Abstract
Background
Damage to olfactory sensory neurons (OSNs), situated within the neuroepithelium of the olfactory cleft, may be associated with anosmia. Although their direct contact with the nasal airspace make OSNs vulnerable to injury and death, multiple mechanisms maintain epithelium integrity and olfactory function. We hypothesized that BMI1, a Polycomb protein found to be enriched in OSNs, may function in neuroprotection. Here, we explored BMI1 function in a mouse model.
Methods
Utilizing a mouse genetic approach to delete Bmi1 selectively in mature OSNs, we investigated changes in OE homeostasis by performing immunohistochemical, biochemical, and functional assays. RT-qPCR, immunostaining, and electro-olfactograms were used to compare gene expression, cell composition, and olfactory function in OSN-specific BMI1 knockout mice (n=3–5) and controls. Chromatin studies were also performed to identify protein-DNA interactions between BMI1 and its target genes (n=3).
Results
OSN-specific BMI1 knockout led to increased neuron death and basal cell activation. Chromatin studies suggested a mechanism of increased neurodegeneration due to de-repression of a pro-apoptosis gene, p19ARF. Despite the increased turnover, we found that olfactory neuroepithelium thickness and olfactory function remained intact. Our studies also revealed the presence of additional Polycomb group proteins that may compensate for the loss of BMI1 in mature OSNs.
Conclusion
The olfactory neuroepithelium employs multiple mechanisms to maintain epithelial homeostasis. Our findings provide evidence that in a mouse model of BMI1 deletion, the overall integrity and function of the olfactory neuroepithelium are not compromised, despite increased neuronal turnover, reflecting a remarkable reparative capacity to sustain a critical sensory system.
Keywords: Olfactory sensory neurons, olfaction, olfactory disorders, anosmia, Polycomb group proteins
INTRODUCTION
The olfactory neuroepithelium (OE) lines the olfactory cleft in the superior-medial region of the nasal cavity, and is the peripheral organ for the sense of smell. Olfactory disorders remain challenging to manage, as treatment options are limited.1 Obstruction or inflammation can contribute to anosmia, however evidence suggests that damage to olfactory sensory neurons (OSNs) in the OE may be a common cause, in situations such as aging or post-viral olfactory disorder.2,3 Therefore, efforts to understand mechanisms involved in adult OSN survival and repair are important for development of treatments for olfactory loss.
In animal models, it is clear that OSNs are vulnerable to experimental injury, requiring mechanisms to protect and replace them to maintain olfactory function.4 One mechanism of protection is a capacity for ongoing neurogenesis due to the presence of multipotent basal cells that are able to differentiate into all the cell types within the OE, including the OSNs.5–7 Other neuroprotective mechanisms have been identified in the OE, including the unfolded protein response in OSNs and the presence of biotransformation enzymes of the cytochrome P450 family within olfactory sustentacular cells.8–10 We recently identified the expression of BMI1 in OSNs,11 and we focus attention here to identifying its potential role in these cells.
In other cell types, such as cortical neurons or auditory hair cells, the expression of BMI1, a Polycomb group protein, has been shown to prevent oxidative damage.12,13 In the aging brain, BMI1 expression is down-regulated, leading to the increased expression of pro-apoptotic, pro-oxidant, and senescence-related genes.14,15 The mechanism by which BMI1 confers neuroprotection in the central nervous system is by acting via the Polycomb repressive complex 1 (PRC1), a multimeric complex that regulates gene expression via post-translational histone modifications and changes to chromatin architecture. As part of PRC1, BMI1 represses the Cdkn2a locus, which encodes p16INK4a and p19ARF, genes that initiate apoptosis and inhibit anti-oxidant activity.
In the adult OE, BMI1 is expressed in globose basal cells and mature OSNs.11,16 Although our lab has previously shown that BMI1 regulates cell proliferation in globose basal cells, the role of BMI1 in the post-mitotic OSNs is unknown. Utilizing a mouse genetic approach to delete Bmi1 selectively in mature OSNs, we investigated changes in OE homeostasis by performing immunohistochemical, biochemical, and functional assays. Since OSN damage and eventual loss appears to be a hallmark common to several olfactory disorders, this work to define the role of BMI1 in mature OSNs will provide insights into mechanisms of OSN neuroprotection and overall OE maintenance.
MATERIALS AND METHODS
Animals
The Institutional Animal Care and Use Committee at the University of Miami approved all procedures. Mouse strains included OMP-Cre17 (Stock# 006668, The Jackson Laboratory) and Bmi1-flox18 strain possessing loxP sites flanking exons 4–8 of the mouse Bmi1 gene on chromosome 2 (Stock# 028572, The Jackson Laboratory).
Tissue processing, staining, and analysis
Two hours before time of sacrifice, animals were administered with 50μg per 1g body weight of 5-bromo-2’-deoxyuridine (BrdU). Animals were put to sleep with a ketamine/xylazine cocktail, exsanguinated, and perfused with phosphate buffered saline (PBS), followed by 4% paraformaldehyde in PBS. The nasal and olfactory bulb tissue was dissected from the head and post-fixed in 4% paraformaldehyde for 2 hours at room temperature. The tissue was cryoprotected and decalcified in PBS containing 30% sucrose and 250mM EDTA at 4°C for 1 week, embedded in OCT compound, placed under vacuum at room temperature for 2 hours, and snap frozen in liquid nitrogen.
Coronal sections 10μm thick were cut using a Leica CM1850 cryostat and mounted onto Superfrost Plus Micro Slides (VWR). Tissue sections were rinsed in PBS and then exposed to an ethanol gradient from 70–95-100–95-70% in order to dehydrate then rehydrate the tissue. Following the required pre-treatments, tissue sections were incubated in 10% horse serum, 5% bovine serum albumin, 4% nonfat dry milk, 0.1% Triton X-100 for 45 minutes at room temperature. Primary antibodies were diluted in this same solution and incubated overnight in a humidified chamber at 4°C. Working dilutions of all primary antibodies are listed in Table 1. Primary antibodies were removed by washing. All washes following primary antibody incubation were done using PBS with 0.3% Triton X-100 for 3×5-minute incubations on a rocker. Fluorescent dye-conjugated secondary antibodies were diluted in block and the sections incubated at room temperature for 45 minutes. Sections were counterstained with 4’,6-daimidino-2-phenylindole (DAPI) and coverslips were mounted using Vectashield (Vector Laboratories) for imaging.
Table 1.
Antibodies used for immunofluorescence staining
| Antibody | Dilution | Source | Identifier |
|---|---|---|---|
| Mouse anti-BMI1 | 1:500 | Cell Signaling Technologies | Cat# ab14389; AB_2065390 |
| Rabbit anti-BMI1 | 1:500 | Abcam | Cat# 6964; AB_10828713 |
| Mouse anti-OMP | 1:1000 | Santa Cruz | Cat# sc-365818; AB_10842164 |
| Rabbit anti-GAP43 | 1:400 | Abcam | Cat# ab75810; AB_1310252 |
| Mouse anti-MEL-18 | 1:250 | Santa Cruz | Cat# sc-515329; AB_2687587 |
| Rabbit anti-H2AK119ub | 1:2000 | Cell Signaling Technologies | Cat# 8240; AB_10891618 |
| Mouse anti-BrdU | 1:10 | Becton Dickinson | Cat# 347580; AB_10015219 |
| Rabbit anti-c-KIT | 1:50 | Cell Signaling Technologies | Cat# 3074; AB_10829442 |
| Rabbit anti-Cleaved Caspase-3 | 1:800 | Cell Signaling Technologies | Cat# 9664; AB_2070042 |
Staining was analyzed on an Olympus IX81 epifluorescent microscope or a Zeiss LSM-710 confocal microscope. Pseudo coloration and brightness/contrast adjustment were performed using ImageJ. Parallel settings for capture and adjustment were used for images included in the same figure.
Images captured were opened in ImageJ/FIJI and the Cell Counts tool was used to quantify cells positive for markers of interest per length of olfactory epithelium. Three to 5 fields across 3 tissue sections were analyzed per side from 3–5 biologic replicates. Comparisons and statistics were performed between control and Bmi1-KO animals in comparable anterior/posterior, dorsal/ventral, and medial/lateral regions.
RNA extraction, cDNA synthesis, and quantitative reverse transcription (RT-qPCR)
Total RNA was extracted with the Quick-RNA MicroPrep kit (Zymo). DNase I on-column digestion was performed. cDNA was generated using 500ng of RNA with the SuperScript IV First-Strand Synthesis System (Invitrogen). RT-qPCR was performed using SsoAdvanced SYBR Green Supermix (Bio-Rad) and run on a Bio-Rad CFX96 Real-Time System. The primers used in the RT-qPCR assays are listed in Table 2. Fifteen nanograms of cDNA was used for each RT-qPCR reaction.
Table 2.
Primer sequences used for RT-qPCR
| Gene | Primer Sequence |
|---|---|
| Bmi1 | F: TGTCCAGGTTCACAAAACCA R: TGCAACTTCTCCTCGGTCT |
| Pcgf3 | F: AGCAAACTACGTGGCCTCAA R: ACAGCAAGTCCATCTTGGGT |
| Pcgf6 | F: CGGACATTTTAAGCCATTGGAA R: CGCTCCAGTAAGTGATCCCC |
| P19ARF | F: GTTCTTGGTCACTGTGAGGATTCAG R: CCATCATCATCACCTGGTCCAG |
| RPO | F: TTCATTGTGGGAGCAGAC R: CAGCAGTTTCTCCAGAGC |
Electro-olfactogram (EOG) recordings
Air-phase EOGs were performed as previously described.19 Briefly, the septum or medial turbinate tissue was exposed and placed in Ringer’s solution with a reference electrode. Recording electrodes were prepared with a 5–10μm diameter tip. Electrodes were advanced using a micromanipulator to contact the mucosal surface. A continuous humidified air stream was applied over the tissue. Amyl acetate was used as the odorant due to its broad stimulus activity. The odor solution was prepared fresh in DMSO at 0.1M, further diluted 50-fold into water, allowed to equilibrate into vapor phase for 30 min prior recording, then injected into the air stream for a 0.1 second pulse using a picospritzer (Parker Hannifin). Recordings were taken from at least 5 stereotyped areas from the medial septum, turbinate II, and turbinate III from control or Bmi1-KO mice. Depolarization responses were recorded and digitized for analysis using the Axograph software. An amplifier (Warner DP-301) in DC mode with 100x gain and a low-pass filter was used, connected to a digitizer (Digidata 1500, Axon Instruments), with output connected to a laptop computer triggering picospritzer. Data from each animal was averaged and the mean±SD peak amplitudes from each group was compared using an unpaired student’s t-test.
Chromatin immunoprecipitation with quantitative polymerase chain reaction (ChIP-qPCR)
Chromatin immunoprecipitation was performed using the ChIP-IT High Sensitivity Kit (Active Motif) according to manufacturer’s instructions. All solutions used were provided in the Active Motif ChIP-IT High Sensitivity Kit unless otherwise specified.
OE tissue was collected from 3–4 animals, washed in Hank’s buffered salt solution (HBSS) and digested in an enzyme solution containing 1mg/ml collagenase (Invitrogen), 2mg/ml Dispase II (Roche), and 0.1 mg/ml DNase I (Sigma) for 25 minutes at 37°C. The tissue was further digested in a 0.125% trypsin-EDTA solution (Gibco) for 2 minutes to yield a single cell suspension. Cells were then strained through a 100μm mesh and crosslinked in 1% formaldehyde for 10 minutes at room temperature. Crosslinking was quenched with Stop Solution for 5 minutes and cells were scraped off culture dishes using a rubber policeman. Pelleted cells were lysed in 5ml of Chromatin Prep Buffer and homogenized by Dounce homogenizer (20–30 strokes). Nuclei were pelleted, resuspended in ChIP Buffer, and sonicated for 25 cycles (1 cycle = 30 seconds on, 30 seconds off) in a Bioruptor Pico (Diagenode). Chromatin was quantified by using Qubit dsDNA HS Assay Kit (Invitrogen).
To immunoprecipitate BMI1, we used 20μg of chromatin and 4μg of mouse anti-BMI1 antibody (Abcam). Antibodies were incubated with chromatin overnight at 4°C. Immunocomplexes were recovered with protein G agarose beads. DNA complexes were de-crosslinked and proteins removed by incubating samples with Proteinase K in Elution Buffer AM4. DNA was then eluted in water using the ChIP DNA Clean and Concentrator kit (Zymo).
Gene enrichment was determined by qPCR using the SsoAdvanced SYBR Green Supermix. The primers used for ChIP-qPCR are shown in Table 3. All fold changes were calculated as cycle number changes compared to negative control and input DNA (12.5ng per reaction). Five microliters of ChIP DNA or 12.5ng of input DNA were used per reaction.
Table 3.
Primer sequences used for ChIP-qPCR
| Gene | Primer Sequence |
|---|---|
| Negative Control | F: gtgcaccatgactccctttt R: ggagcaggagaacagcaaac |
| Prdm14 | F: GTGTCACCCGACTGAAAGGT R: GTTCGTTTTGTTGGGCTGAT |
| Cdkn2a | F: ggcgttttgaacccttagcg R: TAAACAACGTCCTCGGGCTG |
RESULTS
Bmi1 knockout validation, initial characterization of OE changes, and measurement of olfactory function
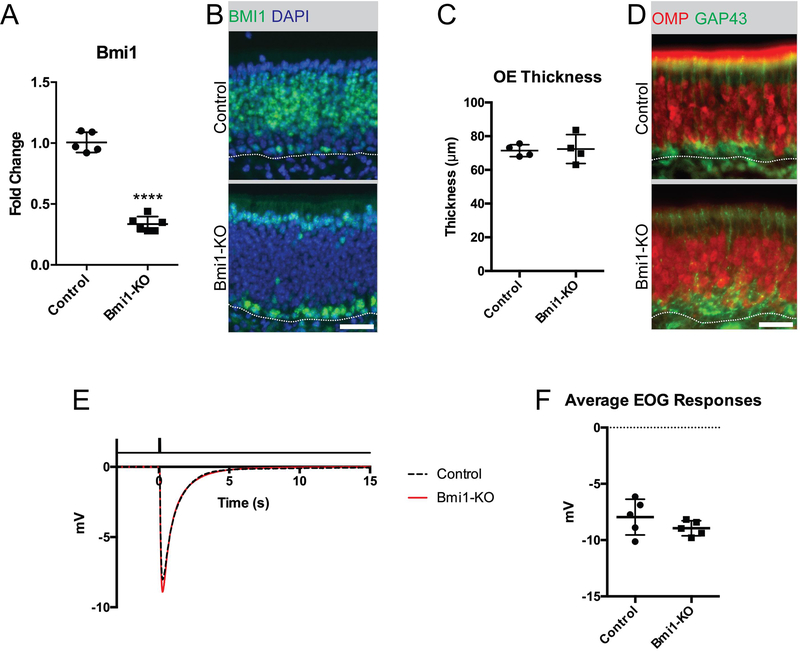
To selectively knock out BMI1 in mature OSNs while leaving the BMI1 expression in other tissues intact, we utilized mice expressing Cre recombinase driven by an olfactory neuron-specific locus, OMP.17 Crossing these mice with Bmi1-flox mice18 resulted in Cre-mediated excision of the Bmi1 gene between exons 4 and 8, which did not yield a protein product. BMI1 knockout (KO) was confirmed by RT-qPCR (Figure 1A, 1.01±0.08 vs. 0.34±.06, mean±SD, unpaired t-test, p<0.0001, n=5–6) and by immunofluorescence (Figure 1B), using antibodies to both the N- and C-terminus. Despite loss of BMI1, the OE did not appear histologically abnormal, compared to controls. OE thickness was normal (Figure 1C, 71.4±3.6μm vs. 72.3±8.6μm, mean±SD, unpaired t-test, p=0.854, n=4) and OMP expression was abundant (Figure 1D). Furthermore, olfactory function remained intact in BMI1 knockout tissue, as measured using electro-olfactogram (EOG) recordings to detect odor responses (Figure 1E and F, −7.95±1.59 vs. −8.95±0.67 mV, mean±SD, unpaired t-test, p=0.235, n=5).
Figure 1. Neuronal organization and olfactory function remain intact in mice with OSN-specific Bmi1-KO.
(A and B) Gene expression analysis and immunofluorescence of BMI1, ****p<0.0001, n=5.
(C) Measurements of OE thickness, p=0.854, n=4.
(D) Representative immunofluorescence of OMP, a marker for mature OSNs, and GAP43, a marker for immature OSNs.
(E) Average traces of electro-olfactogram recordings from all animals in control vs. Bmi1-KO group. Solid black line at the top of the panel indicates the timing of 0.1M amyl acetate stimulation for a duration of 0.1 seconds, n=5.
(F) Average of mean peak EOG responses per animal, p=0.234, n=5.
Dotted lines in B and C mark the basal lamina, bar is 25μm.
Assessment of PRC1 histone marks and other PCGF paralog expression
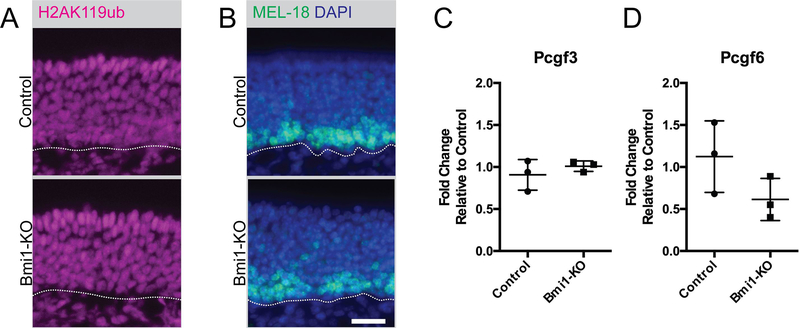
Given the lack of a striking phenotype, we examined for evidence of PRC1 enzymatic activity in Bmi1-KO mice by staining for the monoubiquitylated lysine 119 on histone H2A (H2AK119ub), the post-translational histone mark catalyzed by the RING1 component of PRC1 complexes. Despite an absence of the BMI1 protein in OSNs, we found H2AK119ub marks throughout the OE (Figure 2A). We next investigated whether a paralog of BMI1 might be compensating for its knockout. As part of PRC1, BMI1 is one of six Polycomb group finger (PCGF) paralogs, but only MEL-18 (PCGF2) and BMI1 (PCGF4) are recruited onto canonical PRC1 complexes. MEL-18 and BMI1 have non-overlapping expression patterns in wild type OE 16. Whereas BMI1 is expressed in upstream SOX2+ globose basal cells and mature OSNs (Figure 1), MEL-18 is expressed in neuron progenitors and immature neurons (Figure 2B). Following Bmi1-KO, we found that MEL-18 expression remained unchanged between control and BMI1 knockout animals (Figure 2B). Due to the lack of validated antibodies to the other PCGF paralogs, we determined their expression in OE tissue by RT-qPCR. Pcgf3 and Pcgf6 transcripts were detected in the OE, but there was no significant difference in their expression between control and Bmi1-KO animals (Figure 2C and D).
Figure 2. Intact PRC1 function suggests redundant mechanisms in adult OE.
(A) Representative immunofluorescence of monoubiquitylated lysine 119 of histone H2 (H2AK119ub).
(B) Representative immunofluorescence of a PCGF paralog, MEL-18 (also known as PCGF2).
(C and D) Gene expression analysis of PCGF paralogs, Pcgf3 (p=0.406, n=3) and Pcgf6 (p=0.149, n=3).
Dotted lines in A and B mar the basal lamina, bar is 25 μm.
Basal cell activation following Bmi1 knockout
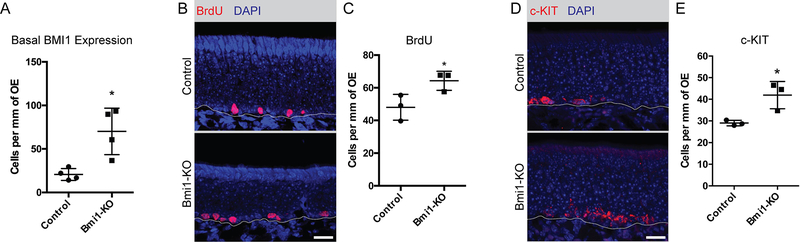
Of interest, the expression of BMI1 protein appeared increased in the basal layers of the epithelium as observed by immunofluorescence (Figure 1). Quantifying the number of BMI1+ nuclei in the basal layers revealed more BMI1+ basal cells in OSN-specific BMI1-deficient mice than controls (Figure 2A; 70.2±26.7 vs 20.6±6.8 cells per mm OE, mean±SD, unpaired t-test, p=0.030, n=4). We further investigated potential changes within the basal layer by quantifying BrdU-incorporation (a measure of mitotically-active cells) and c-KITexpression (a marker for sensory-lineage globose basal cells).20 In agreement with the increase in BMI1 labeling, we observed an increase in BrdU-labeled basal cells (Figure 2B and C; 48.04±7.95 vs. 64.28±5.82 cells per mm OE, mean±SD, unpaired t-test, p=0.046, n=3) as well as increased c-KIT+ cells (Figure 2D and E; 29.01±1.26 vs. 41.91±6.31 cells per mm OE, mean±SD, unpaired t-test, p=0.026, n=3) in BMI1 knockout animals, providing evidence that the basal cells and progenitors of the sensory lineage were activated.
Increased neuronal turnover
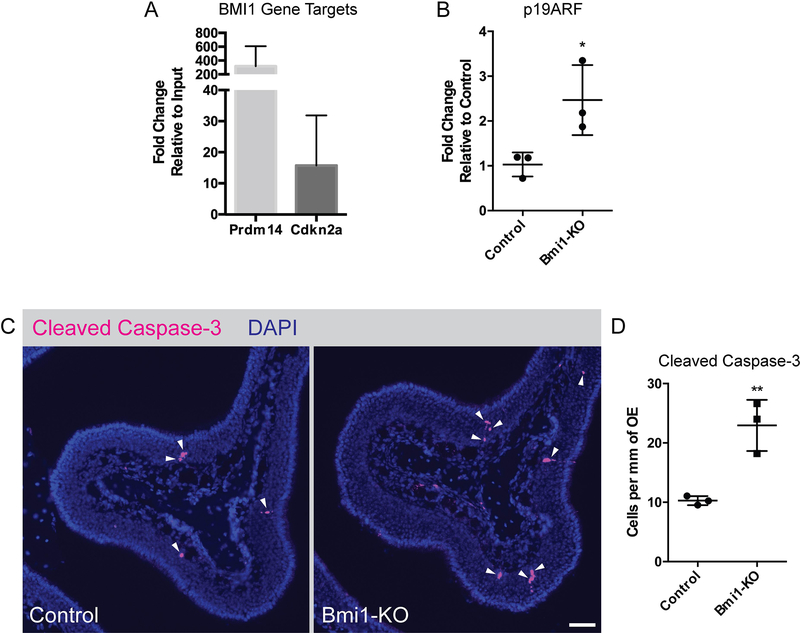
We sought to determine whether the increased basal cell activation was due to neuron injury. In cortical neurons, BMI1 is known to be neuroprotective by repressing the Cdkn2a gene locus, whose gene products p16INK4a and p19ARF can initiate apoptosis. We investigated whether BMI1 in adult OE regulates these same genes by chromatin immunoprecipitation followed by qPCR (ChIP-qPCR). We determined that BMI1 is bound to the Cdkn2a locus in normal adult OE (Figure 4A) and that the loss of BMI1 leads to the de-repression of p19ARF (Figure 4B), but we were not able to detect expression of the alternative gene product p16INK4a. Immunofluorescence experiments probing for the apoptosis marker cleaved caspase-3 revealed that BMI1-KO animals had mildly elevated expression within the neuronal layers, compared to controls (Figure 4C and D; 10.26±0.76 vs. 22.95±4.31 cells per mm OE, mean±SD, unpaired t-test, p=0.007, n=3). These findings suggest that BMI1 may have a subtle role in olfactory neuroprotection, acting via regulation of p19ARF.
Figure 4. De-repression of the Cdkn2a locus leads to increased OSN death.
(A) ChIP-qPCR of BMI1 gene targets, Prdm14 (positive control) and Cdkn2a, n=3.
(B) Gene expression analysis of Cdkn2a gene product p19ARF, p=0.039, n=3.
(C and D) Representative immunofluorescence (bar indicates 50 μm) and gene expression analysis of cleaved caspase-3, **p=0.0074, n=3.
DISCUSSION
Olfactory disorders of sensorineural origin remain a clinical challenge due to our limited understanding of the mechanisms underlying OE homeostasis, particularly those pertaining to neuroprotection and OSN maintenance. In this study, we sought to investigate the role of a Polycomb group protein, BMI1, in mature OSNs. In previous work, our lab identified BMI1 expression in multipotent globose basal cells and mature OSNs of the OE. In those studies, we revealed that BMI1 expression in globose basal cells maintain self-renewal capacity in vitro and give rise to all other cell types in the OE in vivo.11 However, the role of BMI1 in mature OSNs remained unknown. Other groups investigating the role of BMI1 in cortical neurons in the brain provide evidence of its neuroprotective role in the central nervous system.15,21
In this study, we show that, under normal homeostatic conditions, OSN-specific BMI1 knockout leads to a mildly increased neuronal turnover, providing further evidence for the role of BMI1 in regulating OE maintenance. Our analysis of the globose basal cell population in Bmi1-KO animals revealed increased self-renewal and proliferation within the upstream BMI1+ and BrdU+ basal cells, as well as an expanded c-KIT+ population of downstream sensory-lineage progenitors, indicating a likely feedback stimulation activating basal cells in the setting of OSN-specific BMI1 knockout. Feedback mechanisms informing olfactory basal cells of a need to divide and/or differentiate to maintain homeostasis are well-described.22 Although subtle, Bmi1-KO animals did exhibit increased OSN death. However, the increased turnover did not affect olfactory function, reflective of the robust regenerative capacity in the rodent OE. Since OSNs are a highly vulnerable neuron population, having direct contact with the nasal airspace, redundant mechanisms are likely in place to maintain OE integrity.
Importantly, our studies revealed the expression of other PCGF paralogs in the OE. Although there was no change in the expression pattern of MEL-18, the other PCGF paralog recruited into canonical PRC1, the gene expression data of non-canonical PCGFs suggest a possible redundancy within the Polycomb group family of proteins that further compensates for the loss of BMI1 in our knockout animals. In fact, we have found that the expression of a neuron-specific PRC1 component and binding partner of BMI1, CBX8,16 remains unchanged in Bmi1-KO animals (not shown). These findings, together with the lack of change in H2AK119ub marks in the epithelium, suggest intact and functional PRC1 are likely present in the absence of BMI1.
CONCLUSION
Our findings reveal that the loss of BMI1 in mature OSNs leads to mildly increased neuronal cell loss. However, the elevated neuronal death does not compromise the overall health and function of the OE, as determined by OE thickness, abundant OMP expression, and EOG recordings. The lack of a striking phenotype following Bmi1-KO is likely due to redundant neuroprotective and neuroregenerative mechanisms in the olfactory epithelium. Understanding such mechanisms are essential steps towards developing treatment strategies for olfactory damage in humans.
Figure 3. Basal cells and sensory lineage progenitors are activated in Bmi1-KO animals.
(A) Quantification of BMI1+ nuclei in basal cells, *p=0.030, n=4.
(B and C) Representative immunofluorescence and quantification of BrdU-incorporated basal cells, *p=0.046, n=3.
(D and E) Representative immunofluorescence and quantification of c-KIT+ sensory lineage progenitors, *p=0.026, n=3.
Dotted lines in B and D mark the basal lamina, bars are 25μm.
Acknowledgments
DISCLOSURES
Funding sources for the study: National Institute on Deafness and Other Communiction Disorders of the National Institutes of Health (K08DC013556 and R01DC016859 to BJG); Triological Society/American College of Surgeons Clinical Scientist Development Award (BJG)
References
- 1.Choi R, Goldstein BJ. Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig Otolaryngol. 2018;3(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seiden AM. Postviral olfactory loss. Otolaryngologic clinics of North America. 2004;37(6):1159–1166. [DOI] [PubMed] [Google Scholar]
- 3.Paik SI, Lehman MN, Seiden AM, Duncan HJ, Smith DV. Human olfactory biopsy. The influence of age and receptor distribution. Arch Otolaryngol Head Neck Surg. 1992;118(7):731–738. [DOI] [PubMed] [Google Scholar]
- 4.Carr VM, Farbman AI. The dynamics of cell death in the olfactory epithelium. Experimental neurology. 1993;124(2):308–314. [DOI] [PubMed] [Google Scholar]
- 5.Graziadei GA, Graziadei PP. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol. 1979;8(2):197–213. [DOI] [PubMed] [Google Scholar]
- 6.Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10(6):720–726. [DOI] [PubMed] [Google Scholar]
- 7.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. The Journal of comparative neurology. 1998;400(4):469–486. [PubMed] [Google Scholar]
- 8.Sammeta N, McClintock TS. Chemical stress induces the unfolded protein response in olfactory sensory neurons. J Comp Neurol. 2010;518(10):1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Getchell ML, Ding X, Getchell TV. Immunolocalization of two cytochrome P450 isozymes in rat nasal chemosensory tissue. Neuroreport. 1992;3(9):749–752. [DOI] [PubMed] [Google Scholar]
- 10.Dahl AR, Hadley WM, Hahn FF, Benson JM, McClellan RO. Cytochrome P-450-dependent monooxygenases in olfactory epithelium of dogs: possible role in tumorigenicity. Science. 1982;216(4541):57–59. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein BJ, Goss GM, Choi R, et al. Contribution of Polycomb group proteins to olfactory basal stem cell self-renewal in a novel c-KIT+ culture model and in vivo. Development. 2016;143(23):4394–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatoo W, Abdouh M, David J, et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29(2):529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Li L, Ni W, et al. Bmi1 regulates auditory hair cell survival by maintaining redox balance. Cell Death Dis. 2015;6:e1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao G, Gu M, Zhu M, et al. Bmi-1 absence causes premature brain degeneration. PloS one. 2012;7(2):e32015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdouh M, Chatoo W, El Hajjar J, David J, Ferreira J, Bernier G. Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons. PLoS One. 2012;7(2):e31870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein BJ, Choi R, Goss GM. Multiple polycomb epigenetic regulatory proteins are active in normal and regenerating adult olfactory epithelium. Laryngoscope Investig Otolaryngol. 2018;3(5):337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428(6981):393–399. [DOI] [PubMed] [Google Scholar]
- 18.Maynard MA, Ferretti R, Hilgendorf KI, Perret C, Whyte P, Lees JA. Bmi1 is required for tumorigenesis in a mouse model of intestinal cancer. Oncogene. 2014;33(28):3742–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cygnar KD, Stephan AB, Zhao H. Analyzing responses of mouse olfactory sensory neurons using the air-phase electroolfactogram recording. Journal of visualized experiments : JoVE. 2010(37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein BJ, Goss GM, Hatzistergos KE, et al. Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. The Journal of comparative neurology. 2015;523(1):15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flamier A, El Hajjar J, Adjaye J, Fernandes KJ, Abdouh M, Bernier G. Modeling Late-Onset Sporadic Alzheimer’s Disease through BMI1 Deficiency. Cell reports. 2018;23(9):2653–2666. [DOI] [PubMed] [Google Scholar]
- 22.Gokoffski KK, Wu HH, Beites CL, et al. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development. 2011;138(19):4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]