Significance
Collaboration between RecQ helicase and type IA topoisomerase has been found in organisms representing all levels of complexity, including the well-known BLM helicase–Top3α combination found in humans and capable of dissolving DNA double-Holliday junctions. Uniquely, hyperthermophiles have fused these functional units into a chimeric enzyme known as reverse gyrase (RG), which is capable of positively supercoiling DNA using ATP hydrolysis. Single-molecule experimentation allows us to observe at single-molecule resolution the helicase–topoisomerase coupling in RG that leads to the catalytic transitions that make up a complete ATP cycle. Subunit collaboration in RG serves as an entry point to fully understanding the universal cooperation between RecQ and Top IA.
Keywords: DNA topoisomerase, helicase, single molecule, magnetic tweezers
Abstract
Reverse gyrases (RGs) are the only topoisomerases capable of generating positive supercoils in DNA. Members of the type IA family, they do so by generating a single-strand break in substrate DNA and then manipulating the two single strands to generate positive topology. Here, we use single-molecule experimentation to reveal the obligatory succession of steps that make up the catalytic cycle of RG. In the initial state, RG binds to DNA and unwinds ∼2 turns of the double helix in an ATP-independent fashion. Upon nucleotide binding, RG then rewinds ∼1 turn of DNA. Nucleotide hydrolysis and/or product release leads to an increase of 2 units of DNA writhe and resetting of the enzyme, for a net change of topology of +1 turn per cycle. Final dissociation of RG from DNA results in rewinding of the 2 turns of DNA that were initially disrupted. These results show how tight coupling of the helicase and topoisomerase activities allows for induction of positive supercoiling despite opposing torque.
Reverse gyrase (RG) is a unique ATP-consuming topoisomerase that is found only in hyperthermophiles and that can generate positive supercoils in DNA (1–4). The exact role of positive supercoiling in hyperthermophilic life is not fully understood—nor is it fully established. Positive supercoiling could maintain DNA under double-strand form despite elevated temperature, allowing for regulation of gene expression during transcription initiation (5–7). Maintaining DNA in the double-strand form can also make it more resistant to various damage-inducing processes (8–10). Absolutely essential to maintenance in its niche for the hyperthermophilic archaea in which it was discovered (9), RG consists of a single-polypeptide chain that contains two major domains: an N-terminal RecQ-like helicase domain and a C-terminal topoisomerase domain (11, 12). Coupling between these two subunits provides RG the ability to exclusively increase DNA linking number through a strand-passage reaction that relaxes negative supercoils and introduces positive supercoils (3). However, the strength of this coupling can vary: It is weak in the regulated Sulfolobus solfataricus (Sso) reverse gyrase 1 (RG1), which can relax negative supercoils even in the absence of ATP hydrolysis, but it is strong in the constitutive Sso reverse gyrase 2 (RG2), which cannot (13–15).
Despite its importance for genome stability, the detailed molecular mechanisms that enact tight coupling of helicase and topoisomerase activities remain poorly understood. High-resolution single-molecule magnetic trapping experiments have proven useful for the study of topoisomerases as they allow one to easily control and monitor DNA supercoiling (16–21). Although single-molecule methods have recently provided estimates for the turnover rate and torque dependence of a few RG molecules working simultaneously (22, 23), they did not detect individual turnover events and so were unable to parse out the sequence of steps that make up a complete catalytic cycle and enact mechanistic coupling. Here, we analyze Sso RG2 at single-turnover resolution, allowing us to observe discrete substeps in the catalytic cycle and extract from them a mechanistic understanding of the RG’s function (24–29). In addition to the tight coupling existing between the helicase and the topoisomerase domain, RG2 is the most highly processive reverse gyrase and it is able to work well at a temperature as low as 45 °C (14, 15).
Results
Single-Molecule Setup and Manipulation.
As depicted in SI Appendix, Fig. S1A, a single linear DNA molecule is torsionally constrained by the magnetic trap when it is tethered by multiple attachments to a magnetic bead at one end and to a glass surface at the other. The trap allows one to control the rotation and extending force applied to the DNA via the magnetic bead. The position of the bead above the surface can be monitored in real time using videomicroscopy, allowing one to directly observe the end-to-end extension of the DNA resulting from the interplay of DNA topology and extension (30, 31). Rotating the magnets by one full turn allows one to impose a unit change in the DNA linking number, Lk, which is normally constant for a topologically closed system. Lk is the sum of twist (Tw, the number of times the two single strands cross intramolecularly) and writhe (Wr, the number of looped plectonemic supercoils in the molecule): Lk = Tw + Wr (32–34). When DNA is gently extended by a subpiconewton force and supercoiled in the magnetic trap, every additional unit rotation (i.e., a change in DNA linking number ΔLk = 1) causes a unit change in DNA writhe (∆Wr). As a result the bead’s position changes by an amount reflecting the DNA contour length consumed by formation of a plectonemic supercoil (30) (SI Appendix, Fig. S1B). For a constant extending force F = 0.2 pN, this corresponds to a change in DNA extension of ∼60 nm. If for instance a supercoiled DNA comes to be unwound, e.g., as in promoter melting by RNA polymerase (6), this results in a local decrease in Twist (∆Tw = −1) and a concomitant increase, at fixed linking number, of Writhe (∆Wr = +1); this results in an increase in extension for negatively supercoiled DNA but a decrease in extension for positively supercoiled DNA.
Two Types of Events Occur during the Reaction Catalyzed by RG2.
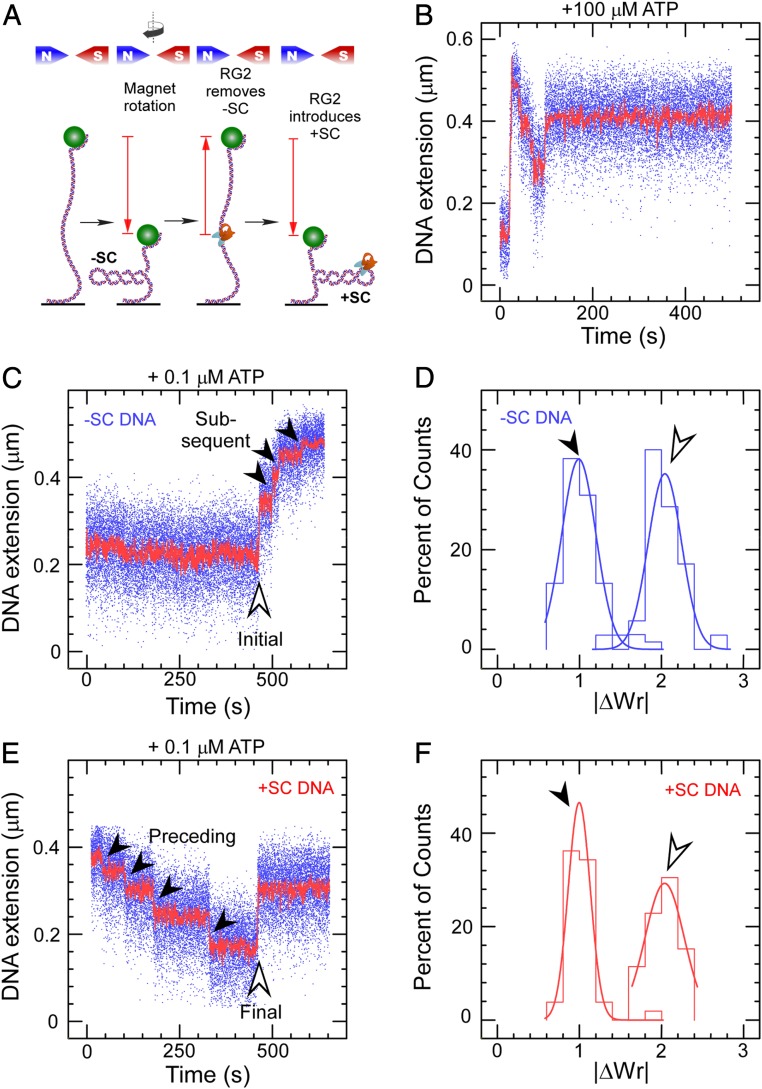
If one provides RG2 with a gently extended, negatively supercoiled DNA substrate and 0.1 mM ATP at 45 °C, the DNA’s extension first rapidly increases and reaches a maximum before slowly decreasing in a stepwise fashion (Fig. 1B). This reflects generation of positive supercoiling by a highly processive enzyme, first rapidly annihilating negative supercoils and then slowly introducing net positive supercoils into the DNA (Fig. 1A and SI Appendix, Fig. S1). The reaction appears to end with dissociation of RG2 from DNA. Discrete steps corresponding to catalytic turnover by a single RG2 are readily observed even at 0.1 µM ATP (Fig. 1 C and E), allowing one to characterize single turnover events more readily for both negatively and positively supercoiled DNA. Due to the very low RG2 concentration used in this experiment (50 pM), only one enzyme is bound to DNA.
Fig. 1.
Catalytic introduction of positive supercoils by RG2. (A) Sketch of the assay showing DNA tethered between a glass surface and a magnetic microsphere, which can be manipulated with a magnetic trap. Clockwise rotation of the magnets (as seen from above) results in negative DNA supercoils, which reduce DNA extension. RG2 first removes the negative supercoils before introducing net positive supercoiling. (B) Time trace for the extension of a negatively supercoiled DNA exposed to 50 pM RG2 and 100 µM ATP. (C) Time trace for the extension of a negatively supercoiled DNA exposed to RG2 and 0.1 µM ATP. The light arrow highlights the initial interaction, and the filled arrows, the subsequent interactions. (D) Histogram of change in DNA extension observed in C, taking into account both the initial and subsequent interaction observed between RG2 and DNA. Data are fit to Gaussian functions, respectively (solid line), with means ∆Wrinitial = 2.04 ± 0.04 (SEM; n = 35) and ΔWrsubsequent = 0.99 ± 0.03 (SEM; n = 68). E and F are as with the prior two panels but for positively supercoiled DNA, and with means ∆Wrpreceding = 1.00 ± 0.02 (SEM; n = 42) and ∆Wrfinal = −2.03 ± 0.07 (SEM; n = 21).
The change in extension of the first step observed on negatively supercoiled DNA is approximately twice as large as that of successive steps (Fig. 1 C and D). Converting step sizes to changes in writhe (plectonemic supercoils, as discussed earlier) indicates the first DNA transaction imposed by RG2 results in ∆Wr = +2, whereas successive transactions on negatively supercoiled DNA are characterized by ∆Wr = +1 (Fig. 1D). Similarly, the change in extension at the last step observed on positively supercoiled DNA is twice as large as that of the preceding steps, although it is of opposite sign (Fig. 1 E and F). Again, the preceding steps are characterized by ∆Wr = +1, while the last DNA transaction imposed by RG2 mirrors the first (∆Wr = −2) (Fig. 1F).
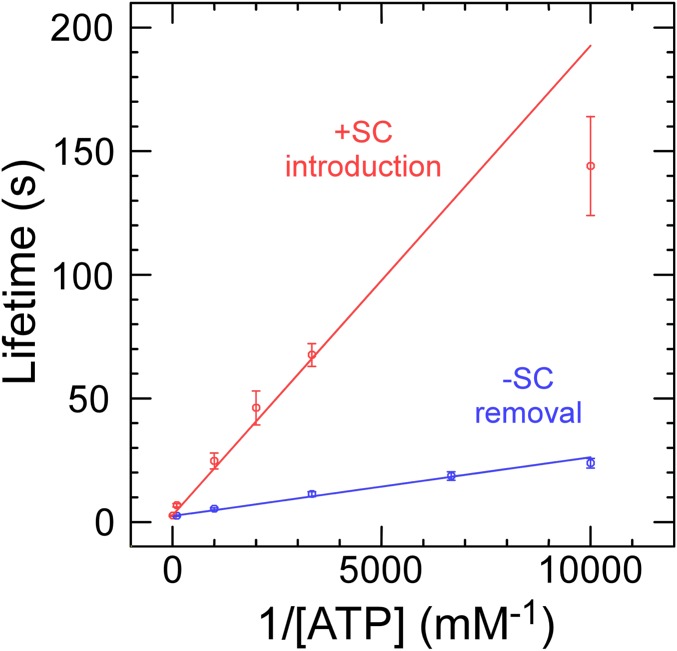
Repeated steps of ∆Wr = +1 are consistent with the expectation from biochemical experiments that RG2 is a type IA topoisomerase that introduces a single unit of positive topology (∆Lk = +1) at each catalytic turnover (2, 35). Dwell time measurements for this state performed at different ATP concentrations allow determination of reaction kinetics as defined by the Michaelis–Menten model (Fig. 2 and SI Appendix, Fig. S2). Comparison of results obtained for negatively or positively supercoiled DNA substrate gives KM = 1.0 ± 0.06 µM (SE) and 1/Vmax = 2.4 ± 0.2 s (SEM) for the former and KM = 7.1 ± 0.5 µM (SE) and 1/Vmax = 2.7 ± 0.2 s (SE) for the latter. The greatest difference between the two reactions thus appears to reside in a roughly sevenfold difference in KM for nucleotide.
Fig. 2.
RG2 catalysis as a function of ATP concentration and sign of supercoiling. Lifetimes reflect average values ± SEM obtained from ∼65 to 269 individual events for removal of negative supercoils (blue) and introduction of net positive supercoils (red). Linear fits are to the Michaelis–Menten model (see text for details), returning for negative supercoiling 1/Vmax = 2.4 ± 0.2 s (SE) and KM = 1.0 ± 0.06 µM (SE) and for positive supercoiling 1/Vmax = 2.7 ± 0.2 s (SE) and KM = 7.1 ± 0.5 µM (SE).
DNA Unwinding in the Absence of ATP.
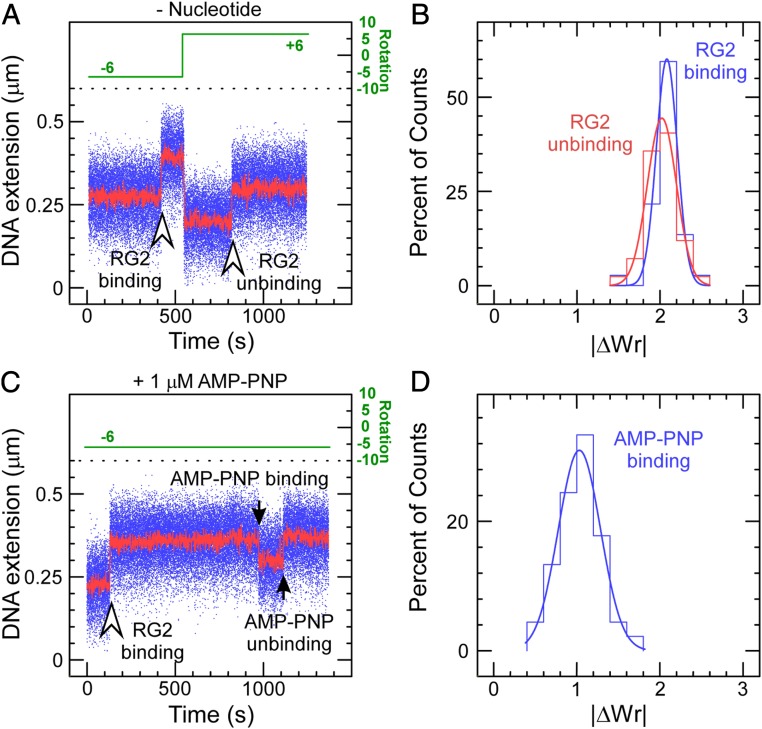
To understand the significance of the initial and final steps of |∆Wr| = 2, we sought to detect binding/unwinding and rewinding/dissociation of RG2 to DNA in the absence of ATP. Binding of RG2 to negatively supercoiled DNA resulted in a stable increase in DNA extension (Fig. 3A), which could be reversed upon positive supercoiling, with amplitude corresponding to |∆Wr| = 2 for binding/unbinding (Fig. 3B). This indicates that the initial step of ∆Wr = +2 reflects the initial DNA transaction imposed by RG2 on DNA prior to ATP usage and processive catalysis by the motor.
Fig. 3.
RG2–DNA interactions in the absence and presence of AMP–PNP. (A) Time trace obtained in the absence of nucleotide cofactor shows RG2 binding to negatively supercoiled DNA results in an increase in DNA extension. This interaction can be reversed upon positive supercoiling, as evidenced by an increase in DNA extension observed in these conditions. (B) Histograms of change in DNA extension observed for negatively and positively supercoiled DNA, corresponding, respectively, to the DNA binding/unwinding step (blue) and DNA rewinding/dissociation step (red). The solid lines are Gaussian fits, indicating that unwinding involves a mean of 2.08 ± 0.02 turns of DNA (SEM; n = 37) and rewinding involves a mean of 2.02 ± 0.03 turns of DNA (SEM; n = 42). (C) Extension time trace obtained in the presence of 1 µM AMP–PNP shows RG2 binding to negatively supercoiled DNA and then binding nucleotide analog. This interaction spontaneously reverses. (D) Histogram of reversible extension changes observed in the presence of AMP–PNP. The solid line is a Gaussian fit giving a mean of 1.03 ± 0.04 (SEM; n = 45).
The changes described above are consistent with formation of a significant unwound DNA region by RG2 in its initial interaction with its substrate. Indeed, unwinding of negatively supercoiled DNA upon/after initial RG2–DNA binding (∆Twbind = −2) will titrate out two negative supercoils (∆Wrbind = +2), resulting in an increase in DNA extension for negatively supercoiled DNA. Rewinding of positively supercoiled DNA prior to/upon RG2 dissociation (∆Twunbind = +2) will titrate out two positive supercoils (∆Wrunbind = −2) and thus result in an increase in DNA extension for positively supercoiled DNA (Fig. 3A). Since the same |∆Wr| value is obtained for binding/unwinding and rewinding/dissociation (Fig. 3B), we conclude that RG2 unwinds the equivalent of 2 turns of DNA upon binding without significant contribution of wrapping or bending. Although a change in DNA topology under similar conditions had been reported previously, the bulk assays used had not provided information on the precise nature or extent of the modification (36).
Unwinding of a DNA Substrate Containing a Premelted Bubble.
Further evidence for the idea that RG2 unwinds DNA in its initial interaction was obtained by providing the enzyme with a negatively supercoiled DNA substrate engineered to contain a mismatched bubble of either 5 or 10 bases. When provided with the 5- or 10-base bubbles but no ATP, RG2 binds and apparently unwinds the DNA (SI Appendix, Fig. S3), albeit to a progressively smaller extent. Analysis of the DNA extension changes indicate that this RG unwinds ∼1.4 turns of DNA when provided with a 5-base bubble (SI Appendix, Fig. S3) and ∼0.9 turn of DNA when provided with a 10-base bubble (SI Appendix, Fig. S3), in a roughly linear relation with respect to what is observed on regular B-DNA. Thus, RG2 acts as a molecular ruler. These results suggest that RG2 unwinds a total of ∼20 bp upon binding DNA and before binding ATP.
Decoupling Nucleotide Binding and Hydrolysis Using AMP–PNP.
To proceed along its reaction pathway, RG2 must next bind nucleotide. We carried out experiments using negatively supercoiled DNA and RG2 in the presence of 1 µM AMP–PNP. We now observe the initial unwinding interaction discussed above, followed by a new transition corresponding to an abrupt decrease in DNA extension (Fig. 3C). Converting into writhe as before indicates the resulting state has WrNTP = +1 relative to the initial state without RG2 (∆WrNTP = −1, where the change is relative to the prior state with RG2 bound to DNA) (Fig. 3D). Because RG2 is not able to hydrolyze this analog, it ultimately will release it back into solution, and this is observed as a return of the DNA extension to the initial state. We find that the dwell time prior to observing the analog-induced transition decreases with analog concentration, although the concentration dependence of the transition rate suggests the analog does not bind as well as ATP and furthermore displays complex association kinetics (SI Appendix, Fig. S4). The overall behavior nevertheless supports the idea that nucleotide binding causes RG2 to rearrange DNA topology as the next step of the catalytic cycle, and we tentatively propose it involves topological rewinding of DNA, leading to a state with one remaining unit of topological unwinding TwNTP = −1 relative to the initial state without RG2 (∆TwNTP = +1, where the change is relative to the prior state with RG2 bound to DNA). The remaining experiments aim to demonstrate this, first by ruling out the possibility of strand passage using a catalytic tyrosine mutant, and second by mixing ATP and AMP–PNP to observe reactions on positively supercoiled DNA.
In the presence of AMP–PNP, the catalytic tyrosine mutant RG2Y903F recapitulated the same initial steps of DNA unwinding (∆Twbind = −2) and nucleotide binding (∆WrNTP = −1)/unbinding (∆WrNTP = +1) (SI Appendix, Fig. S5). As RG2Y903F lacks the ability to cleave DNA, we conclude that the DNA extension changes ∆WrNTP observed upon nucleotide binding are reversible DNA topological transitions independent of DNA strand cleavage and transport, and involve no change in linking number. Thus, also nucleotide binding alone on wild-type enzyme does not permit for even a single round of stable DNA strand passage, which must take place at a later step of the RG2 catalytic cycle. However, because observing AMP–PNP binding to RG2 on positively supercoiled DNA was technically challenging due to rapid dissociation of the topoisomerase, we were not able at this stage to specify the nature of the topological rearrangement imposed on DNA by RG2 in the nucleotide-bound state (e.g., DNA bending/wrapping vs. DNA rewinding).
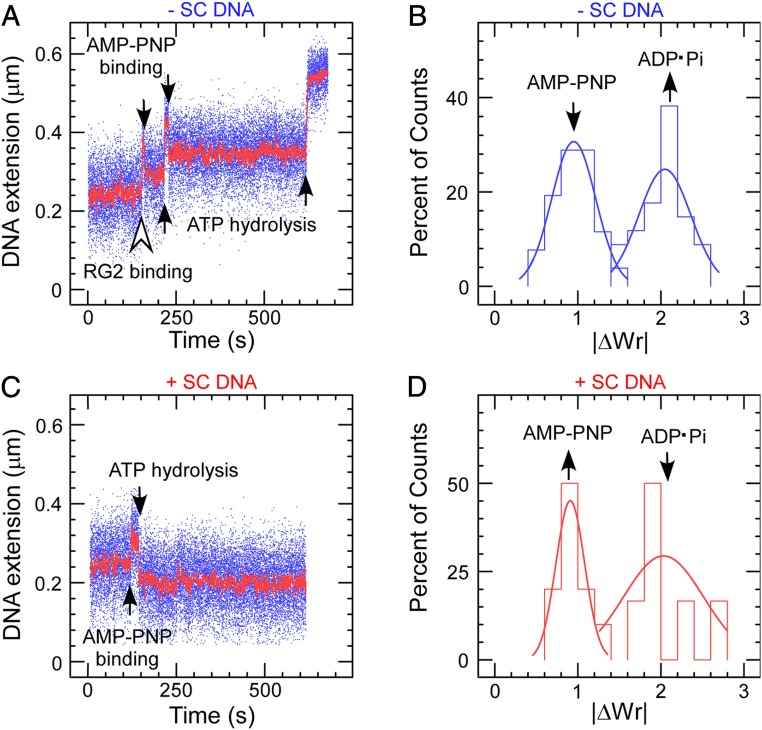
To overcome this difficulty, we next combined AMP–PNP with a very low amount of ATP, increasing the lifetime of the nucleotide-bound state prior to hydrolysis while slowly allowing RG2 to complete its cycle after exchanging AMP–PNP for ATP. After binding to negatively supercoiled DNA (∆Twbind = −2), RG2 generated a slow, multistate staircase pattern clearly displaying a succession of steps alternating between decreases in extension (∆WrNTP = −1 relative to prior state) followed by twofold larger increases in extension (∆WrADP·Pi = +2 relative to prior state) for a net change of ∆Wr = +1 per cycle (Fig. 4 A and B). As per prior results for AMP–PNP binding to RG2 on negatively supercoiled DNA, we propose that the decrease in DNA extension corresponds to nucleotide binding. We further propose that the subsequent increase in writhe ∆WrADP·Pi = +2 reflects ATP hydrolysis, product release, and the strand passage reaction itself. Indeed, control experiments carried out in the presence of RG2 and ADP show that binding/unbinding of this nucleotide causes extension changes similar to those observed with AMP–PNP (SI Appendix, Fig. S6). Because there are no such further extension changes upon cycle completion, we conclude that ADP has already been released by the time this state is formed.
Fig. 4.
RG2–DNA interactions in the presence of a mixture of AMP–PNP and ATP. (A) Time trace obtained on negatively supercoiled DNA. Smaller steps with decrease in DNA extension correspond to AMP–PNP binding and the subsequent extension rebounds relate to ATP hydrolysis and Pi/ADP release. (B) Histogram of extension changes observed on negatively supercoiled DNA. Data are fit to a Gaussian for AMP–PNP binding and hydrolysis/product release, respectively, giving means ∆WrNTP = −0.95 ± 0.04 (SEM; n = 52) and ΔWrADP·Pi = 2.05 ± 0.07 (SEM; n = 34). (C) Time trace showing AMP–PNP binding and ATP hydrolysis/product release of RG2 obtained on positively supercoiled DNA, mirror symmetric with that on negative. The positively supercoiled DNA is obtained via rotating the magnets/magnetic bead immediately after having captured an RG2 binding to negatively supercoiled DNA. (D) Histogram of topological changes observed on positively supercoiled DNA. Data are fit to Gaussian functions (solid line), giving ∆WrNTP = −0.91 ± 0.08 (SEM; n = 10) and ΔWrADP·Pi = 2.03 ± 0.48 (SEM; n = 6).
On positively supercoiled DNA with wild-type RG2, the AMP–PNP plus ATP combination resulted in a symmetric, slow, multistate staircase pattern clearly displaying a succession of steps alternating between increases in extension (∆Wr = −1 relative to prior state) and twofold larger decreases in extension (∆Wr = +2 relative to prior state) for a net change of ∆Wr = +1 per cycle (Fig. 4 C and D).
The mirror symmetry of the time traces obtained on negatively and positively supercoiled DNA implies 1) that the step corresponding to nucleotide binding is the increase in DNA extension observed on positively supercoiled DNA (∆WrNTP = −1 relative to prior state), 2) that this step corresponds to partial rewinding of the RG2:DNA bubble state and not DNA bending/wrapping (∆TwNTP = −∆WrNTP = +1, a consequence of conservation of linking number absent strand transport and cleavage), and 3) that nucleotide hydrolysis is coupled to the strand passage reaction and coupled to an increase in writhe of ∆WrADP·Pi = +2.
Model for the Catalytic Cycle of RG2 and Perspectives.
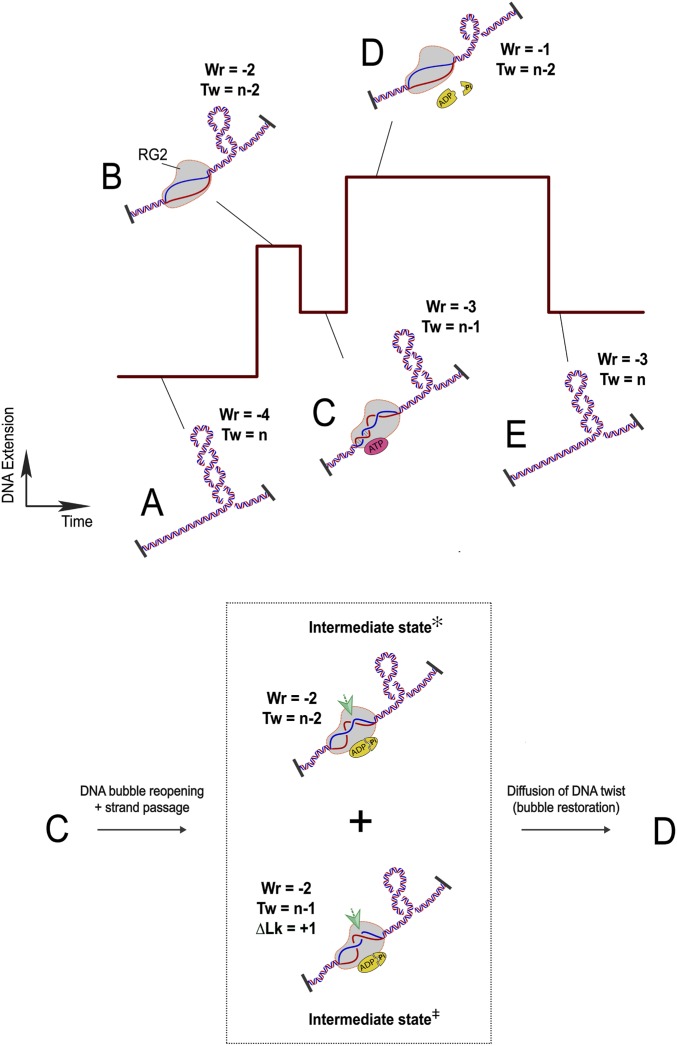
These observations lead to the following model, which considers the different manipulations observed in the DNA (Fig. 5). In regard to the nature of RG2 catalysis, which usually initiates on a negatively supercoiled DNA substrate, we set the starting point of this model to a DNA topological domain with n helical turns and containing four negative supercoils: Tw = n, Wr = −4 (Fig. 5A). Upon binding to DNA, RG2 unwinds the equivalent of 2 turns of DNA, reaching a state with Twbind = n − 2 and Wrbind = −2 relative to the protein-free state. (Fig. 5B). When RG2 binds nucleotide, it partially rewinds the DNA, leading to a state with TwATP = n − 1 and WrATP= −3 (Fig. 5C). After the nucleotide-bound state is formed, ATP hydrolysis and product release transforms the RG2–DNA complex to intermediate states with ∆Wr = +1 (Fig. 5, Inset: intermediate state✻ and state⧧). This transition includes a net increase in DNA linking number (∆LkADP·Pi = +1) via a strand cleavage and transport reaction (Fig. 5, Inset, state‡) and a reunwinding of the previously rewound 10-base DNA bubble (∆TwADP·Pi = −1 and concomitant ∆WrADP·Pi = +1) (Fig. 5, Inset, state✻) allowing the enzyme to restart its catalytic cycle in the nucleotide-free but DNA-bound state with Twbind = −2 (Fig. 5D). Indeed, it is likely that ADP is released during this transition as the RG2 binding states (Fig. 5 B and D) correspond to the ADP-free state in the ADP binding assay (SI Appendix, Fig. S6A). The catalytic cycle of RG ends upon RG2 dissociation, returning DNA to the original state with no twist deformation (Fig. 5E). The DNA linking number and writhe have both increased by 1 unit relative to the prior cycle. Once RG2 has step-by-step relaxed all of the negative supercoils in the DNA substrate, it continues to increase DNA linking number and generates positive DNA supercoils. A sketch of a catalytic cycle of RG2 on the positively supercoiled DNA contains identical topological transitions as described above (Fig. 5 A and E), but the time trace will be mirror symmetric compared with that in Fig. 5, similar as is shown in Fig. 4B.
Fig. 5.
Model for the RG cycle. RG2 is viewed as a topological state machine, manipulating DNA twist/writhe in an ordered reaction, through ATP-regulated helicase–topoisomerase coordination. (A) The starting point of an RG2 catalytic cycle is set with a DNA topological domain containing four negative DNA supercoils (Wr = −4; Tw = n). (B) RG2 binding unwinds 20 bp of DNA (Wr = −2; Tw = n − 2). (C) One-half of the 20-base DNA bubble is rewound due to ATP binding of RG2 (Wr = −3; Tw = n − 1). (Inset) In the subsequent ATP hydrolysis and product release stage (C to D), RG2 performs two steps: RG2 conducts a strand-passage reaction at the intersection point indicated by the green arrows (bottom half, Intermediate state⧧; ΔLk = +1 → ΔWr = +1) and RG2 reopens the rewound DNA (top half, Intermediate state✻; ΔTw = −1 → ΔWr = +1). We do not know the order of the two intermediate steps and also the exact conformation of the base pairs. Together these transitions lead to a +2 unit change in DNA writhe. (D) RG2 finally reaches a nucleotide-free state after ATP hydrolysis and product release with the newly produced DNA twist diffused out of the enzyme and thus titrating out one negative supercoil on the DNA. (E) RG2 dissociation from DNA rewinds the 20-base DNA bubble, which returns two negative plectonemes back to the DNA substrate (Tw = −3; Wr = 0). Therefore, a complete ATP cycle by RG2 adds 1 unit of DNA linking number to the DNA (from A–E).
Finally, these results also indicate that RG2 may be able to positively supercoil DNA until it reaches a natural “set point.” Because the lifetimes shown in Fig. 2 largely reflect the waiting time of RG2 for ATP and ATP hydrolysis itself is rapid (only visible by mixing ATP with AMP–PNP), the KM values in our study describe the ATP binding affinity for RG2. Consequently, the fact that KM for nucleotide is sevenfold lower for positively supercoiled DNA than for negatively supercoiled DNA suggests that the enzyme’s overall reaction rate progressively decreases as positive supercoiling increases. We are not aware of other DNA-processing enzymes for which nucleotide affinity depends on DNA supercoiling. Although the total ATP concentration in vivo may be much higher than the micromolar KM determined here, it should be kept in mind that also in vivo countless enzymes compete for this ATP. The tight affinity of RG2 for ATP means that it is likely to compete effectively against other enzymes for ATP, and the supercoiling dependence of this affinity suggests that a single topoisomerase species may be sufficient to regulate topological homeostasis of the nucleoid.
Discussion
By carrying out single-molecule experiments, we observe well-organized DNA topological transitions imposed by RG2, which couple ATP usage and formation of positive supercoils. This results from the ordered reactions coupling the RecQ helicase and Top IA domains of RG2. On its own, a classical type IA topoisomerase, which does not require ATP, only reduces torsional stress in DNA. It thus relaxes negatively supercoiled DNA and, if provided a ssDNA region, also removes positive supercoils (20, 37, 38). However, by coupling with a RecQ helicase, the Top IA subunit of RG acquires the ability to impose directionality in its strand-passage reaction, which exclusively increases DNA linking number. Indeed, the helicase domain is necessary (but not sufficient) to generate and control the unpaired DNA region, which serves as a substrate for the strand-passage reaction. Otherwise, simply conducting a strand-passage reaction on B-form DNA can only reduce, and never increase, DNA linking number.
Evidence pertaining to protein conformations of RG subunits can be used to explain helicase–topoisomerase coupling. As reported previously, both the helicase and Top IA domains of RG each have two protein conformations.
Thus using single-molecule fluorescence resonance energy transfer, researchers found that a truncated helicase domain of RG switches between an open conformation in the ATP-free state and a closed conformation in the ATP-bound state (39). In addition, ATP-induced closing of the helicase domain dramatically increases its dsDNA binding affinity, whereas affinity to ssDNA is unchanged (39). Both of these results are consistent with our single-molecule observation that ATP binding to RG2 rewinds DNA, converting 10 bases of DNA bubble to dsDNA. Therefore, the open state of the helicase domain is associated with DNA unwinding (Fig. 6 A, C, and D), and the closed state of the helicase domain is associated with DNA rewinding (Fig. 6B). Simultaneously, the coordinated transitions in the Top IA domain reorganize DNA single strands and conduct directional strand passage (Fig. 6 B and C). This is consistent with the 10-base DNA bubble rewinding, which can be considered as byproduct generated and then cleared over the course of an RG2 ATP turnover, and also usefully serves as an indicator for the helicase–topoisomerase coordination.
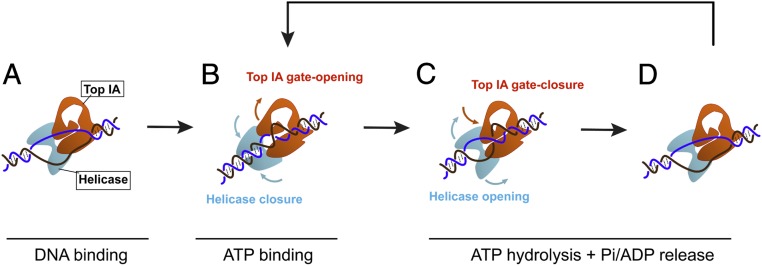
Fig. 6.
Helicase–topoisomerase coupling in RG2 supercoiling. A second model of RG2’s catalytic cycle focuses on the RecQ–Top IA coupling. (A) RG2 unwinds a 20-bp DNA bubble. The open state of the helicase domain is coupled to a closed-gate conformation in the Top IA subunit. (B) ATP binding closes the helicase domain, causing rewinding of 10 bases of the DNA bubble, and opens the Top IA gate to rearrange ssDNA before strand passage. (C) ATP hydrolysis and/or subsequent product release reopens the helicase domain to regenerate the original 20-bp bubble, and this is coordinated with the DNA strand passage through the Top IA gate. (D) The RG2–DNA complex reset to the initial 20-bp DNA unwinding bubble can bind a new ATP and repeat its cycle.
Top IA by itself alternates between an open-gate and a closed-gate conformation to complete a strand-passage reaction (40), as confirmed in recent single-molecule studies (20, 21). Moreover, physical interplay between the two domains of RG has also been reported, whereby a truncated Top IA domain only relaxes negatively supercoiled DNA, but mixing both domains in solution reconstitutes the unique positive supercoiling activity of RG (41, 42). This domain–domain interplay was proposed to be achieved through the latch, a protein insertion in the RG helicase domain important for both helicase–topoisomerase interaction and performance of RG (43–45). These lines of evidence provide insights into the ATP-dependent RecQ–Top IA machinery of RG, in which the opening and closing of the helicase domain tightly gates the conformational transitions of the Top IA domain to carry out strand passage despite opposing torque.
It is worth noting that different levels of helicase–topoisomerase coupling in the RG family have been revealed through the distinct behaviors of family members as observed in the absence of nucleotides. As revealed both in bulk assays (14) and in our single-molecule experiment (SI Appendix, Fig. S7), the Top IA domain in Sso RG1 retains sufficient functional freedom so as to relax negatively supercoiled DNA in the absence of ATP. Similarly, RG from Archaeoglobus fulgidus removes positive supercoils in the absence of ATP when provided a bubble-containing DNA substrate (46). The Sso RG2 studied here is not able to carry out strand passage without NTP (14), which indicates stringent control of Top IA activity by the helicase domain.
Compared with the RecQ–Top IA coupling in RG, the RecQ–Topo III cooperation is presumably achieved in a different way as it results in negative DNA supercoiling. Here, the RecQ helicase first unwinds dsDNA using energy from ATP, providing Topo III with ssDNA to carry out the strand-passage reaction. Although all of the components in the protein complex collaborate with each other, they each retain the freedom to work independently. Thus, when RecQ unwinds DNA, it also generates positive DNA supercoils on the double-strand region of DNA. This situation can further be stabilized by ssDNA binding proteins (47, 48). Topo III then binds and conducts strand passage on the ssDNA, titrating out positive DNA plectonemes. Therefore, plasmid DNA becomes negatively supercoiled after deproteinization and ssDNA renaturation (47, 48).
Compared to the RecQ–Top IA coupling observed in RG, cooperation between RecQ and Topoisomerase III is more universal and has been found in all domains of life. These widespread protein associations are crucial in maintaining genome stability. For example, in eukaryotes, Top3α cooperates with RecQ family helicases to modulate numerous DNA transactions including dissolution of double-Holliday junctions (49–52) and resolution of converging replication forks (53). Defects in any of the three human RecQ family helicases, BLM, WRN, and RecQ4, lead to genetic disorders and diseases such as cancer and premature aging (54–56). Interestingly, a recent study also revealed a similar type of cooperation between human Top3 and the SNF2 family helicase PICH (57), in which they work together to introduce positive DNA supercoiling essential for chromatid separation during anaphase (58, 59). This newly discovered partnership suggests a more widespread existence of cooperation between different helicase and Top IA families.
Materials and Methods
Materials.
All restriction enzymes and DNA ligase were purchased from New England Biolabs; thermostable DNA polymerase and modified nucleotides for attachment of DNA to surfaces were purchased from Roche. Oligonucleotides were from Eurofins Genomics. BSA and Tween 20 were from Roche, and ATP or nonhydroylzable analog AMP–PNP was from Jena Biosciences. All other chemicals (N-mercaptoethanol, glycerol) were purchased from Merck.
Protein Expression and Purification.
RG2, the product of the topR2 gene, was expressed as described previously (14). To create the RG2Y903F catalytic mutant, we used the QuikChange II Site-Directed Mutagenesis Kit (Agilent) to modify a codon-optimized version of the topR2 gene inserted between the NdeI and XhoI sites of the pET28b expression vector (Novagen). The RG2 and RG2Y903F proteins were expressed and purified essentially as described previously (14), except that a gel filtration step was included. The proteins were dialyzed into the same storage buffer as described previously; however, here, the glycerol concentration was 50% (vol/vol). Protein was dispensed into single-use aliquots, snap-frozen in liquid nitrogen, and stored at −80 °C.
Preparation of DNA Tethers for Single-Molecule Experiments.
The 3-kbp DNA tether.
The 3-kbp DNA fragment used in these experiments corresponds to a part of the Thermus aquaticus rpoC gene (seq ID Y19223.3, from position 227 to 3190) cloned into the XbaI and SbfI sites of pUC18. The following oligonucleotides, designed for optimal RG2 binding as per a prior study (60), were then annealed and inserted at the KpnI site of the 3-kbp fragment: 5′-TGTCAGCCCGTGATATTCATTACTTCTTATCCTAAGTAC-3′ and 5′-TTAGGATAAGAAGTAATGAATATCACGGGCTGACAGTAC-3′. The recombinant plasmid was cut with XbaI and SbfI and the 3-kbp fragment of interest purified by gel electrophoresis. The 3-kbp fragment was then ligated to 1-kbp biotin-labeled DNA at one end and 1-kbp digoxigenin-labeled DNA at the other end. These 1-kbp DNA fragments were labeled as described previously (7).
DNA tethers containing a mismatched bubble.
For DNA containing a mismatched 10-bp bubble, we used a 2.2-kbp DNA fragment that corresponds to a part of the T. aquaticus rpoC gene (from position 2103 to 4125) cloned into the XbaI and SbfI sites of pUC18. This DNA fragment was prepared as described above except that, before the ligation with the 1-kbp biotin-labeled or digoxigenin-labeled DNA fragments, the two following oligonucleotides, both bearing 5′ phosphates, were annealed and then ligated into the 2.2-kbp DNA between unique HindIII and SpeI sites: 5′-AGCTGGATACTTACAGCCATATCAGTTACGCCTACTCCATCCCATATG-3′ and 5′-CTAGCATATGGGATGGAGTATCAGCCGTGTATATGGCTGTAAGTATCC-3′ where bases in bold correspond to the 10-bp mismatched bubble region. For DNA containing a mismatched 5-bp bubble, the same procedure was used but with oligonucleotides 5′-AGCTGGATACTTACAGCCATATCAGTTTACTCCATTCCATCCCATATG-3′ and 5′-CTAGCATATGGGATGGAATGGAGTACGTGTATATGGCTGTAAGTATCC-3′.
Assembly in the magnetic trap.
The assembled DNA constructs were attached as described (61) first to streptavidin-coated magnetic beads (MyOne C1; Thermo Fisher), then to anti-digoxigenin–coated capillaries, and finally placed on a temperature-controlled home-built magnetic trap running the PicoTwist software suite (PicoTwist SARL; http://www.picotwist.com).
Single-Molecule DNA Nanomanipulation.
In all experiments, enzymes with or without nucleotides are injected before introducing DNA supercoiling, and it is ensured that the DNA’s mechanical properties are unchanged before and just after injection (i.e., before enzymes begin to act on the DNA).
RG2 catalytic assays.
RG2 catalytic assays were performed at 45 °C in reaction buffer containing 40 mM Na·Hepes, pH 8.0, 100 mM NaCl, 10 mM MgCl2, β-mercaptoethanol, 0.1% Tween 20 (vol/vol), 0.5 mg/mL BSA (wt/vol), and ATP or nonhydrolyzable analog AMP–PNP as indicated. The detailed procedures for manipulation in the magnetic trap have been extensively described elsewhere (31, 61–63). For these experiments, DNA tethers were extended using a 0.2-pN force (1 pN = 10−12 N) and RG2 was at 50 pM. Then, we introduced six negative supercoils into the DNA using the magnetic trap to generate a substrate for RG2. After the reaction was complete, the DNA was once more negatively supercoiled to permit a new reaction round.
RG2-DNA unwinding/rewinding assays.
After addition of 50 pM RG2 but no ATP, we introduced six negative supercoils into the 3-kbp DNA (SI Appendix, Fig. S1). After observing a change in DNA extension mediated by RG2 binding, the magnets were rotated by +12 turns so as to positively supercoil the DNA tethers with ∼6 positive supercoils. Reversal of the RG2-mediated change in DNA extension was observed before regenerating the initial supercoiling state on the DNA with ∼6 negative supercoils.
RG2 unwinding assays on DNA containing a mismatched bubble.
In these assays, the 2.2-kbp DNA tethers containing either a 5-base or a 10-base mismatched bubble were negatively supercoiled by 6 turns and then exposed to 10 pM RG2. After observing the DNA unwinding events on the DNA, RG buffer with 0.1% sodium dodecyl sulfate (SDS) was injected to wash off enzyme bound to DNA. An extra wash step with RG buffer was applied to remove SDS, and then a second round of RG2 addition could be carried out.
RG2/RG2Y903F AMP–PNP binding assays.
Assays for AMP–PNP binding to RG2 and its catalytically inactive mutant RG2Y903F were conducted in the same experimental conditions as the RG2 DNA unwinding/rewinding assays conducted on the 3-kbp DNA tethers and with 1 µM AMP–PNP.
RG2 ADP binding assay.
ADP binding assay of RG2 were performed with the same experimental condition described for the AMP–PNP binding assays, in the presence of 1 µM ADP.
RG2 catalysis in the presence of AMP–PNP and ATP mixes.
In attempts to optimize data collection, we tested a range of ATP:AMP–PNP molar ratios (ranging from 1:1 to 1:20) and concentrations (ATP ranging from 1 to 100 µM), but no clear optimal condition emerged. As a result, we present only step amplitude measurements obtained in the range of conditions explored, and not step dwell-time measurements.
Data Collection and Analysis.
Real-time tracking of DNA extension and data analysis were carried out as described previously using the PicoTwist software package (61, 63). All events for which step–amplitude or step–lifetime measurements were determined were within the linear regime of the extension vs. supercoiling curves as discussed above.
For the step–amplitude measurements, changes of DNA extension (in micrometers) between two unwinding/rewinding or catalytic steps were measured and then converted to changes in DNA writhe (Wr) according to the extension vs. supercoiling curve (SI Appendix, Fig. S1). Histogram plots of step–amplitude distribution were then fitted to a Gaussian or double Gaussian function.
For step–dwell-time measurements (including catalytic step-lifetime and AMP–PNP waiting-time measurements), histograms of dwell-time distribution were fitted to single-exponential functions, yielding average lifetime values and the associated SE. Kinetic analysis was carried out using the linear form of the Michaelis–Menten equation (Eq. 1):
| [1] |
Average lifetimes obtained at different ATP concentrations were further plotted as a function of the inverse of ATP concentration. Points for −SC removal and +SC introduction were fitted separately by linear regression to obtain KM and Vmax. In the AMP–PNP binding analysis, an AMP–PNP concentration vs. association rate scatter plot was created to show the AMP–PNP concentration dependence.
Data Availability.
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
Research was supported in part by the Ligue Nationale Contre le Cancer “Equipe Labellisée” program and by the French Agence Nationale de la Recherche (ANR) (Grant ANR-19-CE11-0001). Further core funding was provided by Institut Jacques Monod, University of Paris Diderot (University of Paris), INSERM, CNRS, and Ecole Normale Supérieure (Paris Sciences et Lettres).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. K.C.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921848117/-/DCSupplemental.
References
- 1.Kikuchi A., Asai K., Reverse gyrase—a topoisomerase which introduces positive superhelical turns into DNA. Nature 309, 677–681 (1984). [DOI] [PubMed] [Google Scholar]
- 2.Forterre P., Mirambeau G., Jaxel C., Nadal M., Duguet M., High positive supercoiling in vitro catalyzed by an ATP and polyethylene glycol-stimulated topoisomerase from Sulfolobus acidocaldarius. EMBO J. 4, 2123–2128 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakasu S., Kikuchi A., Reverse gyrase; ATP-dependent type I topoisomerase from Sulfolobus. EMBO J. 4, 2705–2710 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadal M., Mirambeau G., Forterre P., Reiter W. D., Duguet M., Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 321, 256–258 (1986). [Google Scholar]
- 5.Bell S. D., Jaxel C., Nadal M., Kosa P. F., Jackson S. P., Temperature, template topology, and factor requirements of archaeal transcription. Proc. Natl. Acad. Sci. U.S.A. 95, 15218–15222 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revyakin A., Ebright R. H., Strick T. R., Promoter unwinding and promoter clearance by RNA polymerase: Detection by single-molecule DNA nanomanipulation. Proc. Natl. Acad. Sci. U.S.A. 101, 4776–4780 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revyakin A., Liu C., Ebright R. H., Strick T. R., Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampmann M., Stock D., Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res. 32, 3537–3545 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atomi H., Matsumi R., Imanaka T., Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 186, 4829–4833 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valenti A., Perugino G., Nohmi T., Rossi M., Ciaramella M., Inhibition of translesion DNA polymerase by archaeal reverse gyrase. Nucleic Acids Res. 37, 4287–4295 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nadal M., Reverse gyrase: An insight into the role of DNA-topoisomerases. Biochimie 89, 447–455 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Garnier F., Debat H., Nadal M., Type IA DNA topoisomerases: A universal core and multiple activities. Methods Mol. Biol. 1703, 1–20 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Garnier F., Nadal M., Transcriptional analysis of the two reverse gyrase encoding genes of Sulfolobus solfataricus P2 in relation to the growth phases and temperature conditions. Extremophiles 12, 799–809 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Bizard A., Garnier F., Nadal M., TopR2, the second reverse gyrase of Sulfolobus solfataricus, exhibits unusual properties. J. Mol. Biol. 408, 839–849 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Couturier M., Gadelle D., Forterre P., Nadal M., Garnier F., The reverse gyrase TopR1 is responsible for the homeostatic control of DNA supercoiling in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 269, 27663 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Dekker N. H., et al. , Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 329, 271–282 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Gore J., et al. , Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature 439, 100–104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu A., Schoeffler A. J., Berger J. M., Bryant Z., ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat. Struct. Mol. Biol. 19, 538–546, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szafran M. J., Strick T., Strzałka A., Zakrzewska-Czerwińska J., Jakimowicz D., A highly processive topoisomerase I: Studies at the single-molecule level. Nucleic Acids Res. 42, 7935–7946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunn K. H., Marko J. F., Mondragón A., An orthogonal single-molecule experiment reveals multiple-attempt dynamics of type IA topoisomerases. Nat. Struct. Mol. Biol. 24, 484–490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills M., Tse-Dinh Y.-C., Neuman K. C., Direct observation of topoisomerase IA gate dynamics. Nat. Struct. Mol. Biol. 25, 1111–1118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa T., et al. , Direct observation of DNA overwinding by reverse gyrase. Proc. Natl. Acad. Sci. U.S.A. 112, 7495–7500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa T., Sutoh K., Kikuchi A., Kinosita K. Jr, Torsional stress in DNA limits collaboration among reverse gyrase molecules. FEBS J. 283, 1372–1384 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Strick T. R., Croquette V., Bensimon D., Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 404, 901–904 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Charvin G., Strick T. R., Bensimon D., Croquette V., Topoisomerase IV bends and overtwists DNA upon binding. Biophys. J. 89, 384–392 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charvin G., Vologodskii A., Bensimon D., Croquette V., Braiding DNA: Experiments, simulations, and models. Biophys. J. 88, 4124–4136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charvin G., Strick T. R., Bensimon D., Croquette V., Tracking topoisomerase activity at the single-molecule level. Annu. Rev. Biophys. Biomol. Struct. 34, 201–219 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Koster D. A., Palle K., Bot E. S. M., Bjornsti M.-A., Dekker N. H., Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448, 213–217 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Basu A., et al. , Dynamic coupling between conformations and nucleotide states in DNA gyrase. Nat. Chem. Biol. 14, 565–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strick T. R., Allemand J. F., Bensimon D., Bensimon A., Croquette V., The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Lionnet T., et al. , Single-molecule studies using magnetic traps. Cold Spring Harb. Protoc. 2012, 34–49 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Bauer W., Vinograd J., The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J. Mol. Biol. 33, 141–171 (1968). [DOI] [PubMed] [Google Scholar]
- 33.Fuller F. B., Decomposition of the linking number of a closed ribbon: A problem from molecular biology. Proc. Natl. Acad. Sci. U.S.A. 75, 3557–3561 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White J. H., Bauer W. R., Superhelical DNA with local substructures. A generalization of the topological constraint in terms of the intersection number and the ladder-like correspondence surface. J. Mol. Biol. 195, 205–213 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Nadal M., et al. , Reverse gyrase of Sulfolobus: Purification to homogeneity and characterization. Biochemistry 27, 9102–9108 (1988). [DOI] [PubMed] [Google Scholar]
- 36.Jaxel C., et al. , Reverse gyrase binding to DNA alters the double helix structure and produces single-strand cleavage in the absence of ATP. EMBO J. 8, 3135–3139 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkegaard K., Wang J. C., Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 185, 625–637 (1985). [DOI] [PubMed] [Google Scholar]
- 38.Dekker N. H., et al. , The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 99, 12126–12131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.del Toro Duany Y., Klostermeier D., Nucleotide-driven conformational changes in the reverse gyrase helicase-like domain couple the nucleotide cycle to DNA processing. Phys. Chem. Chem. Phys. 13, 10009–10019 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Viard T., de la Tour C. B., Type IA topoisomerases: A simple puzzle? Biochimie 89, 456–467 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Déclais A.-C., Marsault J., Confalonieri F., de La Tour C. B., Duguet M., Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 275, 19498–19504 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Valenti A., et al. , Dissection of reverse gyrase activities: Insight into the evolution of a thermostable molecular machine. Nucleic Acids Res. 36, 4587–4597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudolph M. G., del Toro Duany Y., Jungblut S. P., Ganguly A., Klostermeier D., Crystal structures of Thermotoga maritima reverse gyrase: Inferences for the mechanism of positive DNA supercoiling. Nucleic Acids Res. 41, 1058–1070 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganguly A., Del Toro Duany Y., Rudolph M. G., Klostermeier D., The latch modulates nucleotide and DNA binding to the helicase-like domain of Thermotoga maritima reverse gyrase and is required for positive DNA supercoiling. Nucleic Acids Res. 39, 1789–1800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganguly A., del Toro Duany Y., Klostermeier D., Reverse gyrase transiently unwinds double-stranded DNA in an ATP-dependent reaction. J. Mol. Biol. 425, 32–40 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Hsieh T.-S., Plank J. L., Reverse gyrase functions as a DNA renaturase: Annealing of complementary single-stranded circles and positive supercoiling of a bubble substrate. J. Biol. Chem. 281, 5640–5647 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Harmon F. G., Brockman J. P., Kowalczykowski S. C., RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J. Biol. Chem. 278, 42668–42678 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Cejka P., Plank J. L., Dombrowski C. C., Kowalczykowski S. C., Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: A mechanism for disentangling chromosomes. Mol. Cell 47, 886–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S.-H., Siaw G. E.-L., Willcox S., Griffith J. D., Hsieh T.-S., Synthesis and dissolution of hemicatenanes by type IA DNA topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 110, E3587–E3594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S. H., Plank J. L., Willcox S., Griffith J. D., Hsieh T.-S., Top3α is required during the convergent migration step of double Holliday junction dissolution. PLoS One 9, e83582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bocquet N., et al. , Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIα and RMI1. Nat. Struct. Mol. Biol. 21, 261–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L., et al. , The Bloom’s syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275, 9636–9644 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Suski C., Marians K. J., Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell 30, 779–789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellis N. A., et al. , The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83, 655–666 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Yu C.-E., et al. , Positional cloning of the Werner’s syndrome gene. Science 272, 258–262 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Kitao S., et al. , Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22, 82–84 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Baumann C., Körner R., Hofmann K., Nigg E. A., PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Baxter J., et al. , Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331, 1328–1332 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Bizard A. H., et al. , PICH and TOP3A cooperate to induce positive DNA supercoiling. Nat. Struct. Mol. Biol. 26, 267–274 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Jaxel C., Duguet M., Nadal M., Analysis of DNA cleavage by reverse gyrase from Sulfolobus shibatae B12. Eur J Biochem. 260, 103–111 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Fan J., Leroux-Coyau M., Savery N. J., Strick T. R., Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature 536, 234–237 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Howan K., et al. , Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature 490, 431–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graves E. T., et al. , A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence. Nat. Struct. Mol. Biol. 22, 452–457 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.