Abstract
Efficient translational bypassing of a 50-nt non-coding gap in a phage T4 topoisomerase subunit gene (gp60) requires several recoding signals. Here we investigate the function of the mRNA stem–loop 5′ of the take-off codon, as well as the importance of ribosome loading density on the mRNA for efficient bypassing. We show that polysomes are less efficient at mediating bypassing than monosomes, both in vitro and in vivo, due to their preventing formation of a stem–loop 5′ of the take-off codon and allowing greater peptidyl-tRNA drop off. A ribosome profiling analysis of phage T4-infected Escherichia coli yielded protected mRNA fragments within the normal size range derived from ribosomes stalled at the take-off codon. However, ribosomes at this position also yielded some 53-nucleotide fragments, 16 longer. These were due to protection of the nucleotides that form the 5′ stem–loop. NMR shows that the 5′ stem–loop is highly dynamic. The importance of different nucleotides in the 5′ stem–loop is revealed by mutagenesis studies. These data highlight the significance of the 5′ stem–loop for the 50-nt bypassing and further enhance appreciation of relevance of the extent of ribosome loading for recoding.
Keywords: NMR spectroscopy, structure, ribosome profiling, translational bypassing, recoding
Abbreviations: ORF, open reading frame; smFRET, single-molecule fluorescence resonance energy transfer; cryo-EM, cryogenic electron microscopy; SD, Shine–Dalgarno; SHAPE, Selective 2′-Hydroxyl Acylation analyzed by Primer Extension; RNase, ribonuclease; NOESY, nuclear Overhauser enhancement spectroscopy; COSY, correlated spectroscopy
Graphical abstract

Highlights
-
•
Monosomes are more efficient than polysome in mediating 50-nt translational bypassing.
-
•
A 5′ mRNA stem–loop facilitates translational bypassing by monosomes.
-
•
Ribosome profiling yields an extra-long, 53-nt, protected fragment of mRNA.
Introduction
The most striking exception known to non-overlapping sequential triplet decoding is in a phage T4 gene next to that used by Crick et al [1]. to establish the general nature of genetic readout. This nearby gene, gene 60, derives from insertion of a mobile DNA cassette consisting of a homing endonuclease gene and an associated separate 50-nt sequence that provides protection against self-cleavage [2]. The insert occurred into an ancestral phage T4 topoisomerase encoding gene. The inserted endonuclease gene split the original gene into two genes. Both genes are functional despite the 3′ gene having a 50-nt insert between codons 46 and 47 of its coding sequence [3]. The insert has stop codons in all frames, suggesting that translation of this sequence would result in a prematurely terminated protein. Studies with plasmid-borne cassettes showed that in Escherichia coli grown on rich media, a substantial proportion of translating ribosomes successfully bypass the 50-nt coding gap to synthesize a single protein from two discontinuous open reading frames (ORFs) [4,5]. Moreover, bypassing can also occur with rare codons in highly expressed genes upon heterologous expression [6], with unassigned codons [7], and even in unstarved cells [8]. Limitation of aminoacyl-tRNA for a “hungry” A-site sense codon induces low-level bypassing [9., 10., 11.]. Interestingly, abundant translational bypassing is productively utilized in mitochondrial decoding in certain yeasts [12,13]. Protein sequence data have shown the absence of amino acids specified by 29 nt within the coding sequence of an adhesion gene of the oral bacterium Prevotella loescheii, and much indirect evidence points to the involvement of translational bypassing [14]. The types of whole (both subunit) ribosome bypassing considered above are very different from that involved in ribosome shunting [15,16].
Gene 60 of bacteriophage T4 is the best-studied example of translational bypassing. The high reading efficiency of these two discontiguous ORFs encoding a single-polypeptide chain is achieved by the presence of a number of recoding signals [17,18]. In WT bypassing, the anticodon of peptidyl-tRNAGly 2 [19] dissociates from codon 46, GGA, the take-off codon, and re-pairs to mRNA at the matched “landing” codon GGA, 5′adjacent to the resume codon 47 [4,20]. Nucleotide position numbers counted 3′ from the take-off codon have the prefix “§” (Figure 1 ). As shown by both single-molecule fluorescence resonance energy transfer (smFRET) studies with zero-mode waveguides, and cryogenic electron microscopy (cryo-EM) studies, when the GGA codon is in the P site, the mRNA in the A site folds into a short stem–loop capped by a UUCG tetraloop (§6–9), termed “A-site SL” (§3–12) [21., 22., 23.]. UUCG has the propensity to form a tetraloop of unusual stability and compactness [24., 25., 26., 27.]. Included in the A-site SL is the third nucleotide, G, of the codon, UAG, 3′ adjacent to the take-off GGA, codon 46. Formation of this stem–loop within the A-site prevents access of release factor 1 and tRNAs near-cognate to the UAG [22] and is consistent with earlier results showing WT levels of release factor 1 do not mediate termination at the UAG [4,20].
Figure 1.
Bacteriophage T4 gene 60 translational bypassing. A compilation of the key regulatory features involved in gene 60 bypassing including: (1) the peptidyl-tRNAGly2 and matched take-off and landing GGA codons (dark green), (2) the UAG stop codon immediately 3′ to the take-off codon (red), (3) an upstream nascent peptide signal (KKYK-LQNNVRRSIKSSS14–30), (4) the 5′ SL, 9-nt 5′ of the take-off codon, (5) the A-site SL with the potential to form an extended version (light green), (6) the stop codons within the coding gap (red underline), (7) a Shine–Dalgarno-like sequence GAG (blue) 6-nt 5′ of the landing codon, (8) the translational resume codon UUA (purple), (9) the nucleotide sequence where ribosomes slow down translation (gold dotted line), (10) the forward slippage barrier, 14-nt 3′ of the landing codon. The common 3′ end of both the large and smaller ribosome profiling fragments obtained at the take-off site is indicated (brown line).
Formation of the A-site SL and the interactions of the nascent peptide with the walls of the polypeptide exit tunnel (see below) facilitate the formation of the non-canonical rotation state of the ribosome, a hyper-rotated state, that initiates bypassing [21,23]. The A-site SL also serves as a tRNA mimic for the recruitment of EF-G and a pseudotranslocation event that initiates bypassing [23]. Following release of the tRNA anticodon from the take-off codon, the ribosome moves over the 5′ part of the coding gap without anticodon scanning of the mRNA for potential complementarity [21,28]. This initial absence of anticodon scanning explains why peptidyl-tRNAGly 2 does not recognize the cognate GGG §9–11 triplet in the A-site SL [28]. Subsequently, continued forward progression toward the 3′ end of the coding gap does involve peptidyl-tRNA anticodon scanning of the mRNA for potential complementarity [21]. [Such scanning makes the mechanism distinct from that described for intact 80s ribosomes resuming translation 3′ of stop codons under stress conditions [29].]
In addition to the A-site SL, another crucial feature for the bypassing is a nascent peptide sequence encoded 5′ of the take-off site [4,20., 21., 22.,30,31]. Changes in the nascent peptide starting from the crucial 14KKYK17 motif resulted in substantial reduction in bypassing efficiency. In vitro studies that systematically scanned the effect of nascent peptide mutations have shown that mutations of residues 14–30 in the nascent peptide sequence reduce bypassing efficiency by 2- to 20-fold [23,31]. The nascent peptide adopts an α-helical conformation and forms multiple interactions with both rRNA and protein components of the interior of the peptide exit tunnel of the ribosome [22]. Nascent peptide-exit tunnel interactions cause progressive ribosome slowing, as the ribosome approaches the take-off codon, enabling ribosomes to adopt an unusual hyper-rotated conformation prior to bypassing [21,23]. The nascent peptide interaction also helps the ribosome to retain peptidyl-tRNA during bypassing [31,32] and serves to increase the accuracy of peptidyl-tRNA re-pairing to mRNA [20,33].
Toward the end of gene 60 bypassing, peptidyl-tRNA re-pairing at the matched landing codon is influenced by the mini Shine–Dalgarno (SD)-like sequence GAG 6-nt 5′ of the landing codon that can pair to the anti-SD sequence in 16S rRNA [28]. Perhaps significantly, this is flanked by A's [34,35]. Landing is also facilitated by an mRNA structure; here termed a forward slippage barrier, 3′ of the landing codon [31]. Translation resumes at the 3′ adjacent “resume codon” (Figure 1) with binding of aa-tRNA to the ribosome in the rotated state [21,23] and then continuing normal translation.
The signals just described conspire to make the initiation of bypassing highly efficient and to overcome the strength of codon:anticodon pairing at the take-off site, which does not affect take-off efficiency [36]. In vivo studies showed that despite the high efficiency of take-off, the overall bypassing efficiency is lower, due to drop-off of peptidyl-tRNA from the ribosome [32]. Quantification in vitro of bypassing efficiency as a function of the gap length indicated that peptidyl-tRNA is progressively lost as the ribosome moves along the non-coding mRNA, but all ribosomes that maintain their P-site tRNA can land [31]. Addition to the 3′ part of the gap of sequence with strong potential to form an SL reduced the amount of product derived from successful bypassing compared to insertion of sequence of the same length without structure potential [31].
The nucleotides that pair to form the A-site stem–loop also pair later to form the initial component of an extended stem–loop (“Extended SL”) that also involves its flanking nucleotides (highlighted in green in Figure 1). Though mutants of its lower part do not affect the early part of bypassing including the rotated state pause [21], it is functionally important for bypassing [28].
A further stem–loop is important for bypassing. It was identified in the course of in vitro studies and because of its location named the 5′ SL (Figure 1) [21,31]. The 5′ SL was suggested to form upon exit from the ribosome mRNA tunnel [31], and there is some evidence for its refolding from the cryo-EM structure [22]. However, mutations of the 5′ SL were not noted as being significant in the assays performed to date in E. coli cells. Interestingly, SL elements are found 5′ of the putative recoding sites in several other cases of recoding, for example, in two Streptomyces phages [37], (O'Loughlin S., et al., unpublished). Similar nucleotide features are present in diverse Streptomyces species [17]. Work in E. coli with a model system has shown that a synthetic SL 5′ of a frameshift site can inhibit − 1 frameshifting and so “backwards” ribosome movement, but promote + 1 frameshifting [38]. The number of nucleotides from the top of the 3′ side of the stem–loop tested to the recode site was much more similar to that of its gene 60 5′SL counterpart, being only 5 nt less, than the equivalent of the hypothesized Streptomyces phage bypassing 5′ SL mentioned above.
Previous in vitro studies clearly identify the 5′ SL as an important facilitator of bypassing, but the question still remains why its effect has not been evident in in vivo studies performed to date. At one time, it was plausible to consider that progression of a bypassing ribosome through the coding gap involved “pushing” by continued translation of a following ribosome. However, in vitro studies showed efficient bypassing under monosome conditions [21,31], where formation of the 5′ SL may have served to favor movement of the initial part of the coding gap through the ribosome [31]. Nevertheless, the bypassing efficiency may be influenced by level of ribosome loading on mRNA; in in vivo studies to date, forward movement could be generated by continuing translation by a following ribosome possibly substituting for the effect of the 5′ SL. The experiments reported here address the nature of the 5′ SL and its significance as well as the relative efficiency of bypassing during high (polysome) and low ribosome loading conditions.
Results
The major take-off site pause facilitates ribosome profiling detection of 5′ SL sequence
To determine the extent of gene 60 mRNA protected by the ribosome, ribosome profiling of T4 infected E. coli was carried out. Micrococcal nuclease (MNase) was used to cleave mRNA unprotected by the ribosome. Because the A-site SL is formed within bypassing ribosomes [22], an increased length of the relevant ribosome protected mRNA fragment might be expected. Monosomes generated by MNase digestion [39,40] were collected from a sucrose gradient. RNAs recovered from the monosomes fraction were size separated by electrophoresis on a polyacrylamide gel. RNAs of 30–40 nt, as well as larger fragments of 45–140 nt, were excised. For only partly understood reasons, in addition to the significant but occasional contribution of interactions involving internal SD sequences [41], protected fragments in bacterial ribosome profiling experiments are often longer and more variable in length than counterparts obtained with larger eukaryotic ribosomes [42].
We performed single read sequencing for the smaller fragments and paired-end sequencing of the longer fragments. As previously reported [43], the T4 bacteriophage almost completely hijacks the host translational machinery. Of the 30- to 40-nt fragments over 1.8 million aligned to E. coli genes and 11 million mapped to T4 genes with 108,000 aligning to gene 60. Around 3.2 million 45- to 145-nt fragments aligned to E. coli genes, 2.4 million of which aligned to tmRNA. One hundred forty-four thousand reads mapped to the genes of T4, with 4311 of these reads aligning to gene 60.
For gene 60, the 30- to 40-nt fragments revealed an extraordinary high density (200 times greater than the gene average) whose 5′ end was 24-nt 5′ of the start of the coding gap. Based on their 5′ ends, it can be inferred that these protected fragments derive from ribosomes whose P-site was within a few nucleotides 5′ of the take-off site (because of the vagaries of cleavage with the micrococcal nuclease used to generate the protected fragments, P-site location could not be precisely predicted). The great majority of these fragments had 13-nt 3′ of the start of the coding gap (U of the UAG stop codon is designated as 0 and counted as 1) consistent with the P-site being located at, or very near, the take-off site (Figure 2(a), top, and Figure 2(b)). Ribosome queuing was not addressed since only monosome-protected mRNA fragments were analyzed (even with high ribosome numbers, ribosome spacing was apparently not tight enough to prevent cleavage). In contrast, the protected fragments detected at the 3′ end of the coding gap were much less numerous. One interpretation of this is that the subsequent re-pairing and the next decoding event are comparatively quick. However, smFRET studies provided direct contrasting evidence [21] raising the possibility that an alternative event, such as increased ribonuclease (RNase) accessibility to the A-site, may be related to the relative paucity of 3′ protected fragments (see Discussion) (Figure 2(b)).
Figure 2.
Gene 60 ribosome profiling of phage T4 infected E. coli. (a) Ribosome protected mRNA fragments (RPFs) isolated from phage T4 infected E. coli cells were fractionated based on size. Boundaries for the smaller sized 30–40 nt RPFs (top) and the larger 45–150 nt RPFs (bottom) are shown. The U nucleotide of the UAG stop codon positioned 3′ of the take-off codon (red on Figure 2(b)) is designated as zero (0), and the 5′ and 3′ boundary of the 50-nt coding gap is indicated (blue dashed lines). (b) The position of the ribosome-protected mRNA fragments recovered in the coding gap, and associated regions of gene 60 are indicated. The smaller RPFs (blue overline) extend 13 nt into the coding gap and include 24-nt 5 ′ of the coding gap. The large RPFs (white text in gray) include 40-nt 5 ′ of the coding gap. It encompasses nucleotides that form the 5′ stem–loop and also extends 13 nt into the coding gap. If MNase digestion occurs in the A-site, then prior to this cleavage, additional 3′ sequence would have been present in the ribosome (3′ sequence shown without highlighting).
Approximately 65% of all of the 45- to 145-nt-long fragments aligning to gene 60 mapped to a single position (and no other single position has many reads mapping to it) (Figure 2(a) bottom). While both the 5′ and 3′ ends of the long fragments aligning at this position display slight variability, most are of one length, 53 nt. This fragment consists of 37-nt 5′ and 13-nt 3′of the (3-nt) take-off site. The 5′ extension incorporates the nucleotide sequence of the 5′ SL (Figure 2(b)). Ribosome profiling of a plasmid-borne gene 60 cassette in E. coli cells uninfected by phage T4 was also performed. The WT gene 60 cassette again yielded just one long protected fragment and similar to the size and sequence as that recovered from phage T4 infected cells (− 41, + 12) (Figure S1).
Figure S1.

Ribosome profiling of E. coli containing a plasmid-borne cassette with the gene 60 sequence. Ribosome-protected mRNA fragments (RPFs) isolated from induced E. coli cells were fractionated based on size. Boundaries for the larger 45–150 nt RPFs are shown. The U nucleotide of the UAG stop codon positioned 3′ of the take-off codon (red) is designated as zero (0) and the 5′ and 3′ boundary of the 50-nt coding gap is indicated (blue dashed lines). The position of the ribosome-protected mRNA fragments recovered in the coding gap and associated regions of gene 60 are indicated. The large RPFs include 41-nt 5′ of the coding gap (green peak). It includes nucleotides that form the 5′ SL and extends 13-nt 3′ into the coding gap (red peak).
5′ SL RNase digestion assays
The secondary structure of the 5′ SL, as deduced from Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE), is composed of two helices, a 4-bp bottom helix and 5-bp upper helix, connected by a 4-nt internal loop and capped by a 6-nt terminal loop (Figure 2(b)) [26]. In this study, the structure of the 5′ SL was explored further through direct measurements in order to understand better how this stem–loop could interact with the translational machinery.
RNase digestion can be a powerful tool for probing secondary structure, because RNases are specific for single- or double-stranded RNA. Thus, paired bases and nucleotides in loops are recognized distinctly. The 5′ SLx construct was designed for RNase digestion assays. 5′ SLx extends three bases 3′ and 5′ of the 5′ SL to include the 34-nt sequence from C96 to U129 with a Cy3 fluorescent label on C96 (Figure S2A). After digestion, only those fragments containing C96 would be detectable on a denaturing gel. Two RNases were chosen for these assays: RNase A, which hydrolyzes the phosphodiester bond 3′ of single-stranded pyrimidine nucleotides, and RNase T1, which targets phosphodiester bonds 3′ of single-stranded guanosines. When 1 ng/μl 5′ SLx was incubated with 0.1 ng/μl RNase A, three fragments were resolved by denaturing gel electrophoresis (Figure S2B). These fragments had approximate lengths of 34, 18, and 15 nt, which represented the full-length 5′ SLx, the product of hydrolysis at U113 or U114, and the product of hydrolysis at C110 or U111, respectively. The absence of other fragments indicated that the remaining pyrimidines in the 5′ SL are protected from RNase A and therefore base-paired.
Figure S2.
RNase digestion assay of 5′ SLx. (A) Secondary structure of 5′ SLx. (B) Denaturing gel electrophoresis of 5′ SLx incubated with RNase A or RNase T1. Cy3-fluorescent RNA fragments appear green. RNA fragments without Cy3 appear blue from SYBR Green II staining. Cleavage points corresponding to the band size relative to the ladder are indicated in the margins.
RNase T1 digestion did not result in fragmentation of the 5′ SLx. One major band was present for the 34-nt full-length RNA oligonucleotide and one minor band at 32 nt represented cleaving after G127 in the 3′ overhang outside of the 5′ SL structure (Figure S2B). Since the 5′ SL SHAPE structure predicted that G121 resides within a loop, digestion 3′ of G121 was expected. Protection from RNase T1 indicated that the 5′ SL adopted a tertiary structure that stacked the internal loop bases and ordered the backbone.
5′ Stem–loop NMR analysis
To build from the RNase secondary structure probes, we endeavored to solve the solution NMR structure of the 5′ SL. NMR has proven to be well-suited for structure determination of oligoribonucleotides [44]. NMR experiments were performed on three RNA constructs: the complete 28-nt 5′ SL sequence, a truncated 5′ SL sequence with the terminal hexaloop and topmost two base pairs replaced by a UUCG tetraloop (5′ SLtrunc), and a 5′ SL sequence with one additional GC base pair at the bottom stem (5′ SLGC) (Figure 3(a)). The 5′ SLtrunc and 5′ SLGC constructs were designed to simplify assignment through signal deconvolution and stabilization of the helical stem, respectively. 5′ SLGC was chosen for isotopic labeling due to its minimal changes from the wild-type sequence and benefits from stabilization of the bottom helix.
Figure 3.
NMR analysis of the 5′ SL. (a) Secondary structures of the three constructs studied by solution NMR. (b) Statistics for the structural constraints used to model the structure of the 5′ SL. (c) Lowest energy model of the 5′ SL. (d) Superpositions of nine of the lowest-energy structures for the bottom stem, internal loop, top stem, and terminal loop regions, highlighting the significant flexibility of the two loop regions.
Exchangeable 1H assignment
The secondary structure of the 5′ SL predicted from SHAPE data included three AU pairs, five GC pairs, and one GU pair. Similarly, 1D exchangeable 1H spectra, which only detected UH3 and GH1 nuclei participating in hydrogen bonds due to base pairing, indicated two AU pairs, five GC pairs, and one GU pair. The missing AU pair corresponding to the terminal AU of the bottom stem was detected from the 5′ SLGC spectra due to addition of the stabilizing terminal GC pair. Assignments of UH3 and GH1 nuclei in the 1D spectra were corroborated by 1H–1H nuclear Overhauser enhancement spectroscopy (NOESY) experiments, which served to connect adjacent UH3 and GH1 in the top and bottom stems. Nucleotides not involved in base pairing, such as those in the purine-rich inner loop and the terminal loop, were not detected in these experiments due to rapid imino 1H exchange in these regions. Thus, the 1D 1H and NOESY spectra completely supported the SHAPE-predicted secondary structure of the 5′ SL.
Nonexchangeable 1H assignment
We next assigned nonexchangeable 1H resonances to model the ribose backbone of the structure. Characteristic A-form RNA helix geometries were leveraged with through-space NOESY experiments to connect 1Hs from adjacent riboses within the top and bottom stems. These connections were rarely found in the flexible internal and terminal loops. NOESY mixing times controlled the maximum internuclear distance that could be detected in the experiment. 50-, 150-, and 300-ms mixing times corresponded to distances of approximately 3, 5, and 6 Å, respectively, so comparisons between NOESY spectra of different mixing times assisted chemical shift assignment. Since the intensity of a NOESY crosspeak inversely correlated with the sixth power of the internuclear distance at shorter mixing times in the absence of spin diffusion, the internuclear distance was indirectly measured by the NOESY experiments.
Two-dimensional through-bond double quantum filtered (DQF)–correlated spectroscopy (COSY) experiments also delivered conformational information in the form of dihedral angles according to Karplus relationships with 3JHH coupling. Ribose sugar pucker conformation was probed through 3JH1′H2′ couplings. Intense COSY signals corresponded to a C3′-endo sugar pucker common in A-form RNA helices, while weak signals indicated a C2′-endo conformation. Within the 5′ SL model, the 18 nt of the top and bottom stems had strong 3JH1′H2′ couplings, which were interpreted as C2′-endo ribose conformations.
To aid 1H resonance assignments, heteronuclear NMR experiments were required, taking advantage of the broader chemical shift dispersion of 13C and 15N resonances. Three-dimensional NOESYHSQC experiments provided an additional heteronuclear dimension with which to distinguish similar 1Hs. 13C chemical shifts distinguished C1′ nuclei from the other ribose carbons, and purine C8 and adenine C2 nuclei were marked by characteristic chemical shifts. 15N chemical shifts also allowed connections to be made between the ribose and nucleobase by observing through-bond energy transfer across purine N9 and pyrimidine N1 nuclei.
Residual dipolar coupling
Residual dipolar coupling was measured by acquiring 1H–13C HSQC spectra without 13C decoupling in the presence and absence of 21 mg/ml Pf1 phage for partial alignment. From a comparison of these spectra, 19 differences in H2-C2 J-coupling, Δ1 J CH values, were measured. Average Δ1 J CH values for the lower helix, internal loop, upper helix, and terminal loop were 18.24 ± 4.69, 23.90 ± 5.72, 23.15 ± 10.87, and 5.03 ± 1.91, respectively. Δ1JCH values provided long-range conformational data used in structural modeling.
Structure modeling
Data from the NMR experiments were used in structural modeling in the following ways. One hundred forty-three internuclear distances were calculated from the NOESY crosspeak intensity and calibrated to the pyrimidine H5–H6 crosspeak intensity and known internuclear distance. Eighteen dihedral angle constraints were collected from COSY 3JHH coupling constants. Eight base-planarity constraints were applied from the strong base pairing data of the exchangeable 1H experiments. The statistics of the modeling constraints are shown in Figure 3(b). The lowest-energy model produced by Xplor-NIH using simulated annealing in the presence of these constraints was refined against a second simulated annealing process including 19 Δ1 J CH residual dipolar coupling restraints measured from the addition of Pf1 phage magnetic resonance cosolvent. The refinement step generated 100 structural models of the 5′ SL construct, and the lowest-energy structure is shown in Figure 3(c). The lower and upper stems were modeled as right-handed A-form helices with an approximate rotation per base pair of 29°, 12.3 bp per turn, rise per base pair of 3 Å, and C3′-endo ribose sugar pucker. The 4-nt internal loop was modeled as a hinge between the two helices such that the loop bases stack nonuniformly to make the helices blend into one semicontinuous helix. The bend of the hinge varied between structures, as indicated by the poor degree of overlapping in the superposition (Figure 3(d)). A non-canonical AG base pair between A104 and G121 was suggested in some structures. The bend followed the right-handedness of the helices, therefore causing the terminal loop to approach the 5′ side of the lower helix. The terminal loop geometry was also extremely heterogeneous, resulting in significant differences between model solutions (Figure 3(d)). Some base stacking was evident for C110, U111, U114, and A115, but nucleobases were often outwardly projected into the solvent.
Comparison of bypassing efficiency with polysomes or monosomes in vivo and in vitro
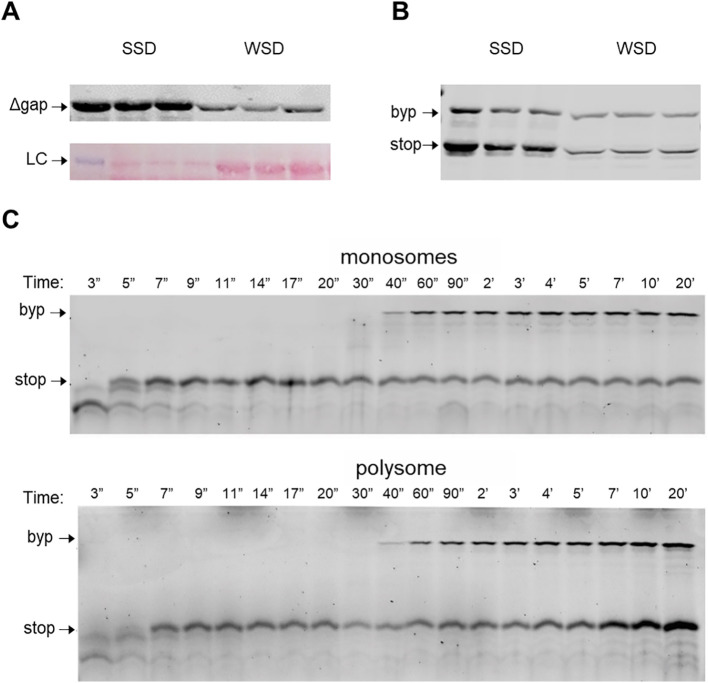
To test whether the level of ribosomal loading has an effect on bypassing efficiency in vivo, an experiment was performed in E. coli cells with a plasmid-borne cassette that permitted either high or low ribosome loading. The former possessed a strong initiating SD sequence, AGGAGG, while the latter had a weaker SD, AGAUGG, designed to lower ribosome loading. There is a 12-fold difference in product level between the two vectors each with a gene 60 cassette lacking the 50-nt coding gap (gene 60 Δgap) (Figure S3A). It was previously confirmed this strong initiating SD vector has a higher ribosome load per message from polysome fractions [45]. With a WT gene 60 cassette in the high ribosome load vector, bypassing efficiency was 26%, whereas in the low ribosome load vector, bypassing efficiency increased to 45% (Figure 4 (a) and Figure S3B).
Figure S3.
Ribosome loading effect on gene 60 bypassing in vivo and invitro. (A) Western blot hybridization of the translation products for the gene 60 Δgap expressed under a strong (left) and weak (right) initiation context in vivo (top). Prior to antibody probing the blot was Ponceau S-stained for total protein normalization (LC, loading control) (bottom). (B) A Western blot hybridization of the gene 60 bypassing translation products under a strong (left) and weak (right) initiation context. (C) Time courses of stop- and byp-product (bypass product) synthesis by monosomes (top) or polysomes (bottom). Protein products were visualized using a fluorescent reporter (Bodipy FL) attached to the N terminus of the peptides.
Figure 4.
Ribosome loading effect on gene 60 bypassing efficacy. (a) Gene 60 bypassing efficiency under weak SD (black) and strong SD (gray) initiation contexts in vivo were quantified from immunoblot assays (Supplementary Figure 1B). (b) The intensities of the stop-product and (c) the bypass product produced by monosomes (white circles) and polysomes (black circles) were quantified and plotted from time courses performed in vitro (Supplementary Figure 1C). (d) Comparison of the bypassing efficiency under mono- and polysome conditions in vitro. (e) Cartoon of the 5′SL including synonymous mutations 5′ and 3′ (f) 5′ side synonymous mutations of the 5′ SL effect on bypassing efficiency under weak SD (left) and strong SD (right) initiation contexts in vivo. (g) Synomonous mutations of the 5′SL to disrupt the secondary structure for the 5′ (white) and 3′ (black) side of the upper and lower stem. Restoration mutations to test for reformation of the secondary structure were performed in parallel (white and black circles). Error bars show the standard deviation for n = 3 replicates **P < .01, ***p < .001 by Student's two-tailed unpaired t test.
Ribosome loading effect on gene 60 bypassing efficacy. (a) Gene 60 bypassing efficiency under weak SD (black) and strong SD (gray) initiation contexts in vivo were quantified from immunoblot assays (Supplementary Figure 1B). (b) The intensities of the stop-product and (c) the bypass product produced by monosomes (white circles) and polysomes (black circles) were quantified and plotted from time courses performed in vitro (Supplementary Figure 1C). (d) Comparison of the bypassing efficiency under mono- and polysome conditions in vitro. (e) Cartoon of the 5′SL including synonymous mutations 5′ and 3′ (f) 5′ side synonymous mutations of the 5′ SL effect on bypassing efficiency under weak SD (left) and strong SD (right) initiation contexts in vivo. (g) Synomonous mutations of the 5′SL to disrupt the secondary structure for the 5′ (white) and 3′ (black) side of the upper and lower stem. Restoration mutations to test for reformation of the secondary structure were performed in parallel (white and black circles). Error bars show the standard deviation for n = 3 replicates **P < .01, ***p < .001 by Student's two-tailed unpaired t test.
Next, a counterpart experiment was performed to see whether one could recapitulate in vitro the ribosome loading effect seen here in vivo. The in vitro translation system [22,23,31] was primed with gene 60 mRNA whose non-bypass product (ORF1 translation product) was 46 amino acids (~ 5 kDa) and the full-length bypassing product was 160 amino acids (~ 17.6 kDa). The set-up for in vitro translation by monosomes involved mixing initiation ribosome complexes (to final concentration 0.016 μM) with the ternary complexes (50 μM). Translation by polysomes was carried out in the same way but in the presence of additional ribosomal subunits, initiation factors and Bpy-Met-tRNAfMet to allow for re-initiation on the same mRNA. Upon translation by a single ribosome, the non-bypass, i.e. ORF1, product accumulates after about 10 s and then decreases, because about 60% of ribosomes bypass and synthesize the bypass product (Figure 4(b) and Figure S3C). A similar accumulation of the ORF1 product is observed for the leading ribosome in the polysome (Figure 4(b) and Figure S3C). In the conditions of high ribosomal loading (polysome), the bypass product is formed slower in comparison with mono ribosome translation (Figure 4(c)), which suggests that even the leading ribosome has difficulties to land. When the leading ribosome leaves the take-off codon, trailing ribosomes in the polysome complete synthesis of the ORF1 product, but cannot complete bypassing, which leads to accumulation of the non-bypass product (Figure 4(b)). Although the level of bypassing product increases in polysomes, (Figure 4(c)), the fraction of ribosomes that complete bypassing is smaller than by a monosome (Figure 4(d)). This suggests that fewer ribosomes reach the landing site when they are arranged in polysomes than in monosomes. The directionality of the change seen corresponds to that of the in vivo analysis. Thus, ribosome loading has a clear effect on gene 60 bypassing both in vivo and in vitro.
Next to determine whether the action of the 5′SL is attenuated by successive ribosomes in a polysome, we examined mutants of the 5′SL under the same high/low ribosome conditions in vivo. The four synonymous substitutions in the 5′ side of the stem, used in the initial in vitro work [31] to preclude stem formation, were tested first (Figure 4(e), Fa1). The in vivo bypassing efficiency of the resulting mutant under high- and low-ribosome loading is 25% and 29%, respectively, which is similar to the bypassing efficiency on WT mRNA at high ribosome loading, 28%, but significantly lower than at low ribosome loading, 42% (Figure 4(f)). This experiment led us to conclude that 5′SL enhances bypassing efficiency in the conditions of low ribosomal density and is consistent with 5′SL formation being precluded or disrupted by the following ribosome in high ribosome conditions (Figure 4(f)). As the only substantial effect for the 5′SL was observed with low-ribosome loading (Fa1, 69% of WT), the remaining mutant constructs were assayed under these conditions. Next, secondary-structure disrupting synonymous changes were made separately to the 3′ side of the stem (Figure 4(e), Fa2), bypassing decreased to 49% of WT. However, when compensatory mutations on the 5′ side were made to restore the secondary structure of the 5′SL (Fa3), bypassing efficiency was only partially restored (64% of WT) (Figure 4(g)).
To further understand the effect of the 5′SL in vivo, we introduced mutations in the lower and upper parts of the 5′SL and measured bypassing in conditions of low ribosome loading (see Materials and Methods). First non-synonymous mutants designed to preclude alternative Watson–Crick pairing were introduced into the lower and upper sections of both sides of the 5′ SL (Figure 5(a)). The mutant of the 5′ side of the lower part of the stem (Fa4) had an efficiency of 73% of WT, and its 3′ side counterpart (Fa5) is 41% of WT. Combining the 5′ and 3′ side mutants in the lower part of the stem to restore base pairing (Fa6) resulted in a partial rescue (68%). The mutant of the 5′ side of the upper part of stem (Fa7) caused bypassing to decrease to 57% of WT. A counterpart on the 3′ side of the upper part of the stem (Fa8), reduced bypassing to 42% of WT. A combination of the mutants that restored complementarity (Fa9) did partially restore bypassing levels (62%) (Figure 5(a)). Together with the earlier in vitro study, these in vivo results support the existence and significance of the 5′SL at conditions of low ribosome loading.
Figure 5.
Assessment of the nucleotide and amino acid effect of the sequence that forms the 5′ SL. (a) Non-synomonous changes that prevent alternative Watson–Crick pairing were made to the upper and lower region of the 5′ and 3′ side of the 5′ SL, alongside with compensatory mutations expected to restore formation of the secondary structure (white text in black). (b) The effect of the encoded amino acids of the 5′SL was also further explored by testing amino acid variants, which permit alternative Watson–Crick pairing to minimize the nucleotide level effect (black, mutations of the left strand; gray, mutations of the right strand; white text, compensatory mutations).
In addition to potential effects of structure formation on bypassing, the encoded amino acid sequence of the 5′SL may also be relevant. To explore potential amino acid level effects, variants with potential alternative Watson–Crick pairing that might minimize nucleotide pairing level effects were tested. Mutating the 5′ side of the stem, GCG (Ala 34), to CGC (Arg) (100–102 GCG-CGC) (Fa10) reduced bypassing efficiency to 63%. Mutating GAC (Asp 41) and GCA (Ala 42) (123–125 CGC-GCG) in combination on the 3′ side to GAG (Glu) and CGA (Arg) (123–125 CGC-GCG and 100–102 GCG-CGC) (Fa11) reduced bypassing efficiency to 53% of WT (Figure 5(b)). A combination of mutated codons 40 and 41 with mutant codon 34 that restored complementarity at the original location (Fa12) yielded WT levels (Figure 5(b)). A complete restoration in this case clearly indicates that base pairing is important in this region of the 5′SL, and not the amino acid identity at these positions, consistent with the lack of specific contacts of Ala34, Asp41, and Ala42 with the exit tunnel [22]. Separately mutating GUC (Val 36) to CUG (Leu) (106 U-C, 108 U-G) (Fa13), reduces bypassing efficiency to 61% of WT. Mutating AUG (Met 39) and ACA (Thr 40) to AUC (Ile) and AGA (Arg), respectively (117 G-C, 119 C-G), on the 3′ side of the stem (Fa14) caused a reduction to 56% of WT (Figure 5(b)). A combination of mutated codons 39 and 40 with mutant codon 36 that restored complementarity at the original location (Fa15), yielded 76% of WT partially rescuing bypassing. A lack of complete restoration could be attributed to the loss of specific interactions of Met39 and Thr40 with the exit tunnel, and in particular the replacement of Thr [40] with Arg may be significant [22]. These results are suggestive of a nucleotide effect on bypassing for both the lower and higher segments of the 5′SL. Disruption of the nucleotide base pairing involved in the formation of the 5′SL without changing the amino acid sequence (Fa1) leads to a 30% reduction in bypassing efficiency. The other compensatory mutations tested cause an amino acid substitution, but nevertheless restore bypassing efficiency, unless the mutations results in a dramatic amino acid substitution, such as Thr to Arg. We note that the observed 30% reduction in 5′SL mutants is not equivalent to the 40% reduction of WT bypassing efficiency in polysome conditions, which indicates that 5′SL unwinding is not the only effect of the trailing ribosome.
Separating the upper and lower parts of the 5′SL is a central region consisting of four purines, AA on the 5′ side and GA on the 3′ side (Figure 6(a)). Extrapolating from the NMR data, one model is that the unpaired nucleotides in this central region act as a molecular hinge providing flexibility to the upper stem. Watson–Crick pairing in the central region connecting the lower and upper parts of the 5′SL into one continuous stem would impede free movement in the central region. This was achieved by changing the 5′ side AA to UC, now complementary to the 3′ side (WT GA) (Fa16), the bypassing efficiency remained at WT levels (100%) (Figure 6(a)). On changing the 5′ side AA in the central region to CC and the 3′ side to UU (i.e. all pyrimidines) (Fa17), bypassing was reduced to 60% of WT. The sequence may be important regardless of the relative flexibility of the upper and lower stems, potentially being involved in some kind of interaction with the ribosome. We also tested the sequence significance of the 6 nt terminal loop (CUAUUA) capping the 5′SL. A minimal effect was observed for loop mutants, e.g., changing the first C nt of the loop to an A reduced bypassing to 80% of WT (Fa18, Fa20). For all other loop mutations, no significant reduction in bypassing was observed (Figure 6(b)).
Figure 6.
Functional potential of the 5′SL loop sequence and flexibility of the adjoining upper stem. (a) Flexibility of the central region of the 5′ SL was explored via creation of one continuous stem by substitutions of the central region permitting Watson–Crick pairing (5′ side UC change to complement the 3′ GA) (white box); connecting both the upper and lower stems together. Purine nucleotides AA on the 5′ side and GA on the 3′ side were changed to pyrimidines CC and UU, respectively (black box). (b) A functional role for the 6-nt loop sequence (CUAUUA) that caps the 5′ SL was examined (loop mutations are highlighted in red).
Discussion
In this paper, the role of the 5′ SL was addressed by ribosome profiling, NMR structural analysis, and examining its effect on bypassing under high (polysome) and low (monosome) ribosome loading conditions. An extraordinarily high proportion of the ribosome-protected fragments in ribosome profiling derived from ribosomes stalled at the take-off codon; in agreement with earlier studies that showed ribosome pausing at the take-off site [21,22]. There were essentially two sizes of protected RNA, 37 and 53 nt. The 37-nt fragment is within the upper part of the size range of standard protected fragments found with ribosomes carrying out canonical translation. However, as extrapolated from the protection pattern of ribosomes in canonical rotation states, the 53-nt fragment is unusually long. If 15-nt 5′ of the P site/A site junction is located in the mRNA channel, the 5′ SL top could form outside the ribosome, whereas the 3′ side of the stem would reside within the mRNA exit channel. On this basis, it is surprising, that the 5′ nt of the protected 53-nt fragment coincides exactly with the 5′ nt of the 5′ SL. However, formation of the SL involving the 3′ side of the stem is highly unlikely, as it is occluded by the mRNA channel, and given the structure of the channel, a double-stranded SL is too bulky to fit into it. Moreover, even if it were somehow to be accommodated in the mRNA exit channel, then either a triple helix or nuclease penetration deep into the exit channel of the hyper-rotated ribosome would apparently be needed to explain the results. Rather, the observed protection could be explained in two possible ways. One possibility is that the 5′SL top and the mRNA region up to the take-off codon is protected in a canonical way due to formation of the SL and protection by the ribosome, respectively, whereas the 5′ part of the 5′ SL is inaccessible for RNase cleavage due to the hyper-rotated conformation of the ribosome. Another possibility is that the parts of the 5′SL that are out of the 30S subunit interact with the ribosome or with the upstream elements of the mRNA, thereby preventing the RNase cleavage. This possibility is supported by the cryo-EM analysis of ribosomes stalled at the take-off codon revealing density for the structured mRNA 5′ of the 30S [22]. Earlier SHAPE analysis showed that the 5′ SL can form in the absence of ribosomes [26]. That work showed that the two bases in the 5′SL closing loop and two bases between the 5′ SL and the extended SL, which were highly accessible to the SHAPE reagent and to Tb3+. These potential cleavage sites in the unstructured regions were not accessible for the MNase in the ribosome profiling experiment, possibly owing to the larger size or the slower cleavage rate of the MNase compared to the SHAPE reagent or Tb3+. Since the 5′SL is dynamic, as indicated by the NMR, the lifetime of the open conformation accessible for cleavage may be too short for the RNase, but sufficient for the chemical probing reagents.
Paradoxically, although we collected and analyzed fragments of up to 150 nt, we did not detect protection of 23-nt 3′ from the take-off codon. Inclusion of 23 nt would be expected if the A-site SL were present in the great majority of the ribosomes paused at the A-site, and if the ribosome hyper-rotated state at take-off, or dynamic properties of the A-site SL does not permit MNase to enter the A-site and cleave the A-site SL structure. While the A-site SL was detected both by cryo-EM analysis and smFRET, the former used temperature trapping and the smFRET analysis showed that the A-site SL only briefly formed [21], with recent work providing consistent results [23]. So it remains possible that the protected fragment being considered derives from ribosomes in which the A-site SL had not yet formed. Though the smFRET study showed that landing/coding resumption is a slow process [21], for unknown reasons possibly related to susceptibility of the mRNA in the vacant A-site in the non-canonical rotated ribosome after the peptidyl-tRNA has paired to the landing site codon, we detect no excess of protected fragments containing the landing site. [Of potentially wider interest, we did detect extra-long protected mRNA fragments in a wide variety of other genes but did not investigate these.]
The NMR analysis shows that the 5′ SL can adopt a compact structure, but remains highly flexible. The top and bottom helices, although rigid and highly stable individually, are weakened by the existence of a connecting loop. This purine-rich internal loop contributes to flexibility not only through fraying of the two helices but also by acting as a hinge that terminal loop nucleobases into the solvent enables intermolecular base pairing, such as that observed in kissing hairpins. The positioning of the 5′ SL during takeoff raises the question whether the ribosome itself could interact with the terminal loop either through protein–RNA or RNA–RNA interactions. However, disruption of terminal loop interactions through synonymous codon mutations of the loop sequence did not affect bypassing. Moreover, it is unclear exactly how much of the 5′ SL sequence is free to fold during takeoff. The upper helix and the terminal loop alone comprise of a modestly stable hairpin. Although mutations designed to restore the 5′ stem–loop do not substantially restore bypassing levels (Figure 4, Figure 5), one possible contributory explanation for this observation could be altered elongation rates at mutant codons with lower codon usage (Table S4). Given the relative instability of the lower stem due to its shorter sequence and rapid fraying detected by NMR, the primary contribution of the lower 5′ SL structure may be to extend the top aspect of the structure to potential interacting partners. The 5′ SL's highly dynamic nature agrees with previous findings that the density for the 5′SL region is disordered in the cryo-EM study of ribosomes paused at the take-off site [22].
Earlier work suggested that as the mRNA exits the ribosome upon bypassing, formation of the 5′ SL may act as either an initial ‘pusher’ of forward bypassing, or as a backstop to prevent backward sliding, or both [21,31]. The 5′ top element, which according to the ribosome profiling data can form when the ribosome resides on the take-off codon, is more stable than the lower stem, and would act as an obstacle for ribosome backward movement even in the absence of the lower part of the structure. While we cannot rule out potential significance of its formation for a contrasting “push” function, it is possible that avoidance, or strict limitation of any potential for a “push” function that could influence the likely key timing of formation of the A-site stem–loop, has limited selection for a stronger backstop, which may anyhow be unnecessary.
As a translating ribosome approaches the coding gap, its leading edge may encounter the 5′SL. There is no evidence that a consequence of this affects the ribosomes changes that occur after the 5′ part of the specific nascent peptide signal coding sequence has been translated, but the possibility has not been ruled out.
Our finding that monosomes are significantly more efficient than polysomes in mediating gene 60 programmed bypassing, both in vivo and in vitro, contrasts with an initial supposition that following translating ribosomes may selectively enhance forward bypassing by a leading ribosome. Gene 60 bypassing starts at codon 46 and this allows a maximum of three ribosomes trailing 5′ behind a ribosome paused at the take-off site. While the context for gene 60 translation initiation is relatively strong [(U)GAGG-6nts-AUG], its initiation level is likely influenced by overlapping translation of an upstream coding sequence and physiological state of the cell at the time of infection [2]. Under polysome conditions, behind the leading ribosome proximity of the mRNA entrance channel of the closest trailing ribosome has potential to prevent nucleotides emerging from the leading ribosome from forming the 5′ SL. While this could account for part of the lower bypassing efficiency under polysome than under monosome conditions, it cannot be the sole reason. Under polysome conditions, the number of ribosomes that complete bypassing is lower than in monosome conditions, due to increased drop-off of bypassing peptidyl-tRNA likely because the 3′SL that facilitates landing [31] does not re-form behind the leading ribosome. It is also possible that the proximity to the bypassing ribosome of its closest trailing ribosome, which is performing continued translation, negatively impacts the pseudotranslocation, e.g., by preventing formation of the hyper-rotated state. The results add to emerging appreciation of the importance of case-specific ribosome loading levels for recoding [45., 46., 47.], as well as elsewhere such as neuronal decoding [48]. This highlights the need, at least in recoding studies, to express cassettes under different loading conditions to avoid missing important stimulatory signals and for ascertaining physiologically relevant efficiencies.
Functional effects of formation of the 5′ SL have formal similarities to one explanation for how a proportion of Cricket Paralysis virus internal IRES-mediated translation initiation involves a 3′ removed site. Formation of an extended IRES in the ribosomal P site may lead to non-adjacent downstream initiation [49]. Further, a synthetically created upstream SL structure 4-nt 5′ of the SARS coronavirus frameshift was shown to attenuate frameshifting [50].
It has become common to regard genetic information as not just codon identity dependent on aminoacyl-tRNA and release factor levels and features, but to also recognize relevant mRNA structure and modification as well as specific nascent peptide sequence as important constituents [51]. Tapes to (protein) shapes are no longer considered an accurate descriptor of the process. Nevertheless, the extent to which aspects other than codon identity contribute to T4 gene 60 decoding is still remarkable. It is fortunate for them and us that Crick et al. [1] performed their pioneering work on the adjacent rII gene and not gene 60!
Materials and Methods
Ribosome profiling
The method for ribosome profiling was described by Ingolia et al. [52] and modified for E. coli [39]. Three 200-ml cultures of E. coli strain MC4100 (OD600 ~ 0.5) were each infected with bacteriophage T4 at a multiplicity of infection of 10 and harvested at different time points (2.5, 4, or 5 min) post-infection. In parallel, a 200-ml culture (uninfected) was also harvested as a control. Cells were collected by fast filtration and immediately frozen in liquid nitrogen. Cells were lysed by mechanical disruption while frozen to prevent further translation elongation during sample preparation. Clarified lysates, corresponding to 20 OD260 (in 100 mM NH4Cl, 10 mM MgCl2, 20 mM Tris (pH 8.0), 0.4% Triton X-100, 0.1% NP40, 5 mM CaCl2, 100 units/ml RNase-free DNase, and 100 μg/ml chloramphenicol), were treated with micrococcal nuclease (MNase) (60 units/OD260) for 1 h at 25 °C. Nuclease digestion was stopped by the addition of EGTA. Lysates were then loaded onto 10%–55% sucrose gradients (gradient buffer: 100 mM NH4Cl, 10 mM MgCl2, 20 mM Tris (pH 8.0), and 100 μg/ml choramphenicol) and centrifuged at 35,000 rpm for 2.5 h at 4 °C. Monosomes fractions were collected and ribosome-protected mRNAs were isolated by acid phenol extraction and isopropanol precipitation. Following dephosphorylation, RNAs (20 μg each) were fractionated on 15% TBE-urea gels and the appropriate size ranges, 30–40 or 45–150 nt, were excised. RNAs were eluted from the gel slices, and sequencing libraries were prepared following the standard protocol [52]. In addition, ribosome profiling was also performed on 200-ml cultures of E. coli containing a plasmid-borne cassette with the gp60 sequence. E. coli cells (OD600 ~ 0.5) were induced for 10 min with IPTG. Following induction, cells were processed and RNA was extracted in the same manner as the T4 infected E. coli as outlined above.
Ribosome profiling data analysis
The adaptor sequence (CTGTAGGCACCATCAATTCGTATGCCGTCTTCTGCTTGAA, for the single-end reads and GATCGTCGGACTGTAGAACTCTGAACGTGTAGATCTCGGTGG or TGGAATTCTCGGGTGCCAAGGAACTCCAGTCACTGAC for the paired end reads) was cleaved with Cutadapt (DOI:10.14806/ej.17.1.200) with parameters (− n 2 --match-read-wildcards –minimum-length = 20). Reads that failed to align to rRNA were aligned to the reference genomes of E. coli sub-strain MG1655 (accession number NC_000913.2) and T4 (NC_000866.4). The single end reads were mapped with Bowtie [19261174] with parameters (− m 1), i.e. allowing for no ambiguous mapped reads. The paired-end reads associated with the longer fragments were aligned with Bowtie2 [22388286] with parameters (− M 10 -I 0 --no-unal --no-discordant --no-mixed –dovetail). Reads were selected to be within the range of 45 to 150 nt in length; however, the majority of fragments were found to be shorter than this. Reads less than 45 nt were discarded. The plots were produced with matplotlib library (https://doi.org/10.1109/MCSE.2007.55). Ribosome profiling data are deposited in the NCBI GEO database (GSE146240).
5′ SL RNase digestion assays
An extended 5' SL sequence, called 5′ SLx and consisting of 34 nt from C96 through U129 with a Cy3 fluorophore chemically attached to C96, was chemically synthesized by Integrated DNA Technologies. 5′ SLx of 1, 10, and 100 ng/μl was digested by 0.1 ng/μl RNase A (Ambion) or RNase T1 (Ambion) in the presence of 1, 10, and 100 ng/μl yeast RNA, respectively, according to recommended protocols. The reactions were incubated at room temperature for 15 min and resolved on a pre-heated 15% TBE-urea denaturing gel at 30 W for 10 min. Cy3 fluorescence was recorded on a Typhoon imager.
5′ SL NMR analysis
Sample preparation
Natural abundance full-length (5′ SL) and truncated 5SL (5′ SLtrunc) RNA constructs chemically synthesized by Integrated DNA Technologies were resuspended individually in RNA Sample Buffer (10 mM sodium phosphates (pH 6.5) with 100 mM NaCl) and purified by size-exclusion chromatography. RNA NMR samples were then exchanged into NMR Sample Buffer (10 mM sodium phosphates (pH 6.5) with 20 mM NaCl) and 10% v/v D2O to final concentrations of 0.8 and 0.7 mM, respectively, for NMR experimentation with exchangeable 1Hs. Samples were then exchanged into NMR Sample Buffer with 99.99% v/v D2O for nonexchangeable 1H acquisition. Uniformly 13C- and 15N-labeled full-length 5′ SL with added terminal GC pair ([U13C,15N]-5′ SLGC) was prepared by in vitro transcription with HiScribe T7 High-Yield RNA Synthesis kits (New England Biolabs) using 2 μM DNA oligomer templates and 7.5 mM [U-13C,15N]-rNTPs (Cambridge Isotopes). The RNA transcript was purified by preparative PAGE followed by size-exclusion chromatography into RNA Sample Buffer [44,53]. The sample was exchanged into NMR Sample Buffer with 10% v/v D2O at a concentration of 0.3 mM for exchangeable spectra, and then into NMR Sample Buffer with 99.99% v/v D2O for nonexchangeable spectra. To measure residual dipolar coupling, Pf1 magnetic resonance cosolvent (ASLA Biotech) was added to 5′ SLGC at a final concentration of 21 mg/ml, and the sample was allowed to equilibrate within the magnetic field overnight.
Spectra acquisition and assignment
Unless indicated otherwise, all NMR experiments were performed on a 800-MHz Agilent VNMRS with 5 mm 1H{13C,15N} cryoprobe using established RNAPack experiments [54]. FIDs were processed in VNMRJ 4.2 and exported to MestreNova (MestreLab Research) and SPARKY [55] programs for analysis and peak assignment. 5′ SLtrunc was analyzed first, and this construct's chemical shifts were used to inform assignment of 5′ SL and 5′ SLGC. For 5′ SL and 5′ SLtrunc, 1D 1H and SSNOESY spectra were acquired on samples in 10% D2O; 1D 1H, TNNOESY, and 1H–13C HSQC spectra were acquired on samples in 99.99% D2O. For 5′ SLGC, 1D 1H, SSNOESY, and HNNCOSY [56] experiments were performed on the sample in 10% D2O; 1D 1H, TNNOESY, 1H–13C HSQC, 1H–15N HSQC, DQF-COSY, HCCH-TOCSY, 3D 1H–15N NOESYHQSC, and 3D 1H–13C NOESYHSQC experiments were performed on the sample in 99.99% D2O. Furthermore, 3D HNCCCH [57] experiments were performed with 5′ SLGC in 99.99% D2O on a 600-MHz Varian INOVA spectrometer (Agilent) with 5 mm 1H{13C,15N} conventional probe. To measure residual dipolar coupling, 1H–13C HSQC experiments were performed on a 500-MHz Bruker AVANCE with 5 mm 1H{13C,15N} cryoprobe with and without 13C decoupling and before and after the addition of 21 mg/ml Pf1 to 5′ SLGC. The chemical shifts for the 5′ SLGC construct are deposited (BMRB ID: 28090).
Structure determination
To model the upper helix and tetraloop of the 5′ SLtrunc construct, 65 NOE distance constraints, 10 dihedral angle constraints, and 8 base-planarity constraints from NMR experiments of 5′ SLtrunc were inputted as parameters into Xplor-NIH 2.44 [58., 59., 60.]. From these parameters, 400 structures were generated from repeated simulated annealing using the RNA-ff1 force field. For the complete 5′ SL sequence model, the same procedure was used incorporating 143 NOE distance constraints, 18 dihedral angle constraints, and 18 base-planarity constraints from NMR experiments of 5′ SL and 5′ SLGC. Two hundred structures were generated, and the 20 structures with the lowest energies were selected. These structures were then refined against 19 Δ1JCH residual dipolar coupling constraints to generate 20 additional structures, from which the two structures with the lowest energies were selected for analysis.
In vitro monosome and polysome formation
Initiation complex was formed by incubating 70S ribosomes (0.5 μM), gene 60 mRNA (1.5 μM), IF1, IF2 and IF3 (0.75 μM each), GTP (1 mM), and fluorescence-labeled Bodipy-[3H]Met-tRNAfMet (Bpy-[3H]Met-tRNAfMet) (0.75 μM) in HiFi buffer for 30 min at 37 °C (22, 31). Resulting initiation complexes were purified by centrifugation through a 1.1 M sucrose cushion in the same buffer. The ternary complex EF-Tu–GTP–aminoacyl-tRNA was prepared by incubating EF-Tu (58 μM) with GTP (1 mM), phosphoenol pyruvate (3 mM), and pyruvate kinase (0.1 mg/ml) for 15 min at 37 °C, then adding purified total aa-tRNA (about 60 μM) and EF-G (2 μM) and incubating for 1 min at 37 °C. In vitro translation by monosomes was started by mixing initiation ribosome complexes (to final concentration 0.016 μM) with the ternary complexes (50 μM). Translation was carried out at 37 °C for different time intervals from 3 to 1200 s. To form the polysome, translation was carried out in the same way but in the presence of additional 30S ribosomal subunits (0.16 μM; 10-fold over the mRNA); 50S ribosomal subunits (0.24 μM), IF1, IF2, and IF3 (0.24 μM each); and Bpy-[3H]Met-tRNAfMet (0.24 μM) to allow for re-initiation on the same mRNA. Translation products were separated by Tris-Tricine gel electrophoresis. Fluorescent peptides were detected in gels using Starion IR/FLA-9000 scanner (FujiFilm) and quantified using the Multi Gauge software. Bypassing efficiency was calculated as a ratio of the density corresponding to the bypass (byp) band to the sum of the byp and stop bands.
In vivo bypassing assays
The E. coli strains DH5α and MG1655 cells were used for plasmid propagation and protein synthesis, respectively. Strains were grown on Luria–Bertani medium (LB) for the gene 60 bypassing assays. Constructs were produced by amplification of complementary oligonucleotides (Integrated DNA Technologies) to produce a full-length sequence containing 5′ Xho1 and 3′ BamHI restriction sites. These were cloned into the vector CRC01 Weak SD vector or CRC01 Strong SD vector. It was previously confirmed the CRC01 Strong SD vector displays a higher ribosome load per message from polysome fractions when compared to the CRC01 Weak SD vector [45]. The low ribosome vector used did not as completely restrict loading as that of another vector whose results are not shown because of artifacts associated with generation of the severe restriction. To distinguish between the stop product and the byp product, the His/Nanoluciferase C-terminal tag is present in the alternative − 1 frame relative to the Firefly luciferase/His N-terminal tag. Both the N-terminal and C-terminal tags are inframe for the ∆ gap control. Overnight cultures of strains containing the appropriate plasmid were diluted 1:100 in LB medium. Each culture was grown in triplicate at 37 °C at 200RPM. Once an OD of 0.4–0.5 was reached, the cultures were induced with 0.1 mM IPTG for 1 h at their respective temperatures. After induction, cultures were incubated on ice for 10 min and the OD was noted. Cells were lysed by resuspension in 2 × laemmli sample buffer (based on OD) and were incubated for a further 30 min on ice. Cells were subsequently centrifuged at 4 °C for 30 min at 20,000g to remove cell debris. Equivalent amounts of proteins were diluted and boiled for 10 min at 95 °C. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane (Protran). Immunoblots were incubated at 4 °C overnight in 5% milk/phosphate buffered saline–Tween containing a 1:5000 dilution of mouse anti-His conjugated to a fluorescently labeled secondary antibody. Immunoreactive bands were detected on membranes after incubation using a LI-COR Odyssey® Infrared Imaging Scanner (LI-COR Biosciences). The amounts of stop and byp products were quantified by densitometry using Image Lite Studio (LI-COR Biosciences). The bypassing efficiency was determined by taking the amount of byp product as a ratio of the total amount of stop plus byp products. Measurements were tabulated from three technical replicates for each sample, and the mean and standard deviations were calculated. Data were represented graphically and statistical analysis was performed (Prism 5).
The following are the supplementary data related to this article.
1H chemical shifts of the 5′ SLGC construct
13C and 15N chemical shifts of the 5′ SLGC construct
1H chemical shifts of the 5′ SLtrunc construct
Codons present in the 5′SL sequence and the substitutions made (frequency per thousand E. coli codons)
Acknowledgments
We thank Sean D. Moore for helpful advice on expression vectors, and him and Angela M. Smith for providing the vectors used in their recent publication; Corey Liu and Stephen Lynch for their help with the NMR analysis; Anna Pfeifer, Olaf Geintzer, Sandra Kappler, Christina Kothe, Theresia Niese, Tanja Wiles, Vanessa Herold, Franziska Hummel, Tessa Hübner, Puyan Nabizadeh-Ardekani, and Michael Zimmermann for expert technical assistance. This work was supported by the following grants: the Deutsche Forschungsgemeinschaft SFB860 to M.V.R., US National Institutes of Health grant GM51266 to J.D.P., and Irish Research Council Advanced Laureate (IRCLA/2019/74) to J.F.A.
Edited by Sarah A. Woodson
References
- 1.Crick F.H., Barnett L., Brenner S., Watts-Tobin R.J. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 2.Bonocora R.P., Zeng Q., Abel E.V., Shub D.A. A homing endonuclease and the 50-nt ribosomal bypass sequence of phage T4 constitute a mobile DNA cassette. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16351–16356. doi: 10.1073/pnas.1107633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang W.M., Ao S.Z., Casjens S., Orlandi R., Zeikus R., Weiss R., Winge D., Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. https://www.jstor.org/stable/1700083 [DOI] [PubMed] [Google Scholar]
- 4.Weiss R.B., Huang W.M., Dunn D.M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maldonado R., Herr A.J. Efficiency of T4 gene 60 translational bypassing. J. Bacteriol. 1998;180:1822–1830. doi: 10.1128/jb.180.7.1822-1830.1998. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC107096/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kane J.F., Violand B.N., Curran D.F., Staten N.R., Duffin K.L., Bogosian G. Novel in-frame two codon translational hop during synthesis of bovine placental lactogen in a recombinant strain of Escherichia coli. Nucleic Acids Res. 1992;20:6707–6712. doi: 10.1093/nar/20.24.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma N.J., Hemez C.F., Barber K.W., Rinehart J., Isaacs F.J. Organisms with alternative genetic codes resolve unassigned codons via mistranslation and ribosomal rescue. eLife. 2018;7:1–23. doi: 10.7554/eLife.34878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsley D., Gallant J., Doneanu C., Bonthuis P., Caldwell S., Fontelera A. Spontaneous ribosome bypassing in growing cells. J. Mol. Biol. 2005;349:261–272. doi: 10.1016/j.jmb.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 9.Weiss R.B., Dunn D.M., Atkins J.F., Gesteland R.F. Slippery runs, shifty stops, backward steps, and forward hops: − 2, − 1, + 1, + 2, + 5, and + 6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 10.Gallant J., Bonthuis P., Lindsley D. Evidence that the bypassing ribosome travels through the coding gap. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13430–13435. doi: 10.1073/pnas.2233745100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wills N.M. In: Translational Bypassing–Peptidyl-tRNA Re-pairing at Non-overlapping Sites. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Atkins J.F., Gesteland R.F., editors. Springer; New York, London: 2010. pp. 365–381. [Google Scholar]
- 12.Lang B.F., Jakubkova M., Hegedusova E., Daoud R., Forget L., Brejova B., Vinar T., Kosa P. Massive programmed translational jumping in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5926–5931. doi: 10.1073/pnas.1322190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nosek J., Tomaska L., Burger G., Lang B.F. Programmed translational bypassing elements in mitochondria: structure, mobility, and evolutionary origin. Trends Genet. 2015;31:187–194. doi: 10.1016/j.tig.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Manch-Citron J.N., Dey A., Schneider R., Nguyen N.Y. The translational hop junction and the 5′ transcriptional start site for the Prevotella loescheii adhesin encoded by plaA. Curr. Microbiol. 1999;38:22–26. doi: 10.1007/PL00006766. [DOI] [PubMed] [Google Scholar]
- 15.Xi Q., Cuesta R., Schneider R.J. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J Virol. 2005;79:5676–5683. doi: 10.1128/JVI.79.9.5676-5683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pooggin M.M., Ryabova L.A. Ribosome shunting, polycistronic translation, and evasion of antiviral defenses in plant pararetroviruses and beyond. Front. Microbiol. 2018;9:644. doi: 10.3389/fmicb.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkins J.F., Loughran G., Bhatt P.R., Firth A.E., Baranov P.V. Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodnina M.V., Korniy N., Klimova M., Karki P., Peng B.Z., Senyushkina T., Belardinelli R., Maracci C. Translational recoding: canonical translation mechanisms reinterpreted. Nucleic Acids Res. 2019;48:1056–1067. doi: 10.1093/nar/gkz783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herr A.J., Atkins J.F., Gesteland R.F. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 1999;18:2886–2896. doi: 10.1093/emboj/18.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herr A.J., Gesteland R.F., Atkins J.F. One protein from two open reading frames: mechanism of a 50 nt translational bypass. EMBO J. 2000;19:2671–2680. doi: 10.1093/emboj/19.11.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Coakley A., O'Connor M., Petrov A., O'Leary S.E., Atkins J.F., Puglisi J.D. Coupling of mRNA structure rearrangement to ribosome movement during bypassing of non-coding regions. Cell. 2015;163:1267–1280. doi: 10.1016/j.cell.2015.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agirrezabala X., Samatova E., Klimova M., Zamora M., Gil-Carton D., Rodnina M.V., Valle M. Ribosome rearrangements at the onset of translational bypassing. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1700147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimova M., Senyushkina T., Samatova E., Peng B.Z., Pearson M., Peske F., Rodnina M.V. EF-G-induced ribosome sliding along the noncoding mRNA. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varani G., Cheong C., Tinoco I., Jr. Structure of an unusually stable RNA hairpin. Biochemistry. 1991;30:3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- 25.Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 26.Todd G.C., Walter N.G. Secondary structure of bacteriophage T4 gene 60 mRNA: implications for translational bypassing. RNA. 2013;19:685–700. doi: 10.1261/rna.037291.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Ascenzo L., Leonarski F., Vicens Q., Auffinger P. Revisiting GNRA and UNCG folds: U-turns versus Z-turns in RNA hairpin loops. RNA. 2017;23:259–269. doi: 10.1261/rna.059097.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wills N.M., O'Connor M., Nelson C.C., Rettberg C.C., Huang W.M., Gesteland R.F., Atkins J.F. Translational bypassing without peptidyl-tRNA anticodon scanning of coding gap mRNA. EMBO J. 2008;27:2533–2544. doi: 10.1038/emboj.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young D.J., Guydosh N.R., Zhang F., Hinnebusch A.G., Green R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3 × UTRs in vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herr A.J., Nelson C.C., Wills N.M., Gesteland R.F., Atkins J.F. Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol. 2001;309:1029–1048. doi: 10.1006/jmbi.2001.4717. [DOI] [PubMed] [Google Scholar]
- 31.Samatova E., Konevega A.L., Wills N.M., Atkins J.F., Rodnina M.V. High-efficiency translational bypassing of non-coding nucleotides specified by mRNA structure and nascent peptide. Nat. Commun. 2014;5:4459. doi: 10.1038/ncomms5459. [DOI] [PubMed] [Google Scholar]
- 32.Herr A.J., Wills N.M., Nelson C.C., Gesteland R.F., Atkins J.F. Drop-off during ribosome hopping. J. Mol. Biol. 2001;311:445–452. doi: 10.1006/jmbi.2001.4899. [DOI] [PubMed] [Google Scholar]
- 33.Herr A.J., Wills N.M., Nelson C.C., Gesteland R.F., Atkins J.F. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. J. Biol. Chem. 2004;279:11081–11087. doi: 10.1074/jbc.M311491200. [DOI] [PubMed] [Google Scholar]
- 34.Baez W.D., Roy B., McNutt Z.A., Shatoff E.A., Chen S., Bundschuh R., Fredrick K. Global analysis of protein synthesis in Flavobacterium johnsoniae reveals the use of Kozak-like sequences in diverse bacteria. Nucleic Acids Res. 2019;47:10477–10488. doi: 10.1093/nar/gkz855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito K., Green R., Buskirk A.R. Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. eLife. 2020;9 doi: 10.7554/eLife.55002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bucklin D.J., Wills N.M., Gesteland R.F., Atkins J.F. P-site pairing subtleties revealed by the effects of different tRNAs on programmed translational bypassing where anticodon re-pairing to mRNA is separated from dissociation. J. Mol. Biol. 2005;345:39–49. doi: 10.1016/j.jmb.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 37.Smith M.C., Hendrix R.W., Dedrick R., Mitchell K., Ko C.C., Russell D., Bell E., Gregory M. Evolutionary relationships among actinophages and a putative adaptation for growth in Streptomyces spp. J. Bacteriol. 2013;195:4924–4935. doi: 10.1128/JB.00618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W.P., Cho C.P., Chang K.Y. mRNA-mediated duplexes play dual roles in the regulation of bidirectional ribosomal frameshifting. Int. J. Mol. Sci. 2018;19:3867. doi: 10.3390/ijms19123867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G.W., Oh E., Weissman J.S. The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor P.B., Li G.W., Weissman J.S., Atkins J.F., Baranov P.V. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013;29:1488–1491. doi: 10.1093/bioinformatics/btt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohammad F., Green R., Buskirk A.R. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife. 2019;8 doi: 10.7554/eLife.42591.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutter E., Kellenberger E., Carlson K., Eddy S., Neitzel J., Messinger L., North J., Guttman B. ASM Press; Washington, DC: 1994. Effects of Bacterial Growth Condition and Physiology on T4 Infection. Molecular Biology of Bacteriophage T4; pp. 406–418. [Google Scholar]
- 44.Puglisi J.D., Wyatt J.R. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–350. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- 45.Smith A.M., Costello M.S., Kettring A.H., Wingo R.J., Moore S.D. Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc. Natl. Acad. Sci. U. S. A. 2019;116:21769–21779. doi: 10.1073/pnas.1910613116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charbonneau J., Gendron K., Ferbeyre G., Brakier-Gingras L. The 5′ UTR of HIV-1 full-length mRNA and the tat viral protein modulate the programmed − 1 ribosomal frameshift that generates HIV-1 enzymes. RNA. 2012;18:519–529. doi: 10.1261/rna.030346.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heyer E.E., Moore M.J. Redefining the translational status of 80S monosomes. Cell. 2016;164:757–769. doi: 10.1016/j.cell.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Biever A., Glock C., Tushev G., Ciirdaeva E., Dalmay T., Langer J.D., Schuman E.M. Monosomes actively translate synaptic mRNAs in neuronal processes. Science. 2020;367 doi: 10.1101/687475. [DOI] [PubMed] [Google Scholar]
- 49.Kerr C.H., Wang Q.S., Moon K.M., Keatings K., Allan D.W., Foster L.J., Jan E. IRES-dependent ribosome repositioning directs translation of a + 1 overlapping ORF that enhances viral infection. Nucleic Acids Res. 2018;46:11952–11967. doi: 10.1093/nar/gky1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu H.T., Lin Y.H., Chang K.Y. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014;42:14070–14082. doi: 10.1093/nar/gku1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J., Grosely R., Prabhakar A., Lapointe C.P., Wang J., Puglisi J.D. How messenger RNA and nascent chain sequences regulate translation elongation. Annu. Rev. Biochem. 2018;87:421–449. doi: 10.1146/annurev-biochem-060815-014818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukavsky P.J., Puglisi J.D. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lukavsky P.J., Puglisi J.D. RNAPack: an integrated NMR approach to RNA structure determination. Methods. 2001;25:316–332. doi: 10.1006/meth.2001.1244. [DOI] [PubMed] [Google Scholar]
- 55.Goddard T.D., Kneller D.G. University of California; San Francisco, CA: 2008. SPARKY 3.https://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 56.Dingley A.J., Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. doi: 10.1021/ja981513x. [DOI] [Google Scholar]
- 57.Simorre J.P., Zimmermann G.R., Pardi A., Farmer B.T., 2nd, Mueller L. Triple resonance HNCCCH experiments for correlating exchangeable and nonexchangeable cytidine and uridine base protons in RNA. J. Biomol. NMR. 1995;6:427–432. doi: 10.1007/bf00197641. [DOI] [PubMed] [Google Scholar]
- 58.Clore G.M., Kuszewski J. Improving the accuracy of NMR structures of RNA by means of conformational database potentials of mean force as assessed by complete dipolar coupling cross-validation. J. Am. Chem. Soc. 2003;125:1518–1525. doi: 10.1021/ja028383j. [DOI] [PubMed] [Google Scholar]
- 59.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. Prog Nucl Mag Res Sp. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 60.Schwieters C.D., Kuszewski J.J., Clore G.M. Using Xplor–NIH for NMR molecular structure determination. Prog Nucl Mag Res Sp. 2006;48:47–62. doi: 10.1016/j.pnmrs.2005.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H chemical shifts of the 5′ SLGC construct
13C and 15N chemical shifts of the 5′ SLGC construct
1H chemical shifts of the 5′ SLtrunc construct
Codons present in the 5′SL sequence and the substitutions made (frequency per thousand E. coli codons)