Abstract
Fighting the current COVID-19 pandemic, we must not forget to prepare for the next. Since elderly and frail people are at high risk, we wish to predict their vulnerability, and intervene if possible. For example, it would take little effort to take additional swabs or dried blood spots. Such minimally-invasive sampling, exemplified here during screening for potential COVID-19 infection, can yield the data to discover biomarkers to better handle this and the next respiratory disease pandemic. Longitudinal outcome data can then be combined with other epidemics and old-age health data, to discover the best biomarkers to predict (i) coping with infection & inflammation and thus hospitalization or intensive care, (ii) long-term health challenges, e.g. deterioration of lung function after intensive care, and (iii) treatment & vaccination response. Further, there are universal triggers of old-age morbidity & mortality, and the elimination of senescent cells improved health in pilot studies in idiopathic lung fibrosis & osteoarthritis patients alike. Biomarker studies are needed to test the hypothesis that resilience of the elderly during a pandemic can be improved by countering chronic inflammation and/or removing senescent cells. Our review suggests that more samples should be taken and saved systematically, following minimum standards, and data be made available, to maximize healthspan & minimize frailty, leading to savings in health care, gains in quality of life, and preparing us better for the next pandemic, all at the same time.
Keywords: COVID-19, Inflammaging, Swabs, Cellular senescence, Biomarker
1. Introduction
In late 2019, clusters of patients with pneumonia of unknown etiology were reported in Wuhan, China. The causative agent was identified as a novel coronavirus, SARS-CoV-2; the disease was named COVID-19. As of March 2020, it has caused many hundreds of thousands of confirmed cases and tens of thousands of deaths worldwide (John Hopkins University CoronavirusMap, https://coronavirus.jhu.edu/map.html). Sepsis is the most frequently observed complication, followed by respiratory failure, acute respiratory deficiency syndrome (ARDS), heart failure and septic shock (Zhou et al., 2020a). Old people, and particularly those affected by one or more comorbidity, are the most vulnerable subjects, where the highest morbidity and mortality has been reported, associated with sequential organ failure and high d-dimer levels (Onder et al., 2020; Shi et al., 2020; Zhou et al., 2020a). Moreover, in some countries like Italy men appear to be more affected and have a higher mortality than women. In order to react effectively to the ongoing and future pandemics, we must improve diagnostics as well as biomarker discovery and validation, for prognosis and for intervention. Moreover, minimum standards are necessary for all sampling procedures. It is not enough to just diagnose; good prognostic and predictive biomarkers are needed.
For long-term gain, we suggest that the most useful biomarkers are the ones that allow us to understand the age-related mechanisms underlying a respiratory infectious disease pandemic such as COVID-19. Here, we can build upon conceptual frameworks developed over the last decade, describing how immune status is influenced by aging processes (see also Section 3). In brief, aging processes such as chronic inflammation and cellular senescence affect the innate as well as the adaptive immune system (Oh et al., 2019). Specifically, older and multi-morbid people are suffering from “inflammaging” (Franceschi et al., 2000, 2018a; Franceschi et al., 2018b), a chronic, low-level inflammation caused by exposure of the immune system to misplaced and/or misfolded self-antigens originating from dying cells, fragments of mitochondria, extracellular vesicles and senescent cells (Franceschi et al., 2017a). The higher mortality from COVID-19 in older people, in subjects with comorbidities and in males can then be explained in part by their higher inflammatory status, including an activation of the innate immune system and the triggering of an inflammatory “storm” that includes IL-6 and other cytokines (Bonafè et al., 2020; Herold et al., 2020; Mehta et al., 2020; Moore and June, 2020; Santesmasses et al., 2020; Storci et al., 2020), as well as inflammatory molecules such as mitochondrial DNA fragments (Pinti et al., 2014), all assumed to be associated with dramatic changes of the peripheral blood lymphocyte subsets (Cossarizza et al., 2020). Moreover, a difference in the amount of inflammaging between men and women (Bonafe et al., 2001) could explain, in part, the higher vulnerability of males to COVID-19 (Marquez et al., 2020). Specifically, Bonafè et al. (2020) suggest that men feature higher levels of subclinical systemic inflammation, connected with a blunted acquired immune system, a blunted type I interferon response, and with accelerated biological aging, as well as downregulation of the ACE2 SARS-CoV-2-receptor, which (paradoxically) triggers inflammation in the aged and comorbid. Thus, there is a strong specific rationale for implicating aging-related processes in COVID-19 mortality, suggesting that interventions into these processes can contribute significantly to better outcomes in this pandemic and the next. Here, we do not just call for action, but we provide a detailed outline of what can be done, and must be done, for maximum health gain for the elderly, in addition to the standard diagnostic tests.
2. Systematic minimally-invasive sampling for immunity/inflammation/aging biomarkers
2.1. Cohort to investigate, and outcomes of interest
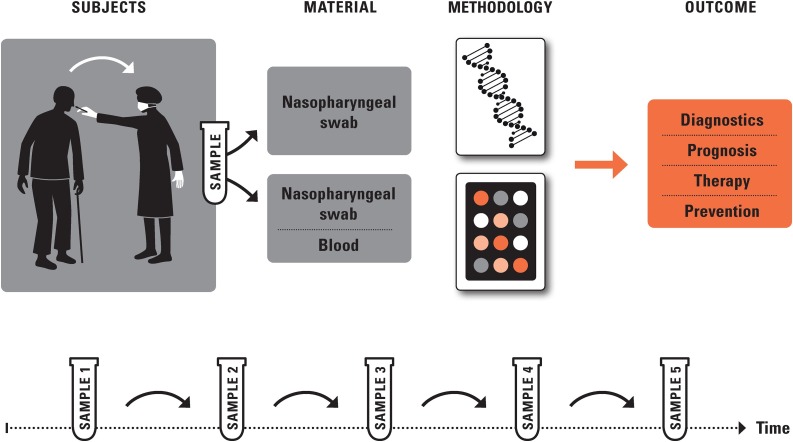
We propose proactive biomarker research as part of ongoing disease screening during a pandemic such as COVID-19 (see Fig. 1 ). The suspected cases being screened are called the “cohort” of probands (or patients). For probands screened during a serious respiratory disease epidemic and found to be positive for the infectious agent, we suggest that the relevant outcomes to predict are disease manifestation, subsequent disease deterioration including hospitalization (and need for intensive care), long-term morbidity after hospital discharge, and mortality. Disease deterioration can be measured in detail by clinical outcomes, see, e.g., (Zhou et al., 2020a). Long-term morbidity, for example in patients that need to undergo intensive care but escape the deadly consequences of the infection, can be measured as newly diagnosed diseases of interest (e.g., related to lung function) and, ultimately, by estimates of biological age, see (Fuellen et al., 2019; Moskalev, 2020).
Fig. 1.
Screening Strategy. We suggest screening, particularly of elderly and frail people, combining routine with minimally-invasive and longitudinal sampling for biomarker research, to test the hypothesis that resilience of the elderly during a pandemic can be improved by countering chronic inflammation (inflammaging) and cellular senescence. Important outcomes are prognosis, occurrence of complications such as lung fibrosis, hints for therapeutic approaches and preventive measures.
2.2. Sample collection and transportation modalities
All sampling we suggest is amending the standard diagnostics. We consider two guiding criteria for selecting the right type of samples: 1) maximum expected usefulness of the samples to identify biomarkers accurately predicting outcomes of interest; 2) minimum effort (time, cost, reliability of supplies, safety) in taking and processing the samples needed. Overall, we suggest that even in the current COVID-19 situation, simple non-invasive or minimally-invasive sampling procedures can easily be implemented. These would be based on using (i) the remainders from the “standard” diagnostic (swab-based) sample, (ii) a double-headed swab (using one head for diagnostics), (iii) in addition to the diagnostic swab, one or more further swabs, (iv) dried capillary blood spot samples, and, with some caveats, (v) venous blood. Overall, sampling methods have to be in agreement with downstream laboratory uses, and of course they have to comply with the corresponding WHO and/or local standards.
The sampling modalities for biomarker identification to be considered at time of pandemic screening are depicted in supplement 1. Nasopharyngeal and throat swabs are the standard for the diagnosis of respiratory infections such as COVID-19. As diagnostic testing will always be performed, we strongly suggest to use remainder material from the diagnostic swab for additional analyses. Of interest, Lopez et al. (2019) elegantly developed a minimally invasive sample collection protocol that allows for multiple diagnostic and research investigations including bacterial culture, viral detection (PCR), cytokine expression (PCR), and 16S ribosomal RNA gene sequencing from a single nasopharyngeal swab. Cutting the tip of the swab also allowed for RNA sequencing. Further, there are a number of additional swab collection options ranging from an additional swab to a double-headed swab. Flocked swabs are most often used and transferred into universal transport medium (UTM) (von Allmen et al., 2019). Dried capillary blood sampling is another “easy” option, routinely applied for neonate screening and drug monitoring. An optimized protocol for the extraction of RNA from dried blood spots yielded similar transcriptome gene counts compared to RNA isolated from venous blood in Tempus tubes (Reust et al., 2018); thus, dried blood spots for transcriptome studies could be a viable option facilitating RNA studies. Blood collection, application onto filter paper, drying, transport and storage can all affect the accuracy of the analysis and comparability with blood serum/plasma; however, handling and transport of the samples is far less problematic (Lim, 2018; Rutstein et al., 2015; Zakaria et al., 2016). We acknowledge that sampling of whole blood may not be practical in times of a health emergency. Nevertheless, specific measurements that require whole (venous) blood (e.g. of circulating antibodies) may still be clinically useful, and then the remaining blood should be used for biomarker analyses, such as transcriptomics based on circulating immune cells. For all samples, for investigating immunity and inflammation, cytokine (or related) protein assays are an obvious first choice; a second choice are modalities allowing unbiased gene expression measurements (transcriptomics). While important, not discussed in this review is the identification of (bacterial) co-infections.
2.3. Towards minimum standards
The most appropriate sampling workflows should be prioritized depending on a number of factors incl. screening location (emergency rooms, specific screening centers, etc.), availability of respective sampling materials, the condition of the proband (asymptomatic, mild, severe) and the objective of the screening. Appropriate pre-analytic sample processing is important particularly for RNA analyses (Stellino et al., 2019), even if standard tubes are used (Lippi et al., 2019). We recommend in the short term the recording of date and time of collection, and of storage and transport conditions, and attention to RNA stability; in the long term, uniform standards must be established.
2.4. Available biomarker analyses
Measuring the host response to infectious pathogens provides a rapid but rather unspecific means of aiding in diagnosis, compared to direct pathogen detection. However, monitoring the host response over time enables evaluation of patient response to therapy. White blood cell counts and single protein or metabolomic biomarkers (such as CRP, procalcitonin and lactate) have historically been used in case of infectious and immunological diseases (Gunsolus et al., 2019; Ross et al., 2019). Moreover, cytokines and other proteins were measured using technologies ranging from ELISA to flow cytometry (Vazquez et al., 2019). Postulating that a broad assessment of host response in terms of gene expression (transcriptomics) will yield superior diagnostic and prognostic accuracy, global changes in gene expression in response to acute infections, trauma, and sepsis were studied intensively (Sweeney and Wong, 2016). Biomarker discovery has initially focused on whole blood or specific blood cell populations (Holcomb et al., 2017; Zaas et al., 2013), but more recently, samples from the respiratory tract have been used more often (Landry and Foxman, 2018). For example, host gene expression profiles in nasopharyngeal (NP) swabs and whole blood samples were investigated during respiratory syncytial virus (RSV) and human rhinovirus (hRV) infection (Do et al., 2017). RSV infection induced strong and persistent innate immune responses and the observed RSV-induced gene expression patterns did not differ much in NP swabs compared to blood. In contrast, hRV infection did not induce expression of innate immunity pathways as strongly, and significant differences were observed between NP swab and blood specimens. With respect to biomarkers for outcome risk stratification, whole blood transcriptomics alongside with clinical data in sepsis patients leads to a significant improvement in the prediction of 30-day mortality (Sweeney et al., 2018), which is useful as a template for investigations in COVID-19 patients.
Moreover, dried capillary blood spots (DBS) provide a minimally-invasive, low-cost option since results often correspond to those derived from gold-standard venous blood samples (McDade et al., 2016). DBS can be used for quantification of virus antigens, RNA (Nguyen et al., 2018) and antibodies (Muzembo et al., 2017), for studies of gene expression (McDade et al., 2016) and age-dependent DNA methylation (Knight et al., 2016). Multiplexing by mass spectrometry is available (Chambers et al., 2015), and global metabolite profiles can be generated, assuming stability at ambient temperature (Drolet et al., 2017). Large numbers of samples can be taken and stored in biobanks (Bjorkesten et al., 2017), and be used to characterize inflammatory responses. For example, multiplex immuno-mass spectrometry can measure acute phase response (inflammatory) proteins from DBS with high precision (Anderson et al., 2019), also allowing longitudinal analyses of inflammation markers, and generating biomarker trajectories of inflammation dynamics. Overall, swabs and dried blood have proven their value, specifically for the identification of infection- and immunity-related biomarkers.
3. Immunity and aging, assessed based on minimally-invasive sampling
We outlined how cytokine assays and transcriptomic estimates of immunity and inflammation in particular are measurable with a minimum of invasion, based on swabs or dried blood, towards predicting age-related outcomes regarding disease manifestation, hospitalization and long-term health deterioration after respiratory infection such as COVID-19. As already pointed out in the introduction, aging-related processes are an important contributor to inferior COVID-19 outcomes. Here, we describe how immunity and aging are related; see supplement 2 for more details. In brief, aging is the primary risk factor for all chronic diseases, and for inferior outcomes in infectious diseases, and COVID-19 is no exception. In people with chronic respiratory disease such as asthma and chronic obstructive pulmonary disease, this risk is attributed to chronic inflammation of the airways, dominated by CD4+ T cells and eosinophils in case of asthma and by CD8+ T lymphocytes, macrophages and neutrophils in case of chronic obstructive pulmonary disease (Cukic et al., 2012), exacerbated by respiratory infection (Barnes et al., 2015; Holgate et al., 2015). Far less is known about COVID-19, and about its chronic long-term consequences. The heterogeneity of responses to infections such as COVID-19 depends not only on the general health condition, predisposition on the genetic level, and the disease history, but in particular on the individual immunobiographies (Franceschi et al., 2017b). Knowledge of the individual life experience, surrounding ecosystem, cultural habit, history of natural infections, vaccinations or allergies should thus additionally be collected. Interestingly, important mechanisms underlying acute severe adult COVID-19 pneumonia cases are a reduction in CD4+ and CD8 + T cells and a decrease in regulatory T cells (Scarpa et al., 2020). As the thymus is a central lymphoid organ responsible for the release of T lymphocytes, inflammaging associated with the absence or a marked reduction of thymopoiesis could be a predisposing condition that also sustains the cytokine release storm observed in many older patients with severe clinical courses (Scarpa et al., 2020). In accordance with this assumption, the clinical manifestations are relatively mild in children. With ageing, antigen stimulation and thymic involution lead to a shift in T cell subset distribution and loss of expression of co-stimulatory molecules such as CD27 and CD28, with increased susceptibility to infections, and a (putatively compensatory) upregulation of cytokines. On the other hand, reduced killing ability by T cells at an early stage after birth could explain susceptibility to SARS-CoV-2 in very young infants (Yuki et al., 2020).
In general, inflammaging, immunosenescence and cellular senescence, all closely related to each other (Fulop et al., 2017), are involved in a deterioration of the performance of the immune system with age, starting with an involution of the thymus (Fagnoni et al., 2000), despite homeostatic mechanisms. Infection with, e.g., cytomegalovirus then leads to repeated reactivation of CD8+ cells, which increases the fraction of specialized T cells and decreases the amount of naive T cells (Vescovini et al., 2010), and to downregulation of CD28, which is an important co-stimulatory signal (Pangrazzi and Weinberger, 2020). Although no cell type of the immune system has been identified that shows all hallmarks of cellular senescence, some immune cells secrete several senescence-related components (cytokines, chemokines, extracellular matrix remodeling proteases) (Callender et al., 2018), indicating that the removal of cells with a senescent like phenotype may prepare elderly people for future infections, despite a protective role also described (Baz-Martinez et al., 2016). In particular, the interference of oncoviruses with senescence pathways suggests that senescence may also be part of the host cell response to fight viruses and that the SASP could stimulate the recruitment of immune cells; some experiments with vesicular stomatitis virus point to this possibility (Baz-Martinez et al., 2016). On the other hand, there are indications that senescent (lung) cells are a host target for SARS-CoV-2 viral infection, possibly due to better conditions for replication such as enhanced protein synthesis, which is also required to produce SASP inflammatory mediators (Sargiacomo et al., 2020). There are clearly many more possible interrelationships between SARS-CoV-2 infection and senescence mechanisms regarding genomic instability, telomere attrition, impaired autophagy, mitochondrial dysfunction, fibrosis, immunosenescence and inflammation, which may help to explain the increased pathophysiological responses to SARS-CoV-2 among older individuals (Mueller et al., 2020; Salimi and Hamlyn, 2020). For example, two suggested host receptors for SARS-CoV-2, CD26 and ACE-2 (angiotensin-converting enzyme 2), are upregulated in senescent cells (Sargiacomo et al., 2020). Moreover, a major cause of immune exhaustion is telomere shortening in viral-specific memory CD8 + T cells (Mueller et al., 2020), which induces cellular senescence and p38 MAPK-dependent SASP (Callender et al., 2018). This is of interest insofar as certain subtelomeric genes are upregulated in aged cells with short telomeres by a mechanism called telomere position effects over long distances (TPE-OLD). One of these TPE-OLD genes, interferon stimulated gene (ISG-15, (Robin et al., 2014)) is the most significantly activated ISG in response to viral infection (Salimi and Hamlyn, 2020). Notably, ISG-15 plays a key role in the innate immune response to viral infection and inhibits viral release by blocking viral assembly (Sadler and Williams, 2008). Therapeutic considerations so far focus at blocking the SASP and viral replication (using substances as rapamycin and doxycycline) or at the removal of infected senescent cells by senolytics (Malavolta et al., 2020; Sargiacomo et al., 2020). In any case, cellular senescence overlaps with immunosenescence and inflammaging, and cytokine assays are a method of choice for discovering biomarkers related to inflammation, immunity, fibrosis, cellular senescence and aging. Whole-genome transcriptomics could enable the further unbiased hypothesis-free exploration of biomarkers.
4. Testing the association of COVID-19 with inflammaging
While the majority of current efforts focus on implementing diagnostic testing capacity, little attention is paid to prognostic testing, e.g. by estimating immune or inflammation status. However, while diagnostic testing is of critical importance to identify and quarantine infected patients, prognostic testing is equally important to allow decisions on the level of care for those that are asymptomatic or show mild symptoms but may deteriorate. As described above, we suggest that the association of COVID-19 with old age may specifically be due to inflammaging- and senescence-associated failures of the immune system, and that it is important to test this hypothesis in more detail, based in particular on molecular biomarker measurements. The human host response to coronaviruses in general was reviewed by (Channappanavar and Perlman, 2017). The coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 can all trigger a strong immune response, characterized by high plasma levels of cytokines and chemokines in intensive care patients (Liu et al., 2020). There are several reports describing extensive lung damage in patients infected in previous SARS pandemics, associated with high initial virus titers (Peiris et al., 2003), increased mononuclear infiltration in the lungs (Nicholls et al., 2003) and elevated levels of serum proinflammatory cytokines and chemokines (Wong et al., 2004). Thus, the clinical deterioration of patients with coronavirus infections appears to result from a combination of direct virus-induced cytopathic effects and immunopathology induced by cytokine activation.
Most biomarker-related studies with respect to COVID-19 that are currently available were done in patients, not in probands undergoing screening, and these studies were done based on sophisticated blood analyses not usually available in the screening situation during a pandemic; we describe some examples because these studies, directly or indirectly, reflect the outcomes we are interested in. Differences in immune responses in patients with severe versus those with mild COVID-19 were investigated by (Liao et al., 2020), characterizing the lung immune microenvironment using bronchoalveolar lavage fluid from 3 severe and 3 mild COVID-19 patients and 8 healthy controls. Through single-cell RNA sequencing combined with TCR-sequencing, monocyte-derived FCN1+ macrophages were identified as the dominant cell population in the lungs of patients with severe disease and ARDS whereas FABP4+ alveolar macrophages and clonal CD8 + T cells predominated in the lungs of patients with mild disease. The predominance of highly inflammatory cells points towards high cytokine activation. Moreover, (Qin et al., 2020) analyzed peripheral lymphocyte subsets from 452 severe and non-severe COVID-19 patients. Severe cases had significantly lower lymphocyte counts, higher leukocyte counts and neutrophil-lymphocyte-ratios, as well as lower percentages of monocytes, eosinophils, and basophils. Most of the severe cases demonstrated elevated levels of infection-related biomarkers and inflammatory cytokines. Finally, for patients with severe COVID-19, monitoring the inflammation and immunity status may suggest treatment options such as immunosuppression by steroids, intravenous immunoglobulin, selective cytokine blockade or JAK inhibition, and specific biomarkers may be increasing ferritin, decreasing platelet counts, erythrocyte sedimentation rate, and the HScore (Mehta et al., 2020; Monteleone et al., 2020). Overall, the role of immune and inflammation-related mechanisms underlying disease protection and progression in COVID-19 need further elucidation, and inflammaging-related processes are a promising focus (see also the Introduction), based on the analysis of local (mucosal) and systemic immune responses, following our suggestions in Section 2 and supplement 1.
5. Discussion and conclusions
5.1. Now is the time to implement a systematic effort
We suggest that all COVID-19 test probands that are able to give informed consent shall participate in systematic surveys as motivated here. It is important to record confounders, and, as in any analysis, potential bias must be considered. Some confounders (time-of-day) can be recorded immediately, while others may need to be established post-hoc (i.e. after sampling) by questionnaires (asking for (co-)morbidity, for anthropomorphic data, etc.). Data analyses shall follow standard procedures for biomarker discovery; since in many cases the data are in a time-to-event format, survival analyses are usually done, as exemplified in some recent reviews, see (Lee and Lim, 2019; Wang et al., 2019). In these analyses, gender must be considered as a confounder, and the immunobiographies of the study participants shall be considered as well. Of note, biological knowledge about the SARS-CoV-2 virus is assembled at a rapid pace, see https://covid.pages.uni.lu/map_curation, can be combined with other knowledge on pathways & gene/protein interaction (Zhou et al., 2020b), and be used to interpret omics data. It is also important to maximize data sharing, fostering open science.
5.2. High-quality data require minimum sampling standards
Tradeoffs are an issue in any biomarker discovery or validation effort; they arise between maximizing the quantity versus the quality of the data, in real-world versus standardized situations, enabling cost-efficient yet safe measurements. As described in Section 2, in any respiratory disease epidemic, and specifically in case of COVID-19, we suggest that with highest priority, swabs and dried blood shall be done based on a simple set of rules, observing minimum standards, to limit the influence of systematic confounders as well as random influences, while still reflecting the real-world situation. Then, if there is sufficient signal for learning biomarkers, these can be expected to generalize well. A specific tradeoff are biosafety issues, see supplement 1.
5.3. Ethical considerations
Any taking of samples for research purposes, as well as the sharing of patient/proband data, must be subject to ethical approval by an institutional review board, and of informed consent from the side of the patient. Nevertheless, while considering all essential data protection aspects, ethical approval and permission for data collection and analysis should primarily enable research promoting health. For the (additional) sampling during screening in times of a health crisis, the initial informed consent should thus be as simple as possible, and be restricted to taking and storing of samples for research purposes, and to processing the data needed to contact the patient/proband electronically or by telephone later. The actual use of the samples may then be consented after a detailed explanation of study purposes and participant rights, including data protection and rights of withdrawal. Such post-hoc informed consent, preferably obtained electronically, also includes the agreements on follow-up exams (see supplement 3).
5.4. Guiding therapeutic intervention by biomarkers
Motivated by recent pilot trials, specifically of intervening into cellular senescence towards improving the health status of idiopathic lung fibrosis (Justice et al., 2019) (and osteoarthritis (UNITY, 2019)) patients, and given that inflammaging can be due in part to senescent cells, we suggest that modulating the immune status of the elderly by reducing inflammation or removing senescent cells (immune cells or lung cells) may improve their health and resilience also in the case of respiratory infections such as COVID-19. The first step towards investigating this hypothesis is the confirmation of biomarkers related to inflammaging or senescence as being predictive for the outcomes considered here, such as hospitalization and long-term deterioration of lung function. The latter is of particular interest in “escapers” that survived serious complications of the disease; if biomarkers could be identified to predict their course of recovery, inferior outcomes may be prevented. More generally, it would be possible to design biomarker-guided intervention trials, where not only the choice of the anti-inflammatory and/or seno-therapeutic intervention(s) shall be guided by the biomarkers found to be most predictive, but patient-specific (“personalized”) biomarker-based treatment is a conceivable option. Moreover, as aging-related processes are at the core of susceptibility to infectious (respiratory) diseases in general, and of the chances to cope with these, any remaining sample material should be used to measure generic aging-related markers. For example, if there are leftovers of human DNA, methylation-based estimates of biological age are possible, and these can help to understand how host age and viral challenge are responsible for the outcomes observed, and specific mechanisms of molecular aging that are related to these outcomes can be confirmed, or discovered anew. In fact, based on DNA from buccal swabs, an epigenetic clock was already established (Eipel et al., 2016). Finally, it was demonstrated in phase-2 studies, although not confirmed in a recent phase-3 study, that influenza vaccination response can be improved by targeting the mTOR pathway by rapalogs (Kaeberlein, 2020). Since mTOR modulates cellular senescence, biomarkers for predicting the reaction of the immune system to mTOR inhibition may be valuable for vaccination research, also for COVID-19.
Funding
GF is supported by the BMBF (FKZ 01ZX1903A) and the European Commission (Aging with elegans, grant agreement 633589). AS is supported by the DFG (RTG 2155 ProMoAge). DQ is supported by the SFI (CÚRAM Research Centre, 13/RC/2073), the European Regional Development Fund and the Dr. Werner Jackstädt-Stiftung. GF and MW are supported by Karls Erdbeerhof, Rövershagen, Germany.
Declaration of Competing Interest
OL is an employee of and holds equity in Inflammatix Inc., developing host response based tests for acute infections and sepsis. Inflammatix does not have a commercial presence anywhere in the world at this time. All other authors: none declared.
Acknowledgments
The authors thank Katja Tränkner (WriteNow, Berlin, Germany) for preparing the drawing.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.arr.2020.101091.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Anderson N.L., Razavi M., Pope M.E., Yip R., Pearson T.W. Multiplexed measurement of protein biomarkers in high-frequency longitudinal dried blood spot (DBS) samples: characterization of inflammatory responses. bioRxiv. 2019 [Google Scholar]
- Barnes P.J., Burney P.G., Silverman E.K., Celli B.R., Vestbo J., Wedzicha J.A., Wouters E.F. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Primers. 2015;1:15076. doi: 10.1038/nrdp.2015.76. [DOI] [PubMed] [Google Scholar]
- Baz-Martinez M., Da Silva-Alvarez S., Rodriguez E., Guerra J., El Motiam A., Vidal A., Garcia-Caballero T., Gonzalez-Barcia M., Sanchez L., Munoz-Fontela C., Collado M., Rivas C. Cell senescence is an antiviral defense mechanism. Sci. Rep. 2016;6:37007. doi: 10.1038/srep37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkesten J., Enroth S., Shen Q., Wik L., Hougaard D.M., Cohen A.S., Sorensen L., Giedraitis V., Ingelsson M., Larsson A., Kamali-Moghaddam M., Landegren U. Stability of proteins in dried blood spot biobanks. Mol. Cell Proteomics. 2017;16:1286–1296. doi: 10.1074/mcp.RA117.000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafe M., Olivieri F., Cavallone L., Giovagnetti S., Mayegiani F., Cardelli M., Pieri C., Marra M., Antonicelli R., Lisa R., Rizzo M.R., Paolisso G., Monti D., Franceschi C. A gender--dependent genetic predisposition to produce high levels of IL-6 is detrimental for longevity. Eur. J. Immunol. 2001;31:2357–2361. doi: 10.1002/1521-4141(200108)31:8<2357::aid-immu2357>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Bonafè M., Prattichizzo F., Giuliani A., Storci G., Sabbatinelli J., Olivieri F. Why older men are the most susceptible to SARS-Cov-2 complicated outcomes. Cytokine Growth Factor Rev. 2020;2020(May 3) doi: 10.1016/j.cytogfr.2020.04.005. S1359-6101(20)30084-30088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender L.A., Carroll E.C., Beal R.W.J., Chambers E.S., Nourshargh S., Akbar A.N., Henson S.M. Human CD8(+) EMRA T cells display a senescence-associated secretory phenotype regulated by p38 MAPK. Aging Cell. 2018:17. doi: 10.1111/acel.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A.G., Percy A.J., Yang J., Borchers C.H. Multiple reaction monitoring enables precise quantification of 97 proteins in dried blood spots. Mol. Cell Proteomics. 2015;14:3094–3104. doi: 10.1074/mcp.O115.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A., De Biasi S., Guaraldi G., Girardis M., Mussini C., Modena Covid-19 Working G. SARS-CoV-2, the virus that causes COVID-19: cytometry and the new challenge for global health. Cytometry A. 2020;97(2020 April (4)):340–343. doi: 10.1002/cyto.a.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukic V., Lovre V., Dragisic D., Ustamujic A. Asthma and chronic obstructive pulmonary disease (COPD) - differences and similarities. Mater. Sociomed. 2012;24:100–105. doi: 10.5455/msm.2012.24.100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do L.A.H., Pellet J., van Doorn H.R., Tran A.T., Nguyen B.H., Tran T.T.L., Tran Q.H., Vo Q.B., Tran Dac N.A., Trinh H.N., Nguyen T.T.H., Le Binh B.T., Nguyen H.M.K., Nguyen M.T., Thai Q.T., Vo T.V., Ngo N.Q.M., Dang T.K.H., Cao N.H., Tran T.V., Ho L.V., De Meulder B., Auffray C., Hofstra J.J., Farrar J., Bryant J.E., de Jong M., Hibberd M.L. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J. Infect. Dis. 2017;217:134–146. doi: 10.1093/infdis/jix519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet J., Tolstikov V., Williams B.A., Greenwood B.P., Hill C., Vishnudas V.K., Sarangarajan R., Narain N.R., Kiebish M.A. Integrated metabolomics assessment of human dried blood spots and urine strips. Metabolites. 2017:7. doi: 10.3390/metabo7030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipel M., Mayer F., Arent T., Ferreira M.R., Birkhofer C., Gerstenmaier U., Costa I.G., Ritz-Timme S., Wagner W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging (Albany NY) 2016;8:1034–1048. doi: 10.18632/aging.100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnoni F.F., Vescovini R., Passeri G., Bologna G., Pedrazzoni M., Lavagetto G., Casti A., Franceschi C., Passeri M., Sansoni P. Shortage of circulating naive CD8(+) T cells provides new insights on immunodeficiency in aging. Blood. 2000;95:2860–2868. [PubMed] [Google Scholar]
- Franceschi C., Bonafe M., Valensin S., Olivieri F., De Luca M., Ottaviani E., De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Vitale G., Capri M., Salvioli S. Inflammaging and’ Garb-aging’. Trends Endocrinol. Metab. 2017;28(2017 March (3)):199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Salvioli S., Garagnani P., de Eguileor M., Monti D., Capri M. Immunobiography and the heterogeneity of immune responses in the elderly: a focus on inflammaging and trained immunity. Front. Immunol. 2017;8:982. doi: 10.3389/fimmu.2017.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Zaikin A., Gordleeva S., Ivanchenko M., Bonifazi F., Storci G., Bonafe M. Inflammaging 2018: an update and a model. Semin. Immunol. 2018;40:1–5. doi: 10.1016/j.smim.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Fuellen G., Jansen L., Cohen A.A., Luyten W., Gogol M., Simm A., Saul N., Cirulli F., Berry A., Antal P., Kohling R., Wouters B., Moller S. Health and aging: unifying concepts, scores, biomarkers and pathways. Aging Dis. 2019;10:883–900. doi: 10.14336/AD.2018.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop T., Larbi A., Dupuis G., Le Page A., Frost E.H., Cohen A.A., Witkowski J.M., Franceschi C. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front. Immunol. 2017;8:1960. doi: 10.3389/fimmu.2017.01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsolus I.L., Sweeney T.E., Liesenfeld O., Ledeboer N.A. Diagnosing and managing Sepsis by probing the host response to infection: advances, opportunities, and challenges. J. Clin. Microbiol. 2019:57. doi: 10.1128/JCM.00425-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold T., Jurinovic V., Arnreich C., Hellmuth J.C., von Bergwelt-Baildon M., Klein M., Weinberger T. Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. medRxiv. 2020 [Google Scholar]
- Holcomb Z.E., Tsalik E.L., Woods C.W., McClain M.T. Host-based peripheral blood gene expression analysis for diagnosis of infectious diseases. J. Clin. Microbiol. 2017;55:360–368. doi: 10.1128/JCM.01057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgate S.T., Wenzel S., Postma D.S., Weiss S.T., Renz H., Sly P.D. Asthma. Nat. Rev. Dis. Primers. 2015;1:15025. doi: 10.1038/nrdp.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice J.N., Nambiar A.M., Tchkonia T., LeBrasseur N.K., Pascual R., Hashmi S.K., Prata L., Masternak M.M., Kritchevsky S.B., Musi N., Kirkland J.L. Senolytics in idiopathic pulmonary fibrosis: results from a first-in-human, open-label, pilot study. EBioMedicine. 2019;40(2019 February):554–563. doi: 10.1016/j.ebiom.2018.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. RTB101 and immune function in the elderly: interpreting an unsuccessful clinical trial. Transl. Med. Aging. 2020;4:32–34. [Google Scholar]
- Knight A.K., Craig J.M., Theda C., Baekvad-Hansen M., Bybjerg-Grauholm J., Hansen C.S., Hollegaard M.V., Hougaard D.M., Mortensen P.B., Weinsheimer S.M., Werge T.M., Brennan P.A., Cubells J.F., Newport D.J., Stowe Z.N., Cheong J.L., Dalach P., Doyle L.W., Loke Y.J., Baccarelli A.A., Just A.C., Wright R.O., Tellez-Rojo M.M., Svensson K., Trevisi L., Kennedy E.M., Binder E.B., Iurato S., Czamara D., Raikkonen K., Lahti J.M., Pesonen A.K., Kajantie E., Villa P.M., Laivuori H., Hamalainen E., Park H.J., Bailey L.B., Parets S.E., Kilaru V., Menon R., Horvath S., Bush N.R., LeWinn K.Z., Tylavsky F.A., Conneely K.N., Smith A.K. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 2016;17:206. doi: 10.1186/s13059-016-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry M.L., Foxman E.F. Antiviral response in the nasopharynx identifies patients with respiratory virus infection. J. Infect. Dis. 2018;217:897–905. doi: 10.1093/infdis/jix648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lim H. Review of statistical methods for survival analysis using genomic data. Genomics Inform. 2019;17:e41. doi: 10.5808/GI.2019.17.4.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Chen L., Li J., Wang X., Wang F., Liu L., Zhang S., Zhang Z. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 [Google Scholar]
- Lim M.D. Dried blood spots for global health diagnostics and surveillance: opportunities and challenges. Am. J. Trop. Med. Hyg. 2018;99:256–265. doi: 10.4269/ajtmh.17-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Betsou F., Cadamuro J., Cornes M., Fleischhacker M., Fruekilde P., Neumaier M., Nybo M., Padoan A., Plebani M., Sciacovelli L., Vermeersch P., von Meyer A., Simundic A.M., Working Group for Preanalytical Phase E.Fo.C.C., Laboratory M. Preanalytical challenges - time for solutions. Clin. Chem. Lab. Med. 2019;57:974–981. doi: 10.1515/cclm-2018-1334. [DOI] [PubMed] [Google Scholar]
- Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez S.M.C., Martin J.M., Johnson M., Kurs-Lasky M., Horne W.T., Marshall C.W., Cooper V.S., Williams J.V., Shaikh N. A method of processing nasopharyngeal swabs to enable multiple testing. Pediatr. Res. 2019;86:651–654. doi: 10.1038/s41390-019-0498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavolta M., Giacconi R., Brunetti D., Provinciali M., Maggi F. Exploring the relevance of senotherapeutics for the current SARS-CoV-2 emergency and similar future global health threats. Cells. 2020;9(2020 April (4)):909. doi: 10.3390/cells9040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez E.J., Chung C.H., Marches R., Rossi R.J., Nehar-Belaid D., Eroglu A., Mellert D.J., Kuchel G.A., Banchereau J., Ucar D. Sexual-dimorphism in human immune system aging. Nat. Commun. 2020;11(2020 February (1)):751. doi: 10.1038/s41467-020-14396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade T.W., K M.R., R L.F., Arevalo J.M., Ma J., Miller G.E., Cole S.W. Genome-wide profiling of RNA from dried blood spots: convergence with bioinformatic results derived from whole venous blood and peripheral blood mononuclear cells. Biodemography Soc. Biol. 2016;62:182–197. doi: 10.1080/19485565.2016.1185600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone G., Sarzi-Puttini P.C., Ardizzone S. Preventing COVID-19-induced pneumonia with anticytokine therapy. Lancet Rheumatol. 2020;2(2020 May (5)):e255–e256. doi: 10.1016/S2665-9913(20)30092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Moskalev A. The challenges of estimating biological age. Elife. 2020;9(February 11) doi: 10.7554/eLife.54969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.L., McNamara M.S., Sinclair D.A. Why does COVID-19 disproportionately affect the elderly? PrePrints. 2020 doi: 10.18632/aging.103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzembo B.A., Mbendi N.C., Nakayama S.F. Systematic review with meta-analysis: performance of dried blood spots for hepatitis C antibodies detection. Public Health. 2017;153:128–136. doi: 10.1016/j.puhe.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Nguyen T.T., Lemee V., Bollore K., Vu H.V., Lacombe K., Thi X.L.T., Luong Q.A., Dubos C., Plantier J.C., Thi H.D., Laureillard D., Lemoine M., Tuaillon E. Confirmation of HCV viremia using HCV RNA and core antigen testing on dried blood spot in HIV infected peoples who inject drugs in Vietnam. BMC Infect. Dis. 2018;18:622. doi: 10.1186/s12879-018-3529-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., Chu C.M., Hui P.K., Mak K.L., Lim W., Yan K.W., Chan K.H., Tsang N.C., Guan Y., Yuen K.Y., Peiris J.S. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Lee J.K., Shin O.S. Aging and the immune system: the impact of immunosenescence on viral infection, immunity and vaccine immunogenicity. Immune Netw. 2019;19:e37. doi: 10.4110/in.2019.19.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020 doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Pangrazzi L., Weinberger B. T cells, aging and senescence. Exp. Gerontol. 2020;134 doi: 10.1016/j.exger.2020.110887. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y., Group H.U.S.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M., Cevenini E., Nasi M., De Biasi S., Salvioli S., Monti D., Benatti S., Gibellini L., Cotichini R., Stazi M.A., Trenti T., Franceschi C., Cossarizza A. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for "inflamm-aging". Eur. J. Immunol. 2014;44:1552–1562. doi: 10.1002/eji.201343921. [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reust M.J., Lee M.H., Xiang J., Zhang W., Xu D., Batson T., Zhang T., Downs J.A., Dupnik K.M. Dried blood spot RNA transcriptomes correlate with transcriptomes derived from whole blood RNA. Am. J. Trop. Med. Hyg. 2018;98:1541–1546. doi: 10.4269/ajtmh.17-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin J.D., Ludlow A.T., Batten K., Magdinier F., Stadler G., Wagner K.R., Shay J.W., Wright W.E. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014;28:2464–2476. doi: 10.1101/gad.251041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.H., Zick B.L., Tsalik E.L. Host-based diagnostics for acute respiratory infections. Clin. Ther. 2019;41:1923–1938. doi: 10.1016/j.clinthera.2019.06.007. [DOI] [PubMed] [Google Scholar]
- Rutstein S.E., Hosseinipour M.C., Kamwendo D., Soko A., Mkandawire M., Biddle A.K., Miller W.C., Weinberger M., Wheeler S.B., Sarr A., Gupta S., Chimbwandira F., Mwenda R., Kamiza S., Hoffman I., Mataya R. Dried blood spots for viral load monitoring in Malawi: feasible and effective. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi S., Hamlyn J.M. COVID-19 and crosstalk between the hallmarks of aging. PrePrints. 2020 doi: 10.1093/gerona/glaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santesmasses D., Castro J.P., Zenin A.A., Shindyapina A.V., Gerashchenko M.V., Zhang B., Kerepesi C., Yim S.H., Fedichev P.O., Gladyshev V.N. COVID-19 is an emergent disease of aging. medRxiv. 2020 doi: 10.1111/acel.13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargiacomo C., Sotgia F., Lisanti M.P. COVID-19 and chronological aging: senolytics and other anti-aging drugs for the treatment or prevention of corona virus infection? Aging (Albany NY) 2020;12:6511–6517. doi: 10.18632/aging.103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa R., Costa L., Del Puente A., Caso F. Role of thymopoiesis and inflamm-aging in COVID-19 phenotype. Pediatr. Neonatol. 2020 doi: 10.1016/j.pedneo.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellino C., Hamot G., Bellora C., Trouet J., Betsou F. Preanalytical robustness of blood collection tubes with RNA stabilizers. Clin. Chem. Lab. Med. 2019;57:1522–1529. doi: 10.1515/cclm-2019-0170. [DOI] [PubMed] [Google Scholar]
- Storci G., Bonifazi F., Garagnani P., Olivieri F., Bonafe M. How studies on inflamm-aging may help to understand and combat COVID-19 pandemic. PrePrints. 2020 [Google Scholar]
- Sweeney T.E., Wong H.R. Risk Stratification and Prognosis in Sepsis: What Have We Learned from Microarrays? Clin. Chest Med. 2016;37:209–218. doi: 10.1016/j.ccm.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.E., Perumal T.M., Henao R., Nichols M., Howrylak J.A., Choi A.M., Bermejo-Martin J.F., Almansa R., Tamayo E., Davenport E.E., Burnham K.L., Hinds C.J., Knight J.C., Woods C.W., Kingsmore S.F., Ginsburg G.S., Wong H.R., Parnell G.P., Tang B., Moldawer L.L., Moore F.E., Omberg L., Khatri P., Tsalik E.L., Mangravite L.M., Langley R.J. A community approach to mortality prediction in sepsis via gene expression analysis. Nat. Commun. 2018;9:694. doi: 10.1038/s41467-018-03078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNITY . 2019. UNITY Biotechnology Reports Promising Topline Data From Phase 1 First-in-human Study of UBX0101 in Patients With Osteoarthritis of the Knee. [Google Scholar]
- Vazquez Y., Gonzalez L., Noguera L., Gonzalez P.A., Riedel C.A., Bertrand P., Bueno S.M. Cytokines in the Respiratory Airway as Biomarkers of Severity and Prognosis for Respiratory Syncytial Virus Infection: An Update. Front. Immunol. 2019;10:1154. doi: 10.3389/fimmu.2019.01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovini R., Biasini C., Telera A.R., Basaglia M., Stella A., Magalini F., Bucci L., Monti D., Lazzarotto T., Dal Monte P., Pedrazzoni M., Medici M.C., Chezzi C., Franceschi C., Fagnoni F.F., Sansoni P. Intense antiextracellular adaptive immune response to human cytomegalovirus in very old subjects with impaired health and cognitive and functional status. J. Immunol. 2010;184:3242–3249. doi: 10.4049/jimmunol.0902890. [DOI] [PubMed] [Google Scholar]
- von Allmen N., Gorzelniak K., Liesenfeld O., Njoya M., Duncan J., Marlowe E.M., Hartel T., Knaust A., Hoppe B., Walter M. Liquid and dry swabs for culture- and PCR-Based detection of colonization with methicillin-resistant Staphylococcus aureus during admission screening. Eur. J. Microbiol. Immunol. (Bp) 2019;9:131–137. doi: 10.1556/1886.2019.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Li Y., Reddy C.K. Machine learning for survival analysis: a survey. ACM Comput. Surv. 2019;51 [Google Scholar]
- Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaas A.K., Burke T., Chen M., McClain M., Nicholson B., Veldman T., Tsalik E.L., Fowler V., Rivers E.P., Otero R., Kingsmore S.F., Voora D., Lucas J., Hero A.O., Carin L., Woods C.W., Ginsburg G.S. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria R., Allen K.J., Koplin J.J., Roche P., Greaves R.F. Advantages and challenges of dried blood spot analysis by mass spectrometry across the total testing process. EJIFCC. 2016;27:288–317. [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.