Abstract
Bacillus thuringiensis (Bt) Cyt1Aa toxin shows toxicity to mosquitoes, to certain coleopteran pests and also to red blood cells (RBC). However, its mode of action in the different target cells is not well defined. This protein is a single α−β domain pore-forming toxin, where a β sheet is wrapped by two α-helices layers. The Cyt1Aa α-helix hairpin in the N-terminal has been proposed to be involved in initial membrane binding and oligomerization, while the β sheet inserts into the membrane to form a pore that lyze the cells. To determine the role of the N-terminal α-helix hairpin region of Cyt1Aa in its mode of action, we characterized different single point mutations located in helices α−1 and α−2. Eight cysteine substitutions in different residues were produced in Bt, and we found that three of them: Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89C, lost insecticidal toxicity against Aedes aegypti larvae but retained similar or increased hemolytic activity towards rabbit RBC. Analysis of toxin binding and oligomerization using Ae. aegypti midgut brush border membrane vesicles showed that the three Cyt1Aa mutants non-toxic to Ae. aegypti were affected in oligomerization. However, these mutants were still hemolytic. Our data shows that oligomerization of Cyt1Aa toxin is essential for its toxicity to Ae. aegypti but not for its toxicity against RBC indicating that the mode of action of Cyt1Aa is different in these distinct target membranes.
Graphical Abstract
1. Introduction
Bacillus thuringiensis subs israelensis (Bti) produces different insecticidal proteins that together show high insecticidal activity against dipteran insects, as Aedes aegypti, vector of different human diseases such as dengue, yellow fever, Zika and Chikungunya (Staples et al., 2009; Kyle and Harris, 2008; Enserink, 2015). Bti produces different toxins, four of them belong to three-domain Cry (3d-Cry) family, Cry4Aa, Cry4Ba, Cry10Aa and Cry11Aa, along with two Cyt proteins, Cyt1Aa and Cyt2Ba (Berry et al., 2002). Both Cyt and Cry toxins are pore-forming toxins (PFT) that lyze larvae midgut cells by osmotic shock. In the case of the Cry toxins, toxicity relies on binding of these proteins to insect gut proteins that facilitate toxin oligomerization and membrane insertion to make a pore that kill the cells (Bravo et al., 2011). In contrast, Cyt toxins directly bind to membrane lipids facilitating its oligomerization and membrane insertion (Soberón et al., 2013). It has been shown that Cyt1Aa toxin synergize the toxicity of Cry11Aa and Cry4Ba by functioning as a receptor, facilitating oligomerization of these Cry toxins and also their membrane insertion (Pérez et al., 2005; Pérez et al., 2007; Cantón et al., 2011). The role of Cyt1Aa as receptor of Cry toxins results in high levels of toxicity and also prevents the evolution of resistance to larvicidal formulations based on Bti, since, the toxicity of Cry11Aa or Cry4 is recovered in the presence of Cyt1Aa when tested against insect populations that have developed resistance to Cry11A or to Cry4 toxins (Wirth et al., 1997; Lacey, 2007 ).
Cyt toxins are present in most Bt strains that show toxicity against dipteran insects (Soberón et al., 2013). However, Cyt1Aa has been shown to also kill certain coleopteran pests, as corn rootworm (CRW), also shows toxicity against different mammalian cell lines and to red blood cells (RBC) (Soberón et al., 2013; Bravo et al., 2018). The mode of action of Cyt toxins is not completely understood. Two different models have been proposed to explain its mechanism of action, a pore formation model and a detergent-like mechanism (Promdonkoy and Ellar, 2000; Manceva et al., 2005). In the pore formation model, it is proposed that β−6, β−7 and β−8 strands insert into the membrane while helices α−1 and α−3 are involved in lipid binding and toxin oligomerization resulting in the formation of high molecular weight oligomers that insert into the membrane forming non-specific lytic pores (Cohen et al., 2011; Promdonkoy and Ellar, 2000; Li et al., 2001, Bravo et al., 2018; López-Díaz et al., 2013). In contrast, the detergent mode of action proposes a non-specific interaction of Cyt toxins with the membrane bilayer inducing a nonspecific aggregation of the toxin on the surface of the membrane leading to lipid bilayer disassembly and cell death (Manceva et., 2005).
Oligomerization is a key step in the toxicity of Cyt toxins as demonstrated by site directed mutagenesis of helices α−1 and α−3 in different Cyt toxins. In the case of Cyt1Aa, certain helix α−3 mutants (V122E and V126E) were shown to be affected in oligomerization with a correlative defect in toxicity to Ae. aegypti larvae and also in the hemolysis of rabbit RBC suggesting that Cyt1Aa oligomerization is necessary for toxicity against different target cells (López-Díaz et al., 2013). However, certain Cyt1Aa helix α−1 mutants (A59C and A61C) were severely affected in hemolysis of RBC but did not show any effect on toxin oligomerization and had increased toxicity against Ae. aegypti and CRW, suggesting that the hemolytic and insecticidal toxicities are independent activities (Bravo et al., 2018). In the case of Cyt2Aa, a single mutant in helix α−1 (A61C) was affected toxin oligomerization and hemolytic activity but this mutant showed no defect on its insecticidal toxicity to mosquito larvae, supporting also that the mode of action of this toxin is different in both cell targets (Promdonkoy et al., 2008). To determine the role of Cyt1Aa helices α−1 and α−2 in binding, oligomerization and toxicity, we performed cysteine substitutions of some residues of these helices and analyzed their toxicity against Ae. aegypti larvae and to rabbit RBC. Our data shows that Cyt1Aa helices α−1 and α−2 are involved in toxin oligomerization required for toxicity against insects while Cyt1Aa oligomerization is not required for toxicity against RBC suggesting a distinct mode of action of Cyt1Aa in different target membranes.
2. Materials and Methods
2.1. Cyt1Aa and Cyt1Aa mutant’s production.
Bt strain 407- acrystalliferous strain (Lereclus et al., 1989) transformed with plasmid pWF45 (Wu et al., 1994) was used for Cyt1Aa production. The Cyt1Aa mutants were also produced in the Bt strain 407- acrystalliferous strain transformed with the corresponding pWF45 plasmid containing the different amino acid substitutions in cyt1Aa gene (see 2.2). All Bt strains were grown on Petri dishes with HCT sporulation medium (Muñoz-Garay et al., 2009) supplemented with 10 μg/mL erythromycin for three to four days at 30 °C until complete sporulation. Suspensions of spores/crystals were washed five times with 0.3 M NaCl, 0.01 M EDTA, pH 8.0 by centrifugation for 10 min at 16,873 xg at 4 °C. A final wash was performed with water containing 0.1 mM PMSF and pellets were stored at −20 °C until used. For crystal solubilization, spore/crystal pellets were suspended in solubilization buffer (50 mM Na2CO3/NaHCO3, pH 10.5) or 50 mM NaOH and incubated with gentle stirring at 37 °C for 2h in solubilization buffer or 2 h at 4 °C for solubilization in NaOH 50 mM. After solubilization the samples were centrifuged at 16,873 xg and the pellets were discarded. The samples solubilized with 50 mM NaOH were then dialyzed in PBS buffer (137 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4, pH 10). Soluble Cyt1Aa protoxins were activated with trypsin using a 1:50 ratio (Trypsin: Cyt1Aa) (Sigma-Aldrich Co., St Louis, MO) w/w for 1 h at 4 °C with agitation at 650 rpm. Protein concentration was determined by Bradford assay and the proteins were analyzed by SDS-PAGE with 15% acrylamide.
2.2. Cyt1Aa site directed mutagenesis
For mutagenesis of cyt1Aa gene, the pWF45 plasmid was used as template. Mutagenic primers for introducing all mutations are described in Table 1. These primers were synthesized in the Institute of Biotechnology, Universidad Nacional Autónoma de México (IBT-UNAM) facilities. Mutants were constructed using the QuickChange mutagenesis kit (Stratagene, La Jolla, CA), following the manufacturer’s instructions. After mutagenesis, the plasmids were transformed into Escherichia coli X-L1 blue strain and transformant strains were growth in LB plates supplemented with ampicillin (100 μg/ml). Plasmids were purified by using a DNA extraction kit (Qiagen, Hilden, Germany) and sequenced at the IBT-UNAM facilities. Plasmids were transformed into E. coli SCS110 (Dam- y Dcm-) for preparing plasmids for Bt transformation. Bt 407 strain was used for transformation of plasmids as described (Lereclus et al., 1989) and transformants were selected in LB supplemented with erythromycin 10 μg/ml at 30 °C.
Table 1.
Sequence of mutagenic oligonucleotides
| Mutation | Location | Oligonucleotide sequence |
|---|---|---|
| I52C | Helix α−1 | 5’-GAA ATT GAT AAT CCG AAT TAT TGT TTG CAA GCA ATT ATG TTA G −3´ |
| L53C | Helix α−1 | 5´-GAT AAT CCG AAT TAT ATA TGT CAA GCA ATT ATG TTA GCA AAT G-3´ |
| Q54C | Helix α−1 | 5´-GAT AAT CCG AAT TAT ATA TTG TGT GCA ATT ATG TTA GCA AAT GC-3´ |
| A55C | Helix α−1 | 5´-CCG AAT TAT ATA TTG CAA TGT ATT ATG TTA GCA AAT GCA TTT C-3´ |
| I56C | Helix α−1 | 5´-CCG AAT TAT ATA TTA CAA GCA TGT ATG TTA GCA AAT GCA TTT C-3´ |
| A65C | Helix α−1 | 5´-GCA AAT GCA TTT CAA AAT TGT TTA GTT CCC ACT TCT ACA-3´ |
| K83C | Helix α−2 | 5´-GCC CTA CGC TTT AGT ATG GCA TGT GGT TTA GAA ATC GCA AAC AC-3´ |
| G84C | Helix α−2 | 5´-CTA CGC TTT AGT ATG GCA AAA TGT TTA GAA ATC GCA AAC AC-3´ |
| L85C | Helix α−2 | 5´-C TTT AGT ATG GCA AAA GGT TGT GAA ATC GCA AAC ACA ATT AC-3´ |
| E86C | Helix α−2 | 5´-GT ATG GCA AAA GGT TTA TGT ATC GCA AAC ACA ATT ACA CC-3´ |
| A88C | Helix α−2 | 5´-GCA AAA GGT TTA GAA ATC TGT AAC ACA ATT ACA CCG ATG-3´ |
| N89C | Helix α−2 | 5´-CAAAAGGTTTAGAAATCGCATGCACAATTACACCGATGGGTGC-3 |
2.3. Insect bioassays and hemolysis assay.
For bioassays, ten Ae. aegypti 4th instar larvae were placed in cups containing 100 ml dechlorinated water and treated with at least five different concentrations of spore/crystals suspensions of Cyt1Aa or Cyt1Aa mutants (100 to 3,000 ng/ml) in triplicate. The larvae viability was scored after 24 h. Controls without toxin were included in the bioassay. Ae. aegypti colony was maintained at the IBT-UNAM facilities at 28 ºC, 75 % humidity, and a 12 h: 12 h, light: dark, photoperiod. The mean lethal concentration (LC50) was determined by Probit analysis (Polo-Plus LeOra Software) using statistical parameters with the data obtained in triplicate from at least three independent assays.
Hemolysis assays were performed as previously described using rabbit RBC (Rodriguez-Almazan et al., 2011) Briefly, RBC were washed three times with buffer 0.1 M dextrose, 0.07 M NaCl, 0.02 M sodium citrate, 0.002 M citrate, pH 7.4 and suspended in the same buffer at 2×108 cells/ml. In a final volume of 200 μl of the same buffer, we add 20 μl of RBC suspension (4×106 RBC) and incubate them with different concentrations of Cyt1Aa or Cyt1Aa mutant toxins (20–6,400 ng) at 37 °C for 30 min in 96 wells microtiter plates. After centrifugation at 1076 xg for 5 min at 4 °C, the supernatants were used to determine absorbance at 405 nm. To obtain 100 % RBC lysis, 20 μl of RBC suspension was incubated with 180 μl of dechlorinated water, then incubated and scored as above. The half maximal effective concentrations (EC50) were estimated from dose response curves adjusting the curves by Sigma plot 10. Three repetitions with different RBC samples were used for t-test analysis by GraphPad Prism program.
2.4. Preparation of brush border membrane vesicles (BBMV).
Ae. aegypti 4th instar larvae were used to dissect the midgut tissue. Midguts were used for BBMV extraction using the MgCl2 differential extraction method previously described (Wolfersberger et al., 1987). BBMV were suspended in 50 mM Na2CO3, pH 9 and stored at −70 °C until used. Protein concentration in BBMV sample preparation was determined by a Lowry DC protein assay (BioRad, Hercules, CA).
2.5. Binding and oligomerization assays
For binding assays of Cyt1A and Cyt1Aa mutants to BBMV, the activated toxins were first labeled with biotin N-hydroxysuccimide ester (Amersham, Litle Chanfont, England) using 20 μl of a biotin solution (25 mg/ml) and 300 μg of Cyt1Aa proteins. The reaction was incubated at room temperature for 1 h and free biotin was discarded using Sephadex-25 column equilibrated and washed with PBS by centrifugation for 2 min at 805 xg on a free angle rotor. Labeled proteins were visualized by separating the labeled Cyt1Aa proteins in 15 % SDS-PAGE and electro blotted to PVDF membrane (Millipore, Burlington, MAS). We used 2 % Tween-20/PBS solution for blocking the membrane for 20 min and washed twice with 0.1 % Tween-20/PBS. HRP-streptavidin (GE Healthcare, Waukesha, WI) was added at 1: 5000 dilution in 1 % Tween 20/PBS and incubated for 1 h at room temperature, washed twice as before and then two times with PBS. The membrane was revealed with SuperSignal reagent (Thermofisher, Waltham, MAS). For binding analysis of the labeled Cyt1Aa toxins to BBMV we incubate 10 μg BBMV protein with 10 nM of biotinylated Cyt1Aa protein for 1 h at room temperature. Precipitation controls were performed without adding BBMV. The membrane fraction was separated by centrifugation (30 min at 117,000 xg) and the membrane pellet was boiled 3 min in loading buffer before loading into 15 % SDS-PAGE gels. Gels were transferred to PVDF membrane (Millipore, Burlington, MAS) for 16 h at 4 °C and 150 mA and biotin-labeled toxins were detected as above.
For oligomerization assays, 200 ng of soluble Cyt1Aa protoxins were incubated with 10 μg of BBMV or 95 μl of RBC suspension (described above) and 10 ng of trypsin (Sigma-Aldrich Co., St Louis, MO) for 1 h at room temperature. Samples were heated at 50 °C for 3 min in loading buffer and loaded onto 15 % SDS-PAGE gels. Gels were transferred to a PVDF membrane (Millipore, Burlington, MAS) for 16 h at 4 °C and 150 mA. The membranes were blocked for 1 h at room temperature with 5 % skimmed milk in 0.1 % Tween-20/PBS. The membranes were washed with 0.1 % Tween-20/PBS. Cyt1Aa was detected by incubating 1 h with a polyclonal anti-Cyt1Aa antibody at 1: 70,000 dilution in 0.1 % Tween-20/PBS. Washing was done as before, followed by incubation with 1: 10,000 HRP labeled anti-rabbit antibody (Sigma-Aldrich Co., St Louis, MO) in 0.1 % Tween-20/PBS. After washing, the membranes were revealed with SuperSignal reagent (Thermofisher, Waltham, MAS).
3. Results
3.1. Characterization of Cyt1Aa helices α−1 and α−2 mutants.
To determine the role of helix α−1 (49PNYILQAIMLANAFQNAL66) in Cyt1Aa mode of action, we previously characterized some helix α−1 residues from the hydrophobic face of the helix by performing cysteine substitutions (L58C, A59C, A61C and F62C) (Bravo et al., 2018). Only Cyt1AaA59C and Cyt1AaA61C mutants produced stable proteins. These mutants retained toxicity to Ae. aegypti or CRW and showed reduced hemolysis of rabbit
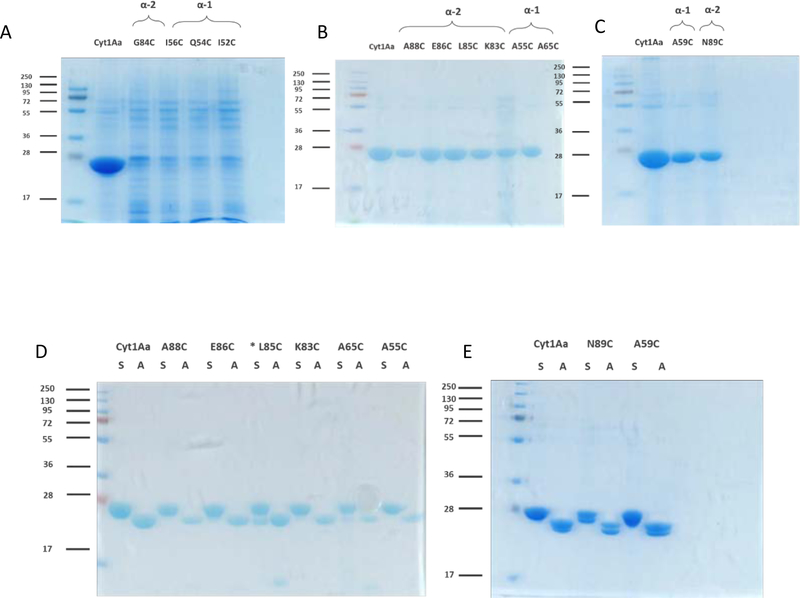
RBC without affecting toxin oligomerization (Bravo et al., 2018). To determine the role of the other helix α−1 and helix α−2 residues (83KGLEIANT90) we constructed six additional helix α−1 mutations (I52C, L53C, Q54C, A55C, I56C and A65C) and six helix α−2 mutations (K83C, G84C, L85C, E86C, A88C and N89C). Cyt1AaL53C mutant could not be obtained after a number of attempts and was not included in further analysis. After transfection of the mutant plasmids into a Bt acrystalliferous strain, the remaining 11 mutants and the helix α−1 A59C mutant previously characterized (Bravo et al., 2018) were produced in sporulation medium as described in Materials and Methods. Figure 1A shows that Cyt1AaI52C, Cyt1AaQ54C, Cyt1AaI56C and Cyt1AaG84C mutant proteins were produced at very low levels and were not further characterized.
Figure 1. Production and activation of Cyt1Aa helices α−1 and α−2 mutants.
A. Analysis of spore/crystal samples of Cyt1Aa and Cyt1AaG84C, Cyt1AaI56C, Cyt1AaQ54C and Cyt1AaI52C. B. Analysis of spore/crystal samples of Cyt1Aa, Cyt1AaA88C, Cyt1AaE86C, Cyt1AaL85C, Cyt1AaK83C, Cyt1AaA55C, Cyt1AaA65C. C. Analysis of spore/crystal samples of Cyt1Aa and Cyt1AaA59C, Cyt1AaN89C after sporulation of Bt strain containing each of these cyt1Aa genes. D. and E. Analysis of soluble Cyt1Aa protoxins and activated Cyt1Aa proteins after activation with trypsin as described in Materials and Methods. D. Cyt1Aa, Cyt1AaA88C, Cyt1AaE86C, Cyt1AaL85C, Cyt1AaK83C, Cyt1AaA65C and Cyt1AaA55C. E. Cyt1Aa, Cyt1AaN89C and Cyt1AaA59C. All Cyt1Aa spore crystal samples were solubilized with 50 mM Na2CO3/NaHCO3 pH 10.5 and treated with trypsin as indicated in Materials and Methods. * Cyt1Aa85C was solubilized with 50 mM NaOH and treated with trypsin as indicated in Materials and Methods. Molecular weight markers are indicated in kDa in the left of each panel.
The rest of the mutants (Fig. 1B, C) were solubilized (Fig. 1D, E). All mutants were solubilized in carbonate buffer pH 10.5 except for Cyt1AaL85C mutant that showed reduced solubilization (data not shown). However, Cyt1AaL85C mutant was efficiently solubilized in 50 mM NaOH (Fig. 1D). Activation of the soluble proteins with trypsin showed that all protoxin mutants, including Cyt1AaL85C, were correctly activated yielding an activated toxin of 24 kDa (Fig. 1D, E). These results indicate that no major structural effects were caused in these proteins since all mutants were correctly processed by trypsin treatment.
3.2. Insecticidal and hemolytic activities of Cyt1Aa helices α−1 and α−2 mutants.
To determine the effect of the different mutations in helices α−1 and α−2 on toxicity we performed bioassays against Ae. aegypti 4th instar larvae. Table 2 shows that Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89C lost toxicity against Ae. aegypti larvae. The other mutants retained toxicity against Ae. aegypti (Table 2). Cyt1AaA59C showed reduced toxicity as previously reported (Bravo et al., 2018) while Cyt1AaA88C showed almost three-fold higher insecticidal toxicity compared to Cyt1Aa (Table 2).
Table 2.
Toxicity of Cyt1Aa α−1 and α−2 mutants against Ae. aegypti
| Toxin | LC50 (ng/ml) |
|---|---|
| Cyt1Aa | 256 (200 ± 312) a |
| Cyt1AaA55C | 619 (486 ± 782) a |
| Cyt1AaA59C | 1531 (1332 ± 1724) a |
| Cyt1AaA65C | >3000 |
| Cyt1AaK83C | 233 (118 ± 343) a |
| Cyt1AaL85C | > 3000 |
| Cyt1AaE86C | 334 (219 ± 445) a |
| Cyt1AaA88C | 92 (64 ± 129) a |
| Cyt1AaN89C | > 3000 |
95% en fiducial limits
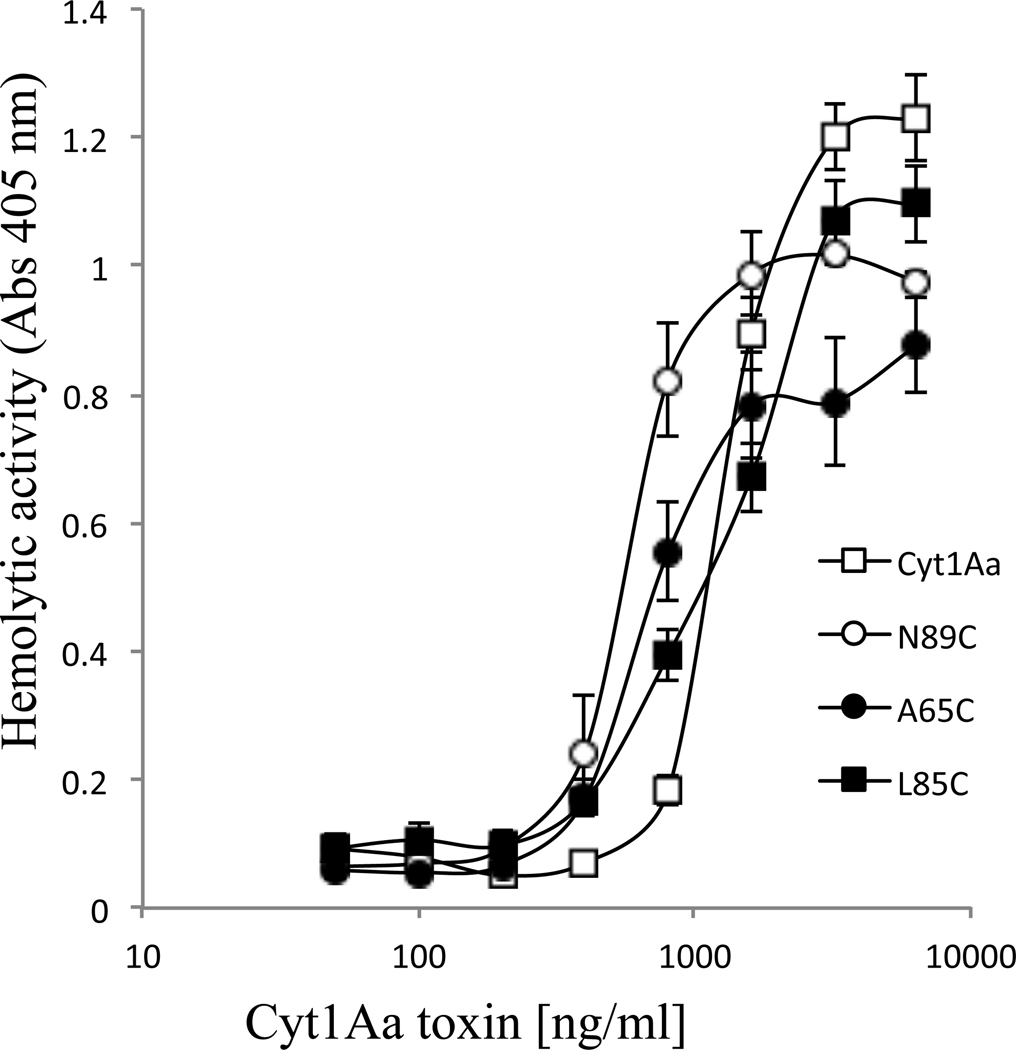
To determine the effect of helices α−1 and α−2 mutations on hemolytic activity, the hemolysis assays against rabbit RBC were performed as described in Materials and Methods. Table 3 shows that, as previously reported, Cyt1AaA59C lost its hemolytic activity (Bravo et al., 2018). In contrast all the other Cyt1Aa mutants, including the non-toxic Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89C, retained significant hemolytic activity. Table 3 also shows that Cyt1AaN89C showed almost 7-fold higher hemolytic activity than Cyt1Aa. Figure 2 shows a dose response curve of the hemolytic activity of Cyt1Aa and the Cyt1Aa mutants that were affected in insecticidal activity.
Table 3.
Hemolytic activity of Cyt1Aa α−1 and α−2 mutants against rabbit RBC
| Toxin | EC50 (ng/ml) |
|---|---|
| Cyt1Aa | 1297 (1293 – 1301.29) a |
| A55C | 813 (806 – 821) a |
| A59C | >5000 |
| A65C | 758 (740 – 780) a |
| K83C | 709 (702 – 718) a |
| L85C | 836 (825 – 851) a |
| E86C | 823 (816 – 831) a |
| A88C | 1626 (1619 – 1633) a |
| N89C | 186 (169 – 208) a |
95% en fiducial limits
Figure 2. Hemolytic activity of activated Cyt1Aa non-toxic mutants.
The hemolytic activity of different protein concentration was analyzed against rabbit RBC as described in Materials and Methods. Hemolysis saturation curves of Cyt1Aa and the non-toxic Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89C are shown.
3.3. Analysis of binding and oligomerization of non-toxic Cyt1Aa mutants
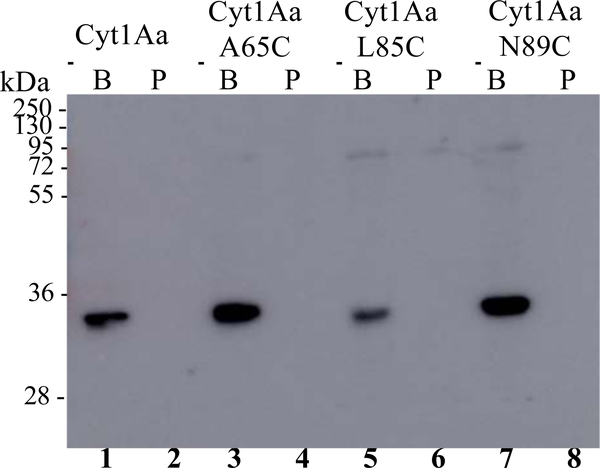
To characterize the three Cyt1Aa mutants that were non-toxic to Ae. aegypti, binding assays of biotinylated activated toxins to BBMV from Ae. aegypti were performed as described in Materials and Methods. Figure 3 shows that Cyt1Aa and Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89 mutant proteins bound to BBMV, although Cyt1AaL85C showed reduced binding compared to Cyt1Aa. Next, we performed oligomerization assays of Cyt1Aa with Ae. aegypti BBMV. For oligomerization assays Cyt1Aa protoxins were incubated simultaneously with BBMV and trypsin and the sample was centrifuged, the pellet and supernatants were separated and moderately heated (50 ºC, 3 min) before loading into SDS-PAGE. Proteins in the gel were transferred to a PVDF membrane and oligomers were detected in western blots using anti-Cyt1Aa antibody as described in Materials and
Figure 3. Binding of Cyt1Aa helices α−1 and α−2 mutants.
Biotin-labeled activated Cyt1Aa proteins were incubated with (lanes labeled as B: 1, 3, 5 and 7) or without (lanes labeled as P: 2, 4, 6, 8) Ae. aegypti BBMV, centrifuged and the pellets were loaded on SDS-PAGE, electro-transferred to PVDF membranes and biotinylated toxins were detected with streptavidin coupled to peroxidase as indicated in Materials and methods. Lanes correspond to, Cyt1Aa (lanes 1 and 2), Cyt1AaA65C (lanes 3 and 4), Cyt1AaL85C (lanes 5 and 6) and Cyt1AaN89C (lanes 7 and 8). Molecular weight markers are indicated in kDa in the left of the figure.
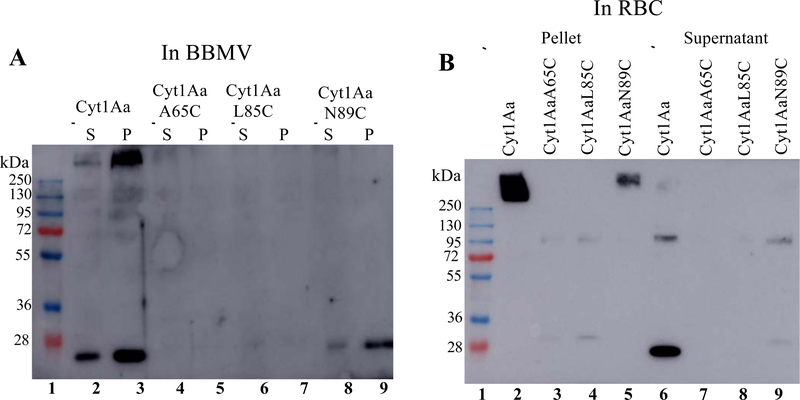
Methods. Figure 4A shows that high molecular weight band oligomers were only observed in the samples of the Cyt1Aa. These data show that the three non-toxic Cyt1Aa mutants were affected in toxin oligomerization in BBMV membranes correlating with their defects in insecticidal activity. Since the three non-toxic mutants retained high hemolytic activity, similar oligomerization assays were performed with rabbit RBC. Figure 4B shows that Cyt1Aa also formed high molecular weight oligomers preferentially in the pellet sample. In contrast, no oligomers were observed in samples from Cyt1AaA65C or Cyt1AaL85C. Interestingly, the Cyt1AaN89C pellet sample revealed high molecular weight oligomers although with substantial lower yield than the Cyt1Aa sample (Fig. 4B).
Figure 4. Oligomerization of Cyt1Aa and Cyt1Aa mutants in BBMV or RBC.
A. Oligomerization of Cyt1Aa and Cyt1Aa mutants with Ae. aegypti BBMV. Soluble Cyt1Aa protoxins were incubated with BBMV and trypsin, the samples were separated by centrifugation and the Cyt1Aa oligomers were detected in the pellets and supernatants after heating the samples (50 °C, 3 min) with loading buffer before loading into SDS-PAGE. The gels were blotted to PVDF membranes and Cyt1Aa oligomers were detected using an anti-Cyt1Aa antibody as indicated in Materials and Methods. Lane 1. Molecular weight marker. Lanes 2 and 3 -Cyt1Aa supernatant (S) or pellet (P) fraction, lanes 4 and 5 - supernatant or pellet fraction of Cyt1AaA65C, lanes 6 and 7 to supernatant or pellet fraction of Cyt1AaL85C, lanes 8 and 9 - supernatant or pellet fraction of Cyt1AaN89C. B. Oligomerization of Cyt1Aa and Cyt1Aa mutants with rabbit RBC. Soluble Cyt1Aa protoxins were incubated with RBC and trypsin, the samples were separated by centrifugation and the Cyt1Aa oligomers were detected in the pellets and supernatants after heating the samples (50 °C, 3 min) with loading buffer before loading into SDS-PAGE. The gels were blotted to PVDF membranes and Cyt1Aa oligomers were detected using an anti-Cyt1Aa antibody as indicated in Materials and Methods. Lane 1. Molecular weight marker. Image shows analysis of pellets (lanes 2, 3, 4 and 5) and supernatant fractions (lanes 6, 7, 8 and 9). Samples of Cyt1Aa (lanes 2 and 6), Cyt1AaA5C (lanes 3 and 7), Cyt1AaL85C (lanes 4 and 8) and Cyt1AaN89C (lanes 5 and 9). Molecular weight markers are indicated in kDa in the left of each panel.
4. Discussion
Two different models for Cyt1Aa toxin mode of action have been proposed based on its interaction with synthetic lipid membranes -a pore formation model and a detergent mode of action (Promdonkoy and Ellar, 2003; Manceva et al., 2005). Also, it has been suggested that both models of Cyt1Aa action are not exclusive and could both be functional depending on toxin concentration (Butko, 2003; Manceva et al., 2005). However, no experimental evidence has been provided regarding the occurrence of both modes of action in insect BBMV. Previous work showed that Cyt1Aa helix α−3 mutants, affected in oligomerization, were affected in toxicity to mosquito larvae and also to rabbit RBC, suggesting that oligomerization is required for toxicity to both cell types and that pore formation was responsible for toxicity in both systems (Lopez-Diaz et al., 2013). However, characterization of two helix α−1 mutants in conserved positions of Cyt1Aa (A59C) or Cyt2Aa (A61C) showed that the insecticidal and hemolytic activities of Cyt proteins could be separated since both mutants Cyt1Aa59C and Cyt2AaA61C lost hemolytic activity but retained insecticidal activity, Cyt1Aa59C against Ae. aegypti and CRW and Cyt2AaA61C against Ae. aegypti and Culex quinquefasciatus mosquitoes (Bravo et al., 2018; Promdonkoy et al., 2008), suggesting that Cyt1Aa may have a different mode of action in insect cells and in RBC. Here, we characterized different Cyt1Aa mutants in helices α−1 and α−2 since it was proposed that these helices were involved toxin oligomerization and in lipid binding to the target membranes. Three Cyt1Aa mutants in these helices (Cyt1AaA65C, Cyt1AaL85C and Cyt1AaN89) were shown to lose toxicity to Ae. aegypti larvae but retained full toxicity to RBC supporting that Cyt proteins display a distinct mode of action in both cell types.
Oligomerization of different PFT has been shown to be essential for their toxicity against their different targets (Abrami et al., 2000; Berube and Bubeck-Wardenburg, 2013). This is also the case for the 3d-Cry toxins that show different insecticidal specificity (Pacheco et al., 2018). Oligomerization assays of the three non-toxic Cyt1Aa mutants with Ae. aegypti BBMV showed that these three Cyt1Aa mutants affected in insecticidal activity were affected in oligomer formation, which correlates with their lack of toxicity against mosquito larvae. Interestingly, oligomerization assays using RBC also confirmed that two Cyt1Aa mutants, Cyt1AaN65C and Cyt1AaL85C, did not form oligomers in contrast to Cyt1Aa while Cyt1AaN89C could form oligomers with reduced efficiency compared to Cyt1Aa. Despite the defect observed in oligomerization, the three mutants showed high hemolytic activity. The different effect of N89C mutation on oligomer formation in both cell types could also indicate a distinct mode of action. Alternatively, the different oligomerization phenotypes of Cyt1AaN89C in both cell membranes could be a matter of oligomerization efficiency in both target membranes, since Cyt1AaN89C showed substantially reduced oligomerization in RBC compared to Cyt1Aa. Nevertheless, these results clearly show that oligomerization is not required for Cyt1Aa toxicity against RBC, indicating also a distinct mode of action in both cell types and that pore formation is likely responsible for toxicity of Cyt1Aa in mosquitoes. What is the mechanism of action of Cyt1Aa in RBC? It is tempting to speculate that the detergent mode of action could be responsible for toxicity to RBS since oligomerization of Cyt1Aa is not involved. The determination of the Cyt1Aa toxin structure in the different target membranes could provide important information of the distinct mode of action of Cyt toxins in its different target cells.
Highlights.
Cyt1Aa shows toxicity to Aedes aegypti larvae and to red blood cells
Cyt1Aa α−1 and α−2 mutants lost toxicity to Ae. aegypti larvae but not to red blood cells
Cyt1Aa α−1 and α−2 mutants affected in toxicity show defects in toxin oligomerization
Cyt1Aa oligomerization is required for toxicity to mosquito larvae but not for red blood cell toxicity
Acknowledgements
To Lizbeth Cabrera for technical assistance. This research was supported in part by DGAPA/UNAM IN202718, CONACyT Fronteras de la Ciencia ID2287 and NIH 2R01 AI066014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrami L, Fivaz M,, van der Goot FG 2000. Adventures of a pore-forming toxin at the target cell surface. Trends Microbiol. 8, 168–172. [DOI] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zarisky A, Parkhill J 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68, 5082–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube BJ, Bubeck-Wardenburg J. 2013. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins (Basel). 5, 1140–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Likitvivatanavong S, Gill SS, Soberón M. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, López-Diaz JA, Yamamoto T, Harding K, Zhao JJ, Mendoza G, Onofre J, Torres MC, Nelson ME, Wu G, Sethi A, Soberón M. 2018. Susceptible and mCry3A resistant corn rootworm larvae killed by a non-hemolytic Bacillus thuringiensis Cyt1Aa mutant. Sci. Rep. 8(1), 17805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko P. 2003. Cytolytic toxin Cyt1Aa and its mechanism of membrane damage: Data and hypotheses. Appl. Environm. Microbiol. 69, 2415–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantón PE, Reyes EZ, Ruiz I, Bravo A, Soberón M, 2011. Binding of Bacillus thuringiensis subsp. israelensis Cry4Ba to Cyt1Aa has an important role in synergism. Peptides. 32, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Albeck S, Ben-Dov E, Cahan R, Firer M, Zaritsky A, Dym O. 2011. Cyt1Aa toxin: crystal structure reveals implications for its membrane-perforating function. J. Mol. Biol. 413, 804–814. [DOI] [PubMed] [Google Scholar]
- Enserink M. 2015. Infectious diseases. An obscure mosquito-borne disease goes global. Science. 350, 1012–1013. [DOI] [PubMed] [Google Scholar]
- Kyle JL, Harris E.. 2008. Global spread and persistence of dengue. Ann. Rev. Microbiol. 62, 71–92. [DOI] [PubMed] [Google Scholar]
- Lacey LA 2007. Bacillus thuringiensis serovar israelensis and Bacillus sphaericus for mosquito control. J. Am. Mosq. Control. Assoc. 23, 133–163. [DOI] [PubMed] [Google Scholar]
- Lereclus D, Arantes O, Chaufaux J, Lecadet MM 1989. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60, 211–218. [DOI] [PubMed] [Google Scholar]
- Li J, Derbyshire DJ, Promdonkoy B, Ellar DJ 2001. Structural implications for the transformation of the Bacillus thuringiensis delta-endotoxins from water-soluble to membrane-inserted forms. Biochem. Soc. Trans. 29, 571–577. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz JA, Canton PE, Gill SS, Soberón M, Bravo A. 2013. Oligomerization is a key step in Cyt1Aa membrane insertion and toxicity but not necessary to synergize Cry11Aa toxicity in Aedes aegypti larvae. Environm. Microbiol. 15, 3030–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceva SD, Pusztai-Carey M, Russo PS, Butko P. 2005. A detergent-like mechanism of action of the cytolytic toxin Cyt1A from Bacillus thuringiensis var. israelensis. Biochem. 44, 589–597. [DOI] [PubMed] [Google Scholar]
- Muñóz-Garay C, Rodriguez-Almazan CR, Aguilar JN, Portugal L, Gómez I, SaabRincon G, Soberón M, Bravo A. 2009. Oligomerization of Cry11Aa from Bacillus thuringiensis has an important role in toxicity against Aedes aegypti. Applied and Environmental Microbiology 75, 7548–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco S, Gómez I, Sánchez J, García-Gómez BI, Czajkowsky DM, Zhang J, Soberón M, Bravo A. 2018. Helix α−3 inter-molecular salt bridges and conformational changes are essential for toxicity of Bacillus thuringiensis 3d-Cry toxin family. Scient. rep. 8, 10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez C, Fernández LE, Sun J, Folch JL, Gill SS, Soberón M, Bravo A. 2005. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 18303–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez C, Muñoz-Garay C, Portugal LC, Sánchez J, Gill SS, Soberón M, Bravo A. 2007. Bacillus thuringiensis ssp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell Microbiol. 9, 2931–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promdonkoy B, Ellar DJ 2000. Membrane pore architecture of a cytolytic toxin from Bacillus thuringiensis. Biochem. J. 350, 275–282. [PMC free article] [PubMed] [Google Scholar]
- Promdonkoy B, Ellar DJ 2003. Investigation of the pore-forming mechanism of a cytolytic delta-endotoxin from Bacillus thuringiensis. Biochem. J. 374, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promdonkoy B, Rungrod A, Promdonkoy P, Pathaichindachote W, Krittanai C, Panyim S. 2008. Amino acid substitutions in alpha A and alpha C of Cyt2Aa2 alter hemolytic activity and mosquito-larvicidal specificity. J. Biotechnol. 133, 287–293. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Almazan C, Ruiz de Escudero I, Cantón PE, Muñoz-Garay C, Pérez C, Gill SS, Soberón M, Bravo A. 2011. The amino- and carboxyl-terminal fragments of the Bacillus thuringiensis Cyt1Aa toxin have differential roles on toxin oligomerization and pore formation. Biochem. 50, 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón M, López-Díaz JA, Bravo A. 2013. Cyt toxins produced by Bacillus thuringiensis: A protein fold conserved in several pathogenic microorganims. Peptides. 41, 87–93. [DOI] [PubMed] [Google Scholar]
- Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49, 942–948. [DOI] [PubMed] [Google Scholar]
- Wirth MC, Georghiou GP, Federeci BA, 1997. CytA enables CryIV endotoxins of Bacillus thuringiensis to overcome high levels of CryIV resistance in the mosquito, Culex. Proc. Natl. Acad. Sci. U.S.A. 94, 10536–10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfersberger M, Lüthy P, Maurer A, Parenti P, Sacchi FV, Giordana B, Hanozet GM 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A, 301–308. [Google Scholar]
- Wu D, Johnson JJ, Federici BA 1994. Synergism of mosquitocidal toxicity between CytA and CryIV proteins using inclusions produced from cloned genes of Bacillus thuringiensis subsp. israelensis. Mol. Microbiol. 13, 965–972. [DOI] [PubMed] [Google Scholar]