Significance
GBM is the most deadly adult brain cancer. The current standard-of-care is surgery followed by RT and temozolomide, which results in a median survival time of only 15 mo. The efficacy of chemotherapies and targeted therapies in GBM is very limited because most of these drugs do not pass the blood−brain barrier (BBB). Ultimately, all patients succumb to the disease. Our study describes radiation-induced cellular plasticity as a resistance mechanism in GBM. We identified a dopamine receptor antagonist as a readily available, FDA-approved drug known to penetrate the BBB, which prevents phenotype conversion of glioma cells into glioma-initiating cells and prolongs survival in mouse models of GBM, thus suggesting that it will improve the efficacy of RT without increasing toxicity.
Keywords: glioblastoma, radiation, dopamine receptor antagonist, dedifferentiation, glioma-initiating cells
Abstract
Glioblastoma (GBM) is the deadliest adult brain cancer, and all patients ultimately succumb to the disease. Radiation therapy (RT) provides survival benefit of 6 mo over surgery alone, but these results have not improved in decades. We report that radiation induces a glioma-initiating cell phenotype, and we have identified trifluoperazine (TFP) as a compound that interferes with this phenotype conversion. TFP causes loss of radiation-induced Nanog mRNA expression, and activation of GSK3 with consecutive posttranslational reduction in p-Akt, Sox2, and β-catenin protein levels. TFP did not alter the intrinsic radiation sensitivity of glioma-initiating cells (GICs). Continuous treatment with TFP and a single dose of radiation reduced the number of GICs in vivo and prolonged survival in syngeneic and patient-derived orthotopic xenograft (PDOX) mouse models of GBM. Our findings suggest that the combination of a dopamine receptor antagonist with radiation enhances the efficacy of RT in GBM by preventing radiation-induced phenotype conversion of radiosensitive non-GICs into treatment-resistant, induced GICs (iGICs).
Glioblastoma (GBM) is among the deadliest cancers in adults, with almost all patients succumbing to the disease. The median survival is only 15 mo (1). The current standard-of-care for GBM patients involves postoperative radiation therapy (RT), which prolongs survival by ∼6 mo over surgery alone (2, 3), and temozolomide treatment, which prolongs progression-free survival (PFS) by another ∼3 mo over postoperative RT alone (1). Addition of classical chemotherapies, antiangiogenic strategies, and novel biologics to the current standard-of-care have all failed to improve the outcome for GBM patients, which has remained largely unchanged since the 1970s (2, 3).
Over the last four decades, RT for GBM underwent several iterations, to whole brain irradiation (WBI) versus gross tumor volume, to standard fractionation schemes versus accelerated hyperfractionated RT, to hypofractionated intensity-modulated RT (4). However, although RT is an indispensable part of the treatment regimen for GBM patients, strategies aimed at improving response to RT have failed to increase PFS. The current standard-of-care in the RT of GBM has its origin in studies showing that escalating the WBI dose from 45 Gy to 60 Gy led to increased survival (3). Yet, dose escalation from 60 Gy to 70, 80, and 90 Gy given to the tumor alone showed no dose–response effect or improved outcome, and the tumors still recurred locally (5).
The almost universal fatal outcome of GBM patients treated by standard-of-care including postoperative RT stands in contrast to the intrinsic radiation sensitivity of GBM cells, which falls into the same range of radiosensitivity seen in other solid tumors frequently cured by total radiation doses of 60 Gy (6). In the above study, the SF2Gy (surviving fraction at 2 Gy) values for glioma cells in vitro were comparable to those of other cancers, including squamous cell carcinoma, colon cancer, and soft tissue sarcoma (6). Likewise, TCD50 (the radiation dose necessary to control 50% of the tumors locally) values were not exceptionally high, and TCD50 and SF2Gy values did not correlate with each other (6). With most systemic agents being ineffective and surgery and RT having reached their maximum potential, treatment for GBM has hit a critical barrier.
A mounting body of evidence points to a hierarchical organization of GBM with a small number of glioma-initiating cells (GICs) at the apex of that hierarchy, able to self-renew and to repopulate a tumor (7, 8). Importantly, GICs were found to be relatively resistant to radiation (9) and chemotherapy (10). A number of marker systems have been used to prospectively identify GICs, including the surface marker CD133 (9) and the lack of proteasome activity (11). A competing stochastic model assumes that every cell in GBM can acquire GIC traits over time. More recent reports indicate that, while the hierarchical model describes the organization of GBM quite accurately, glioma cells exhibit plasticity that allows them to phenotype convert into induced GICs (iGICs) in response to stress (12).
In this study, we identify radiation-induced phenotype conversion of GBM cells into radioresistant iGICs as a facet of GBM treatment resistance. Importantly, we demonstrate that trifluoperazine (TFP), a compound identified in a high-throughput screen for its ability to prevent radiation-induced phenotype conversion of glioma cells into radiation-resistant tumor-initiating cells, can prolong survival in mouse models of GBM.
Materials and Methods
Cell Culture.
Primary human glioma cell lines were established at University of California, Los Angeles (UCLA) as described [Hemmati et al. (7); characteristics of specific gliomasphere lines can be found in Laks et al. (13)]. The GL261 murine glioma cell line was a kind gift of William H. McBride, Department of Radiation Oncology at UCLA. GL261 cells were cultured in log-growth phase in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. Primary GBM cells were propagated as gliomaspheres in serum-free conditions in ultra-low-adhesion plates in DMEM/F12, supplemented with B27, epidermal growth factor (EGF), fibroblast growth factor 2 (bFGF), and heparin as described previously (7, 11, 13). All cells were grown in a humidified atmosphere at 37 °C with 5% CO2. The unique identity of all patient-derived specimens was confirmed by DNA fingerprinting (Laragen). All lines were routinely tested for mycoplasma infection (MycoAlert; Lonza).
ZsGreen−C-terminal degron of murine ornithine decarboxylase (cODC) expressing cells were obtained as described in ref. 11. Briefly, cells were infected with a lentiviral vector coding for a fusion protein between the fluorescent protein ZsGreen and cODC. The latter targets ZsGreen to ubiquitin-independent degradation by the 26S proteasome, thus reporting lack of proteasome function through accumulation of ZsGreen-cODC. We previously reported that cancer cell populations lacking proteasome activity are enriched for tumor-initiating cells in GBM, breast cancer, and cancer of the head and neck region (11, 14, 15), and others have confirmed these findings independently in tumors of the liver, lung, cervix, pancreas, osteosarcoma, and colon (16–21). After infection with the lentivirus, cells expressing the ZsGreen-cODC fusion protein were further selected with G418 for 5 d. Successful infection was verified using the proteasome inhibitor MG132 (Sigma).
Cell Cycle Analysis.
After 5 d of 0- or 8-Gy irradiation, HK-374 ZsGreen-cODC expressing cells were stained with Hoechst 33342 and pyronin Y. Briefly, cells were trypsinized and rinsed with Hanks’ balanced salt solution (HBSS)/1 mM Hepes/10% FBS. DNA was stained with 1 μg/mL Hoechst 33342 in HBSS/1 mM Hepes/10% FBS solution/50 μM Verapamil for 45 min at 37 °C. The cells were rinsed once and then stained for RNA with 4 μM pyronin Y in HBSS/1 mM Hepes/10% FBS solution for 45 min at 37 °C. Finally, cells were rinsed and resuspended in PBS. At least 100,000 cells were analyzed by flow cytometry.
Senescence Analysis.
HK-374 ZsGreen-cODC expressing cells were plated in six-well plates at a density of 50,000 cells per well and, the next day, irradiated with 0 Gy or 8 Gy. Five days after irradiation, the plates were assayed for senescence via the Senescence β-galactosidase Cell Staining Kit (Cell Signaling, Catalog #9860), and the staining was performed as per manufacturer’s protocol.
Animals.
Six- to eight-week-old C57BL/6 mice, or NOD-scid IL2Rgammanull (NSG) originally obtained from The Jackson Laboratories were rederived, bred, and maintained in a pathogen-free environment in the American Association of Laboratory Animal Care-accredited Animal Facilities of Department of Radiation Oncology, UCLA, in accordance with all local and national guidelines for the care of animals. Weights of the animals were recorded every day; 2 × 105 GL261-Luc and 3 × 105 HK-308-Luc or HK-374-Luc cells were implanted into the right striatum of the brains of mice using a stereotactic frame (Kopf Instruments) and a nanoinjector pump (Stoelting). Injection coordinates were 0.5 mm anterior and 2.25 mm lateral to the bregma, at a depth of 3.5 mm from the surface of the brain. Tumors were grown for 3 (HK-374), 7 (GL261) or 21 (HK-308) d, after which successful grafting was confirmed by bioluminescence imaging. Mice that developed neurological deficits requiring euthanasia were killed.
In Vivo Bioluminescent Imaging.
Starting 1 and 3 wk after implantation of xenografts, GL261-Luc-bearing C57BL/6 mice and NSG mice bearing HK-308-Luc or HK-374-Luc tumors were imaged at regular intervals, and the tumor-associated bioluminescent signal was recorded. Prior to imaging, the mice were injected intraperitoneally (i.p.) with 100 µL of D-luciferin (15 mg/mL; Gold Biotechnology). Five minutes later, animals were anesthetized (2% isofluorane gas in O2), and luminescence was recorded (IVIS Spectrum; Perkin-Elmer). Images were analyzed with Living Image Software (Caliper LifeSciences).
Brain Tissue Digestion and Flow Cytometry.
A total of 2 × 105 GL261-Strawberry-Red (StrawRed) and 3 × 105 HK-374- or HK-157-StrawRed cells were implanted into the brains of C57BL/6 or NSG mice, respectively, as described above. Tumors were grown for 3 (HK-374) and 7 (HK-157 and GL261) d for successful grafting. Mice bearing tumors were injected i.p. on a 5-d on/2-d off schedule for 1 or 2 wk (GL261), 4 wk (HK-374), and 6 wk (HK-157) either with TFP or saline. TFP was dissolved in sterile saline at a concentration of 2.5 mg/mL. All animals were treated with 20 mg/kg TFP. At the indicated time points after implantation, the mice were killed, and tumor-bearing brains were dissected for further analysis. Detailed procedure for brain tumor dissociation and flow cytometric analysis is available in SI Appendix, Material and Methods.
For radiation-induced reprogramming experiments in vivo with GL261-StrawRed and GL261-BFP cells, GL261-StrawRed cells were first stained with an anti-Prominin-APC antibody (Miltenyi; Catalog #130-102-197), and the Prominin-positive cells were removed by fluorescence-activated cell sorter (FACS). The Prominin-negative cells were seeded into six-well plates at a density of 50,000 cells per well. Cells were irradiated with 0 or 4 Gy and incubated at 37 °C in a CO2 incubator for 1 d. After 5 d, GL261-StrawRed cells along with GL261-BFP cells were stained with an anti-Prominin-APC antibody to determine the percentage of Prominin-positive cells in the total population. Then, 25,000 cells from each cell population (GL261-StrawRed and GL261-BFP) and from each group (0 or 4 Gy) were mixed together, and a total of 50,000 cells were injected into mice intracranially. The mice were maintained for 3 wk, and, after that, the brains were dissociated as described above for flow cytometric analysis with GL261-StrawRed and GL261-BFP cells stained with anti-Prominin antibody.
Drug Treatment.
TFP was identified as an inhibitor of radiation-induced phenotype conversion in a high-throughput screen described previously (22). After confirming tumor grafting via bioluminescent imaging, mice bearing GL261 tumors were injected i.p. on a 5-d on/2-d off schedule for 3 wk either with TFP or saline. Mice implanted with the HK-374 or HK-308 specimen were injected subcutaneously (s.c.) on a 5-d on/2-d off schedule with TFP or saline until they reached euthanasia endpoints. TFP was dissolved in sterile saline at a concentration of 2.5 mg/mL. All animals were treated with 20 mg/kg TFP, the published (maximum tolerated dose) MTD for mice (23).
Irradiation.
Cells or mice were irradiated at room temperature using an experimental X-ray irradiator (Gulmay Medical Inc.) at a dose rate of 5.519 Gy/min for the time required to apply a prescribed dose. The X-ray beam was operated at 300 kV and hardened using a 4-mm Be, a 3-mm Al, and a 1.5-mm Cu filter and calibrated using National Institute of Standards and Technology-traceable dosimetry. Corresponding controls were sham irradiated.
For the assessment of the effect of TFP in combination with irradiation in vivo, mice were anesthetized prior to irradiation with an i.p. injection of 30 μL of a ketamine (100 mg/mL; Phoenix) and xylazine (20 mg/mL; AnaSed) mixture (4:1) and placed on their sides into an irradiation jig that allows for irradiation of the midbrain while shielding the esophagus, eyes, and the rest of the body. Animals implanted with GL261 cells received a single dose of 10 Gy on day 8 after tumor implantation. Animals injected with the HK-308 glioma specimen received a single dose of 4 or 10 Gy on day 21 after tumor implantation. Animals injected with the HK-374 glioma specimen received a single dose of 10 Gy on day 3 after tumor implantation.
In Vitro Sphere Formation Assay.
For the assessment of self-renewal in vitro, cells were irradiated with 0, 2, 4, 6, or 8 Gy and seeded under serum-free conditions into plates, not treated for tissue culture, in DMEM/F12 media, supplemented with 10 mL/500 mL of B27 (Invitrogen), 0.145 U/mL recombinant insulin (Eli Lilly), 0.68 U/mL heparin (Fresenius Kabi), 20 ng/mL bFGF (Sigma), and 20 ng/mL EGF (Sigma). The number of spheres formed at each dose point was normalized against the nonirradiated control. The resulting data points were fitted using a linear quadratic model.
qRT-PCR.
Total RNA was isolated using TRIZOL Reagent (Invitrogen). Complementary DNA synthesis was carried out using the SuperScript Reverse Transcription III (Invitrogen). Quantitative PCR was performed in the My iQ thermal cycler (Bio-Rad) using the 2× iQ SYBR Green Supermix (Bio-Rad). Ct for each gene was determined after normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or Ribosomal Protein Lateral Stalk Subunit P0 (RPLP0) and ΔΔCt (delta delta Ct) was calculated relative to the designated reference sample. Gene expression values were then set equal to 2−ΔΔCt as described by the manufacturer of the kit (Applied Biosystems). All PCR primers were synthesized by Invitrogen. Primer list and the primer sequences are available in SI Appendix, Table S1.
Chromatin Immunoprecipitation PCR.
Chromatin immunoprecipitation PCR (ChIP-PCR) was performed using SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling, Catalog #9005) by following the manufacturer’s protocol. Briefly, HK-374 cells were plated in 15-cm dishes and cultured until they reached confluency. The plates were irradiated at 0 or 4 Gy, and, 48 h later, these cells were used to perform the ChIP-PCR. Detailed description of the ChIP-PCR is available in SI Appendix, Material and Methods.
Histone Enzyme-Linked Immunosorbent Assay.
HK-374 cells were irradiated with 0 and 4 Gy. Forty-eight hours after irradiation, histone proteins were isolated using EpiQuik Total Histone Extraxtion Kit (Epigentek; Catalog #OP-0006). Detailed description of the method is available in SI Appendix, Material and Methods.
RNA Sequencing.
At 1 and 48 h after 4 Gy irradiation or sham irradiation, RNA was extracted from ZsGreen-cODC−negative HK-374 non-GICs using TRIzol. RNA sequencing (RNA-Seq) analysis was performed by Novogene. Quality and integrity of total RNA was controlled on an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies). The RNA-Seq library was generated using NEBNext Ultra RNA Library Prep Kit (New England Biolabs) according to manufacturer’s protocols. The library concentration was quantified using a Qubit 3.0 fluorometer (Life Technologies), and then diluted to 1 ng/uL before checking insert size on an Agilent Technologies 2100 Bioanalyzer and quantifying to greater accuracy by qPCR (library molarity > 2 nM). The library was sequenced on Illumina NovaSeq6000 with an average of 43.4 million reads per RNA sample. Detailed description of downstream analysis and differential expression analysis is available in SI Appendix, Material and Methods.
Assay for Transposase-Accessible Chromatin Using Sequencing.
Cells were harvested and frozen in culture media containing FBS and 5% dimethyl sulfoxide. Cryopreserved cells were sent to Active Motif to perform the assay for transposases-accessible chromatin using sequencing (ATAC-seq) assay using their standard protocol as described in SI Appendix, Material and Methods.
Small Interfering RNA Treatment.
The 70% confluent HK-374 cell cultures were grown in antibiotic-free culture media overnight. The next day the culture media was replaced with 1 mL of Opti-MEM reduced serum media. To this control or DRD2, small interfering RNA (siRNA)−lipid complex (1:1) was added as per the manufacturer’s protocol. Detailed description of siRNA treatment and protein knockdown validation is available in SI Appendix, Material and Methods.
Western Blotting.
GBM cells were serum starved overnight and, the following day, pretreated with 10 μM TFP for 1 h. Pretreated HK-374 and HK-345 cells were irradiated with a single dose of 8 Gy immediately after a second treatment with 10 μM TFP. One hour after irradiation, the cells were lysed in radioimmuno precipitation assay (RIPA) lysis buffer containing proteinase inhibitor and phosphatase inhibitor. The protein concentration in each sample was determined by bicinchoninic assay (BCA) protein assay, and samples were denaturated in 4× Laemmli sample buffer containing 10% β-mercaptoethanol for 10 min at 95 °C. Western blotting was performed to demonstrate the change in protein expression levels from different treatment conditions.
To analyze for changes in the expression levels of Yamanaka factors in HK-374 cells 5 d after exposing them to 8 Gy irradiation, Western blotting was performed using proteins extracted from FACS-sorted ZsGreen-cODC−negative and ZsGreen-cODC−positive cells treated with 8 Gy. Detailed description of the Western blotting procedure and the protein targets is available in SI Appendix, Material and Methods.
Immunohistochemistry.
Brains were explanted, fixed in formalin for 24 h, and embedded in paraffin. Immunohistochemistry was performed on these paraffin-embedded slides. The method used is available in SI Appendix, Material and Methods.
Statistics.
Unless stated otherwise, all data shown are represented as mean ± SEM of at least three biologically independent experiments. A P value of ≤0.05 in an unpaired two-sided t test indicated a statistically significant difference. Kaplan−Meier estimates were calculated using the GraphPad Prism Software package. For Kaplan−Meier estimates, a P value of 0.05 in a log-rank test indicated a statistically significant difference.
Data Sharing.
All data and methods are included in the manuscript and SI Appendix. The RNA-Seq data are available via GEO accession number GSE166358.
Results
Relative Radiation Sensitivity of Glioma-Initiating Cells.
As described before, the radiation sensitivity of bulk glioma cell populations was found to be in the same range as the radiosensitivity of other solid tumors frequently cured by radiation (6, 24). In light of the known heterogeneity of GBM cell populations and the reported relative radioresistance of GICs compared to their more differentiated progeny (9), we first sought to test whether the radioresistance of GICs differed from that of tumor-initiating cells in breast cancer. Using four primary patient-derived GBM lines, we performed sphere formation assays under serum-free conditions after exposing the cells to radiation doses of 0, 2, 4, 6, or 8 Gy (Fig. 1A). Under these conditions, GICs form gliomaspheres, while more-differentiated glioma cells die from anoikis. As expected, GICs exhibited a range of surviving fractions of 0.4 to 0.63 at 2 Gy. However, a comparison with our published data on the radiation sensitivity of breast cancer-initiating cells (25) did not reveal a more radioresistant phenotype of GICs. Like the radiation sensitivity of bulk cell populations in GBM which falls into the same range of radiation sensitivity reported for other solid cancers (6), the radiation sensitivity of GICs is comparable to that of their equivalent in breast cancer, thus suggesting that the intrinsic radiation resistance of GICs does not explain why GBM cannot be controlled by RT and often recurs within or in proximity to the RT target volume.
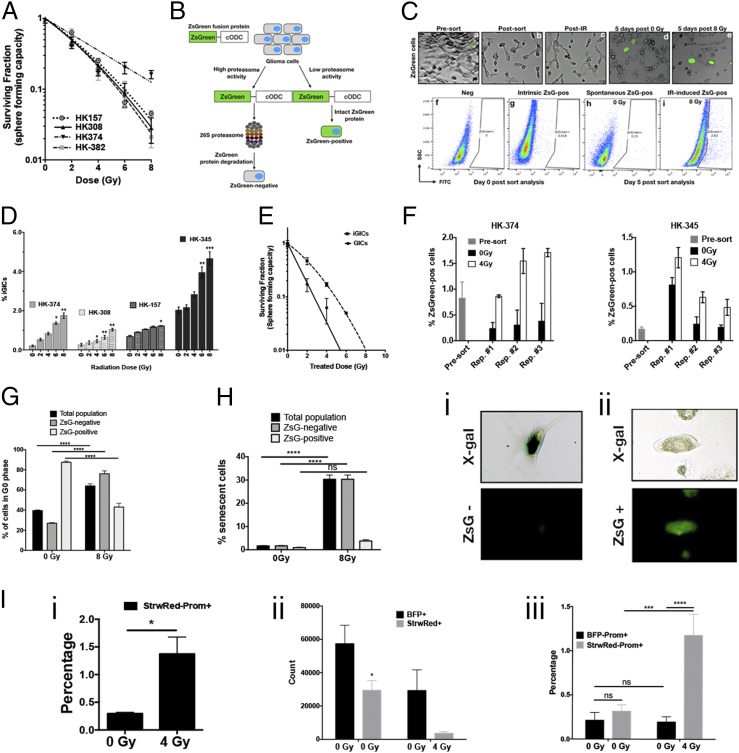
Fig. 1.
Radiation induces phenotype conversion in GBM. (A) Four primary patient-derived GBM lines (parent HK-157, HK-308, HK-374, and HK-382) were seeded as single-cell suspension cultures in 96-well plates containing serum-free medium and exposed to radiation doses of 0, 2, 4, 6, or 8 Gy. The spheres were maintained by feeding with fresh growth factors (10× medium) regularly. The number of spheres formed at each dose point was counted and normalized against their respective nonirradiated control. The final curve was generated using a linear quadratic model. (B) Schematic representation of the ZsGreen-cODC reporter system. (C) Representative images (10×) of HK-374 ZsGreen-cODC infected cells before and after sort (a and b), after irradiation (c), 5 d after 0 Gy irradiation (d), and 5 d post 8-Gy irradiation (e). Flow sort analysis representative images of HK-374−negative cells (f), HK-374 cells before sorting (g), 0-Gy sample 5 d postirradiation (h), and 8-Gy samples 5 d postsorting (i). (D) Four primary patient-derived GBM lines (HK-308, HK-374, HK-157, and HK-345) were infected with ZsGreen-cODC reporter vector and maintained as an adherent culture in a 10% serum-containing medium. After selection, the cells were sorted to deplete the ZsGreen-cODC−positive GICs. The remaining population consisting of differentiated ZsGreen-cODC−negative glioma cells were plated at a density of 50,000 cells per well in a six-well plate and, the following day, irradiated with single doses of 0, 2, 4, 6, or 8 Gy. Five days later, the cells were detached and analyzed for ZsGreen-cODC−positive cells by flow cytometry, using their noninfected parent lines as controls, and represented as percentage iGICs. (E) ZsGreen-cODC vector-expressing HK-374 cells were sorted, and the ZsGreen-cODC−positive cells obtained were seeded in a 96-well plate for sphere formation assay. The remaining population was plated and subjected to irradiation at 0, 2, 4, 6, or 8 Gy. Five days later, the cells were detached and sorted for ZsGreen-cODC−positive cells. These iGICs were then seeded in a 96-well plate for sphere formation assay. The number of spheres formed at each dose point was counted and normalized against the control. The resulting data were fitted using the linear quadratic model. (F) ZsGreen-cODC vector-expressing HK-374 and HK-345 cells were sorted for ZsGreen-cODC−negative cells and irradiated at 4 Gy. Five days later, a small portion of these cells were used for flow cytometry to analyze for percentage of ZsGreen-cODC−positive cells. The remaining cells were sorted for ZsGreen-cODC−negative cells and again irradiated at 4 Gy. This approach was followed two more times. The ZsGreen-cODC−positive cells obtained after each repeat were graphed and presented as percentage ZsGreen-cODC−positive cells. (G) Cell cycle analysis demonstrating percentage of cells in G0 phase of the cell cycle in three different cell populations—total, ZsGreen-cODC−negative, and ZsGreen-cODC−positive cells, after 5 d of irradiation with a single dose of 0- and 8-Gy irradiation. (H) Cellular senescence analysis 5 d post single dose of 0- and 8-Gy irradiation in HK-374 ZsGreen-cODC infected cells demonstrating percentage of senescent cells in three different populations—total, ZsGreen-negative, and ZsGreen-cODC−positive cells. Representative images of a cell showing positive for X-gal but negative for ZsGreen-cODC protein (i); representative image (20×) of a cell showing negative for X-gal but positive for ZsGreen-cODC protein (ii). (I) GL261-StrawRed cells were first sorted for cells negative for Prominin. The sorted cells were then irradiated with a single dose of 4 Gy and incubated at 37 °C in a CO2 incubator for 5 d. Percentage of Prominin+ cells in the cell preparation from the presort (0 Gy) and the sorted, irradiated, and 5-d postincubation (4 Gy) GL261-sorted-irradiated-StrawRed cells were analyzed by performing flow cytometry (i). Next, 1:1 ratio of the sorted GL261-StrawRed cells and the unsorted GL261-BFP cells were intracranially implanted into C57BL/6 mice. Three weeks postimplantation, the brains of the mice were extracted and tumor cells were dissociated and labeled with anti-Prominin antibody. The antibody-labeled cells were subjected to flow cytometric analysis, and the number of either StrawRed or BFP-positive cells obtained from each brain dissociation prep was graphed (ii); the cells positive for both StrawRed and Prominin or BFP and Prominin were also graphed and presented as percentage positive (iii). All experiments in this figure have been performed with at least three biological independent repeats. P values were calculated using unpaired t test. *P value < 0.05, **P value < 0.01, ***P value < 0.001, and ****P value < 0.0001, ns, no significance.
Radiation-Induced Phenotype Conversion in GBM.
We have previously reported that triple-negative and claudin-low breast cancers exhibit high rates of spontaneous and radiation-induced phenotype conversion of nontumorigenic breast cancer cells into breast cancer-initiating cells, while nontumorigenic luminal breast cancer cells show only very low rates of phenotype conversion (26). Likewise, phenotype conversion from CD133neg into CD133pos GICs in response to changes in the tumor microenvironment has been previously reported for GBM (12). Phenotype conversion in irradiated glioma cells has not been investigated, and, therefore, we next tested whether ionizing radiation would also induce this phenomenon in GBM. We utilized our imaging system for tumor-initiating cells (11) to deplete cell populations from ZsGreen-cODC−positive GICs with low proteasome activity by high-speed FACS (Fig. 1B). The system is based on a fusion protein of ZsGreen and cODC, which targets the protein to ubiquitin-independent proteasomal degradation. Cell populations lacking proteasome activity can be identified by accumulation of the fluorescent ZsGreen protein and are enriched for tumor-initiating cells in a large number of solid tumors, including GBM (11, 15, 27). In order to confirm our prior results, we implanted ZsGreen-positive cells into immunodeficient mice and found that they formed more-aggressive tumors in vivo (SI Appendix, Fig. S1). Once GICs were removed by sorting, the remaining populations of differentiated glioma cells were irradiated with single doses of 0, 2, 4, 6, or 8 Gy. Five days after a single dose of radiation, the cells were analyzed by flow cytometry, and the emergence of ZsGreen-cODC−positive cells with low proteasome activity was interpreted as a measure for phenotype conversion. In general, the patient-derived primary GBM samples displayed various degrees of spontaneous (without irradiation treatment) phenotype conversion in the range of 0.21 to 2% (Fig. 1 C and D, 0 Gy). Irradiation with single doses led to a dose-dependent increase of phenotype conversion up to 4.7% of the cells undergoing phenotype conversion after 8 Gy (Fig. 1 C and D). Importantly, iGICs were more resistant to radiation than preexisting GICs in vitro (Fig. 1E).
Next, we tested whether phenotype conversion could be repeatedly induced or whether it rather reflected a population of cells prone to acquiring GIC traits. ZsGreen-cODC−positive cells were purged from the bulk population of HK-374 and HK-345 cells by FACS, and the remaining populations were irradiated with 4 Gy. After 5 d, the number of radiation-induced ZsGreen-cODC−positive cells (iGICs) was assessed, ZsGreen-cODC−positive cells were purged again and the remaining cells reirradiated with 4 Gy and tested for the induction of ZsGreen-cODC−positive iGICs 5 d later. Using this approach, we found that radiation-induced phenotype conversion was not restricted to a single initial irradiation but could be repeatedly observed, thus indicating that the process cannot simply be explained by cell selection or reflect a restriction to a preexisting cell population prone to phenotype conversion (Fig. 1F).
Furthermore, we addressed whether radiation would increase the number of quiescent ZsGreen-cODC−positive cells by staining the RNA and DNA content of the cells. In agreement with the literature, radiation caused cell cycle arrest in the bulk population of ZsGreen-cODC−negative cells. Unirradiated ZsGreen-cODC−positive cells were predominantly in a quiescent state (G0) but were recruited into the cell cycle in response to radiation (Fig. 1G).
Treatment of unsorted bulk tumor cell population with radiation enriches for radioresistant tumor-initiating cells (9). In order to study the cell fate of ZsGreen-cODC−positive and ZsGreen-cODC–negative cells after irradiation, HK-374-ZsGreen-cODC cells were treated with 0 or 4 Gy. Five days later, cells were fixed and stained for beta-galactosidase activity, a marker for cellular senescence (28). Radiation significantly increased the number of senescent ZsGreen-cODC−negative cells but did not significantly increase the number of senescent ZsGreen-cODC−positive cells (Fig. 1H). Together, this indicated that the radiation-induced occurrence of ZsGreen-cODC−positive cells from ZsGreen-cODC−negative cells cannot be explained by induction of senescence or quiescence.
To test whether iGICs contribute to the recurrence of GBM after irradiation, we engineered GL261 cells to constitutively express the fluorescent proteins StrawRed or Blue-fluorescent protein (BFP). Using CD133 as a validated marker for GICs in GL261 (29), we removed CD133-positive cells from StrawRed-expressing cells and irradiated the remaining CD133-negative cells with 0 or 4 Gy. After 5 d in culture, the percentage of CD133-positive cells was assessed by flow cytometry. Radiation induced a phenotype conversion of CD133-negative cells into CD133-positive cells that significantly exceeded the number of preexisting CD133-positive cells in unirradiated GL261 cells (Fig. 1 I, i); 25,000 cells of irradiated StrawRed-expressing CD133-negative and 25,000 unirradiated, unsorted BFP-expressing cells were mixed and injected into the striatum of C57BL/6 mice. After 3 wk, the brains were harvested and analyzed for the number of CD133-positive StrawRed and BFP-expressing cells. Irradiation with 4 Gy reduced the total number of grafting StrawRed-expressing cells, which was in line with the cytotoxic effects of radiation (Fig. 1 I, ii). However, the population of grafting StrawRed cells was significantly enriched for CD133-positive induced GICs, suggesting an in vivo contribution of iGICs to tumor recurrence after in vitro radiation (Fig. 1 I, iii).
Identification of Compounds That Interfere with Phenotype Conversion.
With the exception of temozolomide, previous attempts to prolong survival of glioma patients through pharmacological intervention have largely failed. Clinically, the BBB limits the use of established chemotherapeutic agents in GBM, and GICs have been shown to resist most established systemic therapies (10). Cancer treatment-induced conversion of GBM cells into therapy-resistant GICs could potentially add to this resistance. Therefore, we sought to screen chemical libraries for compounds that interfere with this process. In a high-throughput screen of 83,000 compounds including libraries of Food and Drug Administration (FDA)-approved drugs, we identified the dopamine receptor antagonist TFP as a potent inhibitor of radiation-induced phenotype conversion.
To confirm the presence of TFP’s target on GBM cells, we characterized expression levels of dopamine receptors (DRD1-4 relative to DRD5) in differentiated and matching GIC-enriched GBM cultures from the same GBM specimen via qRT-PCR. The paired specimen from the HK-308, HK-157, and HK-374 lines expressed at least one of the DRD receptors, while HK-345 cells had very low expression levels for all of the DRD receptors relative to DRD5 (Fig. 2A). Reflective of D2-like DRD expression status, TFP prevented radiation-induced phenotype conversion in HK-308, HK-157, and HK-374 specimens, but had no effect on HK-345 cells (Fig. 2B), supporting a role for dopamine receptors in radiation-induced phenotype conversion in GBM.
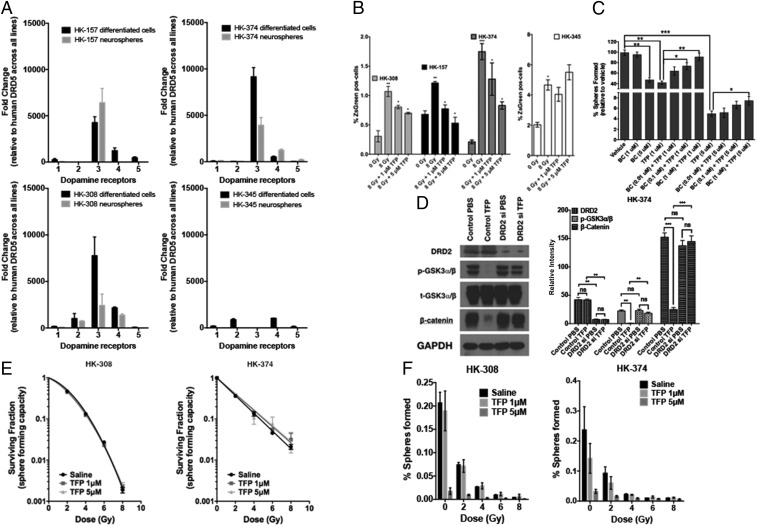
Fig. 2.
TFP prevents radiation-induced phenotype conversion in GBM. (A) Four primary patient-derived GBM lines (HK-157, HK-374, HK-308, and HK-345) were grown as differentiated cells in the presence of 10% serum and as gliomaspheres in serum-free medium. RNA from both cultures was isolated, and qPCR was performed to analyze for the difference in dopamine receptor (DRD1, DRD2, DRD3, DRD4, and DRD5) expression levels. GAPDH was used as the internal control to obtain the dCt (delta Ct) values. The dCt values were normalized to DRD5 across all lines to obtain the ddCt values. The fold change in expression levels of DRDs was calculated by 2^-ddCt method. (B) Sorted ZsGreen-cODC−negative cells from HK-308, HK-157, HK-374, and HK-345 ZsGreen-cODC vector-expressing cells were plated at a density of 50,000 cells per well in a six-well plate and, the following day, pretreated either with TFP (1 and 5 μM) or vehicle (saline) 1 h before irradiation at a single dose of 0 or 8 Gy. Five days later, the cells were detached and analyzed for ZsGreen-cODC−positive cells by flow cytometry, using their noninfected parent lines as controls, and represented as percentage iGICs. (C) Sphere formation assay was performed using the HK-308 parent gliomaspheres plated in a 96-well plate and treating them with different concentrations of TFP (1 and 5 μM) in combination with or without bromocriptine (0.1, 1, and 5 μM). The spheres were fed with growth factors (10×) medium regularly. The number of spheres formed in each condition was counted and normalized against the vehicle control. The resulting data were presented as percentage spheres formed. (D) HK-374 cells were treated with either control siRNA or DRD2-specific siRNA for 72 h, serum-starved for 4 h, and then treated with or without TFP (10 μM). Proteins were extracted and subjected to Western blotting. The membranes were blotted for p-GSK3a/b, t-GSK3a/b, β-catenin, and GAPDH. The intensity of each band was calibrated using ImageJ and presented as density ratio of gene over GAPDH in HK-374 cells. (E and F) Parent HK-308 and HK-374 gliomaspheres seeded in a 96-well plate were pretreated either with TFP (1 or 5 μM) or vehicle (saline) 1 h before irradiation at a single dose of 0, 2, 4, 6, or 8 Gy. This setup was maintained with regular feeding of the spheres with 10× growth factors. The number of spheres formed in each condition was counted and normalized against the respective nonirradiated control. The final curve was generated using a linear quadratic model. All experiments in this figure have been performed with at least three biological independent repeats. P values were calculated using unpaired t test. *P value < 0.05, **P value < 0.01, and ***P value < 0.001.
We further validated DRDs’ interfering with self-renewal by coincubating cells with the DRD agonist, bromocriptine (BC). BC could reverse the inhibitory effect of TFP (Fig. 2C). Next, we knocked down DRD2 expression in HK-374 cells. In wild-type cells, TFP caused a loss of Ser9/21 phosphorylation of GSK3α/β and subsequent degradation of β-catenin. Knockdown of DRD2 prevented TFP-induced loss of Ser9/21 phosphorylation and loss of β-catenin, thus demonstrating dependence of TFP’s effects on β-catenin on the presence of DRD2 (Fig. 2D).
Using sphere-forming assays for HK-308 and HK-374 cells, we further demonstrated that TFP did not act as a classical radiosensitizer, as radiation survival curves of TFP-treated and irradiated samples were identical to their saline-treated corresponding controls (Fig. 2E). However, when combined with radiation, TFP had an additive inhibitory effect on self-renewal capacity, as was assessed in a sphere formation assay (Fig. 2F).
TFP Prevents Radiation-Induced Reexpression of Yamanaka Factors.
We have previously shown that radiation-induced phenotype conversion in breast cancer coincided with the reexpression of the Yamanaka factors Sox2, Oct4, Klf4, and c-Myc and their downstream target Nanog (26). Intrinsic GICs (ZsGreen-cODC−positive cells) showed elevated expression levels of Yamanaka factors when compared to ZsGreen-cODC−negative cells (Fig. 3A, white bars). When patient-derived HK-374 GBM cells, depleted of ZsGreen-cODC−positive cells, were subjected to irradiation, the expression levels of Yamanaka factors, c-Myc, Oct4, Klf4, and Nanog were significantly elevated in the radiation-induced GICs over preexisting GICs (Fig. 3A, checkered bars), and radiation-induced GICs showed elevated protein levels of Sox2, Oct4, Klf4, and Nanog when compared to GBM cells that did not convert their phenotype in response to radiation (Fig. 3B). Similarly, Yamanaka factor expression was up-regulated in iGICs derived from HK-308 cells (Fig. 3C, black bars). Importantly, TFP prevented the radiation-induced up-regulation of the Yamanaka factors and their downstream target, Nanog (Fig. 3C, light gray bars).
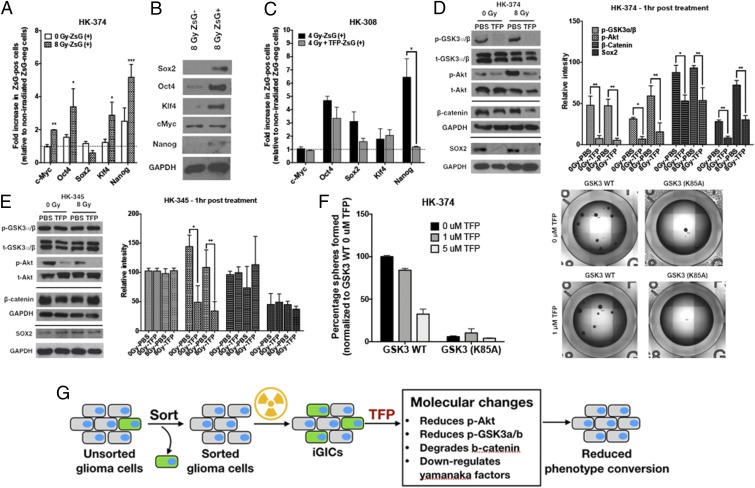
Fig. 3.
TFP reduces stem cell factors at the transcriptional and posttranslational level. ZsGreen-cODC−negative cells were collected after sorting the HK-374 ZsGreen-cODC vector-expressing cells and plated in a six-well plate at a density of 50,000 cells per well. The cells were irradiated at 0 or 8 Gy the following day. Five days after incubation, the cells were detached and resorted to collect both ZsGreen-cODC−positive and ZsGreen-cODC−negative cells. (A) The qPCR was performed using RNA isolated from these samples. GAPDH was used as the internal control. The fold change in expression levels of Yamanaka factors in 0- and 8-Gy ZsGreen-cODC−positive cells was calculated by 2^-ddCt method by normalizing it to nonirradiated ZsGreen-cODC−negative cells. (B) Western blotting was performed with the proteins extracted from 8-Gy ZsGreen-negative and ZsGreen-positive cells, and the membrane was blotted for Sox2, Oct4, Klf4, c-Myc, and Nanog. GAPDH was used as the internal control. (C) ZsGreen-cODC−negative cells were collected after sorting the HK-308 ZsGreen-cODC vector-expressing cells and plated in a six-well plate at a density of 50,000 cells per well. The cells were pretreated with TFP (10 μM) 1 h before irradiating at 0 or 4 Gy the following day. Five days after incubation, the cells were detached and resorted to collect both ZsGreen-cODC−positive and ZsGreen-cODC−negative cells. The qPCR was performed using RNA isolated from these samples. GAPDH was used as the internal control. The fold change in expression levels of Yamanaka factors in 0- and 4-Gy ZsGreen-cODC−positive cells was calculated by 2^-ddCt method by normalizing it to nonirradiated ZsGreen-cODC−negative cells. (D and E) The 90% confluent plates with parent HK-374 and HK-345 cells were serum starved overnight and pretreated with either TFP (10 μM) or vehicle (saline) for 1 h. Just before irradiating these plates with a single dose of 0 or 8 Gy, a second treatment with TFP or saline was performed. Starting from the second treatment, the samples were collected at exactly 1-h time point for protein analysis using Western blotting. The blots were analyzed for p-GSK3a/b, t-GSK3a/b, p-Akt, t-Akt, β-catenin, Sox2, and GAPDH. GAPDH was used as the loading control for β-catenin and Sox2. (D and E) The intensity of each band was calibrated using ImageJ and presented as density ratio of gene over GAPDH in HK-374 and HK-345 cells. (F) HK-374 cells were transduced with lentiviral vector-expressing GSK3 (K85A) mutant gene. HK-374 containing GSK3 WT cells along with HK-374 cells with GSK3 (K85) mutant gene were plated in 96-well plates and treated with TFP (0, 1, and 5 μM). This setup was maintained with regular feeding of the spheres with 10× growth factors. The number of spheres formed in each condition was counted and normalized against the GSK3 WT nonirradiated control. Representative images are of GSK3 WT and GSK3 (K85A) mutant cells at 0 and 5 μM TFP. (G) Schematic representation on how TFP inhibits phenotype conversion. All experiments in this figure have been performed with at least three biological independent repeats. P values were calculated using unpaired t test. *P value < 0.05, **P value < 0.01, and ***P value < 0.001.
TFP Activates GSK3α and Decreases β-Catenin and Sox2 Levels.
GSK3 is a kinase that phosphorylates already phosphorylated proteins to mark them for proteasomal degradation, thus affecting cell signaling in multiple ways (30). Among others, downstream targets include β-catenin, PTEN, p65, HIF1, c-Myc, and Oct4 (30). Active GSK3 lacks phosphorylation on Ser9 or Ser21 and counteracts a stem cell phenotype by phosphorylating key proteins in developmental pathways, such as Oct4 and β-catenin (31), thereby targeting them for degradation. Therefore, inactive GSK3 (phosphorylated on Ser9/21) supports stemness by allowing stem cell factors to escape degradation. In addition, Akt signaling promotes stemness by leading to phosphorylation of Sox2 (32) and phosphorylation of Sox2 requires phosphorylated Akt (32). Phosphorylated Sox2 participates in a transcriptional protein complex composed of XPC, Oct4, Rad23B, and Cent2 that is responsible for initiating transcription of the Nanog gene (33). Signaling through D2-like dopamine receptors leads to phosphorylation of Akt (34) and subsequent inhibitory GSK3 phosphorylation by Akt (35), thus supporting posttranslational stabilization of stem cell factors.
In order to understand how TFP interferes with the acquisition of a stem cell state, HK-374 cells were plated and serum starved overnight and irradiated with 0 or 8 Gy in the presence or absence of TFP. TFP treatment led to a rapid loss of constitutive and radiation-induced phosphorylation of Akt in HK-374 and HK-345 cells at the 1-h time point. Loss of the inhibitory serine-21 phosphorylation of GSK3α was detected in HK-374 cells in which TFP prevented phenotype conversion (Fig. 3D). In contrast, phosphorylation of GSK3α remained unchanged in HK-345 cells (Fig. 3E) in which TFP fails to prevent radiation-induced phenotype conversion (Fig. 2B). Furthermore, TFP treatment reduced constitutive and radiation-induced β-catenin and Sox2 levels at 1 h after TFP treatment in HK-374, but not in HK-345 cells (Fig. 3 D and E).
To show dependence on GSK3, we next overexpressed a dominant-negative kinase-dead form (K85A) of GSK3-beta. Loss of GSK3-beta kinase activity significantly reduced the self-renewal capacity of HK-374 cells measured in a sphere formation assay. This was expected given the multiple known targets of GSK3. Importantly, overexpression of the kinase-dead form of GSK3 rendered the cells unresponsive to TFP in this assay, thus further supporting the role of GSK3 in the response to TFP (Fig. 3F). A graphical summary of these findings is presented in Fig. 3G.
Radiation Induces Developmental Gene Expression Signatures in Glioma Cells.
In order to uncover whether radiation—aside from its DNA-damaging effects and associated cellular responses—also affects developmental programs and to identify additional targets of TFP, we next performed RNA-Seq to map gene expression changes induced in GBM cells by radiation. HK-374 ZsGreen-cODC−negative cells were sorted by FACS, treated with TFP, and irradiated with 4 Gy. At 1 and 48 h after irradiation, total RNA was extracted and subjected to RNA-Seq to study pathways immediately engaged by TFP treatment (1 h) and their downstream consequences for the radiation response (48 h).
Differential gene expression analysis revealed that 1-h treatment with TFP induced changes in the expression levels of 884 genes (Fig. 4 A, Right-Left). In comparison, a single dose of 4 Gy led to 37 differentially expressed genes, with the majority (29/37) of the genes down-regulated (Fig. 4 A, Right-Middle). Combination of TFP treatment with radiation changed the expression of 119 genes (Fig. 4 A, Right-Right). However, at 1 h after drug treatment, no genes were differentially expressed between cells treated with TFP and radiation versus radiation alone (Fig. 4A, Venn diagram, Upper Left). In contrast, 48 h after drug treatment, we identified 2,952 differentially expressed genes when comparing cells treated with TFP and radiation versus radiation alone, creating a distinct gene expression profile in hierarchical clustering (Fig. 4B). We selected the top 9 up-regulated and top 10 down-regulated genes in cells treated with TFP and radiation versus radiation alone and confirmed the expression changes using real-time qRT-PCR (Fig. 4C).
Fig. 4.
TFP interferes with DNA replication and cell cycle progression in GBM. (A) Venn diagram (Upper Left), volcano diagrams (Right), and heat map (Lower Left) of sorted non-GICs from HK-374 cells, 1 h after treatment with radiation (8 Gy), TFP, or TFP and radiation compared to untreated control cells. (B) Venn diagram (Upper Left), volcano diagrams (Right Top and Bottom), and heat map (Lower Left) of sorted non-GICs from HK-374 cells, 48 h after treatment with radiation, TFP, or TFP and radiation compared to untreated control cells. (C) Heat map showing the results of real-time RT-PCR for the top 9 up-regulated and top 10 down-regulated genes in HK-374 non-GICs comparing combination treatment of TFP and radiation with radiation alone, 48 h after treatment. (D) Proteasome subunit and proteasome assembly chaperone mRNA expression 48 h after radiation or TFP treatment. (E) Gene set enrichment analysis was performed on the data obtained by RNA-Seq analysis to identify differentially expressed genes that overlapped with curated gene sets representing expression of gene signatures pertaining to genetic and chemical perturbations.
In agreement with our previous studies (27), radiation led to the down-regulation of proteasome subunit and NF-YB expression at 48 h after irradiation (Fig. 4 D, Left). NF-YB is a subunit of the nuclear transcription factor Y (NF-Y) responsible for the concerted regulation of proteasome subunit expression (36). In contrast, TFP treatment increased proteasome subunit expression (Fig. 4 D, Right). Expression of two inducible beta-type proteasome subunits, PSMB8 (1.68-fold) and PSMB9 (1.47-fold), was up-regulated in cells treated with radiation and TFP compared to radiation alone. These data supported our previous observations (11, 27) that loss of proteasome function is a major event in the acquisition of cancer stem cell traits after irradiation, as it allows for posttranslational stabilization of stem cell factors, otherwise degraded by the proteasome.
In order to study whether radiation engages developmental gene expression programs, we performed a gene set enrichment analysis (37, 38) of our RNA-Seq data, computing the overlap of differentially expressed genes with 3,302 curated gene sets representing expression signatures of genetic and chemical perturbations.
At 48 h after irradiation, differentially expressed genes overlapped with gene sets found up-regulated in neural stem progenitor cells, embryonic stem cells, and induced pluripotent stem cells (Fig. 4E).
Epigenetic Changes in Glioma Cells in Response to Radiation.
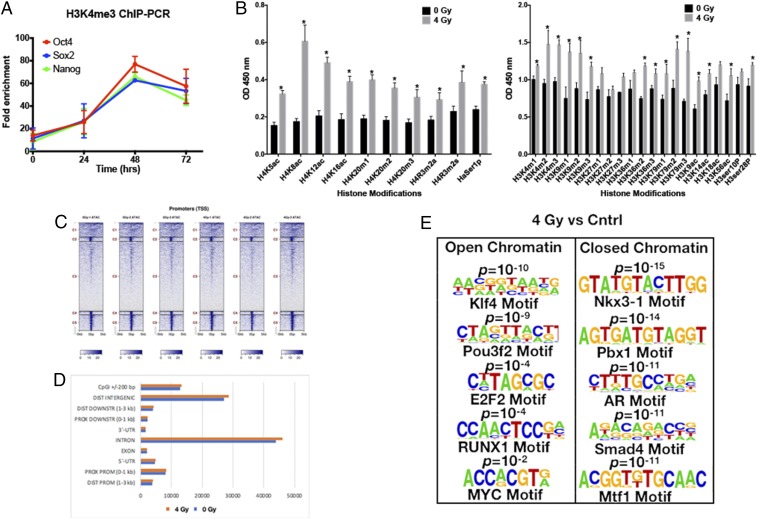
Since we observed reexpression of Yamanaka transcription factors in response to radiation, we next performed ChIP-PCR against the promoter regions of Sox2, Oct4, and Nanog and found radiation-induced acquisition of an open chromatin state for these promoters that peaked at 48 h after irradiation, thus indicating radiation-induced epigenetic remodeling (Fig. 5A). This was further supported by data obtained in a histone enzyme-linked immunosorbent assay (ELISA) that revealed significant global changes in histone acetylation and methylation, 48 h after irradiation with 4 Gy (Fig. 5B). ATAC-seq at the same 48-h time point revealed acquisition of an open chromatin state in 1,785 gene regions and a closed chromatin state in 3,637 gene regions in irradiated cells versus unirradiated cells covering both promoter and nonpromoter regions of the genome (Fig. 5 C and D). A motif search analyzing the differentially opened and closed promoter regions revealed radiation-induced open chromatin with binding motifs for the developmental transcription factors Klf4, Pou3f2, E2F2, RUNX1, and MYC. Conversely, our motif search revealed a radiation-induced closed chromatin state in promoter regions with the binding motifs for Nkx3-1, a prostatic tumor suppressor, Pbx1 and Mtf1 involved in cell fate decisions, the androgen receptor binding motif, and the binding motif for Smad4, a factor correlated with decreased survival in GBM when down-regulated (Fig. 5E).
Fig. 5.
Radiation induces epigenetic changes in GBM. (A) The 70% confluent plates of HK-374 cell cultures were irradiated with 0 or 4 Gy, and, 48 h after irradiation, the cells were extracted for proteins and used to perform ChIP-PCR assay to analyze for H3K4me3 active chromatin interactions with specific proteins such as Sox2, Oct4, and Nanog. (B) The same set of proteins was also used to perform ELISA for H4 and H3 histone modifiers. (C and D) ATAC-seq samples were collected after 48 h of 0 or 4 Gy irradiation in HK-374 cells. These samples were then analyzed for acquisition of an open chromatin and a closed chromatin state in 0- versus 4-Gy samples. (E) A motif search on both samples was also performed to identify the differentially opened or closed promoter regions with binding motifs for the developmental transcription factors such as Klf4, Pou3f2, E2F2, RUNX1, and MYC. P values were calculated using unpaired t test. *P < 0.05.
TFP Delays Tumor Growth in Murine Models of GBM.
Based on the in vitro effect of TFP on the sphere-forming capacity of patient-derived glioma specimen (Fig. 2F), we tested whether TFP also had antitumor activity in a murine model of GBM in vivo. GL261-Luc or GL261-StrawRed murine GBM cells, engineered to express firefly luciferase or StrawRed were implanted into the brains of C57BL/6 mice. On day 7 post tumor implantation, baseline bioluminescence pictures were acquired and animals were randomized and treated with either saline, 20 mg/kg TFP, radiation (10 Gy), or a combination. Mice bearing StrawRed-expressing GL261 tumors were treated with only saline or TFP starting on day 7. The TFP dose of 20 mg/kg, which has previously been reported as the MTD in mice (23), was well tolerated in our experiments. After an initial period of TFP-induced sedation that lasted 2 h to 3 h, the mice did not show any side effects, and TFP-treated animals did not show the tumor-associated weight loss that was observed in the control group (SI Appendix, Fig. S2).
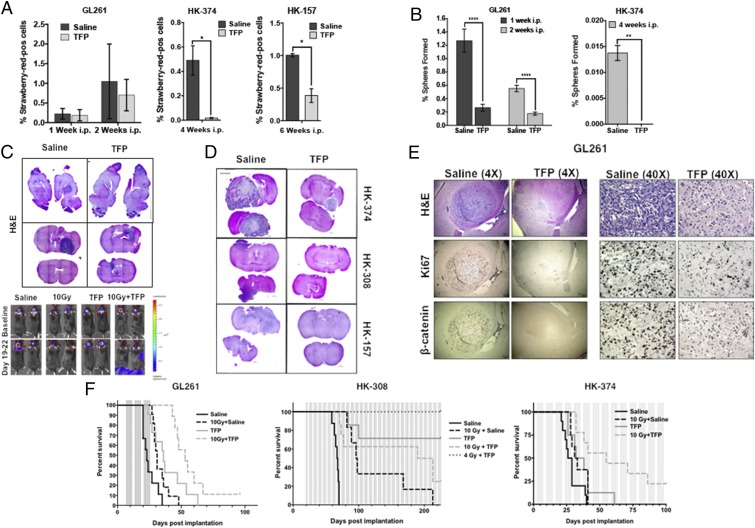
Mice bearing StrawRed-expressing tumors were killed at different time points after TFP treatments. The brains were explanted and digested, and the number of StrawRed-positive cells was quantified and assayed for self-renewal capacity, a functional assay for GICs. In vivo TFP treatment led to a decrease in the total number of tumor cells per brain in the mouse GL261 GBM model and a significant decrease in the total number of tumor cells in human HK-374 and HK-157 patient-derived orthotopic xenografts (PDOXs) (Fig. 6A). Importantly, TFP treatment reduced the sphere formation capacity of the surviving tumor cells (Fig. 6B).
Fig. 6.
TFP in combination with radiation prolongs survival in mouse models of GBM. (A) The 2 × 105 GL261-StrawRed cells were implanted into the brains of the C57BL/6 mice, and 3 × 105 HK-374 and HK-157-StrawRed cells were implanted into the brains of NSG mice. Tumors were grown for 3 d (HK-374) and 7 d (HK-157 and GL261) for successful grafting. Mice bearing tumors were injected i.p. on a 5-d on/2-d off schedule for 1 or 2 wk (GL261), 4 wk (HK-374), and 6 wk (HK-157) either with TFP (20 mg/kg) or saline. At the end of the treatment, brains of these mice were extracted, and tumors were digested and sorted for StrawRed cells. The data from the sort were plotted and presented as percentage of StrawRed-positive cells of the total number of cells in each GBM line. (B) The sorted StrawRed cells from A were plated on 96-well plates in serum-free medium and fed with fresh growth factors every 2 d to 3 d. The number of spheres formed in each condition was counted and normalized against the respective control. (C) Hematoxylin/eosin (H&E) stained sagittal and coronal sections of the brains of C57BL/6 mice intracranially implanted with 2 × 105 GL261 cells. Tumors were grown for 7 d for successful grafting. Mice were injected with TFP or saline i.p. on a 5-d on/2-d off schedule for 3 wk. TFP treatment led to a marked reduction in tumor size. (D) H&E stained coronal sections of the NSG mice brains implanted with HK-374, HK-308, and HK-157-GFP-Luc cells which were treated continuously with either TFP (s.c., 20 mg/kg) or saline until they met the criteria for study endpoint. (E) The 4× and 40× images of coronal sections of brains from C57BL/6 mice implanted with GL261-GFP-Luc cells intracranially and treated with either TFP (20 mg/kg) or saline i.p. for 2 wk and stained for H&E, Ki67, and β-catenin. (F) Survival curves for C57BL/6 mice (Left) implanted intracranially with 2 × 105 GL261-GFP-Luc cells and NSG mice injected intracranially with 3 × 105 HK-308-Luc (Middle) or HK-374-GFP-Luc (Right) cells. Tumors were grown for 3 d (HK-374), 7 d (GL261), and 21 d (HK-308) for successful grafting. Mice were injected with either TFP (20 mg/kg) or saline i.p. for GL261 for 3 wk, and s.c. for HK-308 and HK-374 lines, continuously until they reached the study endpoint. Log-rank (Mantel−Cox) test for comparison of Kaplan−Meier survival curves indicated significant differences in the TFP-treated mice compared to their respective controls. P values in GL261 study: saline vs. TFP (***P value = 0.00014), saline vs. 10 Gy + saline (*P value = 0.0126), saline vs. 10 Gy + TFP (***P value <0.0002), TFP vs. 10 Gy (P value = 0.5320), TFP vs. 10 Gy + TFP (*P value = 0.0324). P values in HK-308 study: saline vs. TFP (***P value = 0.00014), saline vs. 10 Gy + saline (***P value = 0.0004), saline vs. 10 Gy + TFP (****P value 0.0001), saline vs. 4 Gy + TFP (**P value = 0.0011), TFP vs. 10 Gy (*P value = 0.0141), TFP vs. 10 Gy + TFP (P value = 0.1052), TFP vs. 4 Gy + TFP (P value = 0.2137). P values in HK-374 study: saline vs. TFP (*P value = 0.0361), saline vs. 10 Gy + saline (*P value = 0.0413), saline vs. 10 Gy + TFP (***P value = 0.00025), TFP vs. 10 Gy (P value = 0.7217), TFP vs. 10 Gy + TFP (*P value = 0.0273). GBM-StrawRed experiments in this figure have been performed with at least two biological repeats. P values were calculated using unpaired t test. *P value < 0.05, **P value < 0.01, and ****P value < 0.0001.
The loss in total tumor cell numbers and reduction in sphere-forming capacity was accompanied by a reduction in tumor size in tissue sections in GL261 (Fig. 6C), HK-157, HK-308, and HK-374 (Fig. 6D) tumor-bearing mice.
To confirm the effects of TFP on the Wnt pathway in vivo, GL261 cells were implanted into the brains of C57BL/6 mice. After grafting, the animals were administered five daily injections of TFP for 2 wk. Tissue sections of the brains revealed a reduction in tumor size and loss of Ki67+ and β-catenin+ cancer cells (Fig. 6E), which was in line with our in vitro observations (Fig. 3).
Consistent with these observations, the median survival in GL261-bearing animals treated with TFP was 36 d, compared to 23 d in the control group of animals (P = 0.00014). Irradiation with a single dose of 10 Gy increased the median survival from 23 d to 31 d (P = 0.0126). Compared to saline-treated animals, a combination of a single dose of 10 Gy with TFP treatment significantly extended the median survival to 53 d (P = 0.0002) (Fig. 6 F, Left).
In order to assess the effect of TFP on human GBM models, we implanted two different human GBM specimens, the HK-308, a slow-growing specimen, and the HK-374, a fast-growing specimen, into the brains of NSG mice. Tumor take in HK-308-implanted mice was confirmed by bioluminescence imaging, and the animals were randomized into the different treatment groups. Mice were treated with five weekly TFP (s.c.) injections or saline until they reached endpoints for euthanasia. Radiation treatment with a single dose of 10 Gy increased median survival to 97 d, compared to 67.7 d in the control group (P = 0.0004). Remarkably, combining 10 Gy with TFP extended survival to >200 d (P < 0.0001). Combining TFP with a reduced 4-Gy dose of radiation resulted in 100% of the animals surviving to >200 d postimplantation (Fig. 6 F, Middle). The observation that the combined treatment with TFP and 10 Gy was inferior to TFP treatment alone could reflect cental nervous system (CNS) toxicity after 10 Gy in NSG mice, which lack nonhomologous end-joining DNA repair.
Median survival in animals implanted with the more aggressive HK-374 specimen was 27.5 d, and treatment with TFP or 10 Gy extended the median survival to 32 (P = 0.0361) or 35 (P = 0.0413) d, respectively. Animals treated with the combination of TFP and 10 Gy had a median survival of 55 d (P = 0.00025) (Fig. 6 F, Right).
Discussion
Despite decades of research aimed at improving treatment outcome for patients suffering from GBM, the survival rates for this disease remain dismal. The reasons for the failure of RT to control GBM are multiple and include the highly infiltrative nature of GBM (39), which makes complete resection of the tumor nearly impossible; large areas of tumor hypoxia (40) that diminish the efficacy of ionizing radiation (41) and many chemotherapeutic agents (42); and the intrinsic resistance of GICs against established anticancer therapies (9, 10). Yet, although RT does not provide a cure in the GBM setting, it remains one of the few therapies that significantly prolongs the survival of GBM patients (1, 3).
The underlying biology of fractionated RT has been elegantly summarized by Rodney Withers in the 4 Rs of RT. In this model, redistribution within the cell cycle and reoxygenation of hypoxic tumor regions between fractions lead to therapeutic gain, whereas repopulation of the tumor by surviving cells and repair of DNA damage can lead to therapeutic loss (43). The intrinsic radiation sensitivity has been proposed as a fifth R of RT (44); however, this concept is currently less widely accepted, mainly because it will be codetermined by the tumor oxygenation status, position of cancer cells in the cell cycle, and their ability to repair DNA damage, which, altogether, might exaggerate or diminish the intrinsic radiosensitivity that is determined genetically and/or epigenetically.
The 4 (5) Rs of RT consider intratumoral heterogeneity to some extent, but, if interpreted in the context of the cancer stem cell hypothesis and thus intratumoral hierarchy, the model assumes that the differentiation of cancer stem cells during repopulation is unidirectional. Even if one takes into consideration that cancer stem cells are exceptionally radioresistant but rare, this assumption commands that any given tumor can be cured by radiation dose escalation, and the mainstream of research in clinical radiation oncology seeks to enable this through increased precision in dose delivery and reduction in normal tissue dose. However, the infiltrative nature of GBM together with the normal tissue radiation tolerance of the brain limits the success of RT against GBM to the status quo.
The phenotype conversion of nontumorigenic cells into radioresistant GICs through microenvironmental factors such as low pH and hypoxia (12, 45) or through radiation-induced epigenetic changes described in our present study potentially adds reprogramming as a sixth R to the radiobiology of cancer. Our observations that this phenomenon occurs spontaneously but is even more pronounced after irradiation, the fact that it is not restricted to a population of cells prone to phenotype conversion, and the increased self-renewal capacity and radiation resistance of iGICs suggest that it potentially has significant clinical relevance in that the ratio of phenotype conversion to cell killing of existing tumor-initiating cells per fraction determines local control of the tumor. This distinction is highly relevant because it suggests that, unless a therapy given concurrently with radiation interferes with this phenomenon, radiation dose escalation will unlikely lead to local tumor control as long as the rate of phenotype conversion outpaces cell killing of existing tumor-initiating cells.
While generally thought to be associated with neuronal signaling, recent studies have supported a role for the dopamine D2 receptor in gliomagenesis (46, 47). Importantly, in the latter study, dopamine D2 receptor expression was increased in response to temozolomide treatment, elevated in CD133-positive cells, promoted self-renewal, and led to activation of hypoxia-inducible factors 1 and 2 under normoxic conditions (47).
Here we present the dopamine receptor antagonist, TFP, as an already FDA-approved, blood–brain barrier-penetrating drug that not only targets preexisting GICs but also interferes with the process of phenotype conversion of GBM cells into radioresistant iGICs.
Short-term 21-d monotherapy with lower doses (10 mg/kg) of TFP against U87MG xenografts has been previously reported but did not yield a survival benefit (48). A second study reported radiosensitization of U87MG and U251 cells based on increased gamma-H2AX foci formation and inhibition of DNA repair by homologous recombination. In a single survival experiment using a patient-derived specimen, animals were treated with 3 × 5 Gy and TFP at 1 mg/kg. No information about radiation dosimetry was given, and, although the total radiation dose of 15 Gy was substantial, it showed little effect by itself (49).
Our study suggests that ionizing radiation elevates differentiated GBM cells into a state that allows for acquisition of cancer stem cell traits and that this process can also occur spontaneously, although at a much lower frequency. The observation that radiation leads to epigenetic remodeling characterized by an open chromatin state in the promoter region of genes involved in the reprogramming of somatic cells into induced pluripotent stem cells and the promoter regions of their target genes consequently resulting in the activation of developmental gene expression programs indicates the existence of potential new targets to increase the efficacy of RT.
Mechanistically, we were able to connect pathways known to be affected by TFP treatment like Akt signaling to the posttranslational regulation of key molecular mediators of stem cell traits in cancer, like Sox2 and β-catenin. However, our RNA-Seq analysis showed that responses to TFP in combination with radiation are complex and target DNA biosynthesis, cell cycle progression, and cell division in GBM cells.
Although we found that TFP is not a classical radiosensitizer in that it does not affect the intrinsic radiosensitivity of GICs directly, it significantly prolongs the survival in syngeneic and PDOX mouse models of glioma when combined with RT. It is noteworthy that NSG mice are considered radiosensitive with a 50% lethal dose in total body irradiation experiments of 6.5 Gy (50). However, it is known that, when tumors are irradiated in mice carrying the Scid mutation, the radiosensitivity of the microenvironment does not alter tumor responses (51). Furthermore, 10-Gy radiation doses to the mostly postmitotic midbrain were well tolerated in our study, and animals survived for over 200 d without signs of radiation toxicity, thus supporting that NSG mice can be used for radiation studies of GBM PDOXs.
We conclude that a combined treatment with the dopamine receptor antagonist TFP and radiation could be an effective treatment against GBM, as it not only targets GBM cells but prevents the acquisition of a more aggressive phenotype of GBM cells induced by radiation. The data reported in our study indicate the need for continuous drug treatment and the necessity of dose-finding studies for future combination trials, ideally with second-generation dopamine receptor antagonists that exhibit a more favorable side effect profile than TFP.
Supplementary Material
Acknowledgments
F.P. was supported by National Cancer Institute Grants CA200234, CA137110, and CA161294. M.P., P.L.N., T.F.C., L.M.L., H.I.K., and F.P. were supported by National Cancer Institute (UCLA Brain Tumor Specialized Program of Research Excellence) SPORE Grant P50CA211015).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-Seq data are available via the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE146358).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920154117/-/DCSupplemental.
References
- 1.Stupp R., et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups ; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Walker M. D., et al. , Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J. Neurosurg. 49, 333–343 (1978). [DOI] [PubMed] [Google Scholar]
- 3.Walker M. D., Strike T. A., Sheline G. E., An analysis of dose-effect relationship in the radiotherapy of malignant gliomas. Int. J. Radiat. Oncol. Biol. Phys. 5, 1725–1731 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Laperriere N., Zuraw L., Cairncross G.; Cancer Care Ontario Practice Guidelines Initiative Neuro-Oncology Disease Site Group , Radiotherapy for newly diagnosed malignant glioma in adults: A systematic review. Radiother. Oncol. 64, 259–273 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Chan J. L., et al. , Survival and failure patterns of high-grade gliomas after three-dimensional conformal radiotherapy. J. Clin. Oncol. 20, 1635–1642 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Taghian A., et al. , In vivo radiation sensitivity of glioblastoma multiforme. Int. J. Radiat. Oncol. Biol. Phys. 32, 99–104 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Hemmati H. D., et al. , Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 15178–15183 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh S. K., et al. , Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Bao S., et al. , Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Eramo A., et al. , Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 13, 1238–1241 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Vlashi E., et al. , In vivo imaging, tracking, and targeting of cancer stem cells. J. Natl. Cancer Inst. 101, 350–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjelmeland A. B., et al. , Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 18, 829–840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laks D. R., et al. , Large-scale assessment of the gliomasphere model system. Neuro Oncol. 18, 1367–1378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlashi E., et al. , Targeted elimination of breast cancer cells with low proteasome activity is sufficient for tumor regression. Breast Cancer Res. Treat. 141, 197–203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagadec C., et al. , Tumor cells with low proteasome subunit expression predict overall survival in head and neck cancer patients. BMC Cancer 14, 152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adikrisna R., et al. , Identification of pancreatic cancer stem cells and selective toxicity of chemotherapeutic agents. Gastroenterology 143, 234–245.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Pan J., Zhang Q., Wang Y., You M., 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One 5, e13298 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muramatsu S., et al. , Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology 58, 218–228 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Hayashi K., et al. , Visualization and characterization of cancer stem-like cells in cervical cancer. Int. J. Oncol. 45, 2468–2474 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Tamari K., et al. , Identification of chemoradiation-resistant osteosarcoma stem cells using an imaging system for proteasome activity. Int. J. Oncol. 45, 2349–2354 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Munakata K., et al. , Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin. Cancer Res. 22, 5277–5286 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., et al. , Mebendazole potentiates radiation therapy in triple-negative breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 103, 195–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosius S. N., et al. , Combinatorial therapy with tamoxifen and trifluoperazine effectively inhibits malignant peripheral nerve sheath tumor growth by targeting complementary signaling cascades. J. Neuropathol. Exp. Neurol. 73, 1078–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumann M., DuBois W., Pu A., Freeman J., Suit H. D., Response of xenografts of human malignant gliomas and squamous cell carcinomas to fractionated irradiation. Int. J. Radiat. Oncol. Biol. Phys. 23, 803–809 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Lagadec C., Dekmezian C., Bauché L., Pajonk F., Oxygen levels do not determine radiation survival of breast cancer stem cells. PLoS One 7, e34545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagadec C., Vlashi E., Della Donna L., Dekmezian C., Pajonk F., Radiation-induced reprogramming of breast cancer cells. Stem Cells 30, 833–844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagadec C., et al. , The RNA-binding protein Musashi-1 regulates proteasome subunit expression in breast cancer- and glioma-initiating cells. Stem Cells 32, 135–144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debacq-Chainiaux F., Erusalimsky J. D., Campisi J., Toussaint O., Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Wu A., et al. , Persistence of CD133+ cells in human and mouse glioma cell lines: Detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev. 17, 173–184 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Sutherland C., What are the bona fide GSK3 substrates? Int. J. Alzheimers Dis. 2011, 505607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadoya T., et al. , Inhibition of Wnt signaling pathway by a novel axin-binding protein. J. Biol. Chem. 275, 37030–37037 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Jeong C. H., et al. , Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells 28, 2141–2150 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Cattoglio C., et al. , Functional and mechanistic studies of XPC DNA-repair complex as transcriptional coactivator in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E2317–E2326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brami-Cherrier K., et al. , Dopamine induces a PI3-kinase-independent activation of Akt in striatal neurons: A new route to cAMP response element-binding protein phosphorylation. J. Neurosci. 22, 8911–8921 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermida M. A., Dinesh Kumar J., Leslie N. R., GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv. Biol. Regul. 65, 5–15 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Xu H., et al. , The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim. Biophys. Acta 1823, 818–825 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paolillo M., Serra M., Schinelli S., Integrins in glioblastoma: Still an attractive target? Pharmacol. Res. 113, 55–61 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Bell C., et al. , Hypoxia imaging in gliomas with 18F-fluoromisonidazole PET: Toward clinical translation. Semin. Nucl. Med. 45, 136–150 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Semenza G. L., Intratumoral hypoxia, radiation resistance, and HIF-1. Cancer Cell 5, 405–406 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Teicher B. A., Lazo J. S., Sartorelli A. C., Classification of antineoplastic agents by their selective toxicities toward oxygenated and hypoxic tumor cells. Cancer Res. 41, 73–81 (1981). [PubMed] [Google Scholar]
- 43.Withers H. R., Ed., The Four R’s of Radiotherapy (Academic, New York, NY, 1975), vol. 5, pp. 241–271. [Google Scholar]
- 44.Steel G. G., McMillan T. J., Peacock J. H., The 5Rs of radiobiology. Int. J. Radiat. Biol. 56, 1045–1048 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Heddleston J. M., Li Z., McLendon R. E., Hjelmeland A. B., Rich J. N., The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 8, 3274–3284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marisetty A. L., et al. , REST-DRD2 mechanism impacts glioblastoma stem cell-mediated tumorigenesis. Neuro Oncol. 21, 775–785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caragher S. P., et al. , Activation of dopamine receptor 2 prompts transcriptomic and metabolic plasticity in glioblastoma. J. Neurosci. 39, 1982–1993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang S., et al. , Trifluoperazine, a well-known antipsychotic, inhibits glioblastoma invasion by binding to calmodulin and disinhibiting calcium release channel IP3R. Mol. Cancer Ther. 16, 217–227 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Zhang X., et al. , Trifluoperazine, a novel autophagy inhibitor, increases radiosensitivity in glioblastoma by impairing homologous recombination. J. Exp. Clin. Cancer Res. 36, 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller P. H., et al. , Analysis of parameters that affect human hematopoietic cell outputs in mutant c-kit-immunodeficient mice. Exp. Hematol. 48, 41–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Barros M., et al. , Impact of stromal sensitivity on radiation response of tumors implanted in SCID hosts revisited. Cancer Res. 70, 8179–8186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.