Significance
Bacteria utilize sensory signal transduction systems to constantly survey and respond to changes in their environment. The human pathogen Staphylococcus aureus uses two-component signaling systems to sense its surroundings and adapt to hostile stresses of the host in part by shifting its metabolism. Here, we sought to understand how the S. aureus SrrAB two-component system senses changes in its environment during an infection. Using biophysical and biochemical approaches and in vivo experiments, we identified a mechanism by which the SrrB sensor histidine kinase detects a shift in its metabolism that occurs during an infection. Our findings provide general insight into the mechanisms by which bacterial histidine kinases sense environmental cues to regulate signal transduction and pathogenicity.
Keywords: SrrAB two-component system, Staphylococcus aureus, sensor histidine kinase, cysteine disulfide bond
Abstract
Staphylococcus aureus infections can lead to diseases that range from localized skin abscess to life-threatening toxic shock syndrome. The SrrAB two-component system (TCS) is a global regulator of S. aureus virulence and critical for survival under environmental conditions such as hypoxic, oxidative, and nitrosative stress found at sites of infection. Despite the critical role of SrrAB in S. aureus pathogenicity, the mechanism by which the SrrAB TCS senses and responds to these environmental signals remains unknown. Bioinformatics analysis showed that the SrrB histidine kinase contains several domains, including an extracellular Cache domain and a cytoplasmic HAMP-PAS-DHp-CA region. Here, we show that the PAS domain regulates both kinase and phosphatase enzyme activity of SrrB and present the structure of the DHp-CA catalytic core. Importantly, this structure shows a unique intramolecular cysteine disulfide bond in the ATP-binding domain that significantly affects autophosphorylation kinetics. In vitro data show that the redox state of the disulfide bond affects S. aureus biofilm formation and toxic shock syndrome toxin-1 production. Moreover, with the use of the rabbit infective endocarditis model, we demonstrate that the disulfide bond is a critical regulatory element of SrrB function during S. aureus infection. Our data support a model whereby the disulfide bond and PAS domain of SrrB sense and respond to the cellular redox environment to regulate S. aureus survival and pathogenesis.
Staphylococcus aureus is a Gram-positive pathogen that colonizes the mucosal surfaces and skin of ∼30% of humans (1, 2) and contributes to a wide range of infections including soft-tissue abscesses, infective endocarditis, osteomyelitis, necrotizing pneumonia, and toxic shock syndrome (TSS). Infections are mediated by virulence factors whose expression is controlled by various environmental signals, including high concentrations of autoinducing peptides, pH, O2, and CO2, among others (3). A key strategy for S. aureus survival and pathogenicity is its ability to adapt to aerobic and anaerobic conditions found within the host, which vary drastically between different tissues. Oxygen availability plays an important role in S. aureus virulence through the regulation of toxin production and biofilm formation (4). For example, during vaginal colonization, S. aureus is in an anaerobic and low-carbon dioxide environment that represses expression of the TSS toxin-1 (TSST) gene (tstH) (5, 6), thereby reducing the risk of menstrual TSS. Tampon use introduces oxygen, derepressing TSST-1 and increasing the risk of menstrual TSS. Conversely, ica genes involved in cellular adhesion and biofilm formation are stimulated under anaerobic conditions in S. aureus (7, 8).
Several S. aureus two-component systems (TCSs) are involved in the regulation of virulence factor production, including SrrAB, AgrAC, SaeRS, ArlRS, and LytRS (3). Prototypical bacterial TCSs are composed of a sensor histidine kinase (HK) and an effector response regulator (RR) that connect environmental stimuli to the appropriate cellular response. HKs are typically composed of a N-terminal extracellular sensing Cache domain linked by transmembrane helices to a cytoplasmic region that includes a dimerization and phosphotransfer (DHp) domain and an ATP-binding catalytic (CA) domain. Upon stimulation, the HK is autophosphorylated at a conserved histidine residue in the DHp domain by an ATP molecule bound by the CA domain. The phosphorylated HK then transfers the phosphoryl group onto a conserved aspartate residue in the receiver (REC) domain of the cognate RR to promote conformational changes that modify its activity, usually DNA binding and gene regulation. Once the appropriate response is generated and/or the signal is depleted, the RR is dephosphorylated by either autodephosphorylation or phosphatase activity of the HK (9–12).
The S. aureus SrrAB (staphylococcal respiratory response AB) TCS was first identified while investigating the role of tampons in development of menstrual TSS, where TSST-1 production depends on oxygen. SrrAB shares homology to the ResDE TCS in Bacillus subtilis, which regulates aerobic and anaerobic respiration (13). Consistent with this, SrrAB-knockout strains show metabolic and virulence defects, particularly in the regulation of genes involved in aerobic and anaerobic respiration (14, 15). SrrAB regulates S. aureus virulence factors, including TSST-1, RNAIII, and protein A (13), by directly binding to their corresponding promoters (16). A number of studies have established the importance of the SrrAB TCS system in regulation of genes involved in anaerobic metabolism (pflAB, adhE, nrdDG) (17), nitrous oxide detoxification (hmp) (18), cytochrome biosynthesis and assembly (qoxABCD, cydAB, hemeABCX) (19), biofilm formation (7, 8), hydrogen peroxide metabolism (dps, scdA) (20), and programmed cell death (19). In addition, SrrAB controls the up-regulation of fermentation enzymes (lactate dehydrogenase and alcohol dehydrogenase), as well as the down-regulation of tricarboxylic acid cycle enzymes (succinyl-CoA synthetase, aconitase, and fumarase) under anaerobic conditions (3, 14). Finally, several studies have proposed a link between nonrespiratory growth and the activation of the SrrAB TCS (18, 21). However, the mechanism by which the SrrAB TCS senses the shift from respiratory to nonrespiratory growth conditions and the associated redox changes is currently unknown.
Here, we focused on understanding the mechanism of redox sensing and regulation by the SrrAB TCS. Biochemical and structural analysis of the HK catalytic region identified a cysteine disulfide bond that is sensitive to redox status. Disruption of this disulfide bond by mutagenesis affected biofilm formation and TSST-1 production in S. aureus strains. Moreover, the disulfide bond was critical for S. aureus pathogenesis in a rabbit model of infective endocarditis. Together, these data provide insights into the mechanism by which SrrB senses and responds to changes in redox state during S. aureus infection.
Results
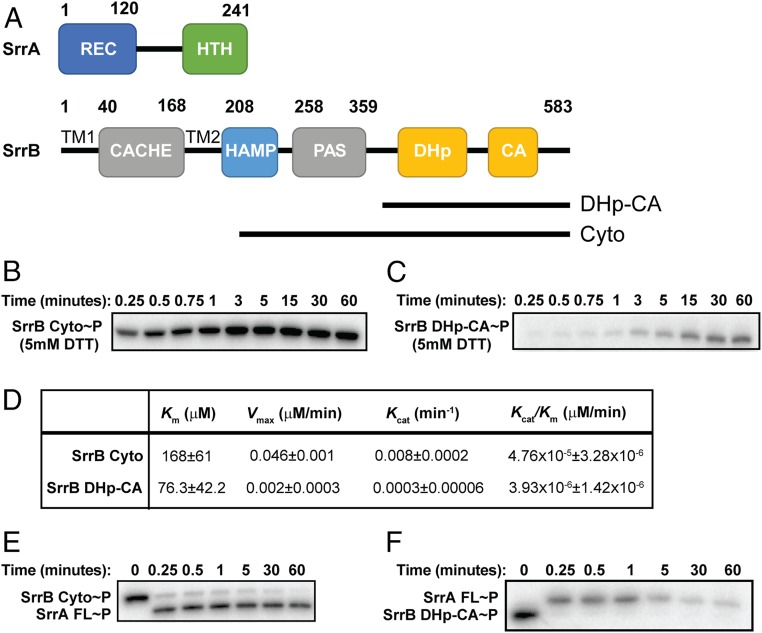
SrrB Is a Kinase/Phosphatase that Regulates the Phosphorylation State of Its Cognate Response Regulator SrrA.
To characterize the kinase/phosphatase activity of SrrB, we performed kinase assays using [γ-32P]-ATP (22). We tested the autophosphorylation and phosphotransfer kinetics of the core catalytic portion of SrrB that includes the DHp and CA domains (DHp-CA) and the entire cytoplasmic region consisting of the catalytic region plus the HAMP and PAS domains (HAMP-PAS-DHp-CA or SrrB Cyto; Fig. 1A). Fig. 1 B and C depict representative autophosphorylation kinetics of SrrB Cyto and DHp-CA, respectively. SrrB Cyto reached 50% phosphorylation (t1/2) at ∼15 s, whereas it took SrrB DHp-CA ∼5 min to reach this level (SI Appendix, Fig. S1). In addition, the Vmax and Kcat values demonstrated that SrrB Cyto has a 10-fold higher rate of autophosphorylation compared to that of the DHp-CA region alone (Fig. 1D). SrrB Cyto and DHp-CA undergo quick (<30 s) phosphotransfer to SrrA (Fig. 1 E and F). However, SrrB phosphatase activity was inhibited in the presence of the HAMP and PAS domains as confirmed by the presence of phosphorylated SrrA throughout the entire 60-min time course (Fig. 1E). In contrast, the DHp-CA region had significant phosphatase activity, as phosphorylated SrrA was lost after 5 min (Fig. 1F). This decrease in SrrA∼P signal was not due to protein instability, as the protein level remained unchanged throughout the experiment (SI Appendix, Fig. S2). We conclude that, like other HK proteins, SrrB is a dual-function kinase and phosphatase enzyme which regulates the phosphorylation state of SrrA.
Fig. 1.
Domain architecture and regulation of SrrB activity by the PAS domain. (A) Domain architecture of SrrA and SrrB proteins. SrrB is a membrane-bound histidine kinase with extracellular Cache domain, transmembrane HAMP domain, intracellular PAS domain, and DHp-CA catalytic domain. SrrA is predicted to be an OmpR-like response regulator with receiver domain and winged helix-turn-helix DNA binding domain. (B) Autophosphorylation of SrrB HAMP-PAS-DHp-CA and (C) DHp-CA. (D) Kinetics of autophosphorylation of SrrB HAMP-PAS-DHp-CA and DHp-CA regions. Km, Vmax, and Kcat values were calculated by fitting the kinetics data to the Michaelis–Menten equation in GraphPad software. The errors represent the error of fitting the model to the data. Phosphotransfer of (E) SrrB HAMP-PAS-DHp-CA and (F) DHp-CA to full-length SrrA. Experiments were performed in triplicate, and representative gels are shown.
Structural Analysis of the Catalytic Region of SrrB Reveals the Presence of a Cysteine Disulfide Bond.
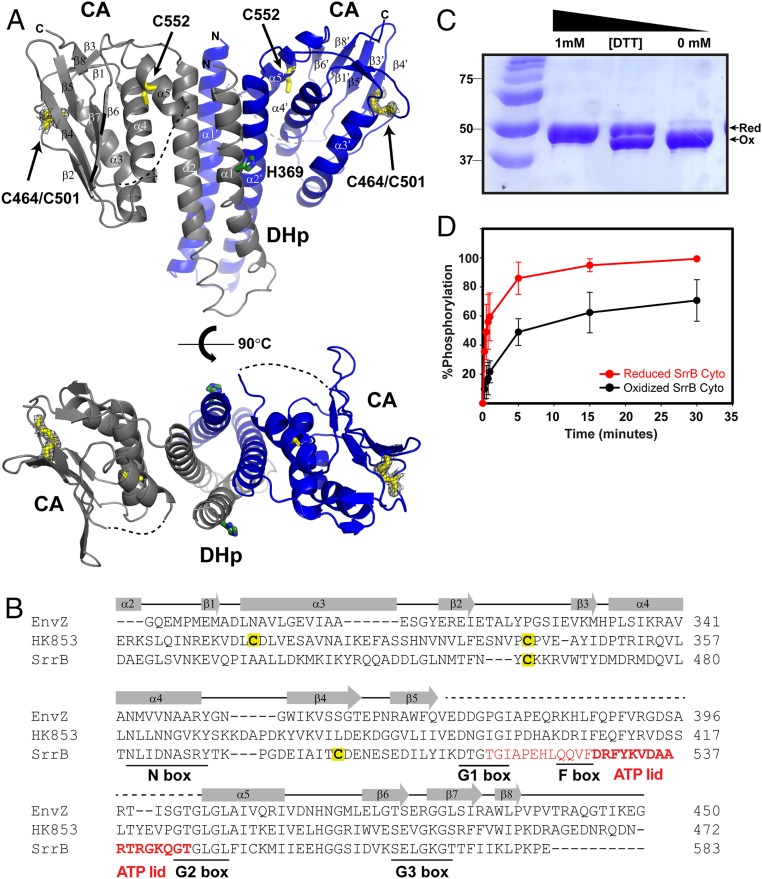
To better understand the catalytic activity of SrrB, we set out to determine the crystal structure of the SrrB catalytic region (DHp-CA) in the presence of MgCl2 and the ATP analog AMPPNP. SrrB belongs to the HisKA (or Subtype 1A) family, characterized by a DHp domain that contains the phosphorylatable histidine residue within an H-box consensus motif (23) (H E X R T/N P, where X is a hydrophobic residue) and a CA domain involved in ATP binding that contains the conserved N-G1-F-G2-G3 box motifs (24). The crystal structure of the DHp-CA region was solved at a resolution of 2.0 Å using multiwavelength anomalous dispersion (MAD) with protein labeled with selenomethionine (SI Appendix, Table S1). The asymmetric unit of the crystal presents two almost identical monomers (rmsd of 1.27 Å for the superimposition of 122 residues) of SrrB organized in a symmetric dimer (Fig. 2A). Size-exclusion chromatography and multiple-angle light-scattering data showed that SrrB DHp-CA and SrrB Cyto form dimers in solution (SI Appendix, Fig. S3). Each monomer showed the characteristic HisKA fold (25–27), consisting of an N‐terminal helical hairpin DHp domain (residues 355 to 426) and a C‐terminal α/β CA domain (residues 433 to 581), connected by a short linker (residues 427 to 432; Fig. 2A). The dimer interface is provided exclusively by the helical‐hairpin domain, burying a surface of around 1,300 Å2 per monomer. The helical‐hairpin DHp domain comprises two antiparallel helices, α1 that includes the His369 phosphorylation site, and α2, connected by a short four‐residue turn (residues 388 to 391). The C‐terminal CA domain assumes an α/β sandwich fold: one layer comprises a mixed five‐strand β sheet (β2, β4–β7), which is nearly parallel to the helical‐hairpin structure, and the other layer consists of three α helices (α3–α5). In addition, this domain contains a pair of short antiparallel β strands (β1 and β3).
Fig. 2.
The SrrB histidine kinase activity is sensitive to the cysteine redox state. (A) The crystal structure of oxidized (-DTT) SrrB catalytic DHp-CA region refined to 2.0 Å (PDB ID code 6PAJ). The phosphorylated histidine residue (H369) is colored in green, and the cysteine residues are in yellow. The missing electron densities for residues 517 to 544 are shown as a dashed line. (Bottom) Top-down view of the DHp-CA region. Each monomer is shown in gray and blue, respectively. The 2Fo-Fc electron density map of the disulfide bond between C464 and C501 is shown in gray wire at contour level 1σ. (B) A structure-based sequence alignment of E. coli EnvZ (PDB ID code 4KP4; NCBI accession no. 584579776), T. maritima HK853 (PDB ID code 3DGE; NCBI accession no. 251836869), and S. aureus SrrB (NCBI accession no. 123003464) is shown with conserved N, G1, F, G2, and G3 box residues underlined. The residues 517 to 544, which are missing in the SrrB crystal structure, are colored in red, and residues forming the ATP-lid are in bold. (C) SDS/PAGE gel of SrrB Cyto showing redox regulation of the intramolecular disulfide bond by DTT. DTT (0 to 1 mM) was added to the reaction buffer containing SrrB Cyto, run on 12% SDS/PAGE gel, and stained with Coomassie blue. (D) Redox-dependent autophosphorylation of SrrB Cyto. Phosphorylated SrrB Cyto was quantified by using ImageJ and plotted in SigmaPlot. The autophosphorylation reactions of reduced and oxidized SrrB Cyto were done in triplicate.
Although the protein was crystallized in the presence of AMPPNP-Mg, no electron density for the nucleotide was observed at the ATP binding site of the CA domain. Similarly, the ATP-lid that covers the nucleotide binding site (residues 516 to 543) is missing electron density, presumably due to enhanced flexibility in the β5–α5 loop in the absence of the nucleotide. Fig. 2B shows a structure-based alignment of the SrrB CA domain with that of Escherichia coli EnvZ (28) and Thermotoga maritima HK853 (29). The alignment delineates the N-G1-F-G2-G3 boxes, highlighting the region not defined in the SrrB structure. A defining feature in the SrrB structure is the presence of two cysteine residues within the CA domain involved in an intramolecular disulfide bond. The electron density of the Cys464 (β2) and Cys501 (β4) side chains unambiguously showed formation of a disulfide bond (Fig. 2 A, Lower).
SrrB Kinase Activity Is Sensitive to Cysteine Redox State.
Cysteine disulfide bonds play structural and functional roles in proteins (30). Previous studies indicate that the SrrAB TCS is redox-sensitive and responds to reduced menaquinones (17, 18, 21). Thus, we hypothesized that the cysteines could be redox-active and functionally relevant. Purified SrrB Cyto reduced with dithiothreitol (DTT) migrates slower than the oxidized form in an SDS/PAGE gel, suggesting that the two forms have distinct conformations (Fig. 2C). To further test the effect of redox on SrrB activity, we performed kinase assays in the presence and absence of DTT. Fig. 2D shows that the level of autophosphorylation is ∼40% higher in the reduced state. We also tested the effect of reduced and oxidized cysteines in phosphotransfer activity of SrrB; however, there was no change in activity with redox state (SI Appendix, Fig. S4).
Our attempts to determine the redox state of SrrB in S. aureus cells were hampered by the lack of an SrrB antibody and the disruption of SrrB function by epitope tags. In order to examine the redox state of SrrB in cells, we overexpressed His-tagged SrrB Cyto in E. coli and probed the oxidized/reduced disulfide population using maleimide-polyethylene glycol to covalently modify the SrrB pool containing reduced sulfhydryl groups (31). SI Appendix, Fig. S5 shows that a pool of both oxidized and reduced SrrB are present in E. coli lysates grown under aerobic conditions, indicating that the disulfide is capable of being formed in cells despite the reducing environment of the cytoplasm.
Conservation of Cysteine Residues in Histidine Kinases.
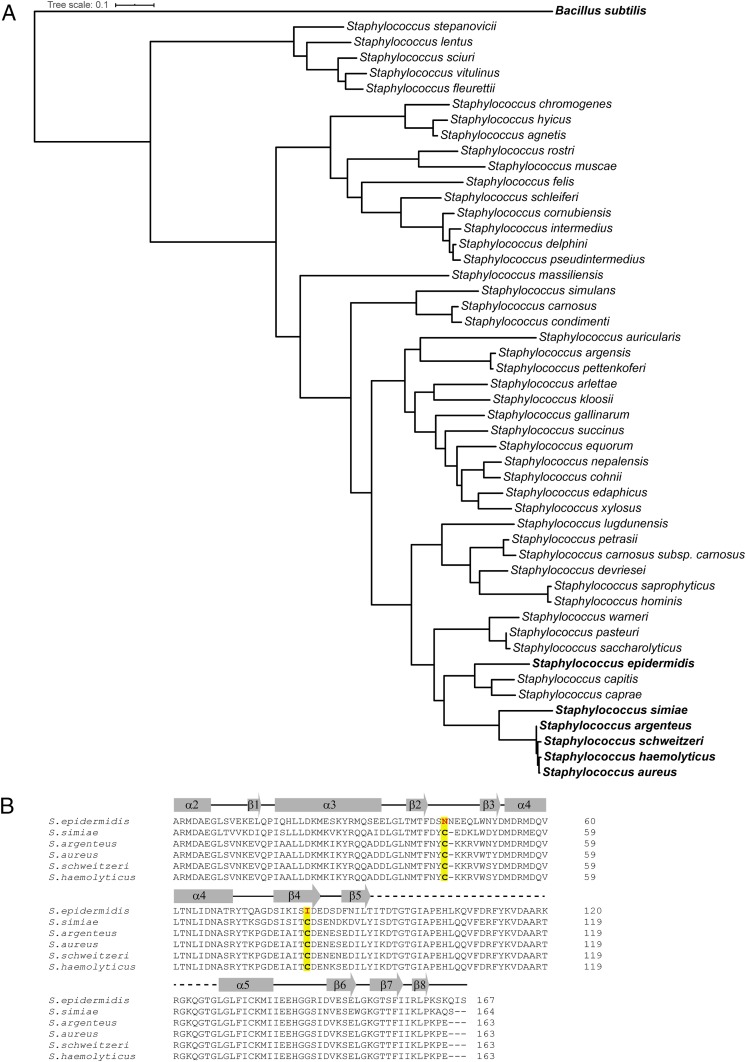
The SrrB cysteine residues are not conserved throughout the Staphylococcus genus (Fig. 3A). Currently, there are 57 known Staphylococcal species, and 49 could be compared phylogenetically based on the srrB gene. Of these, only five Staphylococcus species had cysteine residues at the conserved sites seen in SrrB (Fig. 3B). Fig. 3B shows an alignment of SrrB from these species and includes Staphylococcus epidermidis, which does not contain the disulfide forming cysteine residues, for comparison. Of interest, the cysteines were absolutely conserved in the top 100 S. aureus strains searched in the NIH protein–protein BLAST server, yet not conserved in B. subtilis ResE or S. epidermidis SrrB. In addition, we examined 16 unique HKs deposited in the Protein Data Bank (https://www.rcsb.org/) (32), and, surprisingly, only T. maritima HK853 (PDB ID code 2C2A) (25) had an intramolecular cysteine disulfide bond in the CA domain. Structural comparison of the SrrB and HK853 CA domains showed that the disulfide bond localizes in an almost identical location in both structures in the β-sheet region of the domain. In both structures, one Cys is localized at the end of β2 (Cys464 and Cys359 for SrrB and HK853, respectively), although they are connected in opposite directions to different structural elements: β4 (Cys501) in SrrB and α3 (Cys330) for HK853 (SI Appendix, Fig. S6). Although nothing is known about the role of this disulfide bond in HK853, the spatial conservation with SrrB suggests a biological function. Together, these data suggest that the SrrB and other HKs have evolved a unique intramolecular disulfide bond that is potentially important for function.
Fig. 3.
Alignment of catalytic DHp-CA region of SrrB from various Staphylococcus species. (A) The phylogenetic tree was generated using the full-length SrrB sequence. The five Staphylococcus species containing conserved cysteine residues are in bold. (B) Sequence alignment of the catalytic DHp-CA region of SrrB showing the presence of the conserved cysteine residues. The two conserved cysteines are highlighted in yellow. The S. epidermidis sequence is shown for comparison.
The SrrB Cys464 and Cys501 Residues Are Required for Biofilm Formation and Repression of TSST-1 Expression under Anaerobic and Microaerobic Conditions.
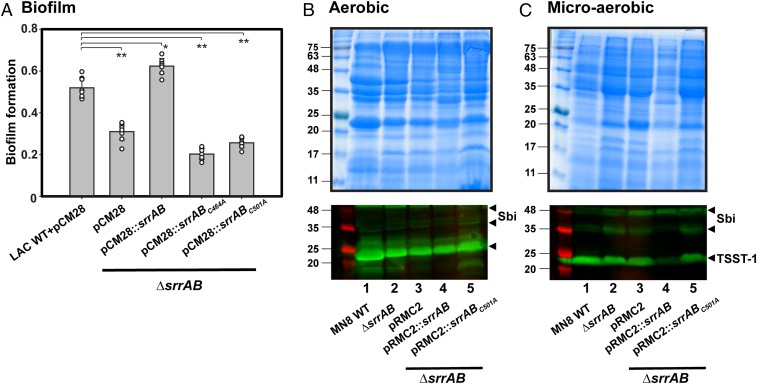
To test the biological significance of the redox state of cysteine residues present in the CA domain of SrrB, we assessed the ability of S. aureus LAC expressing SrrB cysteine to alanine variants (srrBC464A or srrBC501A) to form biofilm. It was previously shown that anaerobic biofilm formation in S. aureus is regulated by SrrAB (21). Fig. 4A shows that wild-type S. aureus LAC forms biofilm under anaerobic conditions and the amount of biofilm was significantly reduced in the LACΔsrrAB strain. This defect could be rescued by genetic complementation with srrAB expressed in trans. In contrast, complementation with srrBC464A or srrBC501A did not restore biofilm formation to wild-type levels (Fig. 4A). Both SrrBC464A and SrrBC501A proteins could be purified from E. coli with WT autophosphorylation and phosphotransfer activity (SI Appendix, Figs. S7 and S8), indicating that the biofilm phenotype was not due to protein misfolding.
Fig. 4.
SrrB cysteine mutations disrupt anaerobic biofilm formation and TSST-1 expression under low O2 conditions. (A) Biofilm formation of the srrAB chromosomal deletion of LAC (ΔsrrAB) and complementation with srrAB mutants. Biofilm formation was quantified as the ratio of the crystal violet absorbance at 570 nm to cell density (absorbance determined at 590 nm). The data summarize six independent experiments (**P < 0.0001; *P < 0.0005). (B and C) Exoprotein profiles (Top) and Western blot analyses of TSST-1 protein expression (Bottom) from S. aureus wild-type MN8, ΔsrrAB mutant, and complemented strains: ΔsrrAB+pRMC2, ΔsrrAB+pRMC2::srrAB, and ΔsrrAB+pRMC2::srrABC501A. Concentrated supernatants from the indicated strains grown in brain heart infusion medium under (B) aerobic conditions for 8 h or (C) low O2 conditions for 24 h were loaded onto 12% SDS/PAGE gels. Molecular mass markers on the left lane are indicated in kilodaltons, and the location of TSST-1 and the second Ig-binding protein (Sbi) are shown by black arrows (Lower). The TSST-1 Western blots were done in duplicate for aerobic conditions and triplicate in low O2 conditions. A representative SDS/PAGE gel and Western blot of TSST-1 is shown.
We also tested the S. aureus MN8 strain for TSST-1 expression, as this clinical isolate contains the tstH gene (33). As shown in Fig. 4 B and C, the wild-type S. aureus MN8 expressed TSST-1 under aerobic (shaking) and microaerobic (static) conditions. Previous studies showed that TSST-1 is induced by a low level of oxygen and negatively regulated by SrrAB (5, 6, 13). As shown in Fig. 4 B and C, Lower, lane 2, and SI Appendix, Fig. S9, the MN8ΔsrrAB deletion strain expresses TSST-1 to a comparable level as the wild-type under both aerobic and microaerobic conditions. The empty vector alone had no effect on TSST-1 expression under aerobic or microaerobic conditions. However, under microaerobic conditions, overexpression of srrAB repressed TSST-1 expression by ∼80% (Fig. 4 C, Lower, lane 4, and SI Appendix, Fig. S9) compared to the wild-type MN8 (Fig. 4 B, Lower, lane 4). Importantly, repression of TSST-1 by overexpressed srrAB was lost in the C501A variant (Fig. 4 C, Lower, lane 5). This effect was not seen under aerobic conditions (Fig. 4B, Lower, compare lanes 4 and 5). These data, consistent with our findings in the LAC strain, show that the cysteine disulfide bond is critical for the cellular activity of SrrB under low oxygen conditions.
SrrB Cysteine Mutations in S. aureus MN8 Increase Rabbit Susceptibility to Endocarditis.
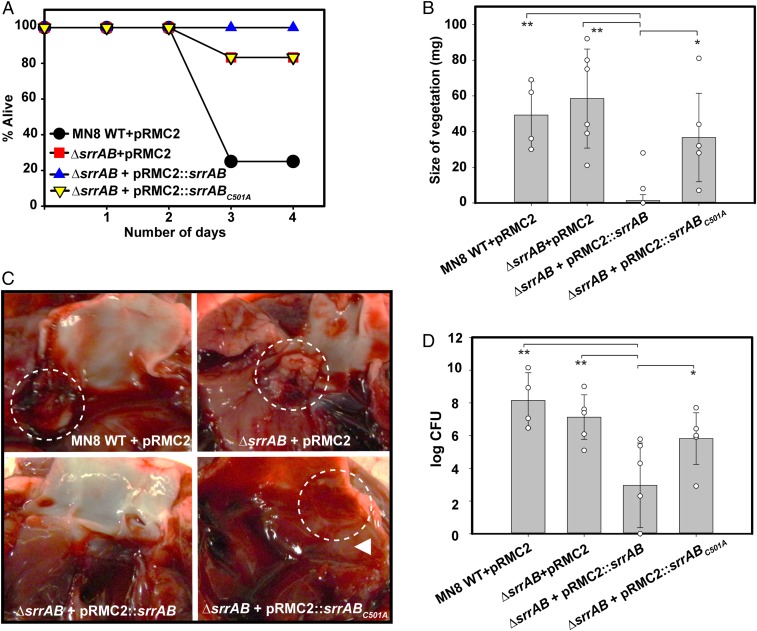
After establishing the importance of the SrrB cysteine disulfide bond in biofilm formation and TSST-1 production in S. aureus strains, we tested its importance for pathogenicity using a rabbit model for infective endocarditis (34). Pragman et al. (16) showed that ectopic overexpression of SrrAB decreases virulence in a rabbit model of infective endocarditis, indicating that overexpressed SrrAB represses some aspects of virulence. We used this same approach by overexpressing wild-type srrAB or srrABC501A on a plasmid in the MN8ΔsrrAB strain background (MN8ΔsrrAB::srrABOE, MN8ΔsrrAB::srrABC501AOE, or MN8srrABKO, respectively). This was done to compare the contribution of the SrrB cysteine disulfide bond in S. aureus virulence in rabbits infected with overexpressed wild-type srrAB versus overexpressed srrABC501A. Fig. 5A shows that 25% of the animals infected with wild-type MN8 (MN8 + pRMC2 vector) strain survived the duration of the experiment, whereas 83% of the animals infected with MN8srrABKO or MN8srrABC501AOE survived. In contrast, 100% of the animals infected with MN8srrABOE survived. The severity of the bacterial infection was quantified by measuring the mass of the vegetations formed in the aortic valve at the site of catheter damage. As expected, animals infected with wild-type MN8 formed large vegetations (Fig. 5B). The animals infected with MN8srrABOE formed essentially no vegetations, consistent with toxin suppression in vivo. In contrast to the animals infected with MN8srrABOE, the animals infected with MN8srrABC501AOE formed large vegetations (Fig. 5B). Remarkably, these animals phenocopied the MN8srrABKO strain. Representative pictures showing vegetations of the dissected aortic valves from animals infected with each of the strains are shown in Fig. 5C. The bacterial count from vegetations extracted from rabbits infected with each strain coincided with vegetation formation (Fig. 5D). Taken together, our data show that the SrrB cysteine disulfide bond is important for SrrAB function in vivo and, in its absence, repression of virulence by the SrrAB TCS is relieved.
Fig. 5.
SrrB cysteine mutation increases susceptibility to endocarditis development and lethality in rabbits infected with S. aureus MN8 strain. (A) The survival curve of rabbits infected with S. aureus MN8 wild-type, ΔsrrAB, and complementation strains: ΔsrrAB + pRMC2, ΔsrrAB + pRMC2::srrAB, and ΔsrrAB + pRMC2::srrABC501A. (B) The size of vegetations excised from dissected aortic valves of animals infected with each MN8 strain. (C) A representative picture of the dissected aortic valve from animals infected with each MN8 strain. Circles show where vegetations were formed. (Original magnification: 2×.) (D) Bacterial count obtained from vegetations of the animals infected with each MN8 strain. Four animals were used for the MN8 wild-type control group, and six animals each were used for the remaining strains. Statistical significance was determined by Student’s unpaired t test (vegetation size and CFU; **P < 0.0005; *P < 0.01).
Discussion
The SrrAB TCS is a major regulator of respiratory growth and virulence in S. aureus. SrrAB is critical for survival under hypoxic conditions and during oxidative and nitrosative stress. How the SrrAB TCS senses these environmental insults and responds accordingly is not known. Here, we show that cysteine residues in the CA domain of SrrB form a redox-sensitive disulfide bond that plays a critical role in virulence and pathogenicity.
The crystal structure of SrrB catalytic DHp-CA region and biochemical experiments identified a redox-sensitive disulfide bond between C464 and C501 in the ATP-binding CA domain. Although we have not determined the crystal structure of SrrB DHp-CA in reduced cysteine form, the structure of the DHp-CA region with oxidized disulfide provides insight into the mechanism by which the disulfide regulates kinase activity. Examination of the SrrB structure with the disulfide suggests that the disulfide bond could affect catalysis by modulating the conformation around the ATP binding pocket of the CA domain. This would readily explain the observation that SrrB Cyto with reduced cysteines is more active the oxidized disulfide form. Consistent with this, the SrrB Cyto C501A mutant lacking the disulfide had similar autophosphorylation kinetics as found in the reduced cysteine state of SrrB Cyto (SI Appendix, Fig. S8).
Cysteine residues at positions 464 and 501 of SrrB were found in only five species closely related to S. aureus, and thus not conserved in the Staphylococcus genus (Fig. 3). Moreover, the cysteines were absolutely conserved in 100 strains of S. aureus but not in S. epidermidis SrrB or B. subtilis ResE. These data suggest a divergent mechanism for SrrB regulation in the B. subtilis or S. epidermidis orthologs, despite overall similar biological function (8, 15, 35). At present, we have not been able to rationalize why S. aureus and related species have these conserved cysteines; however, we speculate that there is an evolutionary advantage to maintaining them. For example, S. aureus, but not B. subtilis and S. epidermidis, is pathogenic in humans, whereas these other organisms are either not pathogenic (B. subtilis) or only weakly pathogenic (S. epidermidis); these latter two organisms lack many of the virulence factors of S. aureus that allow the organism to adapt to different environments found in human infection sites. Interestingly, a disulfide bond with identical spatial localization was present in HK853 HK from T. maritima, suggesting that this redox mechanism of HK regulation may be common.
Over the past ∼20 years, several histidine kinases (ArcB, BvgS, EvgS, RegB, and CikA) have been shown to be regulated by redox-sensitive cysteines or disulfide bonds. The two best examples are E. coli ArcB (31, 36, 37) and Rhodobacter capsulatus RegB (38–41), both of which are regulated wholly or in part by intermolecular disulfide bonds. ArcB has emerged as the paradigm for HK redox regulation. In ArcB, the redox-sensitive cysteine residues are found in the cytoplasmic PAS domain located between the catalytic region and transmembrane domains (31, 36, 42). The prevailing mechanism posits that, under anaerobic conditions, ArcB is autophosphorylated and transfers the phosphoryl group to ArcA, the response regulator (31). Phosphorylated ArcA then regulates several genes, including those involved in energy metabolism (43). Under aerobic conditions, the two redox cysteines from opposite monomers form an intermolecular disulfide bond that endows ArcB with preferential phosphatase activity (44). Additional in vitro and in vivo experiments indicate that oxidized quinones of the respiratory chain promote the disulfide bond and suppress kinase activity (31, 36, 42, 43).
Here, we show the importance of an intramolecular cysteine disulfide bond in an HK involved in S. aureus pathogenicity. Our cellular biofilm experiment showed that the LACΔsrrAB complemented with either srrABC464A or srrABC501A phenocopies the ΔsrrAB strain, resulting in compromised biofilm formation. In MN8ΔsrrAB complemented with srrABC501A, repression of TSST-1 by SrrAB was lost. In addition, loss of the disulfide bond was similar to the deletion of SrrAB, which made the rabbits susceptible to infection. Our studies reinforce that TSST-1 plays a role in vegetation growth and lethality during infective endocarditis, as previously suggested (45–47).
Previous work by Mashruwala et al. showed that decreased respiration results in SrrAB-dependent biofilm formation, likely through the accumulation of reduced menaquinone (21). Similarly, Kinkel et al. showed that the SrrAB TCS senses and responds to nitric oxide and hypoxia through menaquinone (18). It is worth noting that menaquinone is the sole quinone molecule utilized in S. aureus respiration (48). Our in vitro data showed that cysteine-reduced SrrB, which is mimicked by the cysteine mutants, is the more active form. However, oxidized SrrB retains ∼60% activity. Thus, the phenotype observed in cells and in vivo rabbit experiments with the cysteine mutation did not correlate with in vitro redox-dependent activity data. By analogy to ArcB, we expected disruption of the disulfide bond by mutation to activate the SrrAB TCS. However, S. aureus strains complemented with SrrB C464A or C501A mutations were attenuated in anaerobic biofilm formation and TSST-1 repression and were more susceptible to infective endocarditis. Thus, the regulation of SrrB by redox is distinct from other TCS.
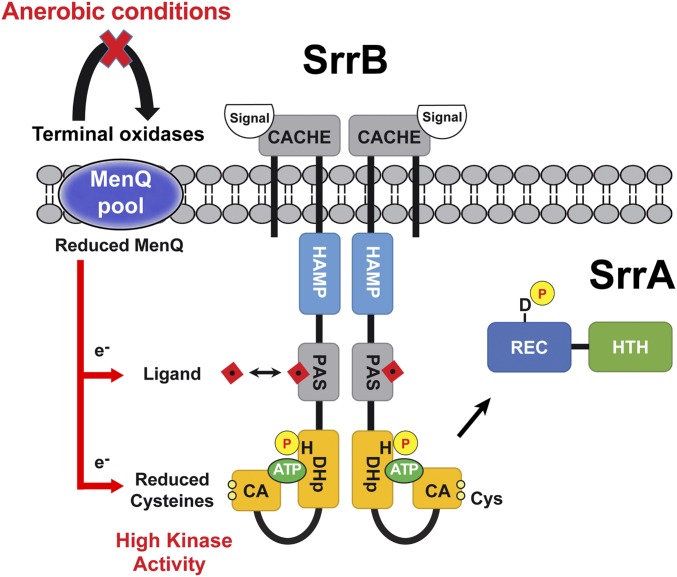
Fig. 6 summarizes our current model where multiple signals, sensed through the extracellular Cache domain, intracellular PAS domain, and redox cysteines, are integrated to regulate SrrB activity, SrrA phosphorylation status, and downstream gene expression. Under aerobic conditions, the menaquinone pool is found oxidized and SrrB has an intact cysteine disulfide bond and steady-state activity. Upon anaerobic or microaerobic conditions (or deletion of terminal oxidases), a reduced menaquinone pool forms to stimulate SrrB activity. Consistent with this idea, our data show that a proportion of the SrrB Cyto expressed in E. coli cells was found in the oxidized disulfide form. It is noteworthy that the regulatory disulfide in E. coli ArcB was also partitioned between the oxidized and reduced states (31). In S. aureus, we propose that the equilibrium state of the disulfide shifts toward the fully oxidized form under the appropriate growth conditions (aerobic) and in the context of full-length and membrane-bound SrrB.
Fig. 6.
Model of SrrAB two-component system regulation by redox. Under oxidizing conditions, terminal oxidases cause accumulation of oxidized menaquinone (MenQ). Under anerobic conditions (or terminal oxidase mutants), there is an accumulation of reduced menaquinone. The pool of reduced MenQ can reduce the SrrB cysteines directly or indirectly via a PAS domain/ligand complex to increase SrrB kinase activity. A putative redox-sensitive ligand binding to the PAS domain is depicted by the red square (see Discussion for details).
Our model suggests that the reduced menaquinone pool could act directly on the cysteine residues located in the CA domain at the C terminus of SrrB or indirectly through the membrane proximal PAS domain (Fig. 6). Thus, the PAS domain might directly bind menaquinone, or the flow of electrons could proceed through an intermediate ligand. In this scenario, a putative redox-active ligand could bind to the PAS domain and act as an intermediary to form a redox relay that reduces the SrrB disulfide bond to stimulate kinase activity. A second scenario allows for the extracellular Cache domain of SrrB to impose another layer of regulation on SrrB beyond that of the redox-active cysteines and a PAS domain/ligand complex. At present, the identity of Cache domain ligand(s) and how it affects SrrB regulation remain unknown. Mutating the cysteines affects the ability of SrrB to respond to these signals in vivo by a mechanism yet to be fully elucidated.
Our in vitro autophosphorylation and phosphotransfer data support a model where the PAS domain is involved in SrrAB regulation. The presence of the intracellular HAMP and PAS domains increase kinase and inhibit phosphatase activity of SrrB. Histidine kinase regulation by a PAS domain has also been shown in several HKs, including the essential S. aureus WalK (49). HAMP domains are predicted to transmit signals from ligand-bound Cache domains to the DHp-CA region by rotational movement (50). PAS domains, on the contrary, are known to sense cellular redox status through a wide variety of bound ligands or cofactors (51–53), including quinones (31, 54). Interestingly, menaquinone analogs have been shown to inhibit TSST-1 production in S. aureus through the SrrAB TCS (55). Despite no exogenous PAS domain ligands being present in our in vitro experiments, the PAS domain itself significantly influenced SrrB kinase and phosphatase activity. We hypothesize that the interdomain loop between the PAS and DHp domain is flexible and may allow for movement of the PAS domain toward the DHp-CA catalytic region to modulate kinase/phosphatase activity. A redox-active ligand binding to the PAS domain may further affect this regulation.
In summary, our studies identified an intramolecular disulfide bond in the SrrB HK that is critical for biological function. Our data suggest a mode of regulation distinct from the E. coli ArcB HK, the current paradigm for HK disulfide bond regulation. We propose a model whereby multiple domains and the redox-sensitive intramolecular disulfide bond in the SrrB HK sense environmental cues that regulate enzymatic activity and gene regulation. Collectively, our data provide insights into the mechanism by which SrrB senses and responds to changes in redox during S. aureus infection. The molecular details of the current model are the focus of ongoing research. The presence of a similar disulfide bond in another HK (HK853) opens the possibility that this strategy could be widely exploited by HKs.
Materials and Methods
Protein Expression and Purification.
Genomic DNA of S. aureus MRSA252 was used to isolate the DNA fragments of SrrB HAMP-PAS-DHp-CA (amino acids 208 to 583), SrrB DHp-CA (amino acids 359 to 583), and full-length SrrA (amino acids 1 to 241). These DNA fragments were cloned into a modified pET21a plasmid (Novagen) using NcoI and XhoI restriction sites. The modified pET21a plasmid has an N-terminal 6×His-tag followed by a recombinant tobacco etch virus (rTEV) protease cleavage site (ENLYFQG). The SrrB HAMP-PAS-DHp-CA C464A and C501A mutants were obtained by oligonucleotide-directed mutagenesis and confirmed by Sanger sequencing at The University of Iowa (Iowa Institute of Human Genetics Genomic Division). All proteins were expressed in E. coli BL21(DE3) (Novagen) grown in Lysogeny Broth (LB) media in the presence of 100 μg/mL ampicillin and induced with 0.5 mM isopropyl-1-thio-D-glactopyranoside (IPTG) at 18 °C overnight. The cells were harvested by centrifugation and resuspended in buffer containing 50 mM Tris⋅HCl, pH 7.5, 250 mM NaCl, and 10 mM imidazole. Proteins were purified by nickel-affinity chromatography (Nickel Sepharose 6 fast flow media; GE Healthcare) followed by incubation with rTEV protease at 4 °C overnight to remove the N-terminal 6×His-tag. Size-exclusion chromatography (Superdex 200; GE Healthcare) was performed as a final step in a buffer containing 25 mM Tris, pH 7.5, 150 mM NaCl, 5% glycerol, and 0.5 mM EDTA.
Kinase and Phosphotransfer Assay.
The radioactivity-based enzyme assays were performed as previously described (56). Briefly, autophosphorylation reactions with 5 μM SrrB HAMP-PAS-DHp-CA or DHp-CA were incubated 1 h with ATP mix (0.25 μM [γ-32P]-ATP and 250 μM cold ATP [Sigma] in 1× kinase buffer [25 mM Tris, pH 7.6, 1 mM MgCl2, 1 mM MnCl2, 1 mM CaCl2, 50 mM KCl, 0 or 5 mM DTT]). The reactions were quenched at the desired time point by addition of SDS/PAGE loading buffer. The samples were then resolved by SDS/PAGE and the bands visualized using a phosphor screen (Typhoon FLA 9500; GE Healthcare).
Phosphotransfer reactions used 5 μM SrrB HAMP-PAS-DHp-CA or DHp-CA that was autophosphorylated for 1 h as described earlier. Autophosphorylated SrrB HAMP-PAS-DHp-CA or DHp-CA were then incubated with 5 μM of full-length SrrA for 1 h. The reactions were quenched at the desired time point by addition of SDS/PAGE loading buffer. The samples were resolved by SDS/PAGE and bands visualized using a phosphor screen.
Sample Preparation and Crystallization.
The catalytic region of S. aureus SrrB, including the DHp and CA domains (DHp-CA), was cloned into plasmid pPROEX-HTb containing an N-terminal 6×His-tag. This plasmid was transformed into the E. coli BL21-CodonPlus(DE3)-RIL (Stratagene), and the expression of SrrB DHp-CA was induced by addition of 1 mM IPTG for 3 h at 37 °C in LB media. Selenomethionine (SeMet)-labeled protein was produced as previously described (57). For protein purification, cells were harvested and resuspended in 50 mM Tris, pH 8, 150 mM NaCl, 5% glycerol, and 1 mM phenylmethanesulfonyl fluoride and lysed using a Soniprep 150 sonicator. After centrifugation at 18,000 × g for 30 min to remove unbroken cells and debris, the supernatant was centrifuged at 90,140 × g for 2 h to remove cell membranes. The supernatant fraction was loaded onto a 5-mL Ni-NTA column (GE Healthcare) preequilibrated with buffer A (50 mM Tris, pH 8, 150 mM NaCl, and 5% glycerol). SrrB DHp-CA was eluted with buffer A containing 250 mM imidazole. The eluted fraction was dialyzed against buffer A overnight at 4 °C with the addition of rTEV protease to cleave the 6×His-tag. The sample was then loaded onto an Ni-NTA column, and the flow through from the column was collected, concentrated, and further purified by size-exclusion chromatography using a Superdex 75 (16/60) column (GE Healthcare) equilibrated with buffer A. The eluted fractions containing SrrB DHp-CA were concentrated and used for crystallization trials.
Purified SrrB DHp-CA at 10 mg/mL supplemented with 5 mM Mg-AMPPNP was used for crystallization. Native and SeMet-derived crystals were grown at 21 °C using the hanging-drop vapor diffusion method: 1 μL protein solution was mixed with 1 μL reservoir solution containing 20% (wt/vol) PEG3350, 15% MPD, 0.1 M sodium citrate, pH 5.5. Crystals appeared within 16 h.
Diffraction Data Processing.
Diffraction datasets used for structure determination were collected at the European Synchrotron Radiation Facility (ESRF) beamline BM-16. The datasets were processed using Mosflm (58) software in space group P21, which suggested the presence of two SrrB DHp-CA monomers in the asymmetric unit (∼73% solvent content). The initial phases were calculated in SOLVE (59) using multiwavelength anomalous dispersion data. The phases were refined and extended to 2-Å resolution using density modification in the CCP4 (60) software suite. The resulting electron density map was of high quality, allowing for a continuous tracing of the main chain. Model building in COOT (61) and refinement using Refmac5 (62) (in CCP4) and CNS (63) gradually improved the model and the fit to the experimental map. The final model yielded Rwork of 19.3% and Rfree of 24.6%. PROCHECK (in CCP4) evaluation of the Ramachandran plot gave 91.6% in favored regions and 0.3% outliers (SI Appendix, Table S1). The coordinates and structure factors of the SrrB catalytic region have been deposited in the PDB under accession code 6PAJ. Figures were generated in PyMOL (Schrödinger).
Phylogenetic Analysis of SrrB Sequences.
The S. aureus SrrB full-length sequence was submitted to the protein–protein BLAST server (64). Unique Staphylococcus species with SrrB sequences were collated and downloaded in FASTA format, and the phylogenetic tree was generated using the Phylogenetic Tree analysis pipeline in the program ETE3 (65).
S. aureus Strains and Plasmid Construction.
SI Appendix, Table S2 lists the strains used in this study. The LAC (21) and MN8 (55) ΔsrrAB strains have been described. MN8 strains containing the multicopy pRMC2 plasmid (66) included the srrAB operon (including 132 bp upstream of srrA and 100 bp downstream of srrB). The srrAB operon was amplified using the PCR from the chromosome of S. aureus MN8 (primers are listed in SI Appendix, Table S3) and cloned into the SacI site of plasmid pRMC2 using E. coli XL1-blue as a cloning host. E. coli SA30B (67) was utilized to methylate DNA prior to electroporation into S. aureus MN8 ΔsrrAB. LAC strains containing the multicopy pCM28 plasmid have been described (21). All plasmid mutations were produced by oligonucleotide-directed mutagenesis and confirmed by nucleotide sequencing.
Biofilm Assays.
S. aureus LAC strains were cultured overnight in tryptic soy broth (TSB) medium containing chloramphenicol before biofilm formation was assessed in microtiter plates after anaerobic culture as previously described (68). Briefly, the cultures were inoculated and passed through an airlock (three vacuum/gas exchange cycles) into an anaerobic chamber (Coy Laboratory Products) with oxygen concentration maintained below 1 ppm. Overnight cultures were diluted into fresh TSB to a final optical density (OD590) of 0.05. Aliquots (200 μL) of diluted cultures were added to the 96-well plate and incubated statically in an anaerobic chamber at 37 °C for 22 h. The optical density (OD590) of the cultures was determined before harvesting the biofilm. The biofilms in 96-well plate were washed twice with water, fixed at 60 °C, and allowed to cool to room temperature. The biofilms were stained with 0.1% crystal violet, washed three times with water, and destained with 33% acetic acid. The absorbance of the eluted stain was measured at 570 nm blanked with acetic acid. The resulting absorbance was normalized to the optical density of the cells prior to harvesting the biofilm. The data for each strain represent six replicates. Statistical analyses was performed, and P values were calculated using the unpaired t test in SigmaPlot (Systat Software).
TSST-1 Western Blot.
Individual MN8 strains were grown overnight and adjusted to an optical density at 600 nm (OD600nm) of 0.05 in 50 mL of brain heart infusion (BHI) medium containing appropriate antibiotics and grown at 37 °C in 100-mL Erlenmeyer flasks for 8 h with shaking (aerobic) or statically in sealed 50-mL conical tubes for 24 h (low O2 conditions). Cells were subsequently pelleted by centrifugation, and cell-free supernatants equivalent to 42 OD600 units of culture were precipitated by using trichloroacetic acid at a final concentration of 6%. Precipitated protein was pelleted, washed with ice-cold acetone, and resuspended in 250 µL 8 M urea and analyzed by SDS/PAGE. For Western blot analysis of TSST-1 expression, parallel protein gels were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) at 100 V for 1 h. The membrane was blocked at room temperature for 2 h with phosphate-buffered saline (PBS) containing 5% skimmed milk, 10% normal horse serum (NHS; Life Technologies), and 10% fetal calf serum (FCS; Wisent). Washed membranes were incubated at 4 °C overnight with rabbit polyclonal anti–TSST-1 antisera (69) diluted 1:1,000 in PBS containing 2.5% skimmed milk, 5% NHS, and 5% FCS. Membranes were washed three times with PBS containing 0.05% Tween 20, followed by incubation with DyLight 800-conjugated donkey anti-rabbit IgG antibody (Rockland) diluted 1:10,000 in PBS containing 2.5% skimmed milk, 5% NHS, and 5% FCS at room temperature for 1 h in the dark. Membranes were washed three times with PBST and imaged with an Odyssey imager (LI-COR Biosciences).
Rabbit Infective Endocarditis and Sepsis.
The infective endocarditis experiments were performed as previously described (34). Male and female New Zealand White rabbits (∼2 kg; Bakkom Rabbitry) were used for the experiments, and males and females responded similarly. Animal care and surgical procedures used were as described in the University of Iowa IACUC protocol number 6121907. Rabbits were anesthetized with ketamine (25 mg/kg) and xylazine (25 mg/kg; Akorn) and administered pain-relieving medications (buprenorphine 0.05 mg/kg twice daily subcutaneous; Reckitt Benckiser Healthcare) throughout the experiment. Each animal’s neck was shaved, and the left carotid artery was exposed by a 5-cm incision. A plastic catheter was inserted through the left carotid artery into each animal until it abutted the aortic valve. The catheter was then tied in place and left for 2 h. After 2 h, the catheter was removed and the animal sutured closed. The animals were injected intravenous through the marginal ear veins with 5 × 108 S. aureus cells (MN8-WT + pRMC2, MN8ΔsrrAB + pRMC2, MN8ΔsrrAB + pRMC2::srrAB, and MN8ΔsrrAB + pRMC2::srrABC501A) grown in 10 μg/mL chloramphenicol. Chloramphenicol (2.5 mg) was injected subcutaneous into the rabbits daily to maintain plasmids. Plasmid stability of MN8 strains in rabbits was verified by plating vegetation homogenates in the presence of chloramphenicol, which showed colony growth indicating retention of plasmids (16, 45, 70). The rabbits were monitored for health status for up to 4 d. During this time, animals that simultaneously failed to exhibit escape behavior and maintain upright positions (a test 100% predictive of lethal infection) were killed with 1 mL/kg of Euthasol (Virbac). The animals were killed after 4 d or prematurely as indicated earlier. Their hearts were removed and vegetation dissected, weighed, and homogenized for colony-forming unit (CFU) determination. Four animals were used for the MN8 wild-type control group, and six animals each were used for remaining strain groups. Statistical analyses of the vegetation size and bacterial counts were performed (P values were calculated using unpaired t test) in SigmaPlot (Systat Software).
Data Availability.
Requests for reagents or further information should be directed to E.J.F. (ernesto-fuentes@uiowa.edu). Coordinates for the model of SrrB DHp-CA region solved by X-ray crystallography have been deposited in the Protein Data Bank with ID code 6PAJ.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (NIH) and National Institute of Allergy and Infectious Diseases (NIAID) to E.J.F. and P.M.S. (NIAID Grant AI135305). J.M.B. was funded by the NIH (NIAID Grant AI139100-01) and US Department of Agriculture Multistate Reseach Fund (Project NE−1028). W.S.-P. was supported by NIH (NIAID Grant AI134692-03). The J.K.M. lab was supported by the Canadian Institutes of Health Research (Grant PJT-166050). A.M. was supported by grant BIO2016-78571-P from the Ministerio de Economia y Competitividad (Spain). Dr. Jonathan Willett is acknowledged for initial observations and experiments with SrrB. Dr. Lokesh Gakhar (University of Iowa Protein and Crystallography Facility) provided scripts to search the Protein Data Bank. We thank the IBV-CSIC Crystallography Facility for protein crystallization screens. The size-exclusion chromatography with in-line with multi-angle scattering data were collected with help by Dr. Srinivas Chakravarthy at the Bio-CAT beamline 18-ID-D at the Advanced Photon Source (Argonne, IL). The synchrotron X-ray diffraction datasets reported in this work were collected in experiments performed at the European Synchrotron Radiation Facility (Grenoble, France). We thank the staff at the beamline for assistance during data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates for the model of SrrB DHp-CA region solved by X-ray crystallography have been deposited in the Protein Data Bank, https://www.rcsb.org/ (ID code 6PAJ).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921307117/-/DCSupplemental.
References
- 1.Nouwen J. L., Fieren M. W. J. A., Snijders S., Verbrugh H. A., van Belkum A., Persistent (not intermittent) nasal carriage of Staphylococcus aureus is the determinant of CPD-related infections. Kidney Int. 67, 1084–1092 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Muthukrishnan G., et al. , Longitudinal genetic analyses of Staphylococcus aureus nasal carriage dynamics in a diverse population. BMC Infect. Dis. 13, 221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronner S., Monteil H., Prévost G., Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol. Rev. 28, 183–200 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Park M. K., Myers R. A., Marzella L., Oxygen tensions and infections: Modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin. Infect. Dis. 14, 720–740 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Ross R. A., Onderdonk A. B., Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect. Immun. 68, 5205–5209 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarwood J. M., Schlievert P. M., Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 38, 1797–1803 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beenken K. E., et al. , Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186, 4665–4684 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrich M., et al. , The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65, 1276–1287 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Stock A. M., Robinson V. L., Goudreau P. N., Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Haag A. F., Bagnoli F., The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. Immunol. 409, 145–198 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Groisman E. A., Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casino P., Rubio V., Marina A., The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 20, 763–771 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Yarwood J. M., McCormick J. K., Schlievert P. M., Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183, 1113–1123 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Throup J. P., et al. , The srhSR gene pair from Staphylococcus aureus: Genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40, 10392–10401 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Wu Y., et al. , Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pragman A. A., Yarwood J. M., Tripp T. J., Schlievert P. M., Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186, 2430–2438 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilde A. D., et al. , Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 11, e1005341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinkel T. L., Roux C. M., Dunman P. M., Fang F. C., The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio 4, e00696-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Windham I. H., Chaudhari S. S., Bose J. L., Thomas V. C., Bayles K. W., SrrAB modulates Staphylococcus aureus cell death through regulation of cidABC transcription. J. Bacteriol. 198, 1114–1122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mashruwala A. A., Boyd J. M., The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS One 12, e0170283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mashruwala A. A., Guchte A. V., Boyd J. M., Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife 6, e23845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willett J. W., Kirby J. R., CrdS and CrdA comprise a two-component system that is cooperatively regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2, e00110-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock J. B., Ninfa A. J., Stock A. M., Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D. J., Forst S., Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147, 1197–1212 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Marina A., Waldburger C. D., Hendrickson W. A., Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24, 4247–4259 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob-Dubuisson F., Mechaly A., Betton J. M., Antoine R., Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 16, 585–593 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Bhate M. P., Molnar K. S., Goulian M., DeGrado W. F., Signal transduction in histidine kinases: Insights from new structures. Structure 23, 981–994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casino P., Miguel-Romero L., Marina A., Visualizing autophosphorylation in histidine kinases. Nat. Commun. 5, 3258 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Casino P., Rubio V., Marina A., Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Sporer A. J., Kahl L. J., Price-Whelan A., Dietrich L. E. P., Redox-based regulation of bacterial development and behavior. Annu. Rev. Biochem. 86, 777–797 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Malpica R., Franco B., Rodriguez C., Kwon O., Georgellis D., Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 13318–13323 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burley S. K., et al. , RCSB protein data bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 47, D464–D474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blomster-Hautamaa D. A., Schlievert P. M., Preparation of toxic shock syndrome toxin-1. Methods Enzymol. 165, 37–43 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Salgado-Pabón W., Schlievert P. M., Aortic valve damage for the study of left-sided, native valve infective endocarditis in rabbits. Methods Mol. Biol. 1396, 73–80 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Härtig E., Jahn D., Regulation of the anaerobic metabolism in Bacillus subtilis. Adv. Microb. Physiol. 61, 195–216 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Alvarez A. F., Rodriguez C., Georgellis D., Ubiquinone and menaquinone electron carriers represent the yin and yang in the redox regulation of the ArcB sensor kinase. J. Bacteriol. 195, 3054–3061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgellis D., Kwon O., Lin E. C., Wong S. M., Akerley B. J., Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J. Bacteriol. 183, 7206–7212 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swem L. R., Gong X., Yu C. A., Bauer C. E., Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 281, 6768–6775 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swem L. R., et al. , Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22, 4699–4708 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J., Bauer C. E., RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. mBio 1, e00272-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J., et al. , RegB kinase activity is repressed by oxidative formation of cysteine sulfenic acid. J. Biol. Chem. 288, 4755–4762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgellis D., Kwon O., Lin E. C., Quinones as the redox signal for the arc two-component system of bacteria. Science 292, 2314–2316 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Malpica R., Sandoval G. R. P., Rodríguez C., Franco B., Georgellis D., Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8, 781–795 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Georgellis D., Kwon O., De Wulf P., Lin E. C., Signal decay through a reverse phosphorelay in the arc two-component signal transduction system. J. Biol. Chem. 273, 32864–32869 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Stach C. S., et al. , Novel tissue level effects of the Staphylococcus aureus enterotoxin gene cluster are essential for infective endocarditis. PLoS One 11, e0154762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung J. W., et al. , Superantigen profiling of Staphylococcus aureus infective endocarditis isolates. Diagn. Microbiol. Infect. Dis. 79, 119–124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nienaber J. J., et al. ; International Collaboration on Endocarditis-Microbiology Investigators , Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J. Infect. Dis. 204, 704–713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bentley R., Meganathan R., Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 46, 241–280 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutu A. D., Wayne K. J., Sham L. T., Winkler M. E., Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: Dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192, 2346–2358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parkinson J. S., Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64, 101–122 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Gilles-Gonzalez M. A., Gonzalez G., Regulation of the kinase activity of heme protein FixL from the two-component system FixL/FixJ of Rhizobium meliloti. J. Biol. Chem. 268, 16293–16297 (1993). [PubMed] [Google Scholar]
- 52.Rebbapragada A., et al. , The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U.S.A. 94, 10541–10546 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delgado-Nixon V. M., Gonzalez G., Gilles-Gonzalez M. A., Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39, 2685–2691 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Sobran M. A., Cotter P. A., The BvgS PAS domain, an independent sensory perception module in the Bordetella bronchiseptica BvgAS phosphorelay. J. Bacteriol. 201, e00286-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlievert P. M., et al. , Menaquinone analogs inhibit growth of bacterial pathogens. Antimicrob. Agents Chemother. 57, 5432–5437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willett J. W., Kirby J. R., Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 8, e1003084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doublie S., Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997). [PubMed] [Google Scholar]
- 58.Leslie A. G., The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Terwilliger T. C., Berendzen J., Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Brünger A. T., et al. , Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 65.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corrigan R. M., Foster T. J., An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61, 126–129 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Monk I. R., Tree J. J., Howden B. P., Stinear T. P., Foster T. J., Complete bypass of restriction systems for major Staphylococcus aureus lineages. MBio 6, e00308-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mashruwala A. A., Gries C. M., Scherr T. D., Kielian T., Boyd J. M., SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect. Immun. 85, IAI.00157-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schlievert P. M., et al. , Alpha and beta chains of hemoglobin inhibit production of Staphylococcus aureus exotoxins. Biochemistry 46, 14349–14358 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salgado-Pabón W., et al. , Superantigens are critical for Staphylococcus aureus Infective endocarditis, sepsis, and acute kidney injury. mBio 4, e00494-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for reagents or further information should be directed to E.J.F. (ernesto-fuentes@uiowa.edu). Coordinates for the model of SrrB DHp-CA region solved by X-ray crystallography have been deposited in the Protein Data Bank with ID code 6PAJ.