Significance
Plants biosynthesize their own proteinogenic aromatic l-amino acids, namely l-phenylalanine, l-tyrosine, and l-tryptophan, not only for building proteins but also for the production of a plethora of aromatic amino acid-derived natural products. Pyridoxal 5′-phosphate (PLP)-dependent aromatic l-amino acid decarboxylase (AAAD) family enzymes play important roles in channeling various aromatic l-amino acids into diverse downstream specialized metabolic pathways. Through comparative structural analysis of four functionally divergent plant AAAD proteins together with biochemical characterization and molecular dynamics simulations, we reveal the structural and mechanistic basis for the rich divergent and convergent evolutionary development within the plant AAAD family. Knowledge learned from this study aids our ability to engineer high-value aromatic l-amino acid-derived natural product biosynthesis in heterologous chassis organisms.
Keywords: enzyme evolution, AAAD, aromatic amino acid metabolism, specialized metabolism
Abstract
Radiation of the plant pyridoxal 5′-phosphate (PLP)-dependent aromatic l-amino acid decarboxylase (AAAD) family has yielded an array of paralogous enzymes exhibiting divergent substrate preferences and catalytic mechanisms. Plant AAADs catalyze either the decarboxylation or decarboxylation-dependent oxidative deamination of aromatic l-amino acids to produce aromatic monoamines or aromatic acetaldehydes, respectively. These compounds serve as key precursors for the biosynthesis of several important classes of plant natural products, including indole alkaloids, benzylisoquinoline alkaloids, hydroxycinnamic acid amides, phenylacetaldehyde-derived floral volatiles, and tyrosol derivatives. Here, we present the crystal structures of four functionally distinct plant AAAD paralogs. Through structural and functional analyses, we identify variable structural features of the substrate-binding pocket that underlie the divergent evolution of substrate selectivity toward indole, phenyl, or hydroxyphenyl amino acids in plant AAADs. Moreover, we describe two mechanistic classes of independently arising mutations in AAAD paralogs leading to the convergent evolution of the derived aldehyde synthase activity. Applying knowledge learned from this study, we successfully engineered a shortened benzylisoquinoline alkaloid pathway to produce (S)-norcoclaurine in yeast. This work highlights the pliability of the AAAD fold that allows change of substrate selectivity and access to alternative catalytic mechanisms with only a few mutations.
Plants are sessile organisms that produce a dazzling array of specialized metabolites as adaptive strategies to mitigate multitudes of abiotic and biotic stressors. Underlying plants’ remarkable chemodiversity is the rapid evolution of the requisite specialized metabolic enzymes and pathways (1). Genome-wide comparative analysis across major green plant lineages has revealed pervasive and progressive expansions of discrete enzyme families mostly involved in specialized metabolism, wherein new enzymes emerge predominantly through gene duplication followed by subfunctionalization or neofunctionalization (2). Newly evolved enzyme functions usually entail altered substrate specificity and/or product diversity without changes in the ancestral catalytic mechanism. In rare cases, adaptive mutations occur in a progenitor protein fold—either catalytic or noncatalytic—that give rise to new enzymatic mechanisms and novel biochemistry (3, 4). Resolving the structural and mechanistic bases for these cases is an important step toward understanding the origin and evolution of functionally disparate enzyme families.
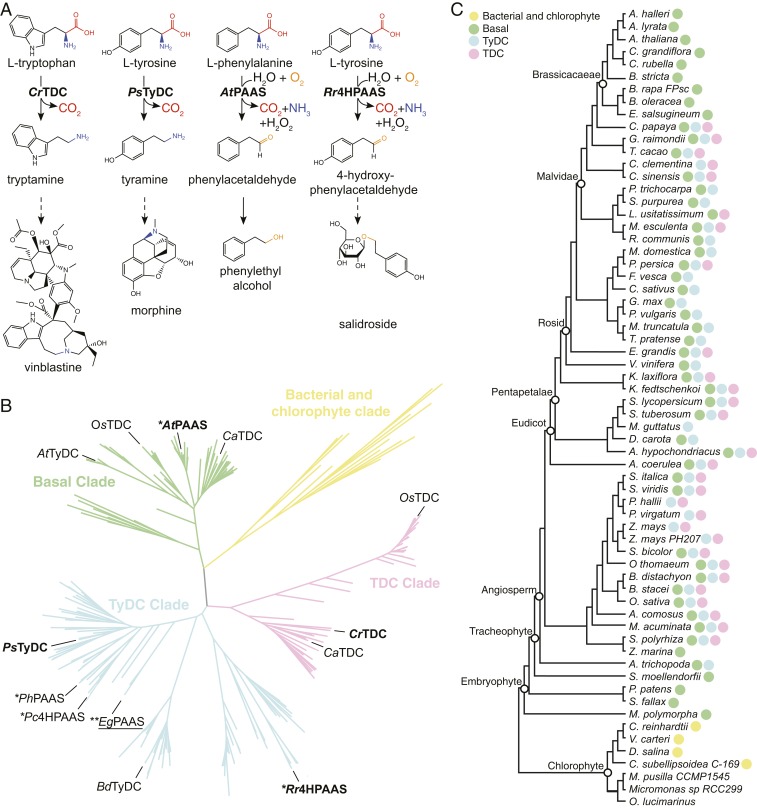
Aromatic amino acid decarboxylases (AAADs) are an ancient group of pyridoxal 5′-phosphate (PLP)-dependent enzymes with primary functions associated with amino acid metabolism. Mammals possess a single AAAD, dopa decarboxylase (DDC), responsible for the synthesis of monoamine neurotransmitters, such as dopamine and serotonin, from their respective aromatic l-amino acid precursors (5). AAAD family enzymes have radiated in plants and insects to yield a number of paralogous enzymes with variation in both substrate preference and catalytic mechanism (Fig. 1 A–C and SI Appendix, Fig. S1) (6). In plants, l-tryptophan decarboxylases (TDCs) and l-tyrosine decarboxylases (TyDCs) are canonical AAADs that supply the aromatic monoamine precursors tryptamine and tyramine for the biosynthesis of monoterpene indole alkaloids (MIAs) and benzylisoquinoline alkaloids (BIAs), respectively (SI Appendix, Fig. S2) (7, 8). In contrast, phenylacetaldehyde synthases (PAASs) and 4-hydroxyphenylacetaldehyde synthases (4HPAASs) are mechanistically divergent aromatic aldehyde synthases (AASs) that catalyze the decarboxylation-dependent oxidative deamination of l-phenylalanine and l-tyrosine, respectively, to produce the corresponding aromatic acetaldehydes necessary for the biosynthesis of floral volatiles and tyrosol-derived natural products (SI Appendix, Fig. S2) (9, 10). Plant AAAD or AAS enzymes thus play a gate-keeping role in channeling primary metabolites into a variety of specialized metabolic pathways.
Fig. 1.
Function, phylogeny, and taxonomic distribution of plant AAADs. (A) Biochemical functions of four representative plant AAADs in the context of their native specialized metabolic pathways. The dashed arrows indicate multiple catalytic steps. (B) A simplified maximum likelihood phylogenetic tree of bacteria, chlorophyte, and plant AAADs. A fully annotated tree is shown in SI Appendix, Fig. S3. The bacterial/chlorophyte, basal, TyDC, and TDC clades are colored in yellow, green, blue, and pink, respectively. Functionally characterized enzymes are labeled at the tree branches. The four AAADs for which their crystal structures were resolved in this study are denoted in bold. The EgPAAS identified and characterized in this study is underlined; * and ** denote two mechanistic classes of AASs that harbor naturally occurring substitutions at the large-loop catalytic tyrosine or the small-loop catalytic histidine, respectively. (C) Taxonomic distribution of plant AAADs across major lineages of green plants. The tree illustrates the phylogenetic relationship among Phytozome V12 species with sequenced genomes. The presence of yellow, green, blue, or pink circles at the tree branches indicates the presence of one or more AAAD sequences from the bacterial/chlorophyte, basal, TyDC, or TDC clades, respectively.
In this work, we seek to understand the evolutionary trajectories that led to the functional divergence and convergence within the plant AAAD family proteins. Through comparative analysis of four representative plant AAAD crystal structures followed by mutant characterization and molecular dynamics (MD) simulation, we identified key structural features that dictate substrate selectivity and the alternative AAAD-versus-AAS catalytic mechanisms. Using these findings, we discovered a group of myrtle plant AASs with a catalytic residue substitution similar to those described in insect AASs but distinct from the substitutions previously described in plant AASs (10). These findings suggest that nature has explored multiple mechanistically distinct trajectories to reconfigure an ancestral AAAD catalytic machinery to catalyze AAS chemistry. Furthermore, we show the feasibility of engineering a shortened BIA biosynthetic pathway in yeast Saccharomyces cerevisiae by harnessing the 4HPAAS activity, highlighting the role of a neofunctionalized enzyme in rewiring ancestral metabolic network during plant specialized metabolic evolution.
Results
The Overall Structures of Four Divergent Plant AAAD Proteins.
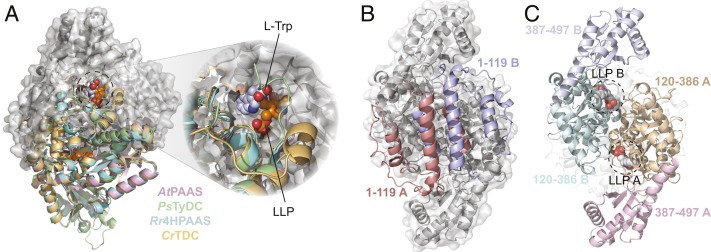
To understand the structural basis for the functional divergence of plant AAAD proteins, we determined the X-ray crystal structures of Catharanthus roseus TDC (CrTDC) in complex with l-tryptophan, Papaver somniferum TyDC (PsTyDC) in complex with l-tyrosine, Arabidopsis thaliana PAAS (AtPAAS) in complex with l-phenylalanine, and unbound Rhodiola rosea 4HPAAS (Rr4HPAAS) (Fig. 2A and SI Appendix, Fig. S3 and Table S1). All four enzymes pack in the crystal lattice as homodimers, and share the typical type II PLP-dependent decarboxylase fold (SI Appendix, Table S2). Each monomer contains three distinct segments (SI Appendix, Fig. S5), which were previously described as domains in the homologous mammalian DDC structure (5). These segments are unlikely to be stable as autonomously folding units, and rather function as stretches of topologically associated constituents necessary for the overall architecture of the α2 dimer. As represented by the l-tryptophan−bound CrTDC structure, the N-terminal segment (CrTDC1-119) comprises three antiparallel helices that interlock with the reciprocal helices of the other monomer to form the primary hydrophobic dimer interface (Fig. 2C and SI Appendix, Fig. S6). The middle (CrTDC120-386) segment harbors a conserved Lys319 with its ζ-amino group covalently linked to the coenzyme PLP (termed as LLP hereafter), and, together with the C-terminal (CrTDC387-497) segment, forms two symmetric active sites at the dimer interface (Fig. 2C).
Fig. 2.
The overall structure of plant AAADs. (A) An overlay of the CrTDC (orange), PsTyDC (green), AtPAAS (pink), and Rr4HPAAS (cyan) structures. All four structures exist as highly similar homodimers, but, for visual simplicity, the cartoon structures were only displayed for the bottom monomers. The top monomer of CrTDC is displayed in gray cartoon and surface representation. The dashed circle highlights the CrTDC active site which contains the l-tryptophan substrate and the prosthetic group LLP. (B) The CrTDC N-terminal segments from the two monomers, one colored in salmon and one in blue, form the hydrophobic dimer interface. The remainder of the homodimer is displayed in gray. (C) The configuration of the CrTDC middle (colored in teal and brown) and C-terminal segments (colored in blue and pink) from the two monomers. The N-terminal segments are displayed in transparent gray cartoons, and the prosthetic LLPs are circled and displayed as spheres. The models exhibited in B and C are in the same orientation, which is rotated 90° around the vertical axis from the view in A.
Structural Features That Control Substrate Selectivity in Plant AAADs.
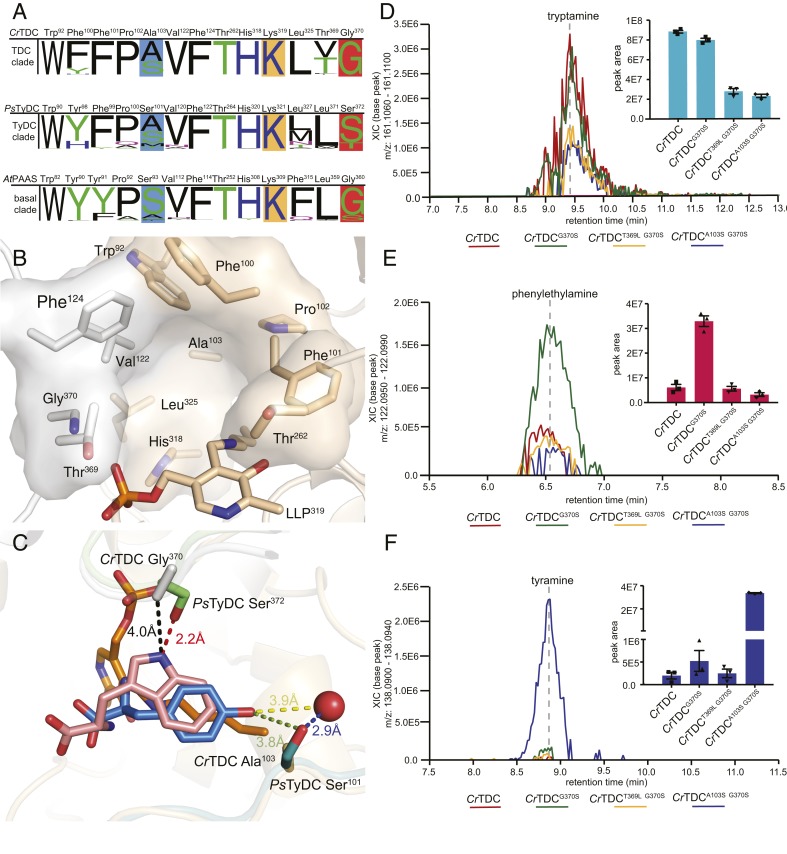
The substrate-binding pocket of plant AAADs is principally composed of conserved aromatic and hydrophobic residues (Trp92, Phe100, Phe101, Pro102, Val122, Phe124, His318, and Leu325 as in CrTDC) in addition to three variable residues (Ala103, Thr369, and Gly370 as in CrTDC) that display sequence divergence across major AAAD clades (Fig. 3 A and B). Comparison of the CrTDC and PsTyDC ligand-bound structures rationalizes the role of Gly-versus-Ser variation at position 370 (numbering according to CrTDC) in dictating the size and shape of the substrate-binding pocket to favor indolic versus phenolic amino acid substrate in CrTDC and PsTyDC, respectively (Fig. 3C) (11). This conclusion is consistent with the previous observation that the CrTDCG370S mutant exhibits enhanced affinity for the nonnative substrate levodopa (l-DOPA) as compared to wild-type CrTDC (11). To test how the G370S mutation would impact CrTDC activity in vivo, we compared the monoamine product profiles of transgenic yeast expressing wild-type CrTDC or CrTDCG370S measured by liquid chromatography high-resolution accurate-mass mass spectrometry (LC-HRAM-MS). Compared to the CrTDC-expressing yeast, the CrTDCG370S-expressing yeast showed little change in tryptamine level (Fig. 3D) but elevated accumulation of phenylethylamine (Fig. 3E), supporting the role of residue at position 370 in gating indolic versus phenolic substrates.
Fig. 3.
Active site pocket composition and residues that dictate substrate selectivity in plant AAADs. (A) Active-site-lining residues from plant AAADs were identified and queried for conservation against all AAAD homologs identified from the 93 Phytozome V12.1 annotated green plant genomes. The height of the residue label displays the relative amino acid frequency, excluding sequence gaps, in the basal, TyDC, and TDC clades. The position of the active site pocket residues from each clade are referenced against the AtPAAS, PsTyDC, and CrTDC sequences. Polar amino acids are colored in green, basic amino acids are colored in blue, acidic amino acids are colored in red, and hydrophobic amino acids are colored in black. Residues highlighted in blue and dark orange boxes denote residues involved in hydroxylated vs. unhydroxylated and phenolic vs. indolic substrate recognition, respectively. The conserved lysine residues, represented as the LLP prosthetic group in several crystal structures, are marked by a light orange box. (B) The CrTDC active-site pocket is composed of residues from both chain A and chain B, colored in beige and white, respectively. The pocket is composed of conserved nonpolar residues (Pro102, Val122, and Leu325), aromatic residues (Trp92, Phe100, Phe101, Phe124, and His318), and a polar residue (Thr262). Additionally, the active site contains three variable residues (Ala103, Thr369, and Gly370), which differ across different AAAD clades. (C) Superimposition of the substrate-complexed CrTDC and PsTyDC structures. CrTDC chain A and chain B are displayed in beige and white, respectively, while relevant portions of the PsTyDC chain A and chain B are displayed in green and deep teal, respectively. The l-tryptophan ligand from the CrTDC structure is colored in pink, while the PsTyDC l-tyrosine ligand is colored in blue. The red sphere represents a PsTyDC active-site water likely involved in substrate recognition. (D−F) Relative in vivo decarboxylation of (D) ʟ-tryptophan, (E) l-phenylalanine, and (F) ʟ-tyrosine catalyzed by wild-type and various mutants of CrTDC as measured in transgenic yeast. The error bars indicate SEM based on biological triplicates while the squares, triangles, and diamonds represent the individual data points.
In the CrTDC structure, the second variable residue Thr369 is adjacent to the indolic-selective Gly370 residue, which is mostly conserved as a leucine in both the basal clade and the TyDC clade but varies more widely as a threonine, valine, or phenylalanine in the TDC clade (Fig. 3A). The variable nature of this Thr369 residue in the TDC clade suggests a potential role of this residue in substrate selectivity. However, transgenic yeast expressing the CrTDCT369L G370S double mutant showed a general decrease in aromatic monoamine production but no significant difference in relative abundance of each monoamine product compared to yeast strain expressing the CrTDCG370S single mutant (Fig. 3 E and F). This observation thus did not support a direct correlation of the identity of this second variable residue with substrate selectivity.
The third variable residue at position 103 (numbering according to CrTDC) is represented only as a serine or alanine in the TDC clade but varies as serine, threonine, alanine, methionine, phenylalanine, or cysteine in the basal and TyDC clades (Fig. 3A). In the l-tyrosine−bound PsTyDC structure, the p-hydroxyl of the tyrosine substrate is coordinated by PsTyDC Ser101 (corresponding to CrTDC Ala103) together with a nearby water through hydrogen bonding (Fig. 3C), suggesting the role of this residue in gating hydroxylated versus unhydroxylated aromatic amino acid substrates. Indeed, transgenic yeast expressing the CrTDCA101S G370S double mutant exhibited an ∼64-fold increase in tyramine level and a significant decrease in phenylethylamine production as compared to the CrTDCG370S-expressing strain (Fig. 3 E and F). The residue variation at this position in the basal and TyDC clades likely plays a role in discerning various phenolic substrates, namely, l-phenylalanine, l-tyrosine, and l-DOPA, with varying ring hydroxylation patterns. Serine or alanine substitution at this position in the TDC clade may likewise distinguish 5-hydroxy-l-tryptophan versus l-tryptophan substrate for entering the plant melatonin and indole alkaloid biosynthetic pathways, respectively. Together, these results suggest that the phylogenetically restricted sequence variations at position 103 and 370 (numbering according to CrTDC) were likely selected to control the respective substrate preferences of the TyDC and TDC clades as they diverged from the basal-clade AAAD progenitors.
The PLP-Dependent Catalytic Center of Plant AAADs.
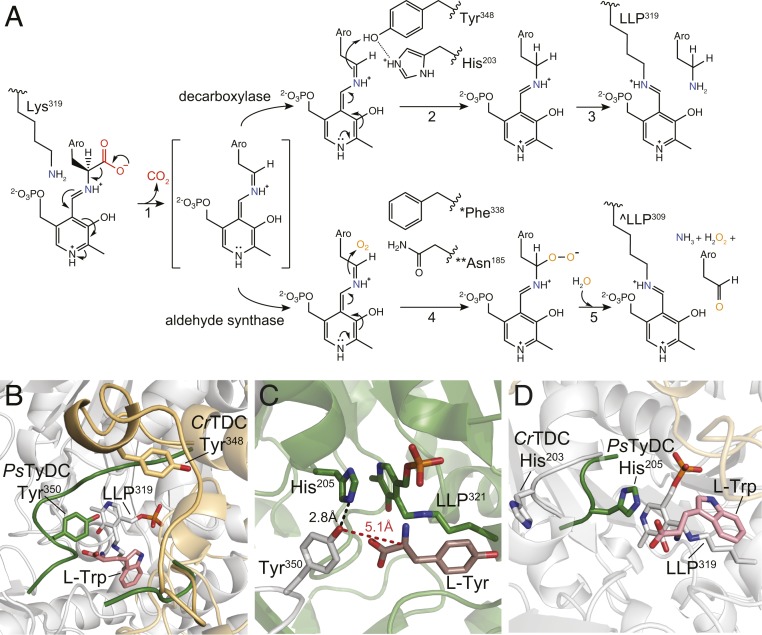
PLP, the active form of vitamin B6, is a versatile coenzyme employed by ∼4% of all known enzymes to catalyze a diverse array of biochemical reactions. The active site of canonical AAADs constrains the reactivity of PLP to specifically catalyze decarboxylation of the α-carbon carboxyl group of aromatic l-amino acids (12). The active site of plant AAADs, as represented in the CrTDC structure, is located at the dimer interface, and features the characteristic prosthetic group LLP in the form of an internal aldimine at resting state (SI Appendix, Fig. S7). The phosphate moiety of LLP is coordinated by Thr167, Ser168, and Thr369, while the pyridine-ring amine forms a salt bridge with the side-chain carboxyl of Asp287, supporting its conserved role in stabilizing the carbanionic intermediate of the PLP external aldimine (SI Appendix, Fig. S7) (13). Evident from the l-tryptophan−bound CrTDC structure, the aromatic l-amino acid substrate is oriented in the active site to present its labile Cα-carboxyl bond perpendicular to the pyridine ring of the internal aldimine LLP (SI Appendix, Fig. S8) as predicted by Dunathan's hypothesis (12). Upon substrate binding, transaldimination then occurs that conjugates the α-amino group of the substrate to PLP through a Schiff base to yield the external aldimine (SI Appendix, Fig. S9), as likely captured by one of the active sites of the l-tyrosine−bound PsTyDC structure (SI Appendix, Fig. S10). The resulting external aldimine subsequently loses the α-carboxyl group as CO2 to generate a quinonoid intermediate (Fig. 4A, reaction step 1). Through an as-yet-unknown mechanism, the nucleophilic carbanion of the quinonoid intermediate is protonated to yield the monoamine product and a regenerated LLP, ready for subsequent rounds of catalysis (Fig. 4A, reaction steps 2 and 3) (14). Despite the availability of several animal DDC crystal structures, several aspects of the full catalytic cycle of the type II PLP decarboxylases remain speculative. Comparative structural analysis of our four plant AAAD structures sheds light on the mechanistic basis for the canonical decarboxylation activity as well as the evolutionarily new decarboxylation-dependent oxidative deamination activity, which are detailed below.
Fig. 4.
Catalytic mechanisms and conformational changes of plant AAAD proteins. (A) The proposed alternative PLP-mediated catalytic mechanisms for the canonical decarboxylase and derived aldehyde synthase in plant AAAD proteins. After transaldimation of the CrTDC internal aldimine to release the active-site Lys319, the PLP amino acid external aldimine loses the α-carboxyl group as CO2 to generate a quinonoid intermediate stabilized by the delocalization of the paired electrons (1). In a canonical decarboxylase (e.g., CrTDC), the carbanion at Cα is protonated by the acidic p-hydroxyl of Tyr348-B located on the large loop, which is facilitated by its neighboring His203-A located on the small loop (2). The CrTDC LLP319 internal aldimine is regenerated, accompanied by the release of the monoamine product (3). In an evolutionarily new aldehyde synthase, the Cα protonation step essential for the canonical decarboxylase activity is impaired when the large-loop catalytic tyrosine is mutated to phenylalanine (* as in AtPAAS), or when the small-loop catalytic histidine is mutated to asparagine (** as in EgPAAS), allowing the Cα carbanion to attack a molecular oxygen to produce a peroxide intermediate (4). This peroxide intermediate decomposes to give aromatic acetaldehyde, ammonia, and hydrogen peroxide products and regenerate the LLP internal aldimine (^ as in AtPAAS) (5). Aro represents the aromatic moiety of an aromatic l-amino acid. (B) An overlay of the PsTyDC structure with its large loop in a closed conformation (green) upon the CrTDC structure with its large loop in an open conformation (beige). (C) The closed conformation of PsTyDC active site displaying the catalytic machinery in a configuration ready to engage catalysis. Chain A is colored in white, chain B is colored in green, and the l-tyrosine substrate is displayed in dark pink. (D) Open and closed small-loop conformations observed in the CrTDC and PsTyDC structures. The PsTyDC small loop with the catalytic histidine is in a closed conformation (green), while the CrTDC structure exhibits its small-loop histidine in an open conformation (white).
Structural Features of Two Catalytic Loops.
Previous structural studies of the mammalian AAADs identified a large loop that harbors a highly conserved tyrosine residue essential for catalysis (15). However, this loop is absent from the electron density map of all of the previously resolved animal DDC structures. We provide direct electron density support for this large loop that adopts different conformations relative to the active site in different plant AAAD crystallographic datasets (Fig. 4B and SI Appendix, Supplementary Note 1). In the CrTDC structure, the large loop (CrTDC342-361) of monomer A adopts an open conformation, revealing a solvent-exposed active site. Conversely, in the PsTyDC structure, a crankshaft rotation puts the large loop over the bound l-tyrosine substrate to seal the active-site pocket. In this closed conformation, the p-hydroxyl of the catalytic tyrosine located on the large loop (Tyr350-A) is in close proximity to the Cα of the l-tyrosine substrate and LLP (Fig. 4C). These structural observations suggest that Tyr350 (as in PsTyDC) serves as the catalytic acid that protonates the nucleophilic carbanion of the quinonoid intermediate at the substrate Cα position, which is preceded by an open-to-closed conformational change by the large loop.
Comparative structural analysis of our four plant AAAD structures identified another small loop (CrTDC200-205-B), which harbors a conserved histidine residue (His203-B as in CrTDC) and cooperatively interacts with the large loop from the opposite monomer. For instance, the small loop in the CrTDC structure is rotated outward from the active-site LLP (Fig. 4D). Conversely, in the AtPAAS, PsTyDC, and Rr4HPAAS structures, the small loop adopts a closed conformation with its histidine imidazole forming pi stacking with the LLP pyridine ring. Such pi stacking between PLP and an active-site aromatic residue has been observed previously as a common feature within AAADs as well as the broader α-aspartate aminotransferase superfamily (16–18). Moreover, in the PsTyDC structure, the τ-nitrogen of the small-loop histidine is in hydrogen-bonding distance of the p-hydroxyl of the large-loop catalytic tyrosine (Fig. 4C), suggesting a potential catalytic role of this histidine. Previous studies of the pH dependence of AAADs suggest that this small-loop histidine cannot function directly as the catalytic acid to protonate the carbanionic quinonoid intermediate (19–21). Based on our structural observations, we propose a catalytic mechanism where the small-loop histidine (CrTDC His203-B) facilitates the proton transfer from the p-hydroxyl of the large-loop tyrosine (CrTDC Tyr348-A) to the quinonoid intermediate (Fig. 4A, reaction steps 2) by serving as a direct or indirect proton source for reprotonation of the Tyr348-A p-hydroxyl. Mechanistically similar Tyr-His side-chain interaction that facilitates protonation of a carbanionic intermediate has also been recently proposed for the C. roseus heteroyohimbine synthase, which is a medium-chain dehydrogenase/reductase-family enzyme (22).
MD Simulations Reveal the Dynamic Nature of the Two Catalytic Loops.
Considering the various alternative conformations observed in our plant AAAD structures, we sought to examine the flexibility and cooperativity of the two loops that harbor the key catalytic residues (CrTDC Tyr348-A and His203-B) by MD simulation. We began with 36 sets of 100-ns simulations on six CrTDC systems with LLP and the substrate l-tryptophan in different protonation states (SI Appendix, Fig. S11 and SI Appendix, Supplementary Note 2). These simulations revealed considerable flexibility of both loops, with one simulation capturing a dramatic closing motion of the open large loop. Upon extending this simulation to 550 ns (SI Appendix, Fig. S12 and Movie S1), the large loop was found to reach a semiclosed state characterized by a minimal Cα rmsd of 4.3 Å with respect to the modeled CrTDC closed-state structure (SI Appendix, Supplementary Note 3). The catalytic Tyr348-A was found to form stacking interactions with His203-B, with a minimal distance of ∼2 Å between the two residues (SI Appendix, Fig. S13). Interestingly, a large-loop helix (CrTDC346-350A) that unfolded at the beginning of this simulation appeared to “unlock” the large loop from its open state. To further examine the correlation between the secondary structure of this helix and the large-loop conformation, we artificially unfolded this short helix and initiated 72 sets of 50-ns simulations from the resulting structure. Similar conformations to that identified in SI Appendix, Fig. S11 were observed in all six systems (SI Appendix, Fig. S14C), with minimal Cα rmsd ranging from ∼5 to 8 Å with respect to the modeled CrTDC closed-state structure. These results suggest that the initial closing motion of the large loop is independent of the coenzyme and substrate protonation states and that the unfolding of the aforementioned helix can significantly accelerate such motion. This is further substantiated by three sets of 600-ns simulations of CrTDC in an apo state with neither PLP nor l-tryptophan present (SI Appendix, Fig. S15 and SI Appendix, Supplementary Note 2). It is noted, however, that the fully closed state of CrTDC was not achieved in our submicrosecond simulations. For instance, in the trajectory shown in SI Appendix, Fig. S11 and Movie S1, l-tryptophan left the active site at around t = 526 ns, shortly after which the simulation was terminated. Overall, while the transition from the semiclosed to the fully closed state can be expected to occur beyond the submicrosecond timescale, the MD results support our hypothesis that Tyr348-A can form close contact with His203-B, readying the latter residue to stabilize pyridine ring resonance and direct proton transfer from the former residue to the carbanionic quinonoid intermediate.
Convergent Evolution of Two Mechanistic Classes of AASs.
Our insights into the structural and dynamic basis for the canonical AAAD catalytic mechanism predicts that mutations to the large-loop tyrosine or the small-loop histidine would likely derail the canonical AAAD catalytic cycle and potentially yield alternative reaction outcomes. Indeed, the canonical large-loop tyrosine is substituted by phenylalanine in AtPAAS and Rr4HPAAS. We propose that absence of the proton-donating p-hydroxyl of the catalytic tyrosine enlarges the active-site cavity to permit molecular oxygen to enter the active site and occupy where the tyrosine p-hydroxyl would be for proton transfer (Fig. 4A, reaction steps 4 and 5, *). Instead of protonation, the nucleophilic carbanion of the quinonoid intermediate attacks molecular oxygen to generate a peroxide intermediate, which subsequently decomposes to yield ammonia, hydrogen peroxide, and the aromatic acetaldehyde product.
The rapid expansion of genomic resources together with our increasing knowledge about the structure−function relationships of disparate enzyme families enable database mining for potentially neofunctionalized enzymes in plant specialized metabolism. Indeed, the identification of the Tyr-to-Phe substitution responsible for the AAS activity in plants (13) served as a “molecular fingerprint” for the functional prediction of unconventional AASs among legume serine decarboxylase-like enzymes (23). Bearing in mind the newly identified functional residues that control substrate selectivity and catalytic mechanism, we queried all plant AAAD homologs identifiable by basic local alignment search tool (BLAST) within National Center for Biotechnology Information (NCBI) and the 1KP databases (24). Interestingly, this search identified a number of TyDC-clade AAAD homologs from the Myrtaceae and Papaveraceae plant families that harbor unusual substitutions at the small-loop histidine (SI Appendix, Fig. S16). Several Myrtaceae plants contain a group of closely related and likely orthologous AAAD proteins with a His-to-Asn substitution at the small-loop histidine. In Papaveraceae, on the other hand, we noticed a His-to-Leu substitution in the previously reported P. somniferum tyrosine decarboxylase 1 (PsTyDC1) (8) and a His-to-Tyr substitution in another Papaver bracteatum AAAD homolog. While the small-loop histidine is typically conserved among type II PLP decarboxylases and the broader type I aspartate aminotransferases, the same His-to-Asn substitution was previously observed in select insect AAAD proteins involved in 3,4-dihydroxyphenylacetaldehyde production necessary for insect soft cuticle formation (25, 26). These sequence observations together with the proposed catalytic role of the small-loop histidine implicate that AAADs with substitutions at this highly conserved residue may contain alternative enzymatic functions.
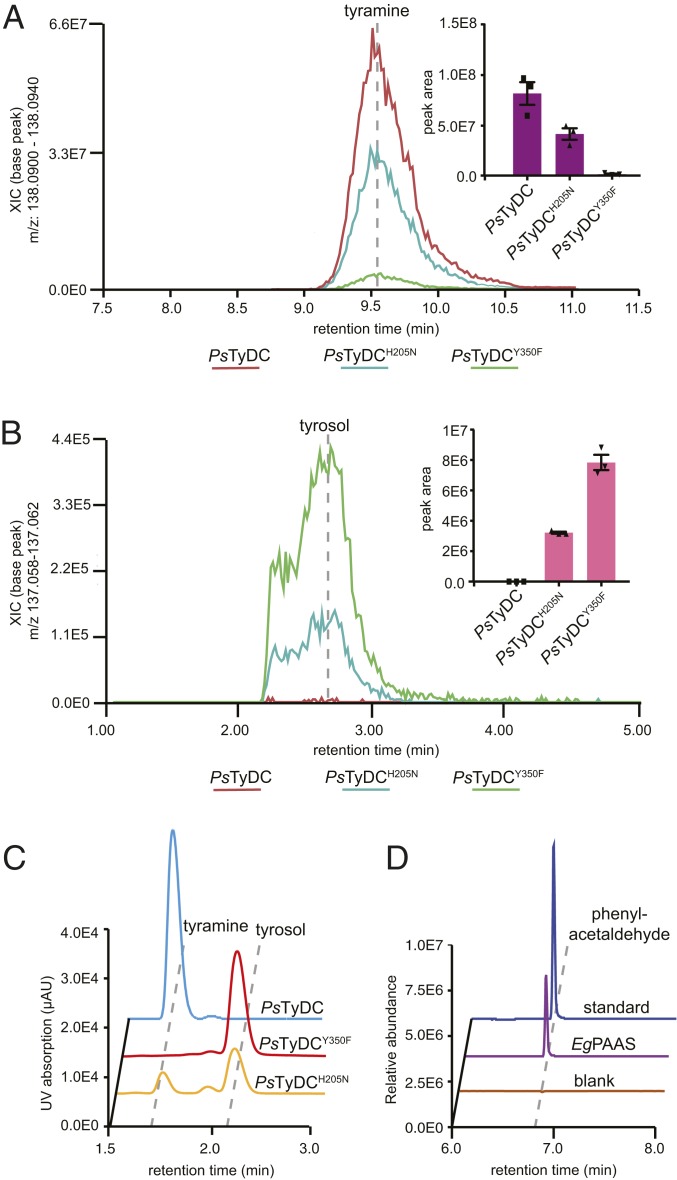
To test this hypothesis, we first generated transgenic yeast strains expressing either wild-type PsTyDC, PsTyDCY350F, or PsTyDCH205N, and assessed their metabolic profiles by LC-HRAM-MS (Fig. 5 A and B). The PsTyDC-expressing yeast exclusively produces tyramine, whereas the PsTyDCY350F-expressing yeast exclusively produces tyrosol (reduced from 4HPAA by yeast endogenous metabolism). This result suggests that mutating the large-loop tyrosine to phenylalanine in a canonical TyDC sequence background is necessary and sufficient to convert it to an AAS. Interestingly, the PsTyDCH205N-expressing yeast produces both tyramine and tyrosol, suggesting that mutating the small-loop histidine to asparagine alone in PsTyDC turned it into an AAAD−AAS bifunctional enzyme. In vitro enzyme assays using recombinant wild-type PsTyDC, PsTyDCY350F, or PsTyDCH205N against l-tyrosine as substrate yielded similar results that corroborated the observations made in transgenic yeast (Fig. 5C). Collectively, these results reinforce the essential role of the large-loop catalytic tyrosine for the canonical AAAD activity, and support the proposed assisting role of the small-loop histidine in quinonoid intermediate protonation for the canonical AAAD activity. The full AAAD catalytic cycle could still proceed, albeit with significantly reduced efficiency, when the small-loop histidine is mutated to asparagine, whereas a significant fraction of the carbanionic quinonoid intermediate undergoes the alternative oxidative deamination chemistry similarly as proposed for AtPAAS (Fig. 4A, reaction steps 4 and 5, **).
Fig. 5.
Two alternative molecular strategies to arrive at aldehyde synthase chemistry from a canonical AAAD progenitor. Transgenic yeast strains expressing PsTyDCH205N and PsTyDCY350F display (A) reduced tyramine production but (B) elevated levels of tyrosol (reduced from 4HPAA by yeast endogenous metabolism) in comparison to transgenic yeast expressing wild-type PsTyDC. Cultures were grown and metabolically profiled in triplicate. The error bars in the bar graphs (Insets) indicate SEM and the squares and triangles represent the individual data points. (C) LC-UV chromatograms showing the relative decarboxylation and aldehyde synthase products produced by purified recombinant PsTyDC, PsTyDCY350F, or PsTyDCH205N enzymes when incubated with 0.5 mM l-tyrosine for 5, 25, and 50 min, respectively. After enzymatic reaction, the 4HPAA aldehyde product was chemically reduced by sodium borohydride to tyrosol prior to detection. (D) Phenylacetaldehyde formation from the incubation of purified recombinant EgPAAS with l-phenylalanine measured by GC-MS.
To assess the function of plant AAAD homologs that naturally contain substitutions at the small-loop histidine, we cloned one of such genes from Eucalyptus grandis (EgPAAS; Fig. 1B), starting from total messenger RNA extracted from the host plant. In vitro enzyme assays conducted using recombinant EgPAAS against a panel of aromatic l-amino acids demonstrated exclusive AAS activity with an apparent substrate preference toward l-phenylalanine (Fig. 5D and SI Appendix, Fig. S17). It is worth noting that phenylacetaldehyde has been previously identified as a major fragrance compound present in the essential oil and honey of numerous Myrtaceae plants (27–29), which may be attributed to the activity of EgPAAS. Unlike the PsTyDCH205N mutant, EgPAAS shows no detectable ancestral AAAD activity in vitro, suggesting that, in addition to the small-loop His-to-Asn substitution, other adaptive mutations must have contributed to the specific PAAS activity. While the AAS activity of EgPAAS was confirmed in transgenic yeast (SI Appendix, Fig. S18), Myrtaceae AAADs from Medinilla magnifica and Lagerstroemia indica (MmAAS and LiAAS), which also contain the unusual small-loop Hist-to-Asn substitution (SI Appendix, Fig. S16), displayed little to no enzymatic activity in this system. Despite our interest in the other two AAAD sequences from Papaveraceae plants harboring alternative substitutions at the small-loop histidine, they could not be directly cloned from their respective host plants, raising the possibility that these genes could potentially be derived from sequencing or assembly errors. Thus, we did not pursue functional characterization of these genes further.
Metabolic Engineering of a Shortened BIA Biosynthetic Pathway in Yeast with 4HPAAS.
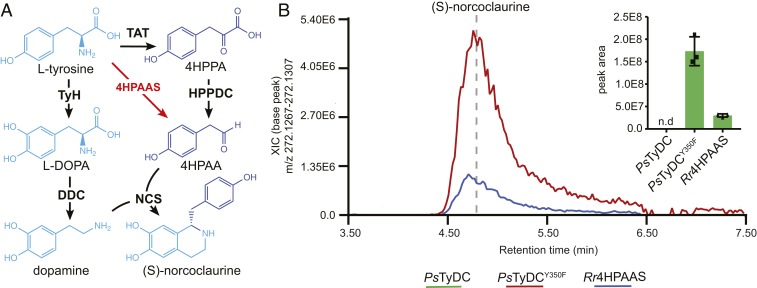
To examine the role of adaptive functional evolution of a single AAAD protein in the larger context of specialized metabolic pathway evolution, we attempted a metabolic engineering exercise to build a shortened BIA biosynthetic pathway in yeast S. cerevisiae. In the proposed plant BIA pathway, the key intermediate 4HPAA is reportedly derived from l-tyrosine through two consecutive enzymatic steps catalyzed by l-tyrosine aminotransferase and an unidentified 4-hydroxyphenylpyruvate decarboxylase (4HPPDC) (30) (Fig. 6A). However, in the R. rosea salidroside biosynthetic pathway, 4HPAA is directly converted from l-tyrosine by a single Rr4HPAAS enzyme (10) (Fig. 6A). We therefore reasoned that 4HPAA activity could be utilized to reroute the 4HPAA branch of the BIA pathway. In our metabolic engineering scheme, either Rr4HPAAS or the PsTyDCY350F mutant was used as the 4HPAAS enzyme, while wild-type PsTyDC was used as a control. Moreover, Pseudomonas putida DDC (PpDDC) and Beta vulgaris l-tyrosine hydroxylase (BvTyH) were used to generate the other key intermediate dopamine from l-tyrosine. Lastly, P. somniferum (S)-norcoclaurine synthase (PsNCS) was added to stereoselectively condense 4HPAA and dopamine to form (S)-norcoclaurine.
Fig. 6.
The utility of 4HPAAS in metabolic engineering of a shortened BIA pathway for (S)-norcoclaurine production in yeast. (A) Canonically proposed (S)-norcoclaurine biosynthetic pathway (black arrows) rerouted by the use of a 4HPAAS (red arrows). (B) Engineering of (S)-norcoclaurine production in yeast using two AAAD proteins with the 4HPAAS activity. All multigene vectors used to transform yeast contain BvTyH, PpDDC, and PsNCS in addition to either PsTyDC, PsTyDCY350F, or Rr4HPAAS. Cultures were grown and measured in triplicate. The error bars in the bar graph indicate SEM whereas the squares and triangles represent the individual data points; n.d., not detected.
Coexpression of PsTyDCY350F with PpDDC, BvTyH, and PsNCS resulted in the highest accumulation of (S)-norcoclaurine among various engineered transgenic yeast strains, whereas replacing PsTyDCY350F with Rr4HPAAS gave ∼sevenfold less (S)-norcoclaurine production (Fig. 6B). Control experiment using wild-type PsTyDC produced tyramine but not (S)-norcoclaurine as expected (Fig. 6B and SI Appendix, Fig. S19). This metabolic engineering exercise illustrates that, when provided with the right metabolic network context, the birth of an evolutionarily new AAS activity (achievable through a single mutation) can lead to rewiring of the ancestral specialized metabolic pathway. Mechanistic understanding of the structure−function relationship within the versatile plant AAAD family adds to the toolset for metabolic engineering of high-value aromatic amino acid-derived natural products in heterologous hosts.
Discussion
Selective pressures unique to plants’ ecological niches have shaped the evolutionary trajectories of the rapidly expanding specialized metabolic enzyme families in a lineage-specific manner. The AAAD family is an ancient PLP-dependent enzyme family ubiquitously present in all domains of life. While AAAD proteins mostly retain highly conserved primary metabolic functions in animals, for example, monoamine neurotransmitter synthesis, they have undergone extensive radiation and functional diversification during land plant evolution. We show that the major monophyletic TDC and TyDC clades of plant AAADs emerged from the basal clade, each acquiring divergent active-site structural features linked to substrate specificity. Moreover, the ancestral AAAD catalytic machinery has also been modified repeatedly through two types of mechanistic mutations to converge on the evolutionarily new AAS activity in numerous AAAD paralogs in plants and insects. The rich evolutionary history of the AAAD family therefore affords a prism for understanding how plants capitalize their ability to biosynthesize proteinogenic aromatic l-amino acids and use them as precursors for further developing taxonomically restricted specialized metabolic pathways (31). Some of these downstream pathways, such as the tryptophan-derived MIA pathway in plants under the order of Gentianales and the tyrosine-derived BIA pathway in plants under the order of Ranunculales, give rise to major classes of bioactive plant natural products critical not only for host fitness but also as a source for important human medicines.
PLP in isolation can catalyze almost all of the reactions known for PLP-dependent enzymes, including transamination, decarboxylation, 0 and y elimination, racemization, and aldol cleavage (32). In the context of PLP-dependent enzymes, PLP reactivity is confined by its encompassing active site to elicit specific catalytic outcomes with defined substrates (32). Evolutionary analysis suggests that major functional classes of PLP-dependent enzymes likely established first, followed by divergence in substrate selectivity (32, 33). Four evolutionarily distinct groups of PLP-dependent decarboxylases exist in nature (33). The largest and most diverse group (Group II) consists of AAADs, histidine decarboxylases, glutamate decarboxylases, aspartate decarboxylases, serine decarboxylases, and cysteine sulfinic acid decarboxylases, implicating a deep divergence of amino acid substrate selectivity among Group II enzymes. Within plant AAADs, the substrate selectivity has continued to evolve to arrive at extant enzymes with exquisite substrate selectivity toward various proteinogenic and nonproteinogenic aromatic l-amino acids. This is largely achieved by fixing specific mutations at the substrate-binding pocket, as revealed in this study.
Oxygenation is one of the most common chemical reactions in plant specialized metabolism. The triplet ground electron state of molecular oxygen necessitates a cofactor to facilitate single-electron transfer, evident from several major oxygenase families involved in plant specialized metabolism, including cytochrome P450 monooxygenases, iron/2-oxoglutarate−dependent oxygenases, and flavin-dependent monooxygenases (34). AASs represent a class of plant oxygenases that exploit a carbanionic intermediate to facilitate oxygenation (Fig. 4A, reaction steps 4 and 5). This catalytic mechanism is reminiscent of the paracatalytic photorespiratory reaction catalyzed by ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), where molecular oxygen instead of CO2 enters the RuBisCO active site and reacts with a ribulose-1,5-bisphosphate−derived carbanionic enediolate intermediate to form a peroxide intermediate that subsequently decomposes to yield 3-phosphoglycerate and 2-phosphoglycerate (35). Carbanion-stabilizing enzymes, including PLP-dependent enzymes DDC, glutamate decarboxylase, and ornithine decarboxylase, are also capable of catalyzing similar paracatalytic oxygenation reactions (35–38). Whereas oxygenation reactions observed in these carbanion-stabilizing enzymes are mostly deemed as paracatalytic activities due to the intrinsic reactivity of the carbanion intermediate, the plant AASs evolved by harnessing such latent paracatalytic activity for dedicated production of aromatic acetaldehydes and their derived secondary metabolites (10, 39–42). We show that, by understanding the structural and mechanistic basis for convergent evolution of two mechanistic classes of AAS within the plant AAAD family, PsTyDC could be converted to a specific 4HPAAS with a single large-loop Tyr-to-Phe mutation, or an AAAD−AAS bifunctional enzyme by the small-loop His-to-Asn mutation. Several aspects of the neofunctionalization of the AAS activities remain unclear and are topics for future investigation. For example, the emergence of AAS activity results in the stoichiometric production of ammonium, hydrogen peroxide, and reactive aldehydes, which may require additional adaptive metabolic and cellular processes for taming their reactivity/toxicity in AAS-expressing cells. While the TDC and TyDC clades appear to clearly function in specialized metabolic pathways, the biochemical functions and physiological roles of the basal clade in plants remain less studied. Furthermore, the exact chemical mechanism underlying decomposition of the peroxide intermediate that yields aromatic acetaldehyde, ammonium, hydrogen peroxide, and the regenerated active-site LLP is yet to be defined.
Coopting progenitor enzymes to synthesize novel and adaptive metabolites is a universal mechanism underscoring metabolic evolution (43). Most specialized metabolic enzymes present in extant plants evolved through the recruitment of malleable ancestral enzyme folds followed by neofunctionalization of substrate specificity, product diversity, or, in much rarer cases, alternative catalytic mechanisms (44–47). The plant AAAD family illustrates all of these evolutionary mechanisms. Applying the learned knowledge about AAAD evolution has further enabled metabolic engineering of a shortened BIA pathway to produce (S)-norcoclaurine in yeast, using a natural or an evolved 4HPAAS. The use of an insect l-DOPA−specific AAS in engineering of tetrahydropapaveroline biosynthesis in Escherichia coli was recently reported (48), highlighting another example of utilizing AAS in metabolic engineering. It is noted that the free aldehydes produced by AASs readily react with amino acids or other free amines to produce iminium conjugation products via nonenzymatic aldehyde−amine condensation chemistry as seen in E. coli (48) or in plant betacyanin and betaxanthin biosynthesis (49). The use of PsNCS that catalyzes the enantioselective Pictet−Spengler condensation of 4HPAA and dopamine (50) therefore helps to channel the reactive 4HPAA toward (S)-norcoclaurine production in engineered yeast. The successful engineering of a shortened (S)-norcoclaurine biosynthetic pathway using 4HPAAS also hints at an alternative hypothesis to the currently unresolved plant BIA pathway regarding the origin of 4HPAA (30).
Materials and Methods
Reagents.
l-tryptophan, tryptamine, l-tyrosine, tyramine, tyrosol, l-phenylalanine, phenylethylamine, phenylacetaldehyde, phenylethyl alcohol, tyrosol, l-DOPA, dopamine, (S)-norcoclaurine, PLP, and sodium borohydride were purchased from Sigma-Aldrich. The 4-hydroxyphenylacetaldehyde was purchased from Santa Cruz Biotechnology.
Multiple Sequence Alignment and Phylogenetic Analysis.
ClustalW2 was used to generate the protein multiple sequence alignments with default settings (51). The phylogenies shown in SI Appendix, Figs. S1 and S3 were inferred using the maximum likelihood method. The bootstrap consensus unrooted trees were inferred from 500 replicates to represent the phylogeny of the analyzed enzyme families. The phylogenetic analysis encompasses AAAD homolog sequences from all sequenced plant genomes available at Phytozome V12 as well as previously characterized AAAD proteins from plants and select bacteria, chordata, and insect sequences (52). All phylogenetic analyses were conducted in MEGA7 (53). ESPript 3.0 was used to display the multiple sequence alignment (54). Conservation of the active-site residues in various AAAD clades was displayed using WebLogo (55).
Plant Materials.
E. grandis seeds were purchased from Horizon Herbs. Seeds were stratified at 4 °C for 3 d, and germinated in potting soil. Plants were grown under a 16-h-light/8-h-dark photoperiod at 23 °C in a local greenhouse.
Molecular Cloning.
Leaf tissue of 70-d-old E. grandis plants was harvested for total RNA extraction using Qiagen’s RNeasy Mini Kit (Qiagen). Total RNAs for A. thaliana, P. somniferum, C. roseus, and R. rosea were extracted as previously described (10, 13). First-strand complementary DNAs (cDNAs) were synthesized by RT-PCR using total RNA as template. The coding sequences of candidate genes were amplified from cDNAs by PCR using gene-specific primers (SI Appendix, Table S3). Gibson assembly was used to ligate the CrTDC, PsTyDC, AtPAAS, and Rr4HPAAS PCR amplicons into pHis8-4, a bacterial expression vector containing an N-terminal 8xHis tag followed by a tobacco etch virus (TEV) cleavage site for recombinant protein production. EgPAAS was alternatively cloned through Gibson assembly into pTYB12, a commercially available N-terminal intein/chitin domain fusion vector designed for affinity chromatography purification.
Recombinant Protein Production and Purification.
BL21(DE3) E. coli containing the pHis8-4 or pTYB12-based constructs were grown in terrific broth at 37 °C to optical density 600 of 0.9 and induced with 0.15 mM isopropyl-β-ᴅ-thiogalactoside. The cultures were cooled to 18 °C and shaken for an additional 20 h. Cells were harvested by centrifugation, washed with phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4), resuspended in 150 mL of lysis buffer (50 mM Tris pH 8.0, 0.5 M NaCl, 20 mM imidazole, and 0.5 mM dithiothreitol [DTT]), and lysed with five passes through an M-110L microfluidizer (Microfluidics). The resulting crude protein lysate from the CrTDC, PsTyDC, AtPAAS, and Rr4HPAAS cultures were clarified by centrifugation prior to Qiagen nickel nitrilotriacetic acid (Ni-NTA) gravity flow chromatographic purification. After loading the clarified lysate, His-tagged recombinant protein-bound Ni-NTA resin was washed with 20 column volumes of lysis buffer, and eluted with 1 column volume of elution buffer (50 mM Tris pH 8.0, 0.5 M NaCl, 250 mM imidazole, and 0.5 mM DTT). One milligram of His-tagged TEV protease was added to the eluted protein, followed by dialysis at 4 °C for 16 h in dialysis buffer (50 mM Tris pH 8.0, 0.1 M NaCl, 20 mM imidazole, and 2 mM DTT). After dialysis, the protein solutions were passed through Ni-NTA resin to remove uncleaved protein and the His-tagged TEV. The EgPAAS enzyme was insoluble when expressed as an N-terminal polyhistidine-tagged protein, and was therefore expressed as a fusion protein with the Intein/Chitin Binding Protein using the pTYB12 vector. EgPAAS-expressing E. coli cell pellets were homogenized in an imidazole-free lysis buffer. The resulting crude protein lysate was then applied to a column packed with chitin beads, washed with 1 L of buffer, and subsequently hydrolyzed under reducing conditions as per the manufacturer's instructions. Recombinant proteins were further purified by gel filtration on a fast protein LC system (GE Healthcare Life Sciences). The principle peaks were collected, verified for molecular weight by sodium dodecyl sulfate polyacrylamide gel electrophoresis, stored in storage buffer (20 mM Tris pH 8.0, 25 mM NaCl, 200 µM PLP, and 0.5 mM DTT) at a protein concentration of 10 mg/mL, and flash frozen for subsequent investigation. Despite the use of a solubilizing domain from the pTYB12 vector in the expression and purification of EgPAAS, this enzyme was ultimately only partially purified, with significant contamination of E. coli chaperone proteins.
Protein Crystallization and Structural Determination.
Crystals for the various plant AAADs were grown at 4 °C by hanging-drop vapor diffusion method with the drop containing 0.9 µL of protein sample and 0.9 µL of reservoir solution at a reservoir solution volume of 500 µL. The crystallization buffer for the AtPAAS contained 0.16 M ammonium sulfate, 0.8 M Hepes:NaOH pH 7.5, and 20% wt/vol polyethylene glycol (PEG) 3350. Crystals were soaked in a well solution containing 15 mM l-phenylalanine for 6 h and cryogenized with an additional 10% wt/vol ethylene glycol. PsTyDC crystals were formed in 1.2 M ammonium sulfate, 0.1 M Bis Tris pH 5.0, and 1% wt/vol PEG 3350. Crystals were soaked in the presence of 4 mM l-tyrosine for 12 h and cryoprotected with an additional 25% wt/vol ethylene glycol. The 0.22 M calcium chloride and 12% wt/vol PEG 3350 formed the CrTDC crystals which were subsequently soaked with 10 mM l-tryptophan for 16 h and then cryogenized with an additional 18% wt/vol ethylene glycol. Finally, to form the Rr4HPAAS crystals, protein solution was mixed with a reservoir buffer of 0.21 M potassium thiocyanate and 22% wt/vol PEG 3350. Ligand soaks for this crystal proved unsuccessful, and, ultimately, the crystals were cryoprotected with an additional 13% wt/vol PEG 3350 in the absence of ligand. The PsTyDC structure was determined first by molecular replacement using the insect DDC structure (56) as the search model in Molrep (57). The resulting model was iteratively refined using Refmac 5.2 (58) and then manually refined in Coot 0.7.1 (59). The CrTDC, AtPAAS, and Rr4HPAAS structures were solved by molecular replacement using the refined PsTyDC structure as the search model, followed by refinement procedure as described above.
Enzyme Assays.
The in vitro decarboxylation and aldehyde synthase activities of the wild-type PsTyDC, PsTyDCH205N, and PsTyDCY350F were assayed in 100 μL of reaction buffer containing 50 mM Tris, pH 8.0, 100 μM PLP, 0.5 mM l-tyrosine, and 20 μg of recombinant enzyme. Reactions were incubated at 30 °C for various time points and subsequently stopped in the linear range of product formation with 200 μL of methanol. After clarification, the soluble fraction was analyzed by LC-MS-ultraviolet (UV). Chromatographic separation and measurement of absorption at 280 nm were performed by an Ultimate 3000 LC system (Dionex), equipped with a 150-mm C18 Column (Kinetex 2.6-µm silica core shell C18 100 Å pore; Phenomenex) and coupled to an UltiMate 3000 diode-array detector in-line UV-Vis spectrophotometer (Dionex). Compounds were separated through the use of an isocratic mobile phase as previously described (10). The reduction of aldehyde products was achieved by the addition of ethanol containing a saturating concentration of sodium borohydride. The EgPAAS enzyme assays were started by adding 2 µg of recombinant protein into 200 µL of reaction buffer containing 50 mM Tris pH 8.0, and 2 mM l-phenylalanine. Reactions were incubated for various time points at 30 °C, and the reactions were stopped with equal volume of 0.8 M formic acid, extracted with 150 µL of ethyl acetate and analyzed by gas chromatography MS (GC-MS) as previously described against an analytical phenylacetaldehyde standard (10). The initial substrate selectivity was measured through the detection of the hydrogen peroxide coproduct using Pierce Quantitative Peroxide Assay Kit (Pierce) and a standard curve of hydrogen peroxide. Reactions were conducted as described using reaction mixtures containing 0.5 mM amino acid substrate concentrations. Triplicate reactions were stopped after 5 min of incubation at 30 °C with an equal volume of 0.8 M formic acid and measured by absorbance at 595 nm.
Metabolic Engineering and Metabolic Profiling of Transgenic Yeast by LC-HRAM-MS.
LiAAS (Phytozome 12: L. indica scaffold RJNQ-2017655) and MmAAS (Phytozome 12: M. magnifica scaffold WWQZ-2007373) were synthesized as gBlocks (IDT) with S. cerevisiae codon optimization. Ectopic expression of various AAADs in S. cerevisiae was achieved through the use of p423TEF, a 2-μm plasmid with the HIS3 auxotrophic growth marker for constitutive expression (60). Fifteen-milliliter cultures of transgenic S. cerevisiae BY4743 strains were grown in 50-mL mini bioreactor tubes for 24 h with shaking at 30 °C. The cultured cells were subsequently pelleted, washed, disrupted, and clarified for LC-HRAM-MS analysis as previously described (61). PpDDC (NP_744697.1), PsNCS2 (AKH61498), and BvTyH (AJD87473) were synthesized as gBlocks (IDT) with S. cerevisiae codon optimization. PCR amplicons or gBlocks were ligated into the entry vector pYTK001 and subsequently assembled into 2-μm pTDH3, tTDH1, and HIS3 multigene vectors for constitutive expression in S. cerevisiae (62). A second multigene vector, containing the S. cerevisiae tyrosine metabolism feedback-resistant mutants ARO4K229L and ARO7G141S, was additionally used to boost tyrosine flux as previously described (10). S. cerevisiae lines were transformed with various multigene vectors to assay ectopic (S)-norcoclaurine production. Here, clarified media extracts from bioreactor cultures were diluted with an equal volume of 100% methanol and analyzed directly by LC-HRAM-MS. Raw MS data were processed and analyzed using MZmine2 (63). Data files were first filtered to only include positive mode ions above the noise filter of 1e5. Shoulder peaks were next removed using the Fourier transform mass spectrometer shoulder peaks filter function. Chromatograms were assembled using the chromatogram builder function and smoothed using the peak smoothing function. Chromatograms were subsequently separated into individual peaks using chromatogram deconvolution. The resulting peak lists were aligned using the join aligner function, and omitted peaks were identified and added using the gap filling function. The project parameters were then grouped by triplicate, and the peak areas were exported for subsequent statistical analysis and graph generation.
MD Simulation and Analysis.
All simulations were performed using GROMACS 5.1.4 (61) and the CHARMM36 force field (64). The nonstandard residue LLP was parameterized using Gaussian (65) and the Force Field Toolkit (66) implemented in VMD (67) based on the initial parameters provided by the CGenFF program (68–71). A number of CrTDC residues buried deeply within the protein or at the monomer−monomer interface were modeled in their neutral forms based on PROPKA (72, 73) calculation results: Asp268-A/B, Asp287-A/B, Asp397-A/B, Lys208-A/B, and Glu169-A. All of the histidines were kept neutral, with a proton placed on the ε-nitrogen, except for His203 and His318, for which the proton was placed on the δ-nitrogen to optimize hydrogen bond network. All simulation systems were constructed as a dimer solvated in a dodecahedron water box with 0.1 M NaCl (SI Appendix, Fig. S11) and a total number of atoms of ∼124,000. Prior to the production runs listed in SI Appendix, Table S4, all systems were subjected to energy minimization followed by a 100-ps constant number of particles, volume, and temperature (NVT) and a 100-ps constant number of particles, pressure, and temperature (NPT) run with the protein heavy atoms constrained. In all simulations, the van der Waals interactions were smoothly switched off from 10 Å to 12 Å. The electrostatic interactions were computed with the Particle Mesh Ewald (PME) method (74) with a grid spacing of 1.5 Å, a PME order of 6, and a cutoff of 12 Å in real space. The system temperature was kept at 300 K using the velocity-rescaling thermostat (75), and the system pressure was kept at 1 bar with the Parrinello−Rahman barostat (76, 77). All bonds were constrained using LINCS (78, 79) to allow an integration time step of 2 fs. The helix-unfolding simulation was performed using the metadynamics method (80) as implemented in PLUMED (81). A 10-ns metadynamics simulation was performed on System 1 by placing Gaussian potentials (height = 35 kJ/mol, sigma = 0.35 rad) every 500 steps on the collective variables, which were chosen as the backbone dihedral angles Ψ and Φ of residues 346 to 350. We should emphasize that this simulation was not intended for an accurate free energy calculation and instead was only used to generate an unfolded structure of the short helix (residues 346 to 350). The resulting unfolded large-loop structure was then used in all systems, each of which was subjected to 12 replicas of 50-ns MD simulations listed in SI Appendix, Table S4. Clustering analysis was performed using gmx cluster over all simulated trajectories with an rmsd cutoff of 3.5 Å. The three-dimensional occupancy maps were created at a resolution of 1 Å3 using the VMD VOLMAP plugin. DSSP calculations (82) were performed with gmx do_dssp implemented in GROMACS. An output of H (ɑ-helix), I (𝜋-helix), or G (310-helix) was considered as a helix, and the corresponding residue was assigned a helical content of 1; otherwise, a helical content of 0 was assigned. Clustering and occupancy analysis as well as the average helical content calculations were performed on the combined trajectories of all simulation replicas for a given CrTDC system. The two monomers of a CrTDC dimer were treated equivalently in these analyses. All simulation figures were made using VMD.
Accession Codes.
The sequences of P. somniferum, C. roseus, and E. grandis genes reported in this article are deposited into NCBI GenBank under the following accession numbers: PsTyDC (MG748690), CrTDC (MG748691), and EgPAAS (MG786260). The atomic coordinates and structure factors for PsTyDC, CrTDC, Rr4HPAAS, and AtPAAS have been deposited in the Protein Data Bank under the accession numbers 6EEM, 6EEW, 6EEQ, and 6EEI, respectively.
Supplementary Material
Acknowledgments
This work was supported by the Pew Scholar Program in the Biomedical Sciences (J.-K.W.), the Searle Scholars Program (J.-K.W.), NSF Grant CHE-1709616 (J.-K.W.), the Keck Foundation (J.-K.W.), and direct grants from The Chinese University of Hong Kong (Y.W.). This work is based on research conducted at the Northeastern Collaborative Access Team (NE-CAT) beam lines, which are funded by the National Institute of General Medical Sciences from NIH Grant P41 GM103403. The Pilatus 6M detector on NE-CAT 24-ID-C beam line is funded by NIH-Office of Research Infrastructure Programs (ORIP) high-end instrumentation (HEI) Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
Competing interest statement: J.-K.W. is a cofounder, a member of the Scientific Advisory Board, and a shareholder of DoubleRainbow Biosciences, which develops biotechnologies related to natural products.
This article is a PNAS Direct Submission.
Data deposition: The sequences of P. somniferum, C. roseus, and E. grandis genes reported in this article are deposited into National Center for Biotechnology Information GenBank under the following accession numbers: PsTyDC (MG748690), CrTDC (MG748691), and EgPAAS (MG786260). The crystal structures of CrTDC in complex with ʟ-tryptophan, PsTyDC in complex with ʟ-tyrosine, AtPAAS in complex with ʟ-phenylalanine, and unbound Rr4HPAAS have been deposited in the Protein Data Bank with the ID codes 6EEW, 6EEM, 6EEI, and 6EEQ, respectively.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920097117/-/DCSupplemental.
References
- 1.Weng J.-K., Philippe R. N., Noel J. P., The rise of chemodiversity in plants. Science 336, 1667–1670 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Chae L., Kim T., Nilo-Poyanco R., Rhee S. Y., Genomic signatures of specialized metabolism in plants. Science 344, 510–513 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Kries H. et al., Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat. Chem. Biol. 12, 6–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaltenbach M. et al., Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol. 14, 548–555 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Burkhard P., Dominici P., Borri-Voltattorni C., Jansonius J. N., Malashkevich V. N., Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 8, 963–967 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Facchini P. J., Huber-Allanach K. L., Tari L. W., Plant aromatic L-amino acid decarboxylases: Evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54, 121–138 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Noé W., Mollenschott C., Berlin J., Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: Purification, molecular and kinetic data of the homogenous protein. Plant Mol. Biol. 3, 281–288 (1984). [DOI] [PubMed] [Google Scholar]
- 8.Facchini P. J., De Luca V., Differential and tissue-specific expression of a gene family for tyrosine/dopa decarboxylase in opium poppy. J. Biol. Chem. 269, 26684–26690 (1994). [PubMed] [Google Scholar]
- 9.Gutensohn M. et al., Role of aromatic aldehyde synthase in wounding/herbivory response and flower scent production in different Arabidopsis ecotypes. Plant J. 66, 591–602 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Torrens-Spence M. P., Pluskal T., Li F.-S., Carballo V., Weng J.-K., Complete pathway elucidation and heterologous reconstitution of rhodiola salidroside biosynthesis. Mol. Plant 11, 205–217 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Torrens-Spence M. P., Lazear M., von Guggenberg R., Ding H., Li J., Investigation of a substrate-specifying residue within Papaver somniferum and Catharanthus roseus aromatic amino acid decarboxylases. Phytochemistry 106, 37–43 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Dunathan H. C., Conformation and reaction specificity in pyridoxal phosphate enzymes. Proc. Natl. Acad. Sci. U.S.A. 55, 712–716 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torrens-Spence M. P. et al., Biochemical evaluation of the decarboxylation and decarboxylation-deamination activities of plant aromatic amino acid decarboxylases. J. Biol. Chem. 288, 2376–2387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansonius J. N., Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 8, 759–769 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Giardina G. et al., Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases. Proc. Natl. Acad. Sci. U.S.A. 108, 20514–20519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vacca R. A., Christen P., Malashkevich V. N., Jansonius J. N., Sandmeier E., Substitution of apolar residues in the active site of aspartate aminotransferase by histidine. Effects on reaction and substrate specificity. Eur. J. Biochem. 227, 481–487 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Mehta P. K., Hale T. I., Christen P., Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214, 549–561 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Zhu H. et al., Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding. Sci. Rep. 6, 27779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii S., Mizuguchi H., Nishino J., Hayashi H., Kagamiyama H., Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J. Biochem. 120, 369–376 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Dominici P., Tancini B., Borri Voltattorni C., Chemical modification of pig kidney 3,4-dihydroxyphenylalanine decarboxylase with diethyl pyrocarbonate. Evidence for an essential histidyl residue. J. Biol. Chem. 260, 10583–10589 (1985). [PubMed] [Google Scholar]
- 21.Hayashi H., Mizuguchi H., Kagamiyama H., Rat liver aromatic L-amino acid decarboxylase: Spectroscopic and kinetic analysis of the coenzyme and reaction intermediates. Biochemistry 32, 812–818 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Stavrinides A. et al., Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat. Commun 7, 12116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torrens-Spence M. P., von Guggenberg R., Lazear M., Ding H., Li J., Diverse functional evolution of serine decarboxylases: Identification of two novel acetaldehyde synthases that uses hydrophobic amino acids as substrates. BMC Plant Biol. 14, 247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.One Thousand Plant Transcriptomes Initiative , One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J., Han Q., Ding H., Li J., Biochemical identification of residues that discriminate between 3,4-dihydroxyphenylalanine decarboxylase and 3,4-dihydroxyphenylacetaldehyde synthase-mediated reactions. Insect Biochem. Mol. Biol. 91, 34–43 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Liao C., Upadhyay A., Liang J., Han Q., Li J., 3,4-Dihydroxyphenylacetaldehyde synthase and cuticle formation in insects. Dev. Comp. Immunol. 83, 44–50 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Pereira M. d C. et al., Chemical composition and antimicrobial activity of the essential oil from Microlicia crenulata. J. Essent. Oil Bear. Plants 18, 18–28 (2015). [Google Scholar]
- 28.Özek T., Demirci B., Baser K. H. C., Chemical composition of Turkish myrtle oil. J. Essent. Oil Res. 12, 541–544 (2000). [Google Scholar]
- 29.D’Arcy B. R., Rintoul G. B., Rowland C. Y., Blackman A. J., Composition of Australian honey extractives. 1. Norisoprenoids, monoterpenes, and other natural volatiles from blue gum (Eucalyptus leucoxylon) and yellow box (Eucalyptus melliodora) honeys. J. Agric. Food Chem. 45, 1834–1843 (1997). [Google Scholar]
- 30.Hagel J. M., Facchini P. J., Benzylisoquinoline alkaloid metabolism: A century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Maeda H., Dudareva N., The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Toney M. D., Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta 1814, 1407–1418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandmeier E., Hale T. I., Christen P., Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221, 997–1002 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Mitchell A. J., Weng J.-K., Unleashing the synthetic power of plant oxygenases: From mechanism to application. Plant Physiol. 179, 813–829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogren W. L., Bowes G., Ribulose diphosphate carboxylase regulates soybean photorespiration. Nat. New Biol. 230, 159–160 (1971). [DOI] [PubMed] [Google Scholar]
- 36.Abell L. M., Schloss J. V., Oxygenase side reactions of acetolactate synthase and other carbanion-forming enzymes. Biochemistry 30, 7883–7887 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Bertoldi M., Dominici P., Moore P. S., Maras B., Voltattorni C. B., Reaction of dopa decarboxylase with α-methyldopa leads to an oxidative deamination producing 3,4-dihydroxyphenylacetone, an active site directed affinity label. Biochemistry 37, 6552–6561 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Bertoldi M., Carbone V., Borri Voltattorni C., Ornithine and glutamate decarboxylases catalyse an oxidative deamination of their alpha-methyl substrates. Biochem. J. 342, 509–512 (1999). [PMC free article] [PubMed] [Google Scholar]
- 39.Torrens-Spence M. P., Liu C.-T., Pluskal T., Chung Y. K., Weng J.-K., Monoamine biosynthesis via a noncanonical calcium-activatable aromatic amino acid decarboxylase in psilocybin mushroom. ACS Chem. Biol. 13, 3343–3353 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Torrens-Spence M. P. et al., Biochemical evaluation of a parsley tyrosine decarboxylase results in a novel 4-hydroxyphenylacetaldehyde synthase enzyme. Biochem. Biophys. Res. Commun. 418, 211–216 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Sakai M. et al., Production of 2-phenylethanol in roses as the dominant floral scent compound from L-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci. Biotechnol. Biochem. 71, 2408–2419 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Kaminaga Y. et al., Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281, 23357–23366 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Weng J.-K., Noel J. P., The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb. Symp. Quant. Biol. 77, 309–320 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Yoshikuni Y., Ferrin T. E., Keasling J. D., Designed divergent evolution of enzyme function. Nature 440, 1078–1082 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Kopycki J. G. et al., Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferases. J. Mol. Biol. 378, 154–164 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Huang R. et al., Enzyme functional evolution through improved catalysis of ancestrally nonpreferred substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 2966–2971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Austin M. B., Bowman M. E., Ferrer J.-L., Schröder J., Noel J. P., An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11, 1179–1194 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Vavricka C. J. et al., Mechanism-based tuning of insect 3,4-dihydroxyphenylacetaldehyde synthase for synthetic bioproduction of benzylisoquinoline alkaloids. Nat. Commun. 10, 2015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schliemann W., Kobayashi N., Strack D., The decisive step in betaxanthin biosynthesis is a spontaneous reaction. Plant Physiol. 119, 1217–1232 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ilari A. et al., Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J. Biol. Chem. 284, 897–904 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Thompson J. D., Gibson T. J., Higgins D. G., Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 00, 2.3.1−2.3.22 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Goodstein D. M. et al., Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouet P., Robert X., Courcelle E., ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Q., Ding H., Robinson H., Christensen B. M., Li J., Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase. PloS One 5, e8826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vagin A., Teplyakov A., MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997). [Google Scholar]
- 58.Murshudov G. N. et al., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mumberg D., Müller R., Funk M., Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Abraham M. J. et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 62.Lee M. E., DeLoache W. C., Cervantes B., Dueber J. E., A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 4, 975–986 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Pluskal T., Castillo S., Villar-Briones A., Oresic M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010).journal [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Best R. B. et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frisch M. J., et al. , Gaussian 09 (Revision D.01, Gaussian, 2016).
- 66.Mayne C. G., Saam J., Schulten K., Tajkhorshid E., Gumbart J. C., Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem. 34, 2757–2770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 27–28, 33–38, (1996). [DOI] [PubMed] [Google Scholar]
- 68.Vanommeslaeghe K., MacKerell A. D. Jr., Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vanommeslaeghe K., Raman E. P., MacKerell A. D. Jr., Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanommeslaeghe K. et al., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu W., He X., Vanommeslaeghe K., MacKerell A. D. Jr., Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 33, 2451–2468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Søndergaard C. R., Olsson M. H. M., Rostkowski M., Jensen J. H., Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Olsson M. H. M., Søndergaard C. R., Rostkowski M., Jensen J. H., PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 75.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 14101 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 77.Nosé S., Klein M. L., Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50, 1055–1076 (1983). [Google Scholar]
- 78.Hess B., Bekker H., Berendsen H. J. C., Johannes G. E., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 79.Hess B., P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Laio A., Parrinello M., Escaping free-energy minima. Proc. Natl. Acad. Sci. U.S.A. 99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tribello G. A., Bonomi M., Branduardi D., Camilloni C., Bussi G., PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014). [Google Scholar]
- 82.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.