To the Editor
Since December 2019, a novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has emerged in Wuhan, Hubei, China, as an etiological agent causing a new infectious respiratory disease, coronavirus disease 2019, which manifests as fever, severe respiratory illness, and pneumonia (Huang et al., 2020). With significant morbidity and mortality, coronavirus disease 2019 has now evolved into a global pandemic (Park, 2020). To date, more than 3 million cases of coronavirus disease 2019 have been reported worldwide, including 252,336 deaths, according to the World Health Organization.
The entry of coronavirus into target cells mainly depends on the binding of the viral spike proteins to cellular receptors (Hoffmann et al., 2020). ACE2 has been identified as a crucial functional receptor of SARS-CoV-2 (Wang et al., 2020). The receptor-binding domain of the SARS-CoV-2 spike protein was reported to have a strong interaction with human ACE2 molecules, which was confirmed in molecular structure (Wrapp et al., 2020, Xu et al., 2020b). These studies suggested that the ACE2-expressing cells were vulnerable to SARS-CoV-2 infection.
The RNA and protein expressions of ACE2 have been widely investigated by bulk samples from the heart, lung, kidney, and other organs (Hamming et al., 2004). Recently, single-cell RNA sequencing (scRNA-seq) was applied to analyze ACE2 mRNA expression in different cell subtypes. High ACE2 expression was identified in type II alveolar cells, bronchial transient secretory cells, small intestinal epithelium cells, and oral epithelial cells in accordance with respiratory clinical manifestations and rare clinical manifestations such as gastrointestinal symptoms, suggesting the respiratory droplet, digestive, and fecal-oral transmission routes of SARS-CoV-2 (Liang et al., 2020, Lukassen et al., 2020, Xu et al., 2020a). Therefore, we hypothesized that the expression and distribution of ACE2 in human organs and tissues could reflect the potential infection routes of SARS-CoV-2. However, scRNA-seq has not yet been applied to examine ACE2 expression in the cells of skin tissues, and the transmission of this virus by percutaneous routes remains unclear.
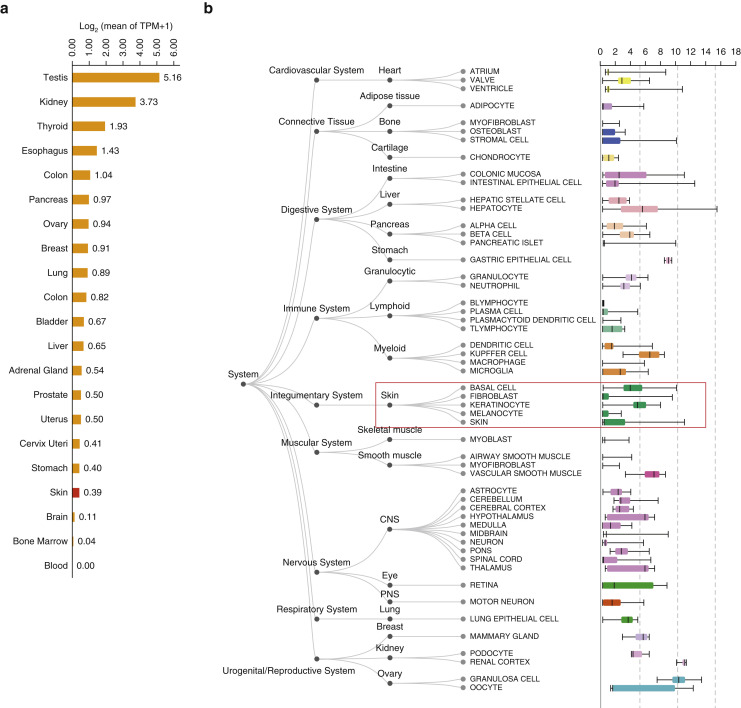
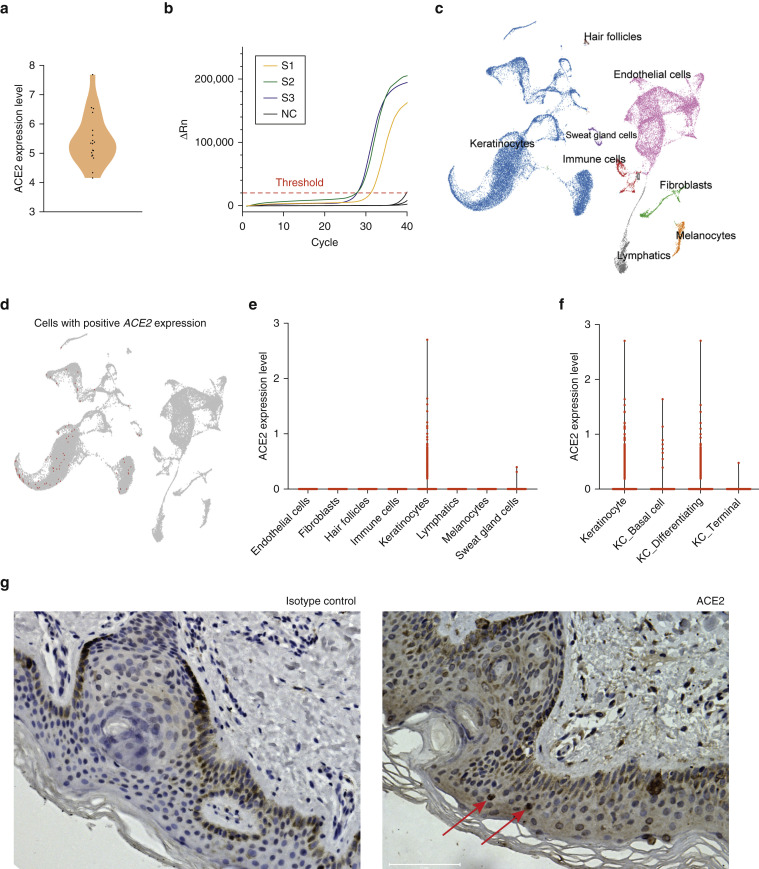
To investigate whether skin was a potential target for SARS-CoV-2 infection, we first analyzed ACE2 mRNA expression and ACE2-positive cell composition in skin tissues based on public databases (GEPIA2 and ARCHS4) and found that ACE2 was expressed in skin tissues in addition to testis, kidney, colon, lung, and so on. The expression of ACE2 was significantly higher in keratinocytes than other cell types in skin tissues, such as fibroblasts and melanocytes (Figure 1 a and b). Moreover, our in-house data of bulk RNA sequencing from 18 skin samples showed ACE2 expression in all samples (Figure 2 a), which was further validated by quantitative real-time RT-PCR (Ct = 28.97 ± 1.91, Figure 2b). We then performed scRNA-seq to evaluate the cell type–specific expression of ACE2 in six skin samples. After data processing, 40,459 cells were acquired and eight cell types were identified based on their canonical markers, including keratinocytes (KRT1 +, KRT5 +, KRT10 +, and KRT14 +), endothelial cells (SELE + and CD93 +), fibroblasts (DCN +, COL6A1 +, and COL6A2 +), hair follicles (SOX9 +), immune cells (PTPRC +, IL32 +, and CD3D +), lymphatic endothelial cells (CCL21 + and LYVE1 +), melanocytes (TYRP1 + and PMEL +), and sweat gland cells (DCD +, KRT19 +, KRT7 +, and AQP5 +) (Figure 2c). We found that 0.19% of skin cells in all six samples were ACE2-positive. Among these positive cells, keratinocytes were most enriched, accounting for 97.37%, and sweat gland cells account for 2.63%, confirming that ACE2 was mainly expressed in keratinocytes (Figure 2d and e). Moreover, we analyzed the ACE2 expression level in keratinocytes from different cell states and found that ACE2 was mainly expressed in differentiating keratinocytes and basal cells (Figure 2f). We did not find ACE2-positive fibroblasts, likely because of the low expression of ACE2 in fibroblasts (Figure 1b). Additionally, the ACE2 expression from eight skin samples by immunohistochemistry was analyzed. The results showed that 0.35% ± 0.07% of cells in the skin samples were ACE2-positive, mainly keratinocytes, consistent with scRNA-seq data. The stratum basale, stratum spinosum, and stratum granulosum of epiderma displayed ACE2-positive keratinocytes (Figure 2g). We also found few ACE2-positive dermal cells in immunohistochemistry, which could not be observed by scRNA-seq, likely because of the limitation of high dropouts and low capture efficiency of scRNA-seq technology (Haque et al., 2017). This study was approved by the institutional review board of Shandong Provincial Hospital for Skin Diseases, Shandong First Medical University, and written informed consent was obtained from all the participants. Detailed study protocols and methods are provided in Supplementary Materials and Methods.
Figure 1.
Expression of ACE2 in skin tissues in public database. (a) Bar plot of ACE2 expression in human normal tissues from GEPIA2 database. Expression level was indicated by log2(mean of TPM + 1). (b) Boxplot of ACE2 in different tissue types from ARCHS4 database. TPM, transcript per million.
Figure 2.
ACE2 RNA and protein expression in skin tissues. (a) Violin plot of ACE2 expression in bulk RNA sequencing of skin tissues. Expression level was indicated by log2(FPKM). (b) Amplification curve of skin samples and negative control from qRT-PCR. (c) Eight primary cell types in skin tissues were identified by cell markers, and cells were clustered by the UMAP method. (d) Cells with positive (red plots) ACE2 expression. (e) Violin plot of ACE2 expression in different cell types. Expression level was indicated by log2(UMI counts). (f) Violin plot of ACE2 expression in different cell subtypes of skin keratinocytes. Expression level was indicated by log2(UMI counts). (g) ACE2 protein expression in human skin tissues (original magnification, ×200). FPKM, fragments per kilobase million; qRT-PCR, quantitative real-time RT-PCR; UMAP, uniform manifold approximation and projection; UMI, unique molecular identifier.
Human skin as a functional physical and immune barrier could prevent the invasion of foreign pathogens, including bacteria, fungi, and viruses. Once the skin barrier is disrupted, humans have an increased susceptibility to microbial colonization and infections (Boguniewicz and Leung, 2011). Recently, the cutaneous manifestations of SARS-CoV-2 infection were reported in 20.4% (18/88) patients and were found to be similar with other viral skin infections (Recalcati et al., 2020). In this study, we systematically analyzed ACE2 expression and ACE2-positive cell composition in skin tissues and found a high expression of ACE2 in keratinocytes, especially in differentiating keratinocytes and basal cells, suggesting that skin might be a potential target of SARS-CoV-2. Eczematoid dermatitis was induced by long-term wearing of protective clothing and contacting disinfectant, which might be a crucial factor to cause percutaneous infection in patients with coronavirus disease 2019 and medical personnel (Yan et al., 2020). Healthcare workers, especially those who worked at the first line, and patients with dermatosis with skin barrier dysfunction may be risk populations for percutaneous infection. In addition, keratinocytes may become infected through hematogenous viral spreading following inoculation of the upper airways (To et al., 2020). Our study provided a viewpoint to the routes of SARS-CoV-2 transmission, which to our knowledge were not reported previously.
In conclusion, the high expression of ACE2 on keratinocytes in human skin indicated that percutaneous transmission might be a potential risk route for SARS-CoV-2 infection, especially in conditions of skin barrier dysfunction. Also, keratinocytes are potential target cells for the viral infection when a patient is in a state of viremia. Currently, SARS-CoV-2 has been pandemic worldwide. The potential risk routes by which SARS-CoV-2 infects keratinocytes and cutaneous manifestations of SARS-CoV-2 infection should be brought to our attention as well.
Data availability statement
The raw sequence data reported in this paper has been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under project PRJCA002557. The accession number is HRA000145. Further information about sequencing data can be found at https://bigd.big.ac.cn/gsa-human/browse/HRA000145.
ORCIDs
Xiaotong Xue: http://orcid.org/0000-0002-2990-0745
Zihao Mi: http://orcid.org/0000-0002-2912-6374
Zhenzhen Wang: https://orcid.org/0000-0001-5927-2471
Zheng Pang: https://orcid.org/0000-0001-7800-1124
Hong Liu: https://orcid.org/0000-0003-4488-0372
Furen Zhang: https://orcid.org/0000-0002-3383-1973
Conflict of Interest
The authors state no conflict of interest.
Acknowledgments
The work was supported by the Academic Promotion Programme of Shandong First Medical University (2019LJ002, 2019RC007), the Youth Technology Innovation Support Project of Shandong Colleges and Universities (2019KJL003), and the Innovation Project of Shandong Academy of Medical Sciences.
Author Contributions
Conceptualization: FZ, HL; Formal Analysis: ZW; Funding Acquisition: FZ; Methodology: HL, XX, ZM; Writing - Original Draft: XX; Writing - Review and Editing: FZ, HL, ZP
Accepted manuscript published online 23 May 2020; corrected proof published online 28 June 2020
Footnotes
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2020.05.087.
Supplementary Materials and Methods
Public database acquisition
The public RNA sequencing (RNA-seq) data of ACE2 expression in various normal tissues were obtained from GEPIA2 (http://gepia2.cancer-pku.cn/#index). The public RNA-seq data of ACE2 expression in various cell types of different tissues were obtained from ARCHS4 (https://amp.pharm.mssm.edu/archs4/data.html), a web source providing most of the published RNA-seq data from human and mouse at the gene and transcript levels (Lachmann et al., 2018).
Samples
A total of 17 skin samples were obtained after written informed consent from all participants and ethical approval from Shandong Provincial Hospital for Skin Diseases, Shandong First Medical University. Six were used for single-cell RNA-seq, and eight were fixed with neutral buffered formalin and embedded in paraffin for immunochemistry. The other three skin samples used for total RNA extraction were stabilized by RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) and stored at −80 °C. The in-house data of bulk RNA-seq of 18 healthy skin samples were available for validation analysis.
Single-cell processing
Fresh skin samples were cut into approximately 3-mm3 pieces and digested using Dispase II (Gibco, Thermo Fisher Scientific, Waltham, MA) to separate dermis and epidermis. Epidermis was minced finely with dissection scissors in 5 ml 1× PBS (Gibco) and was digested in 1 ml 0.25% Trypsin-EDTA (Gibco) for 30 minutes. Subsequently, 6 ml PBS containing 10% fetal bovine serum was added to terminate the digestive reaction. Dermis was minced finely using dissection scissors and was digested in 5 ml digestion buffer (1 mg/ml collagenase P, 100 μg/ml DNase I) (Sigma-Aldrich, St. Louis, MO) for 50 minutes. After that, 5 ml PBS containing 10% fetal bovine serum was added to terminate the digestive reaction. The cell suspension was subsequently passed through a 70-μm cell strainer and centrifuged at 500g for 7 minutes. After washing twice with 1× PBS, the cell pellets were resuspended in 100 μl 1× PBS, and the cells were counted under the microscope using cell counting chamber.
Single-cell RNA-seq
The Chromium instrument and the Single Cell 3’ Reagent kit (v2) were used to prepare individually barcoded single-cell RNA-seq libraries according to the manufacturer’s protocol (10x Genomics, Pleasanton, CA). The libraries were sequenced on an Illumina NovaSeq 6000 System. The FASTQ files were analyzed with the Cell Ranger Single Cell Software Suite (v 3.1; 10x Genomics). The initial processing of all skin cells was performed using the Seurat R Package (v 2.3.4). The mean reads per cell of our single-cell RNA-seq data were 50,000 reads, and about 2,500 genes could be detected per cell. For quality control, we removed the cells with less than 200 genes or larger than 9% mitochondrial reads. The uniform manifold approximation and projection method was used for dimensionality reduction and clustering the cells. The cell types for the analysis were derived from the Human Primary Cell Atlas. Uniform manifold approximation and projection plots and violin plots were generated with Seurat in R.
Quantitative real-time RT-PCR
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instruction and reverse transcribed into cDNA (Applied Biosystems, Foster City, CA). The primers were designed by Primer-BLAST (National Center for Biotechnology Information). Quantitative real-time PCR assays were conducted in triplicate using the SYBR green method (CWBIO) on a StepOnePlus Real-Time PCR Systems (ABI) according to manufacturer’s instructions. The sequence of primers for ACE2 genes were as follows: forward primer, 5′-CGAGTGGCTAATTTGAAACCAAGAA-3′ and reverse primer, 5′-ATTGATACGGCTCCGGGACA-3′ (Zhang et al., 2019).
Immunohistochemistry
The tissue slides were deparaffinized and rehydrated, and the water-bath heating method was used to perform antigen retrieval. After washing with 1× PBS, the endogenous peroxidase was blocked by 0.3% H2O2 for 15 minutes, and nonspecific binding was blocked by 5× BSA for 25 minutes at 37 °C. The slides were incubated with rabbit anti-human ACE2 polyclonal antibody (Proteintech, Rosemont, IL) or rabbit polyclonal IgG isotype control at 1:200 dilution overnight at 4 °C. Next day, the slides were washed with 1× PBS and incubated with horseradish peroxidase–conjugated goat anti-rabbit IgG secondary antibody (Abcam, Cambridge, United Kingdom) for 30 minutes. Sections were mounted by neutral resins and examined under microscopy (EVOS FL Auto 2 Imaging System, Thermo Fisher Scientific).
References
- Boguniewicz M., Leung D.Y. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. 2011;242:233–246. doi: 10.1111/j.1600-065X.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A., Engel J., Teichmann S.A., Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2; coronavirus disease-19) Clin Exp Pediatr. 2020;63:119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recalcati S. Cutaneous manifestations in COVID-19: a first perspective. J Eur Acad Dermatol Venereol. 2020;34:e212–e213. doi: 10.1111/jdv.16387. [DOI] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Chen H., Chen L., Cheng B., Diao P., Dong L. Consensus of Chinese experts on protection of skin and mucous membrane barrier for health-care workers fighting against coronavirus disease 2019 [e-pub ahead of print] Dermatol Ther. 2020 doi: 10.1111/dth.13310. (accessed 13 Mar 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Lu S., Li T., Yu L., Zhang Y., Zeng H. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38:173. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence data reported in this paper has been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under project PRJCA002557. The accession number is HRA000145. Further information about sequencing data can be found at https://bigd.big.ac.cn/gsa-human/browse/HRA000145.