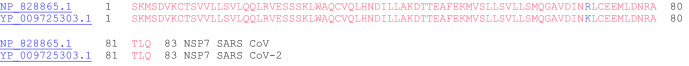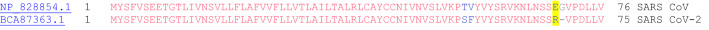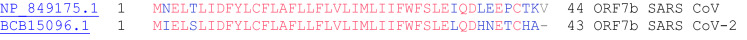Abstract
The devastating effects of the recent global pandemic (termed COVID-19 for “coronavirus disease 2019”) caused by the severe acute respiratory syndrome coronavirus-2 (SARS CoV-2) are paramount with new cases and deaths growing at an exponential rate. In order to provide a better understanding of SARS CoV-2, this article will review the proteins found in the SARS CoV-2 that caused this global pandemic.
Electronic supplementary material
The online version of this article (10.1007/s10930-020-09901-4) contains supplementary material, which is available to authorized users.
Keywords: Proteins, Virus, SARS CoV-2
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS CoV-2) is the virus that caused the global pandemic that was first reported [1] on December 31, 2019 [2]. Taxonomically, SARS CoV-2 belongs to the realm Riboviria, order Nidovirales, suborder Cornidovirineae, family Coronaviridae, subfamily Orthocoronavirinae, genus Betacoronavirus (lineage B), [3] subgenus Sarbecovirus, and the species Severe acute respiratory syndrome-related coronavirus.
The genome of SARS CoV-2 (NCBI Reference Sequence: NC_045512.2) [4] is similar to the genome of the coronavirus that caused the SARS epidemic in 2003 (SARS CoV, NCBI Reference sequence: NC_004718.3) [5, 6]. Much of the understanding of the proteins found in SARS CoV-2 are based on the numerous research studies reported on SARS CoV and other related viruses (e.g. MERS CoV) [7, 8]. However, among the recent coronavirus outbreaks in the new millennium (SARS CoV: 2002–2003, MERS CoV: 2012, SARS CoV-2: 2020), SARS CoV-2 mysteriously had the most devastating global impact. Understanding the proteins present in these viruses enable a more rational approach to designing more effective antiviral drugs [9, 10]. The majority of proteins of SARS CoV have been characterized in detail. The proteins of SARS CoV consist of two large polyproteins: ORF1a and ORF1ab (that proteolytically cleave to form 16 nonstructural proteins), four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N), and eight accessory proteins: ORF3a, ORF3b (NP_828853.1, not present in SARS CoV-2), ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b (NP_828859.1, not present in SARS CoV-2). Although accessory proteins have been viewed as dispensable for viral replication in vitro, some have been shown to play an important role in virus-host interactions in vivo [11]. Similar to SARS CoV, SARS CoV-2 lacks the hemagglutinin esterase gene, which is found in human coronavirus (hCoV) HKU1, a lineage A betacoronavirus [3]. The spike protein, envelope protein, membrane protein, nucleocapsid protein, 3CL protease, papain like protease, RNA polymerase, [10] and helicase protein have been suggested to be viable antiviral drug targets [12]. SARS CoV-2 is an RNA virus and its RNA genome is 30 kb in length. SARS CoV-2 is thought to have originated from its closest relative, BatCov RaTG13 (GenBank: MN996532), [13] which was isolated from horseshoe bats [14].
Discussion: Proteins of SARS CoV-2
SARS CoV-2 (NC_045512.2) has a total of 11 genes with 11 open reading frames (ORFs) (Table 1): ORF1ab, ORF2 (Spike protein), ORF3a, ORF4 (Envelope protein), ORF5 (Membrane protein), ORF6, ORF7a, ORF7b, ORF8, ORF9 (Nucleocapsid protein), and ORF10.
Table 1.
The genes expressed by SARS CoV-2 (NC_045512.2)
| Number(#) | Gene | GeneID | Location | Protein | [LOCUS] |
|---|---|---|---|---|---|
| 1(7,096) | ORF1ab | 43,740,578 | 266–21,555 | ORF1ab polyprotein | [BCB15089.1/BCB97900.1] |
| 1(4,405) | ORF1a | 43,740,578 | 266–13,483 | ORF1a polyprotein | [YP_009725295.1] |
| 2(1,273) | ORF2 (S) | 43,740,568 | 21,563–25,384 | Spike protein (S protein) | [BCA87361.1] |
| 3(275) | ORF3a | 43,740,569 | 25,393–26,220 | ORF3a protein | [BCA87362.1] |
| 4(75) | ORF4 (E) | 43,740,570 | 26,245–26,472 | Envelope protein (E protein) | [BCA87363.1] |
| 5(222) | ORF5 (M) | 43,740,571 | 26,523–27,191 | Membrane protein (M protein) | [BCA87364.1] |
| 6(61) | ORF6 | 43,740,572 | 27,202–27,387 | ORF6 protein | [BCA87365.1] |
| 7(121) | ORF7a | 43,740,573 | 27,394–27,759 | ORF7a protein | [BCA87366.1] |
| 8(43) | ORF7b | 43,740,574 | 27,756–27,887 | ORF7b protein | [BCB15096.1] |
| 9(121) | ORF8 | 43,740,577 | 27,894–28,259 | ORF8 protein | [BCA87367.1] |
| 10(419) | ORF9 (N) | 43,740,575 | 28,274–29,533 | Nucleocapsid phosphoprotein (N protein) | [BCA87368.1] |
| 11(38) | ORF10 | 43,740,576 | 29,558–29,674 | ORF10 protein | [BCA87369.1] |
#Represents the number of amino acids in each gene
Polyprotein Expressed by ORF1ab
The first gene (ORF1ab) expresses a polyprotein. The ORF1ab polyprotein is comprised of 16 nonstructural proteins (NSPs) (Table 2).
Table 2.
The nonstructural proteins (NSPs) found in the polyprotein of SARS CoV-2
| # | Name | Accession | Amino acids | Proposed function |
|---|---|---|---|---|
| (i) | NSP1 | YP_009725297.1 | 180 amino acids | Induce host mRNA (leader protein) cleavage |
| (ii) | NSP2 | YP_009725298.1 | 638 amino acids | Binds to PHBs 1, 2 |
| (iii) | NSP3a | YP_009725299.1 | 1945 amino acids |
Release NSPs 1, 2, 3 (Papain like proteinase) |
| (iv) | NSP4 | YP_009725300.1 | 500 amino acids | Membrane rearrangement |
| (v) | NSP5a | YP_009725301.1 | 306 amino acids | Cleaves at 11 sites of (3C-like proteinase) NSP polyprotein |
| (vi) | NSP6 | YP_009725302.1 | 290 amino acids | Generates autophagosomes |
| (vii) | NSP7 | YP_009725303.1 | 83 amino acids | Dimerizes with NSP8 |
| (viii) | NSP8 | YP_009725304.1 | 198 amino acids | Stimulates NSP12 |
| (ix) | NSP9 | YP_009725305.1 | 113 amino acids | Binds to helicase(?) |
| (x) | NSP10 | YP_009725306.1 | 139 amino acids | Stimulates NSP16(?) |
| (xi) | NSP11 | YP_009725312.1 | 13 amino acids | Unknown |
| (xii) | NSP12a | YP_009725307.1 | 932 amino acids | Copies viral RNA (RNA polymerase) methylation (guanine) |
| (xiii) | NSP13 | YP_009725308.1 | 601 amino acids | Unwinds duplex RNA (Helicase) |
| (xiv) | NSP14 | YP_009725309.1 | 527 amino acids | 5′-cap RNA (3′ to 5′ exonuclease, guanine N7-methyltransferase) |
| (xv) | NSP15a | YP_009725310.1 | 346 amino acids | Degrade RNA to (endoRNAse/endoribonuclease) evade host defense |
| (xvi) | NSP16 | YP_009725311.1 | 298 amino acids | 5′-cap RNA (2′-O-ribose-methyltransferase—potential antiviral drug target) methylation (adenine) |
aIndicates possible targets of antiviral compounds
NSP1 (Leader Protein)
Nonstructural protein 1 (NSP1) is the first protein of the polyprotein of SARS CoV-2 (Fig. 1—sequence alignment of NSP1 for SARS CoV with SARS CoV-2). This protein is also known as the leader protein. This protein is also found in SARS coronavirus and is known to be a potent inhibitor of host gene expression. NSP1 binds to the 40S ribosome of the host cell to inactivate translation and promotes host mRNA degradation selectively, while the viral SARS CoV mRNA remain intact [15]. Figure 1 shows the amino acid sequence alignment for the NSP1 proteins of SARS CoV (from genome: NCBI Reference Sequence: NC_004718.3) and SARS CoV-2.
Fig. 1.
Alignment of the primary amino acid sequence of NSP1 of SARS CoV (top, NP_828860.2) and SARS CoV-2 (YP_009725297.1). Sequence identity: 84.4%. Sequence similarity: 93.9%—determined using LALIGN software (and for subsequent alignments, Figs. 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, and 26, see Supporting Information for output data) [16].
NSP2
Nonstructural protein 2 (NSP2) is the second protein of the polyprotein of SARS CoV-2 (Fig. 2). This protein is conserved in SARS CoV, the related beta coronavirus to SARS CoV-2. In SARS CoV, NSP2 was found to bind to two host proteins: prohibitin 1 and prohibitin 2 (PHB1 and PHB2) [17]. PHB1 and PHB2 proteins are known to play roles in cell cycle progression, cell migration, cellular differentiation, apoptosis, and mitochondrial biogenesis. The binding of NSP2 to PHB1 and PHB2 proteins suggest that NSP2 plays a role in disrupting the host cell environment.
Fig. 2.
The primary amino acid sequence alignment of NSP2 for SARS CoV (NP_828861.2) and SARS CoV-2 (YP_009725298.1). These proteins have 68.3% sequence identity (90.0% similar)
NSP3 (Papain like Proteinase)
NSP3 is the papain-like proteinase protein (Fig. 3). This protein is nearly 200 kDa in size and is the largest protein (not including the polyproteins ORF1a and ORF1ab) encoded by the coronaviruses. With such a long sequence, it possesses several conserved domains: ssRNA binding, ADPr binding, G-quadruplex binding, ssRNA binding, protease (papain-like protease), and NSP4 binding), and transmembrane domain. Among the 16 nonstructural proteins, NSP3, NSP4, and NSP6 have transmembrane domains [18]. The papain like protease 1 (PL1 protease) of alpha coronavirus (alpha CoV) Transmissible Gastroenteritis Virus (TGEV), which is part of NSP3, was shown to cleave the site between NSP2 and NSP3. Furthermore, this papain like protease domain is responsible for the release of NSP1, NSP2, and NSP3 from the N-terminal region of polyproteins 1a and 1ab from coronaviruses [19]. Considering this important protease activity to release essential proteins for viral activity, the inhibition of NSP3 protease activity is an important target for antiviral activity [20]. Tanshinones, a class of natural products have been found to inhibit NSP3 protease activity.
Fig. 3.
The primary amino acid sequence alignment of NSP3 for SARS CoV (NP_828862.2) and SARS CoV-2 (YP_009725299.1). Sequence identity: 76.0%, sequence similarity: 91.8%
NSP4 (Contains Transmembrane Domain 2)
NSP4 interacts with NSP3 and possibly host proteins to confer a role related to membrane rearrangement in SARS CoV. Moreover, the interaction between NSP4 and NSP3 is essential for viral replication [18]. The sequence alignment for NSP4 proteins for SARS CoV and SARS CoV-2 is shown in Fig. 4.
Fig. 4.
The primary amino acid sequence alignment of NSP4 for SARS CoV (NP_904322.1) and SARS CoV-2 (YP_009725300.1). Sequence identity: 80.0%, sequence similarity: 95.0%
NSP5 (3C-like proteinase)
The NSP5 protein based on the Middle East Respiratory Syndrome (MERS) coronavirus has been characterized. NSP5 cleaves at 11 distinct sites to yield mature and intermediate nonstructural proteins (NSPs) [21]. The amino acid sequence alignment for NSP5 of SARS CoV and SARS CoV-2 is shown in Fig. 5.
Fig. 5.
The primary amino acid sequence of NSP5 for SARS CoV (NP_828863.1) and SARS CoV-2 (YP_009725301.1). Sequence identity: 96.1%, sequence similarity: 99.7%
NSP6 (Putative Transmembrane Domain)
The NSP6 protein of the avian coronavirus (infectious bronchitis virus, IBV) was shown to generate autophagosomes from the endoplasmic reticulum (ER) (Fig. 6b shows sequence alignment with SARS CoV-2 NSP6). Autophagosomes facilitate assembly of replicase proteins. Furthermore, NSP6 limited autophagosome/lysosome expansion, which in turn prevents autophagosomes from delivering viral components for degradation in lysosomes [22]. With SARS CoV, NSP6 was shown to induce membrane vesicles [23]. The amino acid sequence alignment for NSP6 of SARS CoV and SARS CoV-2 is shown in Fig. 6.
Fig. 6.
Amino acid sequence alignment between the NSP6 proteins of SARS CoV (top: NP_828864.1) and SARS-CoV-2 (bottom: YP_009725302.1). Sequence identity: 88.2%, sequence similarity: 98.3%
NSP7
NSP7 is required to form a complex with NSP8 (next section) and NSP12 to yield the RNA polymerase activity of NSP8 [24]. The primary amino acid sequence alignment for the NSP8 proteins for SARS CoV and SARS CoV-2 is shown in Fig. 7. Only one amino acid residue is different (arginine vs. lysine) but the charge is conserved at this location.
Fig. 7.
The primary amino acid sequence alignment of NSP7 SARS CoV (NP_828865.1) and SARS CoV-2 (YP_009725303.1). Sequence identity: 98.8%, sequence similarity: 100%
NSP8
NSP8 is a peptide cofactor that makes a heterodimer with NSP7 (the other peptide cofactor), and this NSP7-NSP8 heterodimer complexes with NSP12. In addition to the NSP7-NSP8 heterodimer, an NSP8 monomer unit also complexes with NSP12, which ultimately forms the RNA polymerase complex. The cryo-EM structure of this complex has been solved [25]. The amino acid sequence alignment for NSP8 of SARS CoV and SARS CoV-2 is shown in Fig. 8.
Fig. 8.
The primary amino acid sequence alignment of NSP8 for SARS CoV (NP_828866.1) and SARS CoV-2 (YP_009725304.1). Sequence identity: 97.5%, sequence similarity: 100.0%
NSP9
NSP9 from the porcine reproductive and respiratory syndrome virus (PRRSV) has been found to interact with the DEAD-box RNA helicase 5 (DDX5) cellular protein [26]. This interaction between NSP9 and DDX5 has been shown to be important for viral replication—when the DDX5 gene was silenced in MARC-145 cells, the virus titers were lower by tenfold. Figure 9 shows the amino acid sequence alignment between the two NSP9 proteins from SARS CoV and SARS CoV-2.
Fig. 9.
The primary amino acid sequence alignment of NSP9 for SARS CoV (NP_828868.1) and SARS CoV-2 (YP_009725305.1). Sequence identity: 97.3%, sequence similarity: 99.1%
NSP10
NSP10 has been shown to interact with NSP14 in SARS coronavirus, and this interaction stimulates activity of NSP14. NSP 14 is known to function as an S-adenosylmethionine (SAM)-dependent (guanine-N7) methyl transferase (N7-MTase) [27]. Furthermore, NSP10 has also been shown to stimulate the activity of NSP16, which is a 2′-O-methyltransferase [28]. Figure 10 shows the amino acid sequence alignment between the two NSP10 proteins from SARS CoV and SARS CoV-2.
Fig. 10.
The primary amino acid sequence alignment of NSP10 for SARS CoV (NP_828868.1) and SARS CoV-2 (YP_009725306.1). Sequence identity: 97.1%, sequence similarity: 99.3%
NSP11
The function of NSP11 seems to be unknown. NSP11 is made of thirteen amino acids and the first nine amino acids (sadaqsfln) are identical to the first nine in NSP12. Figure 11 shows the amino acid sequence alignment between the two NSP12 proteins from SARS CoV and SARS CoV-2.
Fig. 11.
The primary amino acid sequence alignment of NSP11 for SARS CoV (NP_904321.1) and SARS CoV-2 (YP_009725312.1). Sequence identity: 84.6%, sequence similarity: 100.0%
NSP12 (RNA Dependent RNA Polymerase)
NSP12 is the RNA-dependent RNA polymerase that copies viral RNA. As mentioned, NSP12 makes a complex with an NSP7-NSP8 heterodimer and an NSP8 monomer to confer processivity of NSP12. NSP12 exhibits poor processivity in RNA synthesis—that is the presence of NSP7 and NSP8 lowers the dissociation rate of NSP12 from RNA [29]. The amino acid sequence alignment between the two NSP12 proteins from SARS CoV and SARS CoV-2 is shown in Fig. 12.
Fig. 12.
The primary amino acid sequence alignment of NSP12 for SARS CoV (NP_828869.1) and SARS CoV-2 (YP_009725307.1). Sequence identity: 96.4%, sequence similarity: 99.4%
NSP13 (Helicase)
SARS CoV was used to characterize the helicase enzyme, NSP13, which unwinds duplex RNA [30]. The crystal structure of NSP13 of SARS CoV has been reported [31]. Furthermore, it has been shown that binding of NSP12 with NSP13 can enhance the helicase activity of NSP13. In addition to its helicase activity, NSP13 of SARS CoV is also known to possess 5′-triphosphatase activity, which is responsible for introducing the 5′-terminal cap of the viral mRNA [32]. Both eukaryotic and most viral mRNA have a 5′-terminal cap structure: m7G(5)ppp(5)N-. This 5′-terminal cap is the site of recognition for translation and plays a role in splicing, nuclear export, translation, and stability of mRNA. This process of incorporating the 5′-terminal cap will be discussed in the next section: (xiv) NSP14. The sequence alignment for NSP13 of SARS CoV and SARS CoV-2 is shown in Fig. 13. Interestingly, only one amino acid residue is different out of the 601 amino acids in these two proteins (isoleucine vs. valine).
Fig. 13.
The primary amino acid sequence of NSP13 SARS CoV (NP_828870.1) and SARS CoV-2 (YP_009725308.1). Sequence identity: 99.8%, sequence similarity: 100.0%
NSP14 (3′ to 5′ Endonuclease, N7-Methyltransferase)
NSP14 from coronavirus is known to have 3′-5′ exoribonuclease activity and N7-methyltransferase activity [33]. The guanine-N7-methyltransferase activity is part of the process for introducing the 5′-cap of the virus, which involves multiple steps: [1] the gamma-phosphate of the 5′end of nascent mRNA is removed by the RNA triphosphatase (NSP13), [32], [2] a GMP moiety derived from a covalent enzyme-GMP intermediate is transferred to the resulting mRNA with a diphosphate end, [3] the GpppA cap is methylated with S-adenosyl-methionine, which is catalyzed by the guanine-N7-methyltransferase (NSP14) to yield the cap-0 structure, [34] and [4] 2′-O-methylation by NSP16 of adenine gives the cap-1 structure [35]. It is currently unknown which enzyme incorporates the GMP group involved in the second step, and it is possible that the virus uses the host guanylyltransferase enzyme [36]. Figure 14 shows the amino acid sequence alignment between the NSP14 proteins of SARS CoV and SARS CoV-2.
Fig. 14.
The primary amino acid sequence alignment of NSP14 of SARS CoV (NP_828871.1) and SARS CoV-2(YP_009725309.1). Sequence identity: 95.1%, sequence similarity: 99.1%
NSP15 (endoRNAse)
NSP15 of SARS coronavirus has been biochemically characterized as an endoribonuclease that cleaves RNA at uridylates at the 3′-position to form a 2′-3′ cyclic phosphodiester product [37]. The NSP15 protein specifically targets and degrades the viral polyuridine sequences to prevent the host immune sensing system from detecting the virus [38]. The crystal structure of NSP15 has been reported for SARS CoV [39] and SARS CoV-2 [40]. NSP15 uses manganese as a cofactor to promote endoribonuclease activity [41]. It has been suggested that NSP15 degrades viral dsRNA to prevent host recognition [42]. The amino acid sequence alignment of NSP15 from SARS CoV and SARS CoV-2 is shown in Fig. 15.
Fig. 15.
The primary amino acid sequence alignment of NSP15 of SARS CoV (NP_828872.1) and SARS CoV-2 (YP_009725310.1). Sequence identity: 88.7%, sequence similarity: 97.7%
NSP16 (2′-O-Ribose-Methyltransferase)
NSP16 for coronavirus has been biochemically [43] (feline coronavirus, FCoV) and structurally [44] (complex of NSP10-NSP16 for SARS CoV) characterized. The viral RNA has a 5′-cap, which protects it from mRNA degradation by 5′-exoribonucleases, promotes mRNA translation, and prevents the viral RNA from being recognized by innate immunity mechanisms [44]. The RNA cap is an N7-methylated guanine nucleotide connected through a 5′-5′ triphosphate bridge to the first transcribed nucleotide (adenine). NSP16 methylates the 2′-hydroxy group of adenine using S-adenosylmethionine as the methyl source. Figure 16 shows the amino acid sequence alignment between the two NSP16 proteins from SARS CoV and SARS CoV-2.
Fig. 16.
The primary amino acid sequence alignment of NSP16 of SARS CoV (NP_828873.2) and SARS CoV-2 (YP_009725311.1). Sequence identity: 93.3%, sequence similarity: 99.0%
Spike Protein (Surface Glycoprotein)
The spike protein (Fig. 17—sequence alignment between SARS CoV and SARS CoV-2) is a glycoprotein, which mediates attachment of the virus to the host cell. The structure of the spike (S) protein has been determined. This protein recognizes the human angiotensin-converting enzyme 2 (ACE2) protein on the host cell surface [45–47]. SARS CoV spike mouse polyclonal antibodies potently inhibited SARS CoV-2 spike protein mediated entry into cells [47]. Interestingly, a furin cleavage site (highlighted in Fig. 17: QTQTNSPRRARSVASQSIIA) was located in the S protein of SARS CoV-2, which was lacking in the S protein of SARS CoV. This difference in site could possibly explain the difference in pathogenicity of these two viruses [47].
Fig. 17.
The primary amino acid sequence alignment of the spike proteins from SARS CoV (NP_828851.1) and SARS CoV-2 (BCA87361.1). Sequence identity: 76.0%, sequence similarity: 91.5%
ORF3a Protein
The ORF3a protein from SARS CoV is an ion channel protein related to NLRP3 inflammasome activation. ORF3a interacts with TRAF3, which in turn activates ASC ubiquitination, and as a result, leads to activation of caspase 1 and IL-1β maturation [48]. The amino acid sequence alignment between the two ORF3a proteins from SARS CoV and SARS CoV-2 is shown in Fig. 18.
Fig. 18.
The primary amino acid sequence alignment of the ORF3a proteins from SARS CoV (NP_828852.2) and SARS CoV-2 (BCA87362.1). Sequence identity: 72.4%, sequence similarity: 90.2%
Envelope Protein
The envelope protein is a small integral membrane protein in coronaviruses, which can oligomerize and create an ion channel [49]. The four structural proteins of coronaviruses are: S protein, M protein, E protein, and N protein [50]. The E protein has been shown to play multiple roles in the viral replication cycle: [1] viral assembly, [51] [2] virion release, [52] and [3] viral pathogenesis [53]. Interestingly, in the sequence alignment of the E proteins from SARS CoV and SARS CoV-2 (Fig. 19), there is a glutamate residue (E69) with a negative charge in SARS CoV that corresponds to a positively charged arginine in SARS CoV-2 (R69).
Fig. 19.
The primary amino acid sequence of the E proteins (ORF4) from SARS CoV (NP_828854.1) and SARS CoV-2 (BCA87363.1). Sequence identity: 94.7%, sequence similarity: 97.4%
Membrane Protein
The SARS coronavirus membrane (M) protein is an integral membrane protein that plays an important role in viral assembly [54]. In addition, the SARS coronavirus M protein has been shown to induce apoptosis [55]. The M protein interacts with the nucleocapsid (N) protein to encapsidate the RNA genome [56]. Figure 20 shows the amino acid sequence alignment of the two ORF5 proteins from SARS CoV and SARS CoV-2.
Fig. 20.
The primary amino acid sequence of the M proteins (ORF5) from SARS CoV (NP_828855.1) and SARS CoV-2 (BCA87364.1). Sequence identity: 90.5%, sequence similarity: 98.2%
ORF6 Protein
The ORF6 protein from SARS coronavirus is an accessory protein that plays an important role in viral pathogenesis [57, 58]. Using a yeast two-hybrid system, ORF6 was shown to interact with NSP8, the nonstructural protein related to promoting RNA polymerase activity [57]. Figure 21 shows the amino acid sequence alignment of the two ORF6 proteins from SARS CoV and SARS CoV-2.
Fig. 21.
The primary amino acid sequence alignment of the ORF6 proteins from SARS CoV (NP_828856.1) and SARS CoV-2 (BCA87365.1). Sequence identity: 68.9%, sequence similarity: 93.4%
ORF7a Protein
ORF7a from SARS coronavirus is an accessory protein that is a type I transmembrane protein and its crystal structure has been determined [59]. Figure 22 shows the amino acid sequence alignment between the two ORF7a proteins of SARS CoV and SARS CoV-2.
Fig. 22.
The primary amino acid sequence of the ORF7a protein from SARS CoV (NP_828857.1) and SARS CoV-2 (BCA87366.1). Sequence identity: 85.2%, sequence similarity: 95.9%
ORF7b Protein
The ORF7b accessory protein from SARS coronavirus is localized in the Golgi compartment [60]. Figure 23 shows the sequence alignment between the two ORF7b proteins of SARS CoV and SARS CoV-2.
Fig. 23.
The primary amino acid sequence of the ORF7b proteins from SARS CoV (NP_849175.1) and SARS CoV-2 (BCB15096.1). Sequence identity: 85.4%, sequence similarity: 97.2%
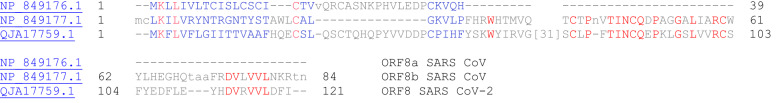
ORF8 Protein
SARS CoV-2 has a single ORF8 protein while SARS CoV has two ORF8 proteins: ORF8a and ORF8b [61]. In SARS CoV, the ORF8b protein binds to the IRF association domain (IAD) region of interferon regulatory factor 3 (IRF3), which in turn inactivates interferon signaling [62]. Interestingly, L84S and S62L missense mutations have been reported in various SARS CoV-2 sequences [5]. Figure 24 shows the alignment between the ORF8 protein of SARS CoV-2 with the ORF8a and ORF8b proteins of SARS CoV.
Fig. 24.
Sequence alignment of ORF8a (NP_849176.1) and ORF8b (NP_849177.1) proteins from SARS CoV (top and middle) with the ORF8 protein (QJA17759.1) from SARS CoV-2 (bottom). Sequence identity and sequence similarity between ORF8a (SARS CoV) and ORF8 (SARS CoV-2): 31.7% and 70.7% in 41 amino acid overlap. Sequence identity and sequence similarity between ORF8b (SARS CoV) and ORF8 (SARS CoV-2): 40.5% and 66.7% in 42 amino acid overlap
Nucleocapsid Protein
The nucleocapsid (N) protein of coronaviruses is a structural protein that binds directly to viral RNA and providing stability [63]. Furthermore, the N protein of SARS CoV-2 (Fig. 24) has been found to antagonize antiviral RNAi [64]. In another study, the nucleocapsid protein of SARS CoV was found to inhibit the activity of cyclin-cyclin-dependent kinase (cyclin-CDK) complex. Inactivation of the cyclin-CDK complex results in hypophosphorylation of the retinoblastoma protein and in turn inhibits S phase (genome replication) progression in the cell cycle [65]. Figure 25 shows the amino acid sequence alignment between the two N proteins of SARS CoV and SARS CoV-2.
Fig. 25.
The primary amino acid sequence of the N protein from SARS CoV (ORF9a, NP_828858.1) and SARS CoV-2 (ORF9, BCA87368.1). Sequence identity: 90.5%, sequence similarity: 97.2%
ORF10 Protein
ORF10 protein from SARS CoV-2 is comprised of 38-amino acids and its function is unknown. Interestingly, SARS CoV possesses an ORF9b protein (NP_828859.1), which is not present in SARS CoV-2. Figure 26 shows the sequence alignment between ORF10 of SARS CoV-2 with ORF9b of SARS CoV. SARS CoV-2 does not have an ORF10 protein. A summary of the sequence identities and similarities of the discussed proteins from SARS CoV and SARS CoV-2 is shown in Table 4.
Fig. 26.
The primary amino acid sequence alignment of the ORF9b protein from SARS CoV and the ORF10 protein from SARS CoV-2 (Accession number: BCA87369.1). Sequence identity: 28.6%, sequence similarity: 52.4%
Table 4.
Sequence identity and similarities between SARS CoV-2 proteins and SARS CoV proteins determined through LALIGN (17) (see Supporting Information)
| Entry | Protein | Amino acid overlap | Sequence identity | Sequence similarity |
|---|---|---|---|---|
| 1 | NSP1 | 180 | 84.4% | 93.4% |
| 2 | NSP2 | 638 | 68.3% | 90.0% |
| 3 | NSP3 | 1,952 | 76.0% | 91.8% |
| 4 | NSP4 | 500 | 80.0% | 95.0% |
| 5 | NSP5 | 306 | 96.1% | 99.7% |
| 6 | NSP6 | 287 | 88.2% | 98.3% |
| 7 | NSP7 | 83 | 98.8% | 100.0% |
| 8 | NSP8 | 198 | 97.5% | 100.0% |
| 9 | NSP9 | 113 | 97.3% | 99.1% |
| 10 | NSP10 | 139 | 97.1% | 99.3% |
| 11 | NSP11 | 13 | 84.6% | 100.0% |
| 12 | NSP12 | 932 | 96.4% | 99.4% |
| 13 | NSP13 | 601 | 99.8% | 100.0% |
| 14 | NSP14 | 527 | 95.1% | 99.1% |
| 15 | NSP15 | 346 | 88.7% | 97.7% |
| 16 | NSP16 | 298 | 93.3% | 99.0% |
| 17 | S protein | 1,277 | 76.0% | 91.5% |
| 18 | ORF3a | 1,381 | 72.4% | 90.2% |
| 19 | E Protein | 76 | 94.7% | 97.4% |
| 20 | M Protein | 222 | 90.5% | 98.2% |
| 21 | ORF6 | 61 | 68.9% | 93.4% |
| 22 | ORF7a | 122 | 85.2% | 95.9% |
| 23 | ORF7b | 41 | 85.4% | 92.7% |
| 24a | (ORF8 vs 8a)a | 41 | 31.7% | 70.7% |
| 24b | (ORF8 vs 8b)a | 42 | 40.5% | 66.7% |
| 25 | N Protein | 422 | 90.5% | 97.2% |
| 26 | (ORF10 vs 9b)a | 21 | 28.6% | 52.4% |
a(SARS CoV-2 protein vs SARS CoV protein). Other reports have also reported amino acid sequence identities using different algorithms (3,67)
Overlapping Genes: ORF9b and Two Proteins with Variation Among SARS CoV-2 Sequences: ORF3b and ORF9c
Overlapping genes in coronavirus have been previously observed [67]. For example, in SARS CoV, the start and end positions in the nucleotide sequence of the N-protein are 28,120 and 29,388 respectively while the ORF9b gene of SARS CoV starts and ends at positions: 28,130 and 28,426 (within the gene sequence of the N-protein) [68]. Similarly, there is a putative ORF9b protein in SARS CoV-2 located within the gene encoding the N-protein, which does not yet have an accession number [4].
In the gene alignment of 2,784 SARS CoV-2 sequences, two variations were recognized in the SARS CoV-2 genome [66]. It was recognized that a premature stop codon at position 14 of ORF3b in SARS CoV-2 in 17.6% of isolates (position E14). Furthermore, there were two mutations that gave rise to premature stop codons in ORF9c (at position Q41 in 0.7% of sequences and at position Q44 in 1.4% of the sequences). The observations of these stop codons suggested that these genes for ORF3b and ORF9c may not be bonafide gene sequences in SARS CoV-2. With the putative SARS CoV-2 ORF3b protein, only 12 out of 57 overlapping amino acid residues were identical (21% sequence identity) to the ORF3b protein of SARS CoV [3]. In the above sections, ORF3b and ORF9c for SARS CoV-2 were not included in the above analysis. Another protein lacking an accession number is ORF14 [69].
Nontranslated (or Untranslated) Regions of SARS CoV-2 Genome
Considering the locations of each gene presented in Table 1, there are regions of the genome that are not translated into proteins, which is related to the non-canonical translational strategy employed by this virus [70]. The nucleotide sequences between the genes are the intergenic regions [71]. For instance, there is a conserved transctiption regulatory sequence (TRS) – a conserved hexanucleotide sequence: (5′-ACGAAC-3′) [71] that could be found in between some of the open reading frames (Table 5, Entries 2, 3, 4, 5, 7, and 9). This particular sequence has previously been identified as the leader-body fusion sites [71]. Furthermore, this sequence is a conserved motif that can be found in subgroup 2b, 2c, and 2d viruses [72]. Another transcriptional regulatory sequence was CUAAAC (e.g. Table 5, Entry 1) [73, 74].
Table 5.
Nontranslated RNA sequence of SARS CoV-2 (NCBI Reference Sequence: NC_045512.2)
| Entry | Location (position) | Sequence |
|---|---|---|
| 1 | Beginning-ORF1ab (1–265) | 1 auuaaagguu uauaccuucc cagguaacaa accaaccaac uuucgaucuc uuguagaucu |
| 61 guucucuaaa cgaacuuuaa aaucugugug gcugucacuc ggcugcaugc uuagugcacu | ||
| 121 cacgcaguau aauuaauaac uaauuacugu cguugacagg acacgaguaa cucgucuauc | ||
| 181 uucugcaggc ugcuuacggu uucguccgug uugcagccga ucaucagcac aucuagguuu | ||
| 241 cguccgggug ugaccgaaag guaag | ||
| 2 | ORF1ab-ORF2 (21,556–21,562) | 1 acgaaca |
| 3 | ORF2-ORF3a (25,385–25,392) | 1 acgaacuu |
| 4 | ORF3a-ORF4 (26,221–26,244) | 1 gcacaagcug augaguacga acu |
| 5 | ORF4-ORF5 (26,473–26,522) | 1 acgaacuaaa uauuauauua guuuuucugu uuggaacuuu aauuuuagcc |
| 6 | ORF5-ORF6 (27,192–27,201) | 1 gugacaacag |
| 7 | ORF6-ORF7a (27,388–27,393) | 1 acgaac |
| 8 | ORF7b-ORF8 (27,888–27,893) | 1 acgaac |
| 9 | ORF8-ORF9 (28,260–28,273) | 1 acgaacaaac uaaa |
| 10 | ORF9-ORF10 (29,534–29,557) | 1 acucaugcag accacacaag gcag |
| 11 | ORF10-end (29,675–29,903) | 1 caaucuuuaa ucagugugua acauuaggga ggacuugaaa gagccaccac auuuucaccg |
| 61 aggccacgcg gaguacgauc gaguguacag ugaacaaugc uagggagagc ugccuauaug | ||
| 121 gaagagcccu aauguguaaa auuaauuuua guagugcuau ccccauguga uuuuaauagc | ||
| 181 uucuuaggag aaugacaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaa |
Exploration of Treatment Options for COVID-19
An intense effort has been put forth to discover potential treatment options for COVID-19, the disease caused by SARS CoV-2 [75–77]. For instance, the FDA approved drug, ivermectin, is known to inhibit nuclear transport, and has been shown to inhibit the replication of SARS CoV-2 [78]. Other drugs have been repurposed and tested against COVID-19 [79, 80]. Remdesivir is a potential antiviral drug originally developed to treat ebola [81] and has been used to treat COVID-19 [82] by inhibiting viral RNA polymerase activity. Hydroxychloroquine [83] and chloroquine [84] have been used to potentially treat COVID-19. However, the use of these drugs has been known to result in cardiotoxicity [85, 86]. In fact, in a recent observational study, it was determined that hydroxychloroquine administration was not associated with a greatly lowered risk of death from COVID-19 [87].
A recent study identified 332 human proteins that interact with SARS CoV-2 proteins [66]. In this report, the predicted SARS CoV-2 proteins (NSPs 1–16 and ORFs) were expressed with 2xstreptavidin affinity tags. These tagged SARS CoV-2 proteins were expressed in human embryonic kidney (HEK)293T/17 cells and isolated the viral protein-(human protein) interactions using affinity purification-mass spectrometry. A total 332 protein–protein interactions (PPIs between SARS CoV-2 proteins and human proteins) were identified. Of these PPIs, 66 of them are targetable by compounds. Table 6 shows a set of compounds that target the identified PPIs based on chemoinformatics (entries 1–28) or expertise knowledge (entries 29–44). From the subset of potential antiviral compounds that were tested, two classes of compounds were found to be effective against viral pathogenesis: [1] protein translation inhibitors (i.e. zotatifin, ternatin-4, and PS3061), and [2] Sigma1 and Sigma2 receptor ligands (i.e. approved drugs: clemastine, cloperastine, and progesterone and PB28, which was ~ 20 times more potent than hydroxychloroquine with an IC90 of 280 nM in the viral titer assay is undergoing pre-clinical trials for anti-cancer [88] activity).
Table 6.
Drugs that potentially target (modulate) proteins that interact with SARS CoV-2 proteins as described in reference [66]
| Entry | Viral Protein-(Human Gene) | Compound Name(s) |
|---|---|---|
| 1 | E protein-(BRD2/4) | JQ1,a RVX-208b |
| 2 | N protein-(CSNK2A2) | Silmitasertib (cancer),c TMCBa |
| 3 | NSP5-(HDAC2) | Apicidin,a Valproic acid (CNS disease, cancer)c |
| 4 | NSP6-(ATP6AP1) | Bafilomycin A1a |
| 5 | NSP6-(SIGMAR1) | E-52862,b PD-144418,a RS-PPCC,a PB28,a |
| Haloperidol (CNS disease)c | ||
| 6 | NSP6-(SLC6A15) | Loratadine (antihistamine)3 |
| 7 | ORF9C-(TMEM97) | PB28,a haloperidol (CNS disease)c |
| 8 | M protein-(ATP6V1A) | Bafilomycin A1a |
| 9 | NSP7-(COMT) | Entacapone (Parkinson’s disease)c |
| 10 | NSP7-(PTGES2) | Indomethacin (inflammation/pain)c |
| 11 | NSP7-(NDUFs) | Metformin (diabetes)c |
| 12 | ORF9C-(NDUFs) | Metforminc |
| 13 | NSP12-(RIPK1) | Ponatinib (cancer)c |
| 14 | NSP13-(PRKACA) | H-89a |
| 15 | NSP14-(IMPDH2) | Merimepodibb |
| 16 | NSP14-(GLA) | Migalastat (Fabry disease)c |
| 17 | NSP14-(IMPDH2) | Mycophenolic acid (organ rejection),3 ribavirin (virus)c |
| 18 | ORF8-(DNMT1) | Azacitidinec |
| 19 | ORF8-(LOX) | CCT 365623a |
| 20 | ORF9b-(MARK2/3) | Midostaurin,3 Ruxolitinibc |
| 21 | ORF9b-(DCTPP1) | ZINC1775962367,a ZINC4326719,a ZINC4511851a |
| 22 | ORF9b/NSP13-(MARK3/TBK1) | ZINC95559591a |
| 23 | ORF9C-(F2RL1) | AC-55541,a AZ8838a |
| 24 | ORF9C-(ABCC1) | Daunorubicinc |
| 25 | ORF9C-(F2RL1) | GB110a |
| 26 | ORF9C-(ABCC1) | S-Verapamil (hypertension)c |
| 27 | ORF9C-(F2RL1) | AZ3451a |
| 28 | M-Protein-(SLC1A3) | UCPH-101a |
| 29 | E protein-(BRD2/4) | ABBV-744,b dBET6,a MZ1,a CPI-0610b |
| 30 | N protein-(LARP1) | Sapanisertib,b Rapamycin (organ rejection)c |
| 31 | NSP2-(FKBP15) | Rapamycinc |
| 32 | ORF8-(FKBP7/10) | Rapamycinc |
| 33 | NSP2-(EIF4E2/H) | Zotatifinb |
| 34 | ORF10-(VCP) | CB5083b |
| 35 | NSP6-(SIGMAR1) | Chloroquine (malaria)c |
| 36 | NSP9-NEK9 | Dabrafenaib (cancer)c |
| 37 | NSP13-CEP250 | WDB002b |
| 38 | NSP14-IMPDH2 | Sanglifehrin Aa |
| 39 | ORF8-(FKBP7) | FK-506 (organ rejection)c |
| 40 | ORF8-(FKBP10) | FK-506c |
| 41 | ORF10-(CUL2) | Pevonedistatb |
| 42 | ORF10-(VCP) | DBeQ, ML240a |
| 43 | ORF8-(PLOD1/2) | Minoxidil (hair loss)c |
| 44 | NSP4/9/ORF6-(NUPs RAE1) | Selinexor (cancer)c |
Entries 1–28 were determined from chemoinformatics. Entries 29–44 were determined from specialist knowledge
aPre-clinical
bClinical trial
cFDA-approved drug. In parentheses after the drug is what the FDA-approved drug is used to treat in the clinic
Moreover, in another collaborative study, a library of 12,000 FDA-approved or clinical-stage drugs were tested against SARS CoV-2 infection in Vero-E6 (African green monkey kidney) cells [89]. Some effective compounds identified in the screen were: PIKfyve kinase inhibitor Apilimod, cysteine protease inhibitors (MDL-28170, Z LVG CHN2, VBY-825, and ONO 5334), and MLN-3897 (a CCR1 antagonist).
Traditional Chinese Medicine (TCM) has also been employed in China to treat COVID-19 [90]. However, due to potential toxic components present in TCM remedies, [91, 92] the use of this strategy should be handled with caution [93]. Ironically, it has been suggested that TCM could have potentially been the cause of COVID-19 [94].
In addition to small molecules, vaccines are also currently being developed against SARS CoV-2, [95] and convalescent plasma transfusions have been used to treat COVID-19 [96]. Nevertheless, more research is needed to develop effective treatments against SARS CoV-2 especially in the context of future outbreaks [97, 98].
Conclusion
Although there is some variation in sequence in the proteins, many of the proteins found in SARS CoV-2 (NC_045512.2) are also found in SARS CoV (AY515512.1 or NC_004718.3) with 77.1% of the protein sequences shared in their proteomes [99]. Thus, previous research on related coronavirus proteins enable a better understanding of how we can approach to understand the current coronavirus (SARS CoV-2) that caused the current global pandemic (COVID-19). The general structures of most of the proteins from SARS CoV-2 can be visualized from homology models [100]. Advances in the knowledge of the structures and functions of the proteins in SARS CoV-2 will enable researchers to design better antiviral drugs that target this virus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Francis K. Yoshimoto, PhD holds a Voelcker Fund Young Investigator Award from the MAX AND MINNIE TOMERLIN VOELCKER FUND.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li X, Zai J, Wang X, Li Y. Potential of large “first generation” human-to-human transmission of 2019-nCoV. J Med Virol. 2020;92:448–454. doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, Yuen K-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Liu Z, Chen Z, Huang X, Xu M, He T, Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khailany RA, Safdar M, Ozaslan M. Genomic characterisation of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–455. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y-H, Hu C-Y, Wu N-P, Yao H-P, Li L-J. Molecular characteristics, functions, and related pathogenicity of MERS-CoV proteins. Engineering. 2019;5:940–947. doi: 10.1016/j.eng.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. From SARS to MERS thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calligari P, Bobone S, Ricci G, Bocedi A. Molecular investigation of SARS-CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu DX, Fung TS, Chong KK-L, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prajapa M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, Kumar S, Bhattacharyya A, Kumar H, Bansal S, Medhi B. Drug targets for corona virus: a systematic review. Indian J Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cagliani R, Forni D, Clerici M, Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus, SARS-CoV-2. J Virol. 2020 doi: 10.1128/JVI.00411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornillez-Ty CT, Liao L, Yates JR, Kuhn P, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai Y, Kawachi K, Terada Y, Omori H, Matsuura Y, Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baez-Santos YM, St. John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function, and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomar S, Johnston ML, St. John SE, Osswald HL, Nyalapatla PR, Paul LN, Ghosh AK, Denison MR, Mesecar AD. Ligand-induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro) implications For nsp5 Regulation And The Development Of Antivirals. J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;13:e00524–e1513. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.te Velthuis AJ, van de Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Ge X, Wang X, Liu A, Guo X, Zhou L, Yu K, Yang H. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductiv e and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. 2015;195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Wu L, Shaw N, Gao Y, Wang J, Sun Y, Lou Z, Yan L, Zhang R, Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Sun Y, Wu A, Xu S, Pan R, Zeng C, Jin X, Ge X, Shi Z, Ahola T, Guo D. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J Virol. 2015;89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, Snijder EJ, Canard B, Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang K-J, Jeong S, Kang DY, Sp N, Yang YM, Kim D-E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case JB, Ashbrook AW, Dermody TS, Denison MR. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J Virol. 2016;90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin X, Chen Y, Sun Y, Zeng C, Wang Y, Tao J, Wu A, Yu X, Zhang Z, Tian J, Guo D. Characterization of the guanine-N7 methyltransferase activity of coronavirus nsp14 on nucleotide GTP. Virus Res. 2013;176:45–52. doi: 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa K, Lokugamage KG, Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci USA. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj K, Palaninathan S, Ortiz Alcantara JM, Li Yi L, Guarino L, Sacchettini JC, Cheng Kao C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, Michalska K, Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78:12218. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng X, Baker SC. An “Old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163. doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B. Coronavirus nonstructural protein 16 Is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. Crystal structure and functional analysis of the sars-coronavirus RNA cap 2’-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5s. [DOI] [PubMed] [Google Scholar]
- 46.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Vessler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siu K-L, Yuen K-S, Castano-Rodriguez C, Ye Z-W, Yeung M-L, Fung S-Y, Yuan S, Chan C-P, Yuen K-Y, Enjuanes L, Jin D-Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdia-Baguena C, Nieto-Torres JL, Alcaraz A, DeDiego ML, Torres J, Aguilella VM, Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim KP, Liu DX. The Missing Link in Coronavirus Assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-golgi compartments and physical interaction between the envelope and membrane proteins. J Biol Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsoi H, Li L, Chen ZS, Lau K-F, Tsui SKW, Chan HYE. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1-PKB/Akt signalling. Biochem J. 2014;464:439–447. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- 56.Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, Tsao SW, Nicholls JM, Altmeyer R, Peiris JSM, Bruzzone R, Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus re required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar P, Gunalan V, Liu B, Chow VTK, Druce J, Birch C, Catton M, Fielding BC, Tan Y-J, Lal SK. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366:293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao J, Falcon A, Zhou H, Netlan J, Enjuanes L, Brena PP, Perlman S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J Virol. 2009;83:2368–2373. doi: 10.1128/JVI.02371-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson CA, Pekosz A, Lee CA, Diamond MS, Fremont DH. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le TM, Wong HH, Tay FPL, Fang S, Keng C-T, Tan YJ, Liu DX. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007;274:4211–4222. doi: 10.1111/j.1742-4658.2007.05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grunewald ME, Fehr AR, Athmer J, Perlman S. The coronavirus nucleocapsid protein is ADP-ribosylated. Virology. 2018;517:62–68. doi: 10.1016/j.virol.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y, Qiu Y, Zhou X. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 2020;63:10. doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Surjit M, Liu B, Chow VTK, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Huettenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGrego MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang X-P, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu D, Wang H-Y, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, Garcia-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shukla A, Hilgenfeld R. Acquisition of new protein domains by coronaviruses: analysis of overlapping genes coding for proteins N and 9b in SARS coronavirus. Virus Genes. 2015;50:29–38. doi: 10.1007/s11262-014-1139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hussain S, Pan J, Chen Y, Yang Y, Xu J, Peng Y, Wu Y, Li Z, Zhu Y, Tien P, Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quan P-L, Firth C, Street C, Henriquez JA, Petrosov A, Tashumukhamedova A, Hutchison SK, Egholm M, Osinubi MOV, Niezgoda M, Ogunkoya AB, Briese T, Rupprecht CE, Lipkin WI. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MmBio. 2010;1:00208–00210. doi: 10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang D, Leibowitz JL. The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jean S-S, Lee P-I, Hsueh P-R. Treatment options for COVID-19: The reality and challenges. J Microbiol. 2020 doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr Med Chem. 2020;27:32297571. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 77.Zeng Q-L, Yu Z-J, Gou J-J, Li G-M, Ma S-H, Zhang G-F, Xu J-H, Lin W-B, Cui G-L, Zhang M-M, Li C, Wang Z-S, Zhang Z-H, Liu Z-S. Effect of convalescent plasma therapy on viral shedding and survival in COVID-19 patients. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esposito S, Noviello S, Pagliano P. Update on treatment of COVID-19: ongoing studies between promising and disappointing results. Le Infezioni Med. 2020;2:198–211. [PubMed] [Google Scholar]
- 80.Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 81.Siegel D, Doerffler E, Clarke MO, Chun K, Zhang L, Neville S, Carra E, Lew W, Ross B, Wang Q, Wolfe L, Jordan R, Soloveva V, Knox J, Perry J, Perron M, Stray KM, Barauskas O, Feng JY, Xu Y, Lee G, Rheingold AL, Ray AS, Bannister R, Strickley R, Swaminathan S, Lee WA, Bavari S, Cihlar T, Lo MK, Warren TK, Mackman RL. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f ][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J Med Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 82.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020 doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyerowitz EA, Vannier AGL, Friesen MGN, Schoenfeld S, Gelfand JA, Callahan MV, Kim AY, Reeves PM, Poznansky MC. Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB J. 2020;34(5):6027–6037. doi: 10.1096/fj.202000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Touret F, de Lamballerie X. Of Chloroquine and COVID-19. Antiviral Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong YK, Yang J, He Y. Caution and clarity required in the use of chloroquine for COVID-19. Lancet Rehumatol. 2020;2:255. doi: 10.1016/S2665-9913(20)30093-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Joyce E, Fabre A, Mahon N. Hydroxychloroquine cardiotoxicity presenting as a rapidly evolving biventricular cardiomyopathy: key diagnostic features and literature review. Eur Heart J. 2012;2:77–83. doi: 10.1177/2048872612471215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geleris J, Sun Y, Platt J, Zucker J, Baldwin M, Hripcsak G, Labella A, Manson D, Kubin C, Barr RG, Sobieszczyk ME, Schluger NW. Obersvational study of hydroxychloroquine in hospitalized patients with Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pati ML, Hornick JR, Niso M, Berardi F, Spitzer D, Abate C, Hawkins W. Sigma-2 receptor agonist derivatives of 1-cyclohexyl-4-[3-(5-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl)propyl]piperazine (PB28) induce cell death via mitochondrial superoxide production and caspase activation in pancreatic cancer. BMC Cancer. 2017;17:51. doi: 10.1186/s12885-016-3040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Burgstaller-Muehlbacher S, Pache L, De Jesus PP, Hull MV, Chang M, Chan JF-W, Cao J, Poon VK-M, Herbert K, Nguyen T-T, Pu Y, Nguyen C, Rubanov A, Martinez-Sobrido L, Liu W-C, Miorin L, White KM, Johnson JR, Benner C, Sun R, Schultz PG, Su A, Garcia-Sastre A, Chatterjee AK, Yuen K-Y, Chanda SK. A Large-scale drug repositioning survey for SARS-CoV-2 antivirals. bioRxiv. 2020 doi: 10.1101/2020.04.16.044016. [DOI] [Google Scholar]
- 90.Xu J, Zhang Y. Traditional Chinese Medicine treatment of COVID-19. Complement Ther Clin Pract. 2020;39:101165. doi: 10.1016/j.ctcp.2020.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C-H, Dickman KG, Moriya M, Zavadil J, Sidorenko VS, Edwards KL, Gnatenko DV, Wu L, Turesky RJ, Wu X-R, Pu Y-S, Grollman AP. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci USA. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duan L, Guo L, Wang L, Yin Q, Zhang C-M, et al. Application of metabolomics in toxicity evaluation of traditional Chinese medicines. Chin Med. 2018;13:60. doi: 10.1186/s13020-018-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lv W, Piao J-H, Jiang J-G. Typical toxic components in traditional Chinese medicine. Expert Opin Drug Saf. 2012;11:985–1002. doi: 10.1517/14740338.2012.726610. [DOI] [PubMed] [Google Scholar]
- 94.Wassenaar TM, Zou Y. 2019_nCoV/SARS-CoV-2: rapid classification of betacorona viruses and identification of Traditional Chinese Medicine as potential origin of zoonotic coronaviruses. Lett Appl Microbiol. 2020;70:342–348. doi: 10.1111/lam.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lurie N, Saville M, Hatchett R, Halton J. Developing Covid-19 vaccines at pandemic speed. N Engl J Med. 2020 doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 96.Brown BL, McCullough J. Treatment for emerging viruses: convalescent plasma and COVID-19. Transfus Apher Sci. 2020 doi: 10.1016/j.transci.2020.102790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu S, Li Y. Beware of the second wave of COVID-19. Lancet. 2020;395:1321–1322. doi: 10.1016/S0140-6736(20)30845-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leung K, Wu JT, Liu D, Leung GM. First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. Lancet. 2020;395:1382–1393. doi: 10.1016/S0140-6736(20)30746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.https://swissmodel.expasy.org/repository/species/2697049. Accessed 26 Apr 2020, 6:05 AM
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.