Significance
This work shows that the genome of a multipartite virus, brome mosaic virus (BMV), is highly disordered and relatively unconstrained within its virion, suggesting a different method of assembly for multipartite viruses compared with well-studied monopartite viruses like MS2 and Qβ with genomes that have been shown to be highly ordered within their capsids. This can be understood in the context of their associated viral lifecycles as BMV must package separately each of several different RNA molecules and has been shown to replicate and package them in specially formed replication factories largely isolated from the rest of the host cell cytoplasm, whereas MS2 and Qβ exploit capsid protein binding “packaging signals” throughout viral RNA to selectively package their monopartite genomes.
Keywords: cryoelectron microscopy, virus, single-stranded RNA
Abstract
We report the asymmetric reconstruction of the single-stranded RNA (ssRNA) content in one of the three otherwise identical virions of a multipartite RNA virus, brome mosaic virus (BMV). We exploit a sample consisting exclusively of particles with the same RNA content—specifically, RNAs 3 and 4—assembled in planta by agrobacterium-mediated transient expression. We find that the interior of the particle is nearly empty, with most of the RNA genome situated at the capsid shell. However, this density is disordered in the sense that the RNA is not associated with any particular structure but rather, with an ensemble of secondary/tertiary structures that interact with the capsid protein. Our results illustrate a fundamental difference between the ssRNA organization in the multipartite BMV viral capsid and the monopartite bacteriophages MS2 and Qβ for which a dominant RNA conformation is found inside the assembled viral capsids, with RNA density conserved even at the center of the particle. This can be understood in the context of the differing demands on their respective lifecycles: BMV must package separately each of several different RNA molecules and has been shown to replicate and package them in isolated, membrane-bound, cytoplasmic complexes, whereas the bacteriophages exploit sequence-specific “packaging signals” throughout the viral RNA to package their monopartite genomes.
Single-stranded RNA (ssRNA) viruses pathogenic to a wide range of prokaryotic and eukaryotic cells assemble into infectious virions with incredible precision. These particles can be as simple as an RNA genome packaged inside a capsid—a shell composed of a specific number of copies of a single gene product, the capsid protein (CP). Most such viruses form icosahedrally symmetric structures involving 12 pentamers and 10(T −1) hexamers of CP, with triangulation number T = 1, 3, 4, or 7 (1). Since about 30 y ago, the detailed structures of the capsids of a number of viruses have been determined by X-ray crystallography to near-atomic resolution (2 to 5 Å) (2), notable among them the leviviruses MS2 (3) and Qβ (4) and the bromoviruses brome mosaic virus (BMV) (5) and cowpea chlorotic mottle virus (CCMV) (6, 7). CCMV and BMV have T = 3 icosahedral capsids 30 nm in diameter consisting of 90 dimers of a single CP. The capsids of the MS2 and Qβ bacteriophages are similar to those of BMV and CCMV in that they have T = 3 structures and are 30 nm in diameter, but they contain only 89 protein dimers, with 1 dimer being replaced by a single maturation protein—called the A protein.
While their capsid structures are quite similar, viruses in the Leviviridae and Bromoviridae families display marked differences in the organization of their RNAs and in the interactions between their RNAs and their CPs. As is the case with 30 to 40% of plant viruses, the bromoviruses are multipartite (8). Specifically, bromovirus genomes are composed of three molecules that are not all packaged in the same virion: RNA1 (3.2 kilobases [kb]) and RNA2 (2.8 kb) are each packaged separately into individual virions, whereas genomic RNA3 (2.1 kb) and a subgenomic RNA4 (0.8 kb) are copackaged into a third virion (9, 10), which makes up 55 to 80%—dependent on the host—of the virion population in a wild-type infection (11). The three virions have indistinguishable capsid structures (12) and almost identical overall mass densities (13), complicating their physical separation into individual virion types. In contrast, MS2 and Qβ are monopartite ssRNA viruses, with all of their genetic information—single-molecule RNA genomes consisting of 3.6 kb for MS2 and 4.2 kb for Qβ—packaged into a single capsid (14).
The CPs of these simple spherical RNA viruses have been shown to spontaneously self-assemble around their RNA in vitro and have, therefore, been used as model systems to study viral assembly. Among them, the bromoviruses are notable because infectious virions can be assembled in vitro from their pure components, CP and RNA (15); in contrast, in vitro assembly of MS2 and Qβ is possible only in the absence of the maturation protein, yielding a noninfectious form of the virus consisting of 90 CP dimers (16). A variety of in vitro assembly studies has shown that the packaging of RNA by leviviral CP is dependent on the presence of “packaging signals,” essential stem loops organized in a specific arrangement along the primary sequence (17) that interact strongly with the CP (18, 19). In addition, the maturation protein has been shown to bind both the 5′ and the 3′ ends of the RNA genome and to be involved in delivering it to the host cell during infection. Moreover, numerous experimental analyses have shown that the bromovirus CPs interact nonspecifically with the RNA genome through electrostatic attractions with their positively charged N termini (20–23). Previous studies (24, 25) have indicated that a 16-nucleotide stem loop in the 3′ tRNA-like sequence present in each of the BMV genomic RNAs plays a role in packaging the viral RNA in vivo. It is significant that, in vivo, BMV packages its own RNA with extremely high fidelity, with fewer than 0.1% of the RNA molecules representing nonviral (cellular) RNA (11).
Recently, important insights into the role played in viral assembly by the interaction between CP and the genome have been made possible by major advances in cryoelectron microscopy (cryoEM). As shown by recent investigations of MS2 (26, 27) and Qβ (28), it is now possible to resolve much of the structure of the RNA genome inside the capsid and determine details of its interactions with the capsid. As much as 80% of the RNA can be traced, displaying a well-defined organization in which specific stem loops are found to interact with CP. Furthermore, these studies illustrate the power of asymmetric cryoEM reconstruction, which does not rely on imposing icosahedral symmetry on the dataset—but requires rather a much larger dataset—to resolve asymmetric features of the genome.
The focus of the present work is to use cryoEM to examine the organization of the RNA genome and its interaction with CP in the multipartite BMV. Previous structural studies have been performed on the naturally occurring mixtures of the three types of virions (5, 13, 29). In this study, we examine a subset of the virions, prepared in planta, with unique RNA content—in this case, one copy each of RNA3 and RNA4. Utilizing this pure BMV particle (BMV3 + 4) and employing asymmetric cryoEM reconstruction, we avoid both averaging over different RNA contents and averaging out asymmetric portions of the virion, in principle allowing for visualization of the organization of the structured portions of the RNA. We find in BMV3 + 4 virions that the RNA is only organized at the capsid surface and that there is not a single dominant conformation for the RNA inside the virus but rather, an ensemble of RNA structures that interact with the CP. These findings demonstrate that, while some monopartite viruses (like MS2) utilize specific protein–RNA interactions to drive virion assembly and result in their having highly organized RNA genomes, multipartite viruses (like BMV) take advantage of nonspecific electrostatic interactions between CP and RNA to ensure the separate but simultaneous packaging of two or more molecules of the multipartite genome, resulting in significantly less order of the genome.
Results
In Vivo Assembly of Pure BMV3 + 4 Virions.
Characteristic properties of the plasmid agroconstructs, pB1 (coding for RNA1), pB2 (RNA2), and pB3 (RNA3), engineered to express biologically active full-length BMV genomic RNAs have been described previously (30); RNA1 and RNA2 transcribed from pB1 and pB2 code for the replicase proteins that bind and amplify both of those molecules and also RNA3. The agroconstructs p1a and p2a, on the other hand, are transcribed to yield mRNAs that code for the same replicase proteins, but these RNAs lack the 5′ and 3′ noncoding sequences needed for their amplification; as a consequence, only RNA3 is amplified (31). Since genome packaging in BMV is coupled to replication (30–32) and because the interaction between CP and replicase protein p2a is sufficient to dictate packaging specificity, only virions copackaging genomic RNA3 and subgenomic RNA4 (BMV3 + 4 virions) are assembled in Nicotiana benthamiana as described in Materials and Methods. Negative stain electron microscopy and the northern blot analysis data shown in Fig. 1L confirm the purity and the genetic identity of BMV3 + 4. Additionally, in contrast to the induction of local and systemic infection in Chenopodium quinoa by a purified virion preparation of wild-type control (33), inoculation with purified virions of BMV3 + 4 failed to induce any visible local or systemic infection, further attesting to the purity of BMV3 + 4 virions.
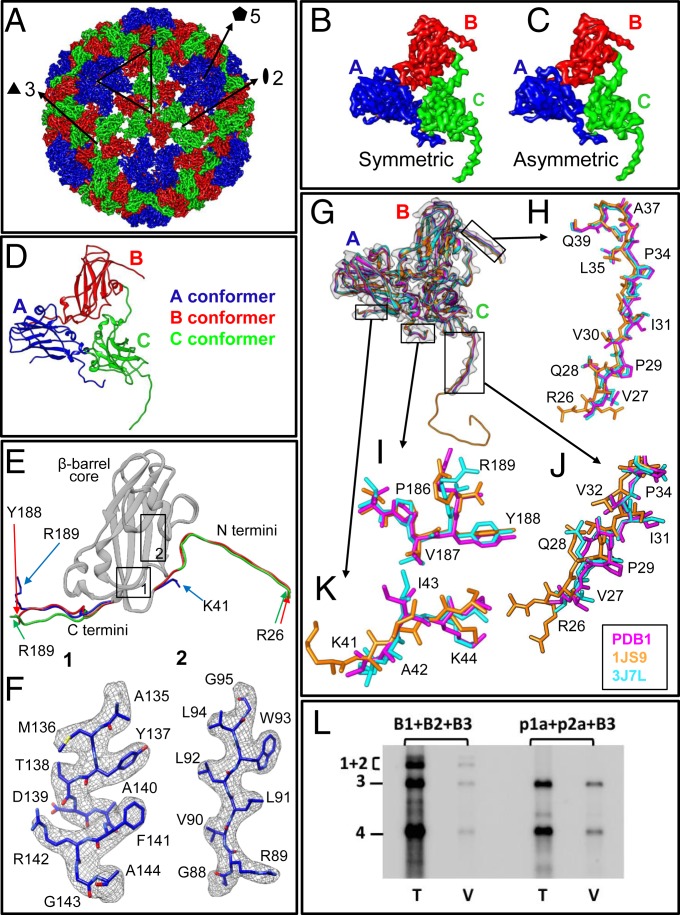
Fig. 1.
Atomic structure of the BMV3 + 4 pure virions. (A) Exterior of the 3.1-Å resolution symmetric reconstruction of BMV3 + 4, highlighting the highly symmetric capsid, in close agreement with previous structures determined for wild-type BMV (5, 29). (B and C) CryoEM density of the asymmetric unit of the capsid showing that both symmetric (B) and asymmetric (C) reconstruction result in the same CP density. (D) Ribbon depiction of the atomic model of the asymmetric unit. (E) Superposition of the atomic model of each of the three asymmetric CP subunits A, B, and C. Note that B and C are nearly identical, while A differs at both the N and C termini. The rectangles labeled 1 and 2 represent the regions of the CP shown in F. (F) Close-up view of the CP structure. The cryoEM density (mesh) is superposed with our atomic model (sticks). (G–K) Comparison of our atomic model of the asymmetric unit with those determined by X-ray crystallography (5) and a previous cryoEM study (29); G depicts the three models overlaid on top of one another within our cryoEM density, while H–K show the boxed regions from G, emphasizing the similarity between the three models at the level of the amino acid side chains. (L) Demonstration of purity of BMV3 + 4 virions by northern blot analysis of either total nucleic acid preparations (T) or RNA extracted from virions (V) isolated from leaves infiltrated with a mixture of agroplasmids containing either B1 + B2 + B3 (lanes 1 and 2) or p1a + p2a + B3 (lanes 3 and 4); Materials and Methods has details. For reference, the position of the four BMV RNAs is shown to the left.
The RNA Is Disordered but Is Located Preferentially near the Two- and Threefold Symmetry Axes.
We obtained 2,943 high-resolution electron-counted movies (SI Appendix, Fig. S1 and Movies S1–S5) containing 79,140 frozen hydrated BMV3 +4 particles and determined high-resolution symmetric and asymmetric reconstructions. The resolution of the symmetric reconstruction (Fig. 1 A and B) is 3.1 Å (SI Appendix, Fig. S6A) and that of the asymmetric reconstruction (Fig. 1C) is 3.9 Å (SI Appendix, Fig. S6B) based on “gold standard” Fourier shell correlation (FSC) curves. The symmetric reconstruction shown in Fig. 1A is colored by CP conformation, depicting each of the A (blue), B (red), and C (green) conformers distinctly. Atomic models of the three CP conformers were built de novo based on side chain densities (Fig. 1 D and E) clearly visible in both the symmetric and asymmetric reconstructions. Fig. 1F shows the cryoEM density (mesh) overlaid on top of the atomic model, illustrating the quality of the dataset as individual amino acid side chains are easily resolved.
The symmetric and asymmetric reconstructions shown in Fig. 1 B and C are essentially identical, with both structures lacking features at the interior of the capsid (SI Appendix, Fig. S2). This similarity in both the capsid and RNA structure is attributed to the dominance of the CP signal in the reconstruction, which obscures that of the RNA genome. Both capsid structures agree well up to the level of amino acid side chain orientations with those determined previously for wild-type BMV—which involves a mixture of all three virions, with 3 + 4 particles in the majority—using cryoEM symmetric reconstruction (29) and X-ray crystallography (5) (Fig. 1 G–K). We resolve residues 41 to 189 of the A conformer, residues 26 to 188 of the B conformer, and residues 26 to 189 of the C conformer. While the study performed by Lucas et al. (5) modeled the first 25 amino acids of the N terminus of the C conformer as a simple polyalanine chain, our results suggest that this density is likely composed of a mixture of RNA and the flexible N terminus as we see density at both the B and C conformers that comprise the capsid hexamers.
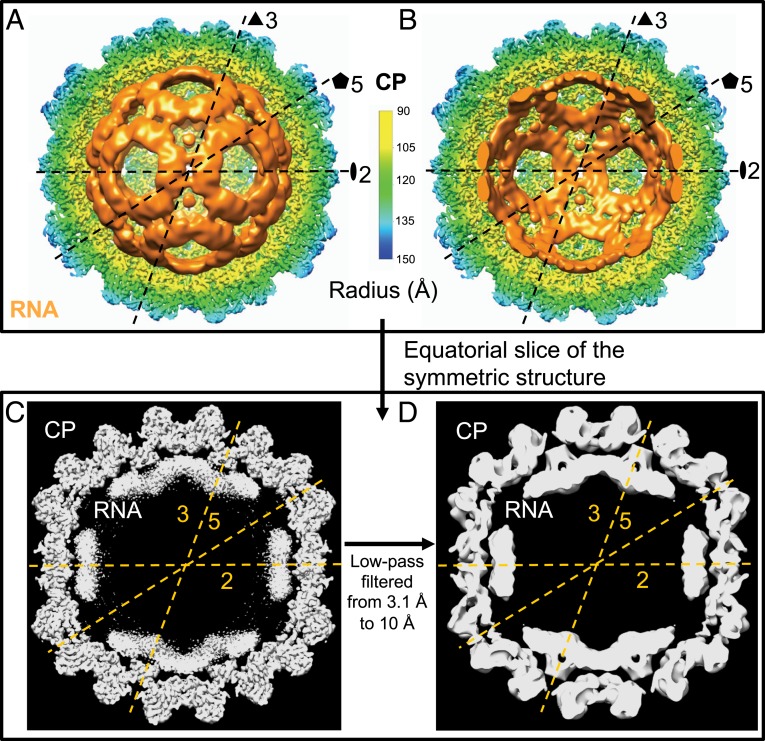
Fig. 2 A and B shows the internal density of the ssRNA genome (colored orange) that survives icosahedral averaging during the structure determination, making clear its interaction with the N termini of the hexameric capsomers (Fig. 2 C and D). The RNA density in BMV3 + 4 virions is found predominantly at the hexamers near the two- and threefold symmetry axes and forms rings around the pentamers at the fivefold symmetry axes. In fact, the resolved density suggests that the N termini of the CP hexamers at the threefold symmetry axes bind the RNA and order it between the two hexameric capsomers. Notably, the interior of the particle appears empty, indicating that there is little ordered RNA here. The RNA density resolved in our structure is similar to that from previous work, although in the prior work—due to the presence of a mixture of three different RNA-containing particles (7, 13, 34)—the internal density cannot be interpreted. Unfortunately, it is problematic to associate this RNA density with a particular structure (e.g., a dsRNA hairpin loop). In this context, an attempt to fit a model 23-base pair dsRNA hairpin loop [the HIV-1 A-rich hairpin loop (35)] into our structure made clear that the density that we see is significantly flatter than that expected for dsRNA. It is interesting, however, that we see strong density attributable to the RNA genome at these locations on the capsid; the fact that this density lacks a particular structure implies that no specific RNA–protein motif is dominant but rather, that there are nonspecific interactions between CP and RNA at this position. Furthermore, if we assume that the density seen is in fact an ensemble of RNA duplexes with varying sequence and coordinates, then the amount of RNA resolved is roughly equivalent to 1.5 kb—or 50% of the total.
Fig. 2.
Symmetric reconstruction showing partially ordered RNA. (A and B) Interior radially colored view of the symmetric reconstruction showing the back half of the CP shell and either the entire (A) or the back half of (B) the RNA genome filtered to 10 Å and colored orange. The capsid symmetry axes are shown on the RNA genome for guidance. Notice that, in A and B, the RNA sits near the two- and threefold axes but not near the fivefold. No RNA density is resolved at the center of the particle. (C and D) Equatorial slab of the symmetric reconstruction without (C) and with (D) low-pass filtering to 10 Å. The two-, three-, and fivefold symmetry axes have been indicated, and it is clear that the RNA is situated preferentially near the two- and threefold axes and away from the fivefold axes. In fact, in the 10-Å structure, we see that the only resolved RNA–CP contact occurs between the RNA at the threefold axes and the hexameric N termini at these positions.
The RNA Adopts an Ensemble of Secondary/Tertiary Structures inside the Capsid.
While the symmetric structure (Fig. 2) shows segments of the internal RNA genome that survive icosahedral averaging, visualization of the rest of the RNA is in principle possible by carrying out an asymmetric reconstruction (26–28). However, the asymmetric reconstruction of BMV shows that simply using the raw data is not sufficient for visualizing the asymmetric genome, suggesting that the symmetrical capsid dominates the signal and interferes in the orientation search for the asymmetric components attributable to the RNA (SI Appendix, Fig. S2).
This problem is similar to that faced in the recent cryoEM studies of the genomic structures in dsRNA viruses (36, 37) and was addressed by subtracting the icosahedrally symmetric contribution to the raw data (SI Appendix, Fig. S4). We have thus followed this procedure to generate a new dataset that only contains the RNA and internal CP density (i.e., the disordered N termini) that was not resolved in the symmetric reconstruction. This subtracted dataset is then used to obtain RNA-dominant structures. We obtain 70 similar but distinct three-dimensional (3D) structures—in fact, as many as we ask for, all of which have an identical capsid structure with variations in the RNA density (four examples are in SI Appendix, Fig. S3). Each of these structures represents a roughly equivalent portion of the dataset, and notably, the capsid structure is resolved to high resolution (around 4 Å); however, the RNA genome shows little ordered density at the interior of the particle, but instead, it is found to be disordered and preferentially near the capsid surface.
Among the many distinct classes, each corresponds to a different organization of the RNA and can be characterized as a shell of RNA density (surface RNA in SI Appendix, Fig. S3) consistently concentrated along the two- and threefold symmetry axes and often having low density at the fivefold axes. These common features of RNA organization agree well with both our own symmetric reconstruction and previous structural works on wild-type BMV (5, 13, 29). Additionally, the low resolution to which these structures are determined indicates that each of these structures themselves represents an average of many RNA secondary structures. In fact, Relion is capable of sorting each of the classes shown above into as many roughly equally populated classes as we ask for (38). These results strongly suggest that the RNA inside of BMV3 + 4 particles is associated with an ensemble of secondary/tertiary structures.
Only the N Termini of the Hexameric CPs Contact the RNA.
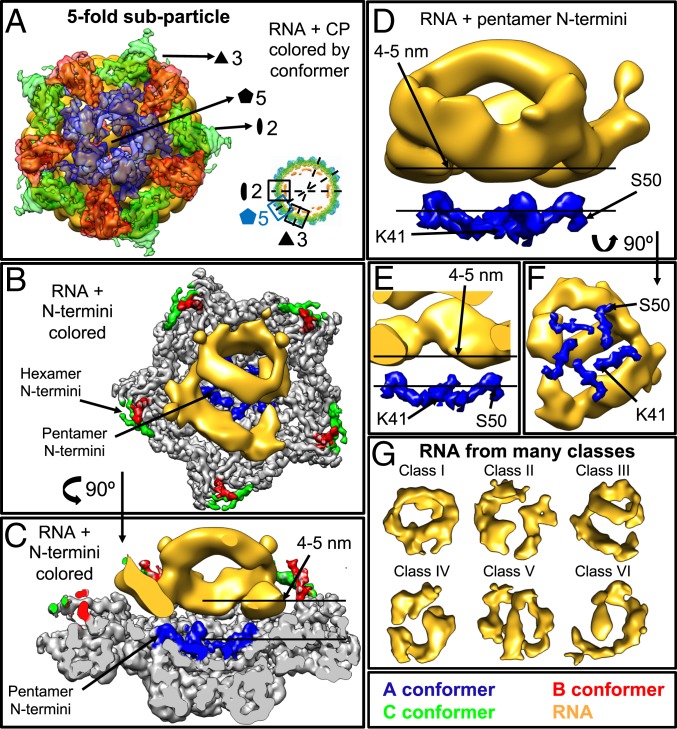
While more of the genome was resolved through capsid subtraction, RNA–CP contacts were still not resolved. We attempted to resolve these contacts by following a subparticle reconstruction protocol, which focuses in on each CP symmetry axis. More explicitly, three-, five-, and twofold “subparticles” (Figs. 3–5) were extracted from specified positions; then, 3D classification was carried out to generate 20 classes, and six representative classes were selected and reconstructed (SI Appendix, Fig. S5). In other words, subparticles in the three- (Fig. 3), five- (Fig. 4), or twofold (Fig. 5) symmetry groups are classified, and the resulting subclasses are asymmetrically reconstructed, allowing for only the signal in these regions to contribute to their classification and final reconstruction.
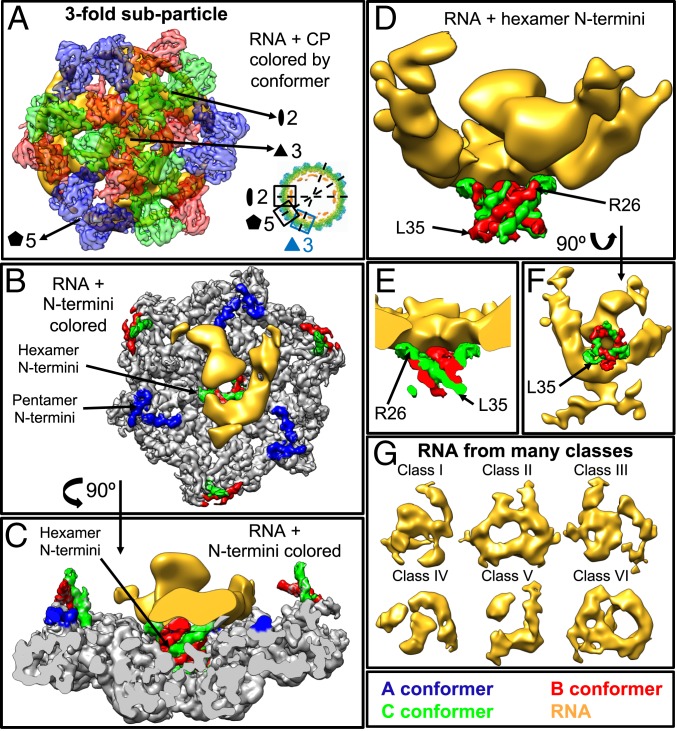
Fig. 3.
Subparticle reconstruction of the threefold axis showing that only the N termini of the hexameric CP contact the RNA. (A) Exterior view of the density from a representative class generated by threefold subparticle reconstruction. The CP has been colored by conformer, and the two-, three-, and fivefold symmetry axes are indicated. Inset shows an equatorial slab of the 3.1-Å symmetric reconstruction indicating the locations of the two-, three-, and fivefold subparticle reconstructions. (B and C) Interior (B) and side (C) views of the density shown in A, with most of the CP colored gray and only the N termini colored by conformer. The RNA has been low-pass filtered to 10-Å resolution. (D and E) Side view (D) and half-side view (E) of the density shown in B and C with only the N termini of the hexameric B and C conformers shown interacting with the RNA. (F) Exterior view of the RNA shown at the interior of the CP N termini colored by conformer. Note that, in B–F, the RNA density is interacting with the hexameric B and C conformers. This is the only RNA–CP contact that is seen across all of the reconstructions generated by subparticle reconstruction. (G) Representative structures of the internal density associated with the CP N termini and the RNA genome generated by threefold subparticle reconstruction and low-pass filtered to 10-Å resolution. Many RNA conformations are generated, but in all, the RNA preferentially sits near the two- and threefold axes and not near the fivefold.
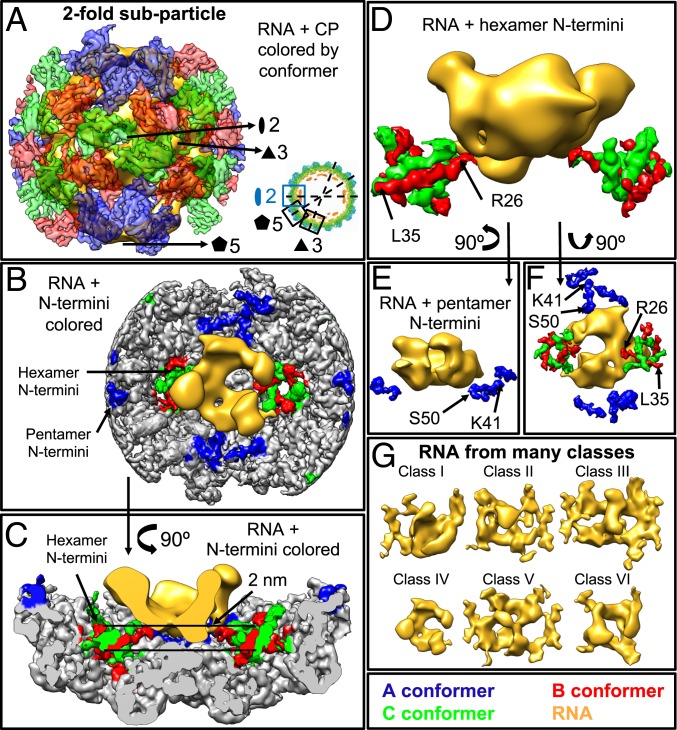
Fig. 5.
Subparticle reconstruction of the twofold axis showing that only the N termini of the hexameric CP contact the RNA. (A) Exterior view of the density from a representative class generated by twofold subparticle reconstruction. The color schemes and map resolutions are described in Fig. 3. (B and C) Interior (B) and side (C) views of the density shown in A. (D and E) Side view as shown in C with (D) only the N termini of the hexameric B and C conformers shown interacting with the RNA or (E) only the pentameric A conformers shown away from the RNA. (F) Exterior view of the RNA shown at the interior of the CP N termini colored by conformer. Note that, in B–F, the RNA density is preferentially near the hexameric B and C conformers. (G) Representative structures of the internal density in the twofold subparticle reconstruction.
Fig. 4.
Subparticle reconstruction of the fivefold axis showing that the N termini of the pentameric CP do not contact the RNA. (A) Exterior view of the density from a representative class generated by fivefold subparticle reconstruction. The color schemes and map resolutions are described in Fig. 3. (B and C) Interior (B) and side (C) views of the density shown in A. (D and E) Side view (D) and half-side view (E) of the density shown in B and C with only the N termini of the pentameric A conformers shown, which are 4 to 5 nm away from the nearest RNA. (F) Exterior view of the RNA shown at the interior of the CP N termini colored by conformer. Note that, in B–F, the RNA density is relatively far away from the pentameric N termini and that little RNA density is resolved at the center of the fivefold symmetry axes. (G) Representative structures of the internal density in the fivefold subparticle reconstruction.
Each of Figs. 3–5 shows subparticle reconstructions of one of the three (three-, five-, and twofold) symmetry axes, and each figure, therefore, presents the information about each symmetry axis in a similar way. Figs. 3A, 4A, and 5A show the exterior of the subparticle reconstruction to illustrate the CPs that have been reconstructed, while Figs. 3 B and C, 4 B and C, and 5 B and C show the interior (Figs. 3B, 4B, and 5B) and side (Figs. 3C, 4C, and 5C) views of a representative density map with the CP N termini colored by conformer, highlighting where the RNA density sits within the subparticle region. Figs. 3 D–F, 4 D–F, and 5 D–F show only the RNA density and the CP N termini since the N terminus is the RNA binding motif. Finally, Figs. 3G, 4G, and 5G show the six unique RNA secondary/tertiary structures generated for each subparticle axis.
Once again, classification generated almost equally populated classes as shown in SI Appendix, Figs. S4 and S5, and if a particular class is chosen and allowed to be separated into more subclasses, then the number of subclasses asked for is again generated with roughly equal populations. While these analyses show as before that an ensemble of RNA–CP conformations exists within the BMV3 + 4 virion (Figs. 3G, 4G, and 5G and SI Appendix, Fig. S3), they also provide additional information about the RNA–CP contacts in both the hexameric and pentameric capsomers.
More explicitly, we find that the β-barrel core at the threefold axes (Fig. 3) acts as the dominant RNA binding domain (Fig. 3 D and E), with the N termini of the B and C conformers extending from the core into the interior of the particle in close proximity to—in fact, often touching—the RNA. Fig. 3 D–F highlights the last nine amino acid residues (between R26 and L35) built into the protein atomic model, showing that our inability to resolve the first 25 amino acids in the atomic model is likely because these residues are strongly bound to the RNA.
In contrast, the fivefold axes (Fig. 4) have little to no RNA density near them—in fact, the RNA is as far as 4 to 5 nm away from the capsid at this point (Fig. 4 C–E)—and the RNA does not contact the pentameric capsomers in any of the maps generated. The A conformer, which makes up the fivefold capsomers, is also exceptional in that the first 40 amino acid residues cannot be resolved. One possible explanation for the lack of RNA density resolved at the fivefold axes is that the N termini of the A conformers are structurally dynamic, resulting in little order at these positions. In this scenario, it is possible that the RNA and N termini are binding to one another, but we are unable to see either of them because they are fluctuating significantly from particle to particle.
The twofold axes (Fig. 5) are unique in that all three protein conformers (A, B, and C) are present. Fig. 5C shows that the RNA is about 2 nm away from the capsid at the twofold axes, and we find that the RNA density contacts various amino acid residues within this region. However, none of the observed contacts are conserved across the many classes that were reconstructed, indicating that they are weaker interactions than those seen between the hexameric N termini and the RNA. Reconstructing this axis confirms the results obtained for the three- and fivefold axes, namely that the RNA density is predominantly found near the threefold axes and away from the fivefold axes. Fig. 5 D and F shows that the N termini of the B and C conformers (amino acids R26 to L35) are contacting the RNA density, while Fig. 5 E and F shows that the N termini of the A conformers (amino acids K41 to S50) are located away from the RNA density.
X-ray crystallography of CCMV has shown that residues 34, 42, 45, 47, 48, 82, 85, 87, 89, 140, 143, 179, 181, and 184 interact with density that was attributed to RNA (7), while recent studies using Clip-Seq analysis (13, 39)—in which the binding footprints of CP for RNA and vice versa are investigated by cross-linking capsids with different RNA cargo—have found that over 60% of the CP interacts with RNA. We find that residues 34, 42 to 48, 82 to 89, and 140 to 143 in BMV (Figs. 3–5), all of which are on the interior surface of the capsid near the hexameric N termini, make contact with the RNA. While some RNA density is resolved near residues 179 to 184, the RNA signal is significantly weaker, indicating a weaker interaction between these residues and the RNA.
In summary, we find that the only strong points of contact between the CP and the RNA—those that are seen in many of the generated classes—occur at the N termini of the hexameric CPs at the threefold symmetry axis (Figs. 3 D–F and 5 D–F). In fact, in every structure generated for the hexameric capsomer, at least one of the N termini and often several interact with the RNA, while no other points of CP–RNA contact are seen in more than one of the generated structures. To illustrate this point further, we counted the number of CP–RNA contacts observed at each of the three symmetry axes for the six classes and found the number of RNA–CP contacts at the three-, two-, and fivefold axes to be 32, 2, and 4, respectively. These results agree with the low pass-filtered symmetric reconstruction shown in Fig. 2D, where the only RNA–CP contact is again seen near the threefold symmetry axis. Furthermore, little to no contact between pentameric CPs and the RNA is resolved (Figs. 4 D–F and 5 D–F), which agrees with the lack of RNA density seen near pentameric capsomers in the symmetric and asymmetric reconstructions and also, agrees well with previous symmetric reconstructions of wild-type BMV (5, 13).
Discussion
We resolve the organization of the ssRNA genome inside a multipartite, symmetric virus particle with pure RNA content. We find that most of the RNA density is organized into a shell at the capsid surface, indicating that many different portions of the RNA genome are interacting with the CP in a variety of ways. Additionally, we show that many RNA conformations exist within BMV virions—an ensemble of RNA structures—all of which are associated with a shell of RNA interacting with the capsid.
Our results indicate that the multipartite plant virus BMV and the monopartite (14) bacteriophage MS2 differ dramatically in the way that their genomes are organized in virions. It has been shown that a single RNA secondary structure is present in each MS2 particle and that the RNA density is conserved even at the center of the particle (26, 27), while in BMV, we do not resolve a high-resolution RNA secondary structure and only see RNA density at the capsid surface. This result provides further evidence that these viruses assemble differently, with BMV utilizing nonspecific electrostatic interactions to drive assembly and MS2 using highly specific CP–RNA contacts—packaging signals. Intermediate situations have been observed in cases where the viral genome is a single molecule with packaging signals that are weaker and less broadly distributed than in MS2. More explicitly, for satellite tobacco mosaic virus, double-helical portions of genomic RNA have been shown to be ordered along the inner icosahedral edges of the T = 1 capsid (40–42), while for satellite tobacco necrosis virus, specific single-stranded tetraloop sequences are identified that interact strongly with N-terminal CP (43). In the case of tomato bushy stunt virus with its T = 3 capsid, on the other hand, the N termini are less ordered, and a smaller portion of the RNA genome density survives icosahedral averaging (44). Studies of other ssRNA virus structures are expected to point to further instances of intermediate behavior for the nature and extent of genome ordering in viral capsids.
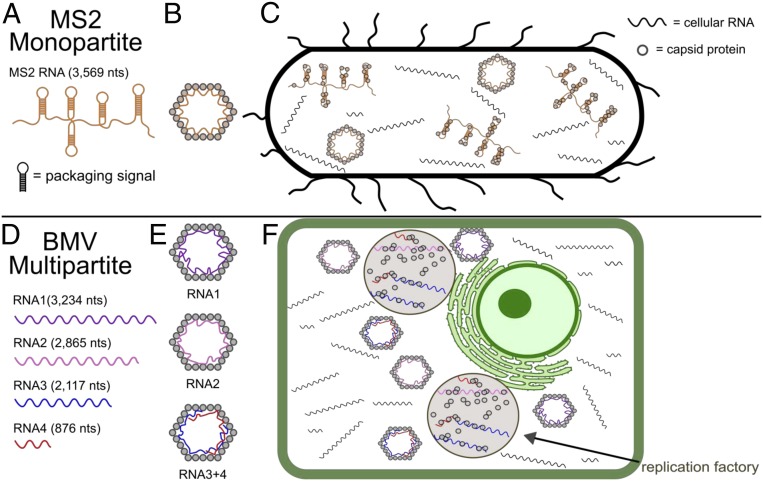
We hypothesize that BMV utilizes nonspecific interactions for virion assembly to enable its multipartite lifestyle in which it packages four RNAs into three otherwise-identical particles. The ensemble of RNA secondary/tertiary structures allowed by the CP enables the virus more flexibility in packaging its many genomic RNAs. This result also emphasizes a difference in the lifecycle of these two classes of viruses (Fig. 6). More explicitly, multipartite viruses often modify host cell membranes to create isolated vesicles in which viral replication and genome packaging can occur (45, 46). These “replication factories” allow the viruses to package directly their own newly synthesized genomes without needing the sequence-specific interactions utilized by MS2 and Qβ to exclude nonviral RNAs.
Fig. 6.
Cartoon illustrating the differences between (A–C) a monopartite virus (MS2) and (D–F) multipartite virus (BMV). (A) The monopartite genome of MS2 with its sequence-specific packaging signals. (B) The monopartite MS2 particle with its RNA strongly interacting with particular sites of the MS2 CP. (C) The viral lifecycle of a monopartite virus occurs in the cytosol, and the virus relies on sequence-specific interactions to preferentially package its own RNA. (D) The multipartite BMV genome has four RNAs of varying length and sequence. (E) The multipartite BMV is composed of a mixture of three different particles, one containing RNA1, another containing RNA2, and a third containing both RNAs 3 and 4. Additionally, the RNA interacts nonspecifically with the CP. (F) The multipartite BMV replicates and assembles in replication factories—separate lipid vesicles (gray regions) created by the viral infection, allowing it to preferentially package its own genome.
We are unable to ascertain to what extent the packaging of two molecules (RNAs 3 and 4) rather than a single one (RNA1 or RNA2) affects the extent to which RNA is ordered in the capsid. Recent experiments involving trypsin-protease digestion of B1, B2, and B3 + 4 virions analyzed by both western blot analysis and MALDI-TOF show that B1 and B2 virions exhibit a distinct protease digestion profile from B3 + 4 virions, suggesting that the packaged RNA affects capsid dynamics (47). Accordingly, we plan to carry out high-resolution reconstructions, similar to those reported here, with in planta-prepared BMV1 and BMV2 virions to determine differences in RNA ordering when a single RNA molecule is inside.
Materials and Methods
Buffers.
The following buffers were employed. Virus suspension buffer for imaging (VSB) was 50 mM Tris⋅HCl, pH 7.2, 50 mM NaCl, 10 mM KCl, and 5 mM MgCl2. Virus purification buffer was 50 mM sodium acetate, pH 4.5, and 8 mM magnesium acetate.
Strategy for In Vivo Production of Pure BMV3 + 4 Virions and Transmission Electron Microscopy.
The strategy used to produce in vivo pure BMV virions exclusively containing RNA3 + 4 is as follows. Nicotiana benthamiana plants were agroinfiltrated with an inoculum containing a mixture of three agroplasmids: p1a and p2a (nonreplicating constructs engineered to transiently express replicase protein 1a and 2a, respectively) (48) as well as pB3 (engineered to express a replication-competent full-length BMV RNA3) (30). Following infiltration, a functional replicase complex is assembled from the transiently expressed p1a and p2a that directs the complete replication of B3 RNA followed by the translation of CP subunits from replication-derived subgenomic RNA4 (B4). Since RNA packaging in BMV is functionally coupled to replication (31, 32) and no RNA1 or RNA2 is replicated, only RNA3 + 4 containing virions are produced. BMV3 + 4 virions are then purified from agroinfiltrated leaves using protocols as described by Annamalai and Rao (30). Virions purified from plants infiltrated with a mixture of agrotransformants of all three wild-type BMV RNAs (i.e., pB1 + pB2 + pB3) served as positive controls. The structural integrity of the virions is verified by negative stain electron microscopy—as described previously by Cadena-Nava et al. (49), while the genetic purity of the virions is confirmed by both northern blot hybridization (30) and infectivity assays in C. quinoa, a local lesion host for BMV (50).
Northern Blot Analysis.
Approximately 5 µg of total nucleic acid preparation and 100 ng of virion RNA were denatured with formamide/formaldehyde and subjected to 1.2% agarose gel electrophoresis prior to vacuum blotting onto a nylon membrane. The blot was hybridized with 32P-labeled riboprobes complementary to the homologous 3′ noncoding region present on all four BMV RNAs (30).
Cryoelectron Microscopy.
After purification from plants, virus was washed three times with VSB through a 100-kDa Amicon filter and concentrated to a final concentration of 5 mg/mL. CryoEM grids were prepared as described previously (51). CryoEM imaging was performed with an FEI Titan Krios equipped with a Gatan imaging filter (GIF) and post-GIF K2 direct electron detector. The microscope was operated at 300 kV, and the GIF slit was set to 20 eV. Movies were recorded as dose-fractionated frames using Leginon (52) at a 130-kx nominal magnification in superresolution mode (corresponding to a calibrated pixel size of 0.535 Å on the specimen level). Each movie was recorded for 6 s at an electron dose rate of 8 electrons per 1 Å2 per second and fractionated equally into a stack of 30 frames, resulting in an accumulated dose of 48 electrons per 1 Å2 on the specimen level; 2,943 movies were collected over a 3-d session.
The movies were aligned using MotionCor2 (53) with subframe motion correction (5 × 5 array) and binned two times to a final pixel size of 1.07 Å to generate both dose-weighted—used for final reconstruction—and dose-unweighted averaged micrographs—used for manual screening, CTF determination, and particle picking. The defocus values of the micrographs were determined with CTFFIND4 (54). Micrographs with ice contamination or a defocus value outside the range −0.8 to −3 µm were discarded. A total of 79,140 particles were picked from manually selected micrographs using Ethan (55).
Symmetric and Asymmetric Reconstruction.
All reconstruction and classification methods were carried out using Relion (38). The 79,140 selected particles were extracted followed by 3D classification to generate three classes with icosahedral symmetry using a Gaussian ball as the initial template; 70,470 particles were selected from the “good” class as indicated in SI Appendix, Fig. S4, and those particles were subjected to 3D refinement with icosahedral symmetry, resulting in a 3.1-Å resolution icosahedrally symmetric reconstruction as shown in Fig. 1 A and B. The same particles were refined with C1 symmetry (i.e., no symmetry) using the symmetric map as the initial model, resulting in a 3.9-Å resolution asymmetric reconstruction shown in Fig. 1C and SI Appendix, Fig. S2B. During refinement, particles were divided into two random halves and refined separately. The resolution was then determined by the gold standard FSC = 0.143 (56) (SI Appendix, Fig. S6). The final density map was generated by merging the two half datasets. The atomic model of the CP was built with Coot (57) using the Protein Data Bank ID code 3J7L as the reference, refined with the real-space refinement modules in Phenix (58), and visualized with UCSF Chimera (59) following the protocol described previously (60).
Capsid Subtraction and RNA-Focused Asymmetric Reconstruction.
As described in Results, in order to focus on the RNA density we used the capsid subtraction method described below. The capsid signal in the raw images was subtracted using projections of the CP (zeroing the density in the inner shell of the icosahedral map). The Relion command “relion_particle_symmetry_expand” (38) was used with the icosahedral symmetry option (I2) to expand each icosahedral orientation entry in the subtracted particles’ data STAR file into 60 icosahedrally related entries in an expanded data STAR file. This expanded STAR file was then used to run Relion 3D classification with the option “–skip_align” (38) with a Tau number of three by requesting 70 classes (SI Appendix, Fig. S4). Four representative classes were selected, and the raw (i.e., unsubtracted) particles corresponding to the subtracted particles in each class were used to generate four full-capsid maps with the Relion command “relion_reconstruct” with a maximum resolution of 4 Å. In this way, full-virus asymmetric maps were generated but with their C1 orientations sorted out from 60 icosahedrally related orientations by the internal genome.
Subparticle Reconstruction.
As described in Results, in order to resolve important CP–RNA contacts near each CP symmetry axis, we used a subparticle reconstruction workflow (61–66) as illustrated in SI Appendix, Fig. S5. Briefly, the Relion command "relion_particle_symmetry_expand" (38) was used with the icosahedral symmetry option (I2) to expand the icosahedral orientation entry in the data file generated from icosahedral reconstruction into 60 icosahedrally oriented entries in a new data STAR file. This data STAR file was then used to reextract subparticle regions from specified subparticle positions from each virus particle image. For subparticles at the icosahedral two-, three-, and fivefold symmetry axes, the specified positions in the icosahedral (I2) map are (x = 90, y = 0, and z = 0 pixels); (x = 0, y = 34, and z = 90 pixels); and (x = 0, y = 80, and z = 48 pixels), respectively. For each symmetry axis, 3D classification was then carried out in Relion with the option "–skip_align" by requesting 20 classes as shown in SI Appendix, Fig. S5. Six representative classes were selected, and the particles in each class were used to generate six maps using the "relion_reconstruct" command with a maximum resolution of 4 Å.
Data Deposition.
All data are available in the manuscript or SI Appendix. Accession numbers listed here are also shown in SI Appendix, Table S1: 1 is in the Protein Data Bank, which is ID code 6VOC, and 24 are in EMDB, which are accession numbers EMD-21260, EMD-21261, and EMD-21279 to EMD-21300.
Supplementary Material
Acknowledgments
We thank Ivo Atanasov for technical assistance with the electron microscopy, Wei Liu for help generating animations, Cheylene Tanimoto for help with illustrations, and Robijn Bruinsma for discussions related to RNA genome packaging. This research was supported by the University of California Multi-Campus Research Programs Initiative (A.L.N.R. and W.M.G.); NIH Grants AI094386 (to Z.H.Z.), DE025567 (to Z.H.Z.), DE028583 (to Z.H.Z.), and GM071940 (to Z.H.Z.); and NSF Grant Macrosystems Biology (MCB) 1716925 (to W.M.G.). We acknowledge the use of resources at the Electron Imaging Center for Nanomachines supported by the University of California, Los Angeles and by NIH Instrumentation Grants 1S10RR23057, 1S10OD018111, and U24GM116792 and NSF Instrumentation Grants DMR-1548924 and DBI-1338135.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Twenty-four cryoEM structures have been deposited in the Electron Microscopy Data Bank (EMDB) (accession nos. EMD-21260, EMD-21261, and EMD-21279–EMD-21300), and one atomic model has been deposited in the Protein Data Bank (ID code 6VOC).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915078117/-/DCSupplemental.
References
- 1.Caspar D. L. D., Klug A., Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962). [DOI] [PubMed] [Google Scholar]
- 2.Ho P. T., et al. , VIPERdb: A tool for virus research. Annu. Rev. Virol. 5, 477–488 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Valegård K., Liljas L., Fridborg K., Unge T., The three-dimensional structure of the bacterial virus MS2. Nature 345, 36–41 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Golmohammadi R., Fridborg K., Bundule M., Valegård K., Liljas L., The crystal structure of bacteriophage Q beta at 3.5 A resolution. Structure 4, 543–554 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Lucas R. W., Larson S. B., McPherson A., The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317, 95–108 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Rossmann M. G., Smiley I. E., Wagner M. A., Crystalline cowpea chlorotic mottle virus. J. Mol. Biol. 74, 255–256 (1973). [DOI] [PubMed] [Google Scholar]
- 7.Speir J. A., Munshi S., Wang G., Baker T. S., Johnson J. E., Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3, 63–78 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sicard A., Michalakis Y., Gutiérrez S., Blanc S., The strange lifestyle of multipartite viruses. PLoS Pathog. 12, e1005819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahlquist P., Dasgupta R., Kaesberg P., Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J. Mol. Biol. 172, 369–383 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Ahlquist P., Luckow V., Kaesberg P., Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 153, 23–38 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Shrestha N., et al. , Next generation sequencing reveals packaging of host RNAs by brome mosaic virus. Virus Res. 252, 82–90 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Fox J. M., et al. , Comparison of the native CCMV virion with in vitro assembled CCMV virions by cryoelectron microscopy and image reconstruction. Virology 244, 212–218 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Hoover H. S., et al. , Phosphorylation of the brome mosaic virus capsid regulates the timing of viral infection. J. Virol. 90, 7748–7760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiers W., et al. , Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene. Nature 260, 500–507 (1976). [DOI] [PubMed] [Google Scholar]
- 15.Bancroft J. B., Hiebert E., Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology 32, 354–356 (1967). [DOI] [PubMed] [Google Scholar]
- 16.Hung P. P., Ling C. M., Overby L. R., Self-assembly of Q-beta and MS2 phage particles: Possible function of initiation complexes. Science 166, 1638–1640 (1969). [DOI] [PubMed] [Google Scholar]
- 17.Dykeman E. C., Stockley P. G., Twarock R., Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. J. Mol. Biol. 425, 3235–3249 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Basnak G., et al. , Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid. J. Mol. Biol. 395, 924–936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolfsson Ó., et al. , Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 428, 431–448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayer T. S., Booth L. N., Knudsen S. M., Ellington A. D., Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11, 1848–1857 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calhoun S. L., Rao A. L., Functional analysis of brome mosaic virus coat protein RNA-interacting domains. Arch. Virol. 153, 231–245 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Ni P., et al. , An examination of the electrostatic interactions between the N-terminal tail of the Brome Mosaic Virus coat protein and encapsidated RNAs. J. Mol. Biol. 419, 284–300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., Konecny R., Baker N. A., McCammon J. A., Electrostatic interaction between RNA and protein capsid in cowpea chlorotic mottle virus simulated by a coarse-grain RNA model and a Monte Carlo approach. Biopolymers 75, 325–337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y. G., Dreher T. W., Rao A. L., tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. U.S.A. 99, 655–660 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs A., et al. , The intrinsically disordered N-terminal arm of the brome mosaic virus coat protein specifically recognizes the RNA motif that directs the initiation of viral RNA replication. Nucleic Acids Res. 46, 324–335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai X., et al. , In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 541, 112–116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koning R. I., et al. , Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat. Commun. 7, 12524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorzelnik K. V., et al. , Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ. Proc. Natl. Acad. Sci. U.S.A. 113, 11519–11524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., et al. , An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 5, 4808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annamalai P., Rao A. L., Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: A functional role for viral replicase in RNA packaging. Virology 338, 96–111 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Seo J. K., Kwon S. J., Rao A. L., A physical interaction between viral replicase and capsid protein is required for genome-packaging specificity in an RNA virus. J. Virol. 86, 6210–6221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annamalai P., Rao A. L. N., Delivery and expression of functional viral RNA genomes in planta by agroinfiltration. Curr. Protoc. Microbiol. 16, 16B.2.1–16B.2.15 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Choi Y. G., Grantham G. L., Rao A. L., Molecular studies on bromovirus capsid protein. Virology 270, 377–385 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Konecny R., et al. , Electrostatic properties of cowpea chlorotic mottle virus and cucumber mosaic virus capsids. Biopolymers 82, 106–120 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglisi E. V., Puglisi J. D., HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 5, 1033–1036 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Liu H., Cheng L., Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 349, 1347–1350 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., et al. , In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527, 531–534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni P., Vaughan R. C., Tragesser B., Hoover H., Kao C. C., The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J. Mol. Biol. 426, 1061–1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larson S. B., Day J. S., McPherson A., Satellite tobacco mosaic virus refined to 1.4 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 70, 2316–2330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larson S. B., et al. , Double-helical RNA in satellite tobacco mosaic virus. Nature 361, 179–182 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Larson S. B., McPherson A., Satellite tobacco mosaic virus RNA: Structure and implications for assembly. Curr. Opin. Struct. Biol. 11, 59–65 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Ford R. J., et al. , Sequence-specific, RNA-protein interactions overcome electrostatic barriers preventing assembly of satellite tobacco necrosis virus coat protein. J. Mol. Biol. 425, 1050–1064 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmins P. A., Wild D., Witz J., The three-dimensional distribution of RNA and protein in the interior of tomato bushy stunt virus: A neutron low-resolution single-crystal diffraction study. Structure 2, 1191–1201 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Ertel K. J., et al. , Cryo-electron tomography reveals novel features of a viral RNA replication compartment. eLife 6, e25940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Restrepo-Hartwig M. A., Ahlquist P., Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70, 8908–8916 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarty A., Reddy V. S., Rao A. L. N., Unravelling the stability and capsid dynamics of the three virions of brome mosaic virus assembled autonomously in vivo. J. Virol., 10.1128/JVI.01794-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaturvedi S., Rao A. L. N., Live cell imaging of interactions between replicase and capsid protein of Brome mosaic virus using Bimolecular Fluorescence Complementation: Implications for replication and genome packaging. Virology 464–465, 67–75 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Cadena-Nava R. D., et al. , Self-assembly of viral capsid protein and RNA molecules of different sizes: Requirement for a specific high protein/RNA mass ratio. J. Virol. 86, 3318–3326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao A. L., Grantham G. L., Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 211, 42–52 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Cui Y, et al. , pH-dependent gating mechanism of the Helicobacter pylori urea channel revealed by cryo-EM. Sci. Adv. 5, eaav8423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C., et al. , Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kivioja T., Ravantti J., Verkhovsky A., Ukkonen E., Bamford D., Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 131, 126–134 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen E. F., et al. , UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Yu I., et al. , Building atomic models based on near atomic resolution cryoEM maps with existing tools. J. Struct. Biol. 204, 313–318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilca S. L., et al. , Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Z., et al. , Double-stranded RNA virus outer shell assembly by bona fide domain-swapping. Nat. Commun. 8, 14814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui Y., Zhang Y., Zhou K., Sun J., Zhou Z. H., Conservative transcription in three steps visualized in a double-stranded RNA virus. Nat. Struct. Mol. Biol. 26, 1023–1034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y., et al. , Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes. Nat. Commun. 10, 5346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Y., et al. , In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating. Proc. Natl. Acad. Sci. U.S.A. 116, 16535–16540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ilca S. L., et al. , Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature 570, 252–256 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.