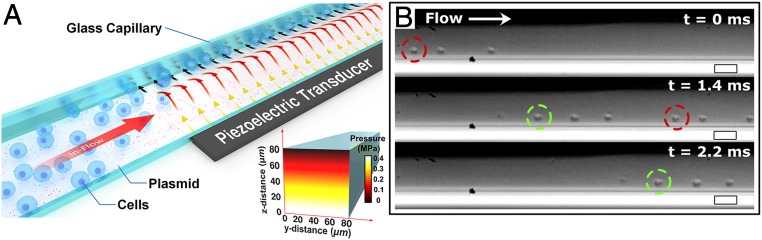
Acoustofluidic devices were designed to transduce acoustic pressure waves into a square glass microcapillary, physically shearing cell membranes against the capillary wall (Fig. 1A). These waves are generated by applying a continuous sinusoidal electrical potential to a lead zirconate titanate piezoelectric (PZT) transducer at a resonance frequency (3.3 MHz) predetermined with a network analyzer. At this frequency, the inner width of the capillary is smaller than a quarter wavelength of the generated acoustic waves in the capillary medium, which results in a minimum Gor’kov potential farthest from the acoustic source (30). Simulations of the generated acoustic pressure coincide with this potential minimum, with an amplitude gradient that decreases moving away from the PZT transducer (SI Appendix, Fig. S1A). This acoustic pressure yields predictable cell movement toward the capillary wall opposite the transducer (Fig. 1B).
Fig. 1.
(A) Schematic of the device components and application, where target cells undergo acoustofluidic treatment via flow through a glass capillary over a piezoelectric transducer and are collected at the outlet. (Inset) Simulated acoustic pressure amplitude of the aqueous medium in the glass capillary showing minimum pressure presents at the wall farthest from the piezoelectric transducer at an excitation frequency of 3.3 MHz. (B) Sequential images taken with a high-speed camera at 0, 1.4, and 2.2 ms. Jurkat cells are observed to localize against a capillary wall and are pushed forward by laminar flow. Colored circles are used to track cells moving through the capillary. (Scale bars, 50 μm.)
To observe cell behavior within the cross-section of the glass capillary, acoustofluidic devices were vertically aligned with the optical path of a microscope (SI Appendix, Fig. S1A). Images of Jurkat cells under acoustofluidic treatment (without flow) confirmed cell displacement away from the PZT transducer, pressing cells against the capillary wall due to the acoustic radiation force (SI Appendix, Fig. S1B). Additional analyses of tracking cell displacement to the capillary wall with respect to time (SI Appendix, Fig. S1C) enabled acoustic energy density measurements. This displacement analysis provided estimates for the maximum pressure amplitude (0.48 ± 0.04 MPa) (Fig. 1 A, Inset) and the acoustic radiation force (43 ± 10 pN) when applying an input voltage of 40 V peak-to-peak.
Cell movement under flow conditions (192 μL/min) was studied with a high-speed camera, enabling us to observe mobile Jurkat cells under acoustofluidic treatment (Fig. 1C and Movie S1). Under these conditions, the cells experience a combination of forces that lead to sonoporation, including the shearing force induced by microscale acoustic streaming (31) and the acoustic radiation force that pushes the cells to the microcapillary wall. We also note that cavitation is not completely suppressed in our acoustofluidic platform as the calculated acoustic pressures fall within Krasovitski’s theory of intramembranous cavitation (0.2–0.8 MPa), where membrane leaflets cyclically expand and contract, which results in increased cellular deformation, pore formation, and thus membrane permeability (32).
Intracellular Delivery with Fluorescently Labeled DNA.
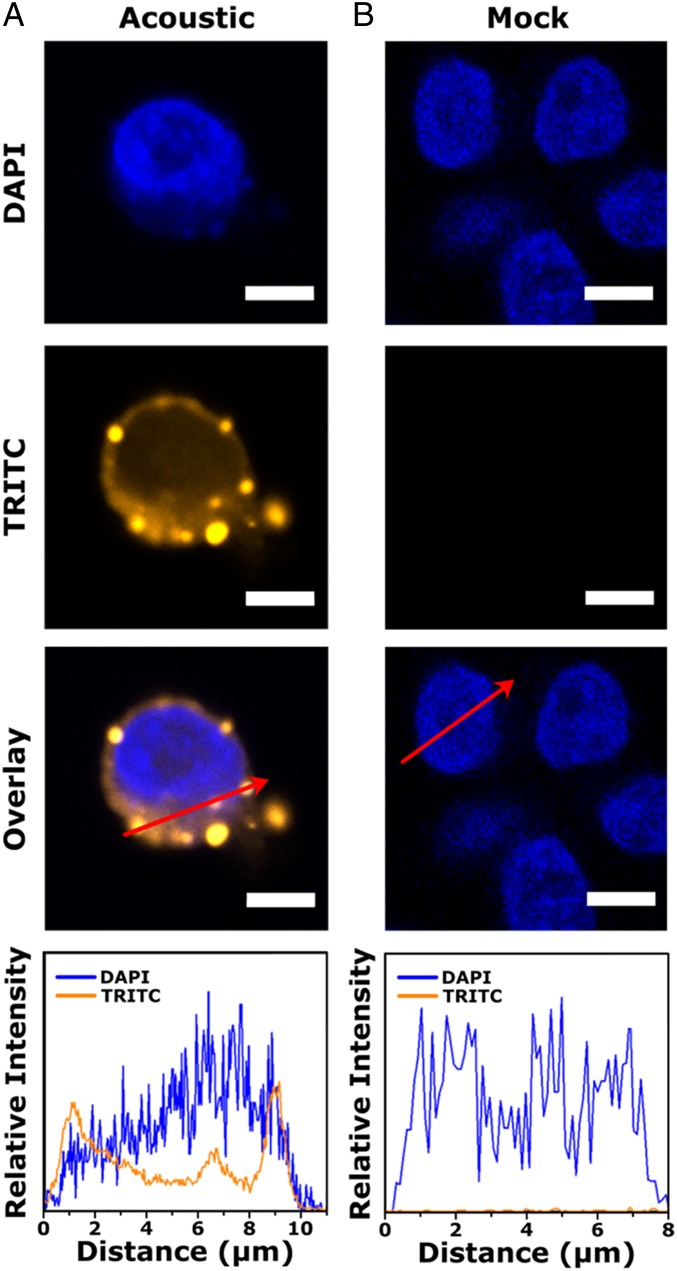
To examine whether intracellular delivery is achieved through cell membrane shearing, fluorescently labeled DNA (Cy3-DNA) was electrostatically tethered to the glass capillary by prefunctionalizing the surface with (3-aminopropyl)triethoxysilane (APTES). The Cy3-DNA was introduced into the glass capillary with a zone-loading technique using a three-way valve to prevent air from entering the capillary. Jurkat cells were flowed into the glass capillary at 192 μL/min and exposed to acoustic waves. Postacoustofluidic treatment, cells were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI), enabling observations of acoustofluidic-mediated delivery by visualizing the distribution of Cy3-DNA around the cell nucleus with confocal laser scanning microscopy. Micrographs of acoustofluidic-treated cells indicated delivery of Cy3-DNA into the cell cytosol, into the nucleus, and on the cell membrane (Fig. 2 and SI Appendix, Fig. S2). We characterized the Cy3-DNA distribution by plotting relative fluorescence intensity profiles across the red arrows and observed peak intensity maximums at the plasma membrane and nuclear membrane of the cell. These bright spots are postulated to represent formations of DNA aggregates that correspond to endosomal trafficking from the plasma membrane into the cell (33, 34).
Fig. 2.
Confocal laser scanning micrographs for (A) acoustic-treated and (B) untreated Jurkat cells. Line profiles (red arrows) of 4′,6-diamidino-2-phenylindole (DAPI) and TRITC channels show fluorescence signal of Cy3-labeled DNA at the cell membrane, cytosol, and nucleus for acoustic-treated cells. The overlays show the two images above, combined with ImageJ software. Scale bars are 10 μm.
Device Optimization with Model Cells.
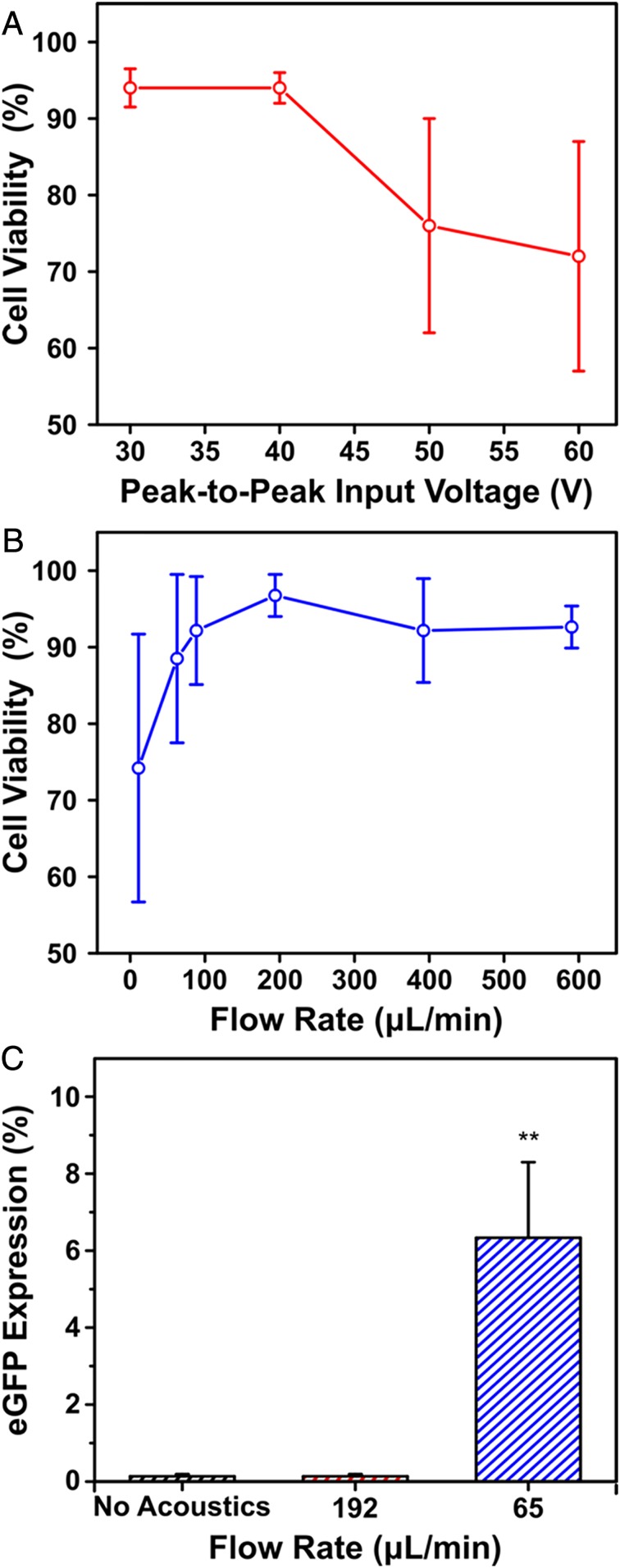
Given the successful delivery of Cy3-DNA to Jurkat cells, the input voltage to the PZT transducer and the flow rate were optimized for both cell viability and gene expression. When applying an input voltage up to 40 V peak-to-peak, we observed cell viabilities that exceeded 90% (Fig. 3A). This voltage was subsequently used as the standard input voltage. The effect of flow rate on cell viability was then studied over a range of 15 μL/min to 576 μL/min. Viability was found to exceed 90% at flow rates >192 μL/min (Fig. 3B), which is inversely related to the exposure time of acoustofluidic treatment. Following these observations, an eGFP expression plasmid driven by a cytomegalovirus (CMV) promoter was tethered to the sidewalls of the channel and coflowed with Jurkat cells (3 million cells/mL) to examine eGFP expression under different flow rates and acoustofluidic treatment. When reducing the flow rate to 65 μL/min, eGFP expression was observed after 24 h of incubation (Fig. 3C). This density (3 million cells/mL) and flow rate (65 μL/min) established a processing throughput of 200,000 cells/min, which can meet the clinical throughput for pediatric patients within 2 h using a single channel. We also conducted additional viability experiments with an annexin V-propidium iodide assay and flow cytometry using these device parameters. This assay showed ∼77% live cells with <10% apoptotic cells 72 h after acoustofluidic treatment (SI Appendix, Fig. S3). To exclude the possibility of endocytotic processes that could lead to transfection, cells were flowed through a control device consisting of a glass capillary mounted on a glass scaffold with a tubing inlet and outlet. These control samples showed no evidence of eGFP expression and viabilities equivalent to untreated cells, thereby establishing optimized peak-to-peak input voltage (40 V peak-to-peak) and flow rate (65 μL/min) that were used for the rest of the work. Successful eGFP expression in Jurkat cells also provides evidence of plasmid diffusion into the cell nucleus, since it is known that cytoplasmic nucleases can degrade free DNA, resulting in low gene expression (35, 36).
Fig. 3.
(A) Cell viability as a function of the applied peak-to-peak input voltage to the piezoelectric transducer with a constant flow rate of 192 μL/min. (B) Cell viability as a function of flow rate through the glass capillary with a constant input voltage of 40 V peak-to-peak. (C) eGFP in Jurkat cells 24 h postacoustofluidic delivery of an eGFP-expression plasmid. Protein expression is plotted as a function of flow rate and compared to a no acoustics control flowed at 65 μL/min. All cell viability measurements were assessed through trypan blue staining. Data are expressed as mean and SD for n = 3. Significance is determined using a one-way ANOVA and a Tukey means comparison test (**P < 0.01).
Nuclear Membrane Rupture Induced by Acoustofluidics.
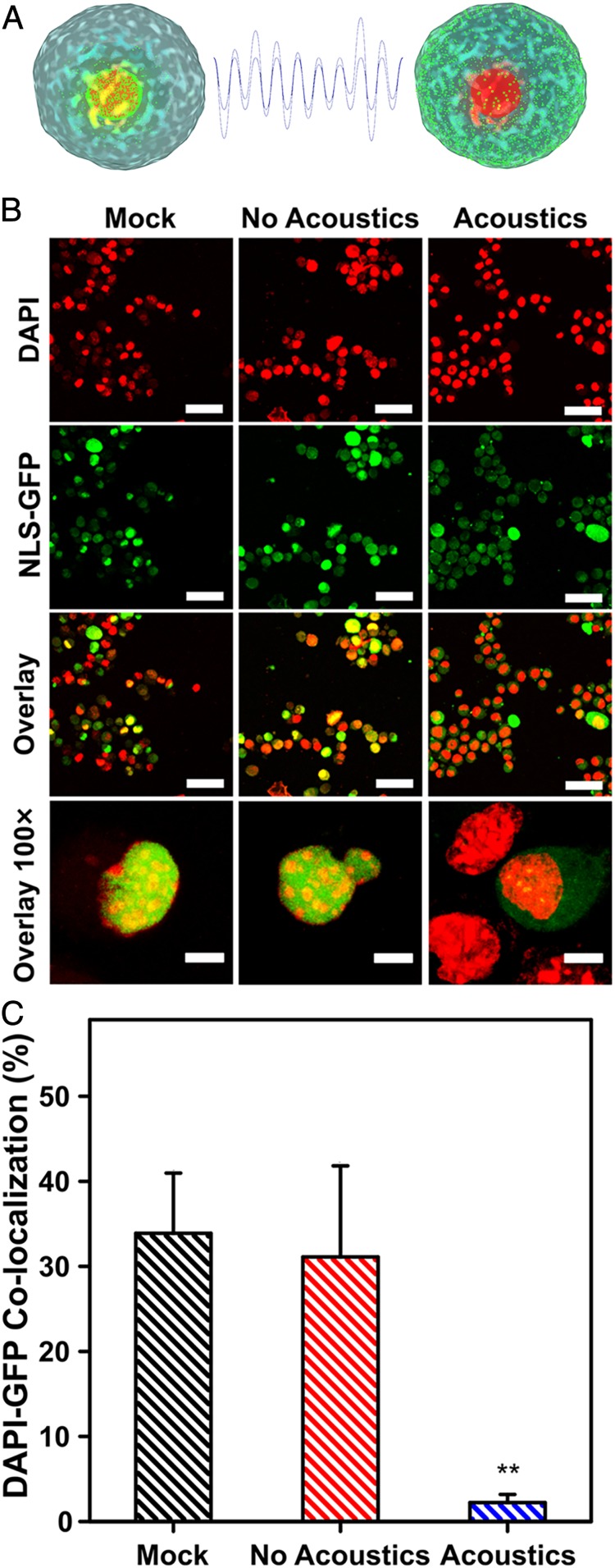
To determine the effects of acoustofluidic treatment (using the aforementioned parameters of 65 μL/min and 40 V peak-to-peak) on cell nuclei, we investigated nuclear membrane ruptures in MEFs (Fig. 3A). Cells were first virally transduced with an NLS-GFP reporter (green fluorescent protein fused to a nuclear localization signal) (37); these cells constitutively express green fluorescent protein that localizes to the nuclear envelope and enables observations of perturbations on cell nuclei. These NLS-GFP MEFs were fixed and stained with DAPI immediately after acoustofluidic treatment for confocal laser scanning microscopy (Fig. 4). A significant difference in the percentage of cells with nuclear membrane rupture was observed in acoustofluidic-treated cells compared with mock and no-acoustics controls, quantified by the numbers of cells with colocalization of DAPI and GFP signals (Fig. 4B and SI Appendix, Fig. S4). Notably, cells that underwent acoustofluidic treatment showed dispersed GFP throughout the cytosol and decreased nuclear fluorescence intensity compared with untreated control (mock) and samples flowed through the control device (no acoustics). The displacement of nuclear-localized GFP into the cytosol via acoustofluidic treatment provides evidence of nuclear envelope ruptures, which could enable diffusion of plasmid DNA into the nucleus. These results demonstrate the potential of the acoustofluidic platform as a tool to examine both cell membrane and nuclear membrane repair mechanics and for development of reagents that increase the entry of DNA into the nucleus. It is also possible that rupture of the nuclear envelope could be a driver for genomic instability and DNA damage (38). Thus, we took an immunohistochemical approach to analyze DNA damage by staining NLS-MEFs with DAPI and a γH2AX antibody immediately after acoustofluidic treatment. We determined highly damaged cells by counting cells that had >7 labeled DNA foci and found insignificant amounts of DNA damage when compared to untreated cells (mock) (SI Appendix, Fig. S5). The finding of nuclear membrane ruptures with insignificant DNA damage will make it possible to elucidate potential effects of acoustofluidic treatment on the genome, a relevant consideration for gene-editing applications (39).
Fig. 4.
(A) A schematic of MEFs green fluorescent protein fused to a nuclear localization signal (NLS-GFP) and acoustofluidic treatment inducing nuclear membrane rupture, resulting in dispersed NLS-GFP throughout the cell cytosol. (B) Confocal laser scanning micrographs of MEFs showing colocalization events of NLS-GFP and DAPI signals in the untreated (mock), no acoustics, and acoustics-treated samples. The MEFs are virally transduced to express GFP at their nuclei (green) and are stained postacoustofluidic treatment with DAPI to label the cell nuclei (red). Colocalization of GFP and DAPI signals (yellow) are shown in the overlay, and lack thereof is evidence of nuclear membrane rupture. (Scale bars, upper three rows, 50 μm; bottom row, 10 μm.) (C) Quantification of colocalized DAPI and GFP signals for mock, no acoustic, and acoustic-treated cells. Data are expressed as mean and SD for n = 3 and significance is determined using a one-way ANOVA and Tukey’s mean comparison test (**P < 0.01). Colocalization % of DAPI and GFP signals are normalized to 60 cells for each condition.
Gene Delivery to Primary Cells.
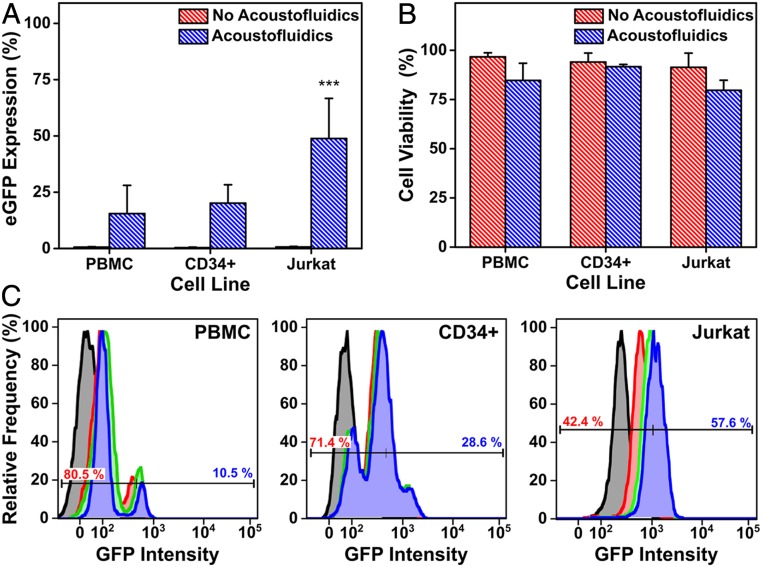
We explored the potential of this platform using the optimized device parameters (3 million cells/mL, 65 μL/min, and 40 V peak-to-peak) for therapeutic applications, in which an eGFP plasmid was delivered to Jurkat, PBMCs, and CD34+ HSPCs. Protein expression was monitored at 24-, 48-, and 72-h time points with flow cytometry (assessing continued eGFP expression). Delivery to Jurkat cells revealed 6% eGFP expression after 24 h, increasing to 62% after 72 h. Primary human cells (mixed donor CD34+ HSPCs derived from umbilical cord blood, PBMCs) demonstrated similar increases in transfection efficiency over time, with 15% and 20% eGFP expression and 85% and 92% viability, respectively, after 72 h (Fig. 5). We also observed positive eGFP expression in PBMCs 12 h postacoustofluidic delivery (SI Appendix, Fig. S7). Since there are a limited amount of cells in a batch of mixed donor HSPCs, the cell density of CD34+ HSPCs was reduced to 500,000 cells/mL in order to obtain data in triplicate. Fluorescence microscopy of CD34+ HSPCs confirmed eGFP expression, revealing bright fluorescence intensity compared to the “no acoustofluidics” control (SI Appendix, Fig. S6). Furthermore, the acoustofluidic treatment of Jurkat cells yielded comparable efficiencies to electroporation-mediated eGFP plasmid delivery (40). While delivery to primary cell types showed lower efficiency, we expect that optimization of device parameters and delivery media for each cell type will enhance levels of gene expression (41).
Fig. 5.
eGFP expression (A) and cell viability (B) 72 h postacoustofluidic delivery of an eGFP-expression plasmid to Jurkat, PBMC, and CD34+ hematopoietic stem and progenitor cells (CD34+). (C) Flow cytometry quantification of eGFP expression over a 72-h period postacoustofluidic delivery of an eGFP-expression plasmid. Histograms show relative frequency of detected eGFP events with time points defined as 0 h (black), 24 h (red), 48 h (green), and 72 h (blue). A representative bisector gate is overlaid on each histogram to show the flow cytometry gating for each cell type, with negative GFP populations in red text and positive in blue. Data are expressed as mean and SD for n = 5 for Jurkat and n = 3 for PBMCs and CD34+. Statistical significance is determined using a Student’s t test (***P < 0.001).
The high viabilities observed here indicate that cells could be circulated through multiple rounds of acoustofluidic treatment in order to boost efficiency further. Likewise, multiple parallel channels can be driven by a single acoustic source in order to increase throughput substantially. Avoiding viral methods increases safety, adds flexibility to the biomolecular cargo delivered, and should significantly reduce the cost for clinical and research applications of gene delivery.
Conclusions and Prospects
We developed a gene-delivery platform that utilizes acoustofluidic-mediated sonoporation of target cells to facilitate DNA uptake across plasma membranes. With optimization of our device, we demonstrated plasmid delivery from model cells (Jurkat) to clinically relevant cell types (PBMCs, CD34+ HSPCs) with throughputs of 200,000 cells/min and viabilities exceeding 80%. This device employs a facile and cost-effective design, taking advantage of a commercially available square glass capillary as the microfluidic channel, thereby circumventing the need for specialized facilities and complex microfluidic geometries. These data indicate scalable and economical acoustofluidic strategies for applications involving disease treatment. For example, successful eGFP expression in PBMCs suggests a strong potential to manufacture cells expressing chimeric antigen receptors for cancer immunotherapies. Furthermore, analyses of intracellular delivery revealed disruption of the cell membrane and the nuclear membranes of Jurkat and mouse embryonic fibroblasts, respectively. Further investigation of membrane disruption with our acoustofluidic platform will make it possible to examine membrane rupture, repair, and membrane mechanics in a variety of cell types. These studies, along with prospective applications in the delivery of CRISPR-Cas9 and other targeted nuclease systems, are important steps for the clinical application of the acoustofluidic platform for gene editing.
Materials and Methods
Surface Functionalization of Glass Microcapillaries.
Square glass microcapillaries (Vitrocom) with 5 cm × 80 μm × 80 μm in internal dimensions were cleaned in piranha solution (3:1 concentrated sulfuric acid and 30% hydrogen peroxide) for 30 min to remove organic molecules while adding hydroxyl functionalities to the glass surface. Next, the capillaries were rinsed and sonicated in 18-MΩ deionized water (Millipore) for five cycles of 5 min and placed in a drying oven at 110 °C for 6 h. The dried capillaries were then dipped in a 5% (vol/vol) ethanolic solution of APTES (Sigma Aldrich) and placed in an oven at 60 °C for 5 min followed by three cycles of sonication in ethanol for 5 min to remove any passively adsorbed APTES molecules from the channel walls. Clean functionalized capillaries were stored in ethanol until device assembly.
Device Fabrication.
The acoustofluidic devices are comprised of a piezoelectric lead zirconate titanate (PZT) transducer (SMPL26W16T07111, StemInc), a functionalized glass microcapillary, and a glass slide that provides a supporting substrate. The PZT transducers were mounted onto the glass slide with a thin layer of Devcon 5-min epoxy adhesive (300007-392, VWR) after soldering 30-gauge wire to the front and back electrodes of the PZT transducer. A functionalized glass microcapillary was attached onto the transducer with adhesive and cured for 30 min. Polyethylene tubing (PE-50, Instech) was connected to both ends of the microcapillary and sealed with small drops of epoxy. After curing, the tubing was secured to the glass slide with double-sided tape and tested for leaks. The resonant frequency for each device was determined with a vector network analyzer (VNA-120, Array Solutions).
Operation.
Fabricated acoustofluidic devices were vertically aligned in a custom-built stage that aligned the cross-section of the microfluidic channel within the optical path of a Nikon TE300 optical microscope. Tubing was connected to a syringe by inserting a 23-gauge needle adapter, and the flow rate was controlled with a syringe pump (Fusion 4000, Chemyx). The PZT transducers were excited with a sinusoidal wave at the desired frequency and an amplitude of 40 Vp-p with a signal generator (81150A, Agilent) and a broadband amplifier (25A250B, Amplifier Research).
DNA and Plasmid Delivery.
The APTES-treated glass capillaries were prerinsed with 5 mL of 70% ethanol, followed by 3 mL of 1× phosphate-buffered saline (PBS) solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, Gibco) before introducing plasmid DNA. The eGFP expression vector (pCMV-GFP, Plasmid 11153, Addgene) or Cy3-labeled DNA (Integrated DNA Technologies) was diluted to 50 ng/mL in 1× PBS and was zone-loaded with a flow a rate of 3.33 μL/min for 30 min with a three-way valve (CMA 110, Harvard Apparatus) connected to the acoustofluidic device. Cells were then dispersed at a density of 3 million cells/mL (except for CD34+ HSPCs) in a delivery medium consisting of 1% (vol/vol) Pluronic F-68 (Gibco), 1× PBS, and 0.1 mg/mL of eGFP-expressing plasmid and collected into a 1-mL syringe. Cells were introduced into the glass capillary at the designated flow rate, and acoustofluidic-treated cells were collected into a sterile tube. The peak-to-peak input voltage was applied to the PZT transducer after the initial drop of solution was collected to ensure that cells were under continuous acoustofluidic treatment while passing through the device. Additional PBS was flowed through the device for 3 min after the cell syringe reached depletion to collect any remaining cells in the dead volume of the device. Upon collection, cells were incubated in the delivery medium for 10 min to facilitate membrane recovery and biomolecule diffusion. Cells were then centrifuged at 500 × g for 5 min and dispersed in their respective culture media. Viability was determined using a Cell Countess II (Invitrogen) and 0.4% trypan blue (Invitrogen).
Confocal Laser Scanning Microscopy.
Cells were initially fixed via incubation in 0.5% paraformaldehyde (Sigma Aldrich) in 1× PBS solution. The cells were then spun down, dispersed in 1× PBS at a density of at least 1,000,000 cells/mL, plated on microscope slides (Denville) in a 2:7 mixture of cells to ProLong diamond antifade mounting solution (Thermo Fisher Scientific), and mixed thoroughly using a P100 pipette (Gilson). A coverslip was carefully placed on top of the cell mixture and allowed to dry at room temperature for 1 h with foil covering the slides. Slides were stored at 4 °C and imaged within 1 wk. Confocal laser scanning microscopy was performed with a confocal microscope (Zeiss, LSM 700) with Plan Apochromat 10×/0.45 and 20×/0.8 objectives. Z-stacks were acquired, and a maximum intensity projection was applied to each stack using Zeiss Zen Blue software.
Cell Culture.
Jurkat cells (American Type Culture Collection, Inc., ATCC) were cultured in 1× RPMI 1640 with l-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (10,000 units/mL penicillin and 10 mg/mL streptomycin) (Gibco). PBMCs were sourced from healthy donors and isolated by the University of California, Los Angeles (UCLA) Virology core and cultured with the aforementioned medium supplemented with 10 ng/mL recombinant human IL-2 (Peprotech). Mouse embryonic fibroblasts with an NLS-GFP reporter were cultured and derived as described previously (42). Staining with the γH2AX antibody to identify DNA damage was described previously by Chen et al. (43). Mixed donor umbilical cord CD34+ Hematopoietic stem and progenitor cells (Allcells Inc.) were thawed and prestimulated as described by Hoban et al. (44). Postacoustofluidic treatment, HSPCs were cultured in X-VIVO 15 supplemented with 50 ng/μL recombinant human stem cell factor (SCF) (Peprotech), 50 ng/μL human recombinant Flt3-ligand (Peprotech), and 50 ng/μL recombinant human TPO (Peprotech) in a 12-well plate at a density of 400,000 cells/mL for 24 h. The HSPCs were then spun down at 500 × g for 5 min and transferred to Iscove’s Modified Dulbecco’s Medium (Thermo Fisher Scientific) supplemented with 50 ng/μL recombinant human IL-3 (Peprotech), 50 ng/μL recombinant human IL-6 (Peprotech), 50 ng/μL human recombinant SCF (Peprotech), 1.5% BSA (Sigma Aldrich), 20% FBS, and 1% l-glutamine/penicillin/streptomycin (Gemini). For all cell types, cell culture media were changed every 2 d.
High-Speed Imaging.
The high-speed imaging setup was described in detail in a previous work (45). The frame rate of the camera is limited by the chosen pixel resolution (512 × 512 pixels resolution here). The resulting frame rate was 21,000 per s with an exposure time of 0.25 μs.
Flow Cytometry.
Flow cytometry data were acquired and processed using an LSR Fortessa cytometer (BD Biosciences). Data analyses were performed using FlowJo software (FlowJo, LLC). Fluorescence emission was stimulated using a 488-nm, 50-mW laser with a 505-nm long-pass filter and 515/20-nm bandpass filters for detecting green fluorescent protein.
Materials and Data Availability.
All original data are available in the manuscript and SI Appendix. Materials are available either commercially or upon request.
See SI Appendix, SI Materials and Methods for more details on the methods we used.
Supplementary Material
Acknowledgments
We thank Dr. Dino Di Carlo (UCLA) and Hector Munoz for the use of a high-speed camera for visualizing cells in flow conditions; Geoffrey Pronovost, Alexander Sercel, and Dr. Thomas Fung for their insights on fluorescence analyses; the UCLA Center for AIDS Research (CFAR) virology core (National Institute of Health Grant 5P30 AI028697) for isolating peripheral blood mononuclear cells; Dr. Donald Kohn (UCLA), Dr. Zulema Romero, and Suzanne Said for sharing hematopoietic stem cell transfection protocols and cell culture reagents. This work was supported by NIH Grants U54HL119893, R01GM132603, R33CA223908, and R01GM127714, and NIH National Center for Advancing Translational Sciences (NCATS) UCLA Clinical and Translational Science Institute (CTSI) Grant Number KL2TR001882 and UL1TR001881 through the University of California (UC) Center for Accelerated Innovation. J.N.B. thanks the NIH for a Predoctoral Fellowship; research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the NIH under Grant F31HL149356. A.M.M. thanks the National Science Foundation for a Graduate Research Fellowship (Grant DGE‑1144087). Y.G. thanks the UCLA chemistry department for funding through the S.G. Fellowship. L.S. thanks the American Italian Cancer Foundation for a Postdoctoral Fellowship. S.D.O. acknowledges support from National Cancer Institute (NCI) Grant K23CA222659, American Society of Hematology Scholar Award. P.S.W. and S.J.J. acknowledge support from the UCLA Innovation Fund MedTech Innovator Award and seed funding provided through a UCLA David Geffen School of Medicine Regenerative Medicine Theme Award. S.J.J. is supported by NIH Common Fund through a NIH Director’s Early Independence Award co-funded by the National Institute of Dental and Craniofacial Research and Office of the Director, NIH Grant DP5OD028181. S.J.J. also acknowledges Young Investigator Award funds from the Alex’s Lemonade Stand Foundation for Childhood Cancer Research, the Hyundai Hope on Wheels Foundation for Pediatric Cancer Research, and the Tower Cancer Research Foundation. We acknowledge the facilities and thank the staff of the UCLA Broad Stem Cell Research Center Flow Cytometry and Microscopy Cores.
Footnotes
Competing interest statement: P.S.W., S.J.J., A.Z.S., and J.N.B. are inventors on US and international patent applications filed by the Regents of the University of California relating to the acoustofluidic platform.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917125117/-/DCSupplemental.
References
- 1.Hoban M. D., Bauer D. E., A genome editing primer for the hematologist. Blood 127, 2525–2535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunbar C. E., et al. , Gene therapy comes of age. Science 359, eaan4672 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Thomas C. E., Ehrhardt A., Kay M. A., Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rui Y., Wilson D. R., Green J. J., Non-viral delivery to enable genome editing. Trends Biotechnol. 37, 281–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J. H., Davis K. M., Liu D. R., Chemical biology approaches to genome editing: Understanding, controlling, and delivering programmable nucleases. Cell Chem. Biol. 23, 57–73 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rees H. A., et al. , Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzalone A. V., et al. , Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart M. P., et al. , In vitro and ex vivo strategies for intracellular delivery. Nature 538, 183–192 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Stewart M. P., Langer R., Jensen K. F., Intracellular delivery by membrane disruption: Mechanisms, strategies, and concepts. Chem. Rev. 118, 7409–7531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharifi Tabar M., et al. , Evaluating electroporation and lipofectamine approaches for transient and stable transgene expressions in human fibroblasts and embryonic stem cells. Cell J. 17, 438–450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mout R., et al. , Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11, 2452–2458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q., “Review of microinjection systems” in Micromachines for Biological Micromanipulation, Moldvai M., Hal B., Eds. (Springer, Cham, 2018), pp. 15–47. [Google Scholar]
- 14.Morgan R. A., Gray D., Lomova A., Kohn D. B., Hematopoietic stem cell gene therapy: Progress and lessons learned. Cell Stem Cell 21, 574–590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw K. L., et al. , Clinical efficacy of gene-modified stem cells in adenosine deaminase-deficient immunodeficiency. J. Clin. Invest. 127, 1689–1699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharei A., et al. , A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. U.S.A. 110, 2082–2087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X., et al. , CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 1, e1500454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding X., et al. , High-throughput nuclear delivery and rapid expression of DNA via mechanical and electrical cell-membrane disruption. Nat. Biomed. Eng. 1, 0039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiTommaso T., et al. , Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 115, E10907–E10914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fechheimer M., et al. , Transfection of mammalian cells with plasmid DNA by scrape loading and sonication loading. Proc. Natl. Acad. Sci. U.S.A. 84, 8463–8467 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao S., Thrall B. D., Miller D. L., Transfection of a reporter plasmid into cultured cells by sonoporation in vitro. Ultrasound Med. Biol. 23, 953–959 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Helfield B., Chen X., Watkins S. C., Villanueva F. S., Biophysical insight into mechanisms of sonoporation. Proc. Natl. Acad. Sci. U.S.A. 113, 9983–9988 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller D. L., Pislaru S. V., Greenleaf J. E., Sonoporation: Mechanical DNA delivery by ultrasonic cavitation. Somat. Cell Mol. Genet. 27, 115–134 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Ozcelik A., et al. , Acoustic tweezers for the life sciences. Nat. Methods 15, 1021–1028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodamporn S., Harris N. R., Beeby S. P., Boltryk R. J., Sanchez-Elsner T., HeLa cell transfection using a novel sonoporation system. IEEE Trans. Biomed. Eng. 58, 927–934 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Carugo D., et al. , Contrast agent-free sonoporation: The use of an ultrasonic standing wave microfluidic system for the delivery of pharmaceutical agents. Biomicrofluidics 5, 44108–4410815 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramesan S., Rezk A. R., Dekiwadia C., Cortez-Jugo C., Yeo L. Y., Acoustically-mediated intracellular delivery. Nanoscale 10, 13165–13178 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Ramesan S., Rezk A. R., Yeo L. Y., High frequency acoustic permeabilisation of drugs through tissue for localised mucosal delivery. Lab Chip 18, 3272–3284 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z., et al. , Hypersonic poration: A new vesatile cell poration method to enhance cellular uptake using a piezoelectric nano-electromechanical device. Small 13, 1602962 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Guo F., et al. , Three-dimensional manipulation of single cells using surface acoustic waves. Proc. Natl. Acad. Sci. U.S.A. 113, 1522–1527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lentacker I., De Cock I., Deckers R., De Smedt S. C., Moonen C. T. W., Understanding ultrasound induced sonoporation: Definitions and underlying mechanisms. Adv. Drug Deliv. Rev. 72, 49–64 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Krasovitski B., Frenkel V., Shoham S., Kimmel E., Intramembrane cavitation as a unifying mechanism for ultrasound-induced bioeffects. Proc. Natl. Acad. Sci. U.S.A. 108, 3258–3263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delalande A., Kotopoulis S., Postema M., Midoux P., Pichon C., Sonoporation: Mechanistic insights and ongoing challenges for gene transfer. Gene 525, 191–199 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Delalande A., Leduc C., Midoux P., Postema M., Pichon C., Efficient gene delivery by sonoporation is associated with microbubble entry into cells and the clathrin-dependent endocytosis pathway. Ultrasound Med. Biol. 41, 1913–1926 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Shah P., Wolf K., Lammerding J., Bursting the bubble–Nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. 27, 546–555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean D. A., Strong D. D., Zimmer W. E., Nuclear entry of nonviral vectors. Gene Ther. 12, 881–890 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denais C. M., et al. , Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox D. B. T., Platt R. J., Zhang F., Therapeutic genome editing: Prospects and challenges. Nat. Med. 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim S., Quinton R. J., Ganem N. J., Nuclear envelope rupture drives genome instability in cancer. Mol. Biol. Cell 27, 3210–3213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L. H., et al. , Highly efficient, large volume flow electroporation. Technol. Cancer Res. Treat. 1, 341–350 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Jordan E. T., Collins M., Terefe J., Ugozzoli L., Rubio T., Optimizing electroporation conditions in primary and other difficult-to-transfect cells. J. Biomol. Tech. 19, 328–334 (2008). [PMC free article] [PubMed] [Google Scholar]
- 42.Chen N. Y., et al. , Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. U.S.A. 115, 10100–10105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen N. Y., et al. , An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl. Acad. Sci. U.S.A. 116, 25870–25879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoban M. D., et al. , Delivery of genome editing reagents to mematopoietic stem/progenitor cells. Curr. Prot. Stem Cell Biol. 36, 5B.4.1–5B.4.10 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Gossett D. R., et al. , Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. U.S.A. 109, 7630–7635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.