Abstract
The type VI secretion system (T6SS) is a multiprotein weapon that kills eukaryotic predators or prokaryotic competitors by delivering toxic effectors. Despite the importance of T6SS in bacterial environmental adaptation, it is still challenging to systematically identify T6SS effectors because of their high diversity and lack of conserved domains. In this report, we discovered a putative effector gene, U876-17730, in the whole genome of Aeromonas hydrophila NJ-35 based on the reported conservative domain DUF4123 (domain of unknown function), with two cognate immunity proteins encoded downstream. Phylogenetic tree analysis of amino acids indicates that AH17730 belongs to the Tle1 (type VI lipase effector) family, and therefore was named Tle1AH. The deletion of tle1AH resulted in significantly decreased biofilm formation, antibacterial competition ability and virulence in zebrafish (Danio rerio) when compared to the wild-type strain. Only when the two immunity proteins coexist can bacteria protect themselves from the toxicity of Tle1AH. Further study shows that Tle1AH is a kind of phospholipase that possesses a conserved lipase motif, Gly-X-Ser-X-Gly (X is for any amino acid). Tle1AH is secreted by T6SS, and this secretion requires its interaction with an associated VgrG (valine-glycine repeat protein G). In conclusion, we identified a T6SS effector-immunity pair and verified its function, which lays the foundation for future research on the role of T6SS in the pathogenic mechanism of A. hydrophila.
Introduction
Aeromonas hydrophila is a prevalent agent of aquatic infections, mainly causing motile aeromonad septicemia (MAS). As a food-borne pathogen, this organism represents a public health concern and causes soft-tissue wound infection and diarrhea [1]. Many virulence factors of A. hydrophila have been investigated, including motility [2], toxins [3], tissue-destructive enzymes [4] and S-layer [5]. Protein secretion systems are also essential for virulence and competition with nearby microorganisms. To date, nine types of bacterial secretion systems, from type I to type IX secretion systems (T1SS to T9SS), have been reported [6–8]. Among them, the type VI secretion system (T6SS) is one of the most commonly described secretion systems in Gram-negative bacteria.
T6SS was discovered as early as 2006 in Pseudomonas aeruginosa [9] and Vibrio cholerae [10], and approximately 25% of all Gram-negative bacteria have highly conserved T6SS gene clusters based on bioinformatics analysis [11]. T6SS is the main machinery that delivers antagonistic effector molecules into the environment, eukaryotic hosts and prokaryotic competitors for pathogenesis in a contact-dependent manner [12, 13]. The T6SS apparatus is composed of thirteen highly conserved “core” proteins and is believed to resemble the structure of the contractile tail of Escherichia coli bacteriophage T4 [14, 15]. Among the core components, TssB and TssC make up the bacteriophage contractile sheathe, which promotes the injection of a puncturing tube structure that comprises Hcp (hemolysin co-regulated protein) rings topped by a spike-like trimer of VgrG (valine-glycine repeat protein G). A previous study from Shneider et al. [16] showed that PAAR (proline-alanine-alanine-arginine) repeat-containing proteins bind to the ends of endogenous VgrG proteins. Another important component, ClpV, represents the central energy source for T6SS and mediates remodeling of the VipA/VipB cogwheel-like protein complex, which is conserved and essential for T6SS function [17]. Among these “core” proteins, Hcp and VgrG are the most studied components. Additional copies of these two proteins can also be found outside of the main T6SS cluster, generally around genes encoding putative effector proteins, and are used as chaperones for the secretion of effectors [18, 19]. Effector proteins are delivered through the T6SS and injected into target cells in a one-step manner. The known T6SS effectors could be transported either fused to structural components (specialized effectors) or via direct interactions with one protein of the core complex (cargo effectors) [20]. Meanwhile, there are corresponding antagonistic immunity genes located downstream of each effector gene to neutralize toxic effectors, preventing self-killing or sibling-intoxication, and thus a new toxin-antitoxin module called the effector-immunity (E-I) pair was constituted [21–23]. Although diverse approaches have been used to identify T6SS effectors, the number of identified effectors remains limited. The most common method to identify effectors is comparing the secretomes of WT (wild type) and T6SS mutants by transcriptomics, mutant library screening or proteomics-based methods [24]. In recent years, bioinformatics analysis has been used more frequently to predict effector genes by identifying highly conservative domains, such as the common N-terminal MIX motif, and DUF4123 and DUF2169 proteins [25–27]. However, due to the diversity of bacteria and the complexity of effector proteins, systematic identification of T6SS effectors is still a challenge.
In this study, we report the identification of a novel T6SS effector, Tle1AH (type VI lipase effectors of A. hydrophila), based on the conserved domain DUF4123, that plays a critical role in the successful colonization of A. hydrophila in the host. Two immune proteins work together to protect bacteria from the toxicity of Tle1AH. Further study found that the secretion of Tle1AH is associated with an interaction with VgrG but not Hcp. The present findings will provide valuable insights into the role of T6SS in A. hydrophila.
Materials and methods
Ethics statement
Zebrafish (Danio rerio) and crucian carp (Carassius auratus) were obtained from the Pearl River Fishery Research Institute, Chinese Academy of Fishery Science. The animal experiments were performed according to the Ethical Committee for Animal Experiments of Nanjing Agricultural University, China [permit number: SYXK (SU).2017-0007]. All operations are in line with the guidelines of the Animal Welfare Council of China.
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown at 28 °C or 37 °C in Luria–Bertani (LB; 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) media. When required, the final concentrations of antibiotics in growth media were as follows: 34 µg/mL chloramphenicol (Cm, in absolute ethanol), 100 μg/mL ampicillin (Amp, in water), 50 μg/mL kanamycin (Kan, in water) and 20 µg/mL gentamicin (Gm, in water). All antibiotics were stored at 4 °C.
Table 1.
Strains used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strain | ||
| A. hydrophila NJ-35 | Wilde-type, isolated from diseased crucian carp, in China, Ampr | Laboratory stock |
| A. hydrophila J-1 | Wilde-type, isolated from diseased crucian carp, in China, Ampr | Laboratory stock |
| A. hydrophila NJ-3 | Wilde-type, isolated from pond water, in China | Laboratory stock |
| A. hydrophilaATCC 7966 | Wilde-type, isolated from fishy milk, in USA | Laboratory stock |
| A. salmonicida CS-2 | Wilde-type, isolated from pond water, in China | Laboratory stock |
| A. media NJ-8 | Wilde-type, isolated from pond water, in China | Laboratory stock |
| A. veronii XH-14 | Wilde-type, isolated from diseased Chinese bream, in China | Laboratory stock |
| V. parahaemolyticus RIMD 2210633 | Wilde-type, isolated from a patient suffering from diarrhoea, in Japan | Laboratory stock |
| E. coli SM10 | λpir+, Kanr | [28] |
| E. coli BL21 | E. coli strain, competent invitrogen cells | CWBIO |
| E. coli TOP10 | E. coli strain, competent invitrogen cells | CWBIO |
| ∆tle1AH | tle1AH gene deletion mutant from NJ-35 | This study |
| ΔclpV | clpV gene deletion mutant from NJ-35 | [32] |
| ∆tle1-tli1tli2AH | tle1-tli1tli2AH gene deletion mutant from NJ-35 | This study |
| C∆tle1AH | ∆tle1AH with the vector pMMB-tle1AH | This study |
| C∆tli1AH | ∆tle1-tli1tli2AH with the vector pMMB-tli1AH | This study |
| C∆tli2AH | ∆tle1-tli1tli2AH with the vector pMMB-tli2AH | This study |
| C∆ tli1tli2AH | ∆ tle1-tli1tli2AH with the vector pMMB-tli1tli2AH | This study |
| Plasmid | ||
| pYAK1 | R6K-ori suicide vector, SacB+, Cmr | [27] |
| pYAK1-tle1AH | pYAK1 carrying the flanking sequence of tle1AH, Cmr | This study |
| pYAK1-tle1-tli1tli2AH | pYAK1 carrying the flanking sequence of tle1−tli1tli2AH, Cmr | This study |
| pMMB207 | Low-copy-number vector, Cmr | [29] |
| pMMB-tle1AH | pMMB207 carrying the tle1AH coding region, Cmr | This study |
| pMMB-tli1AH | pMMB207 carrying the tli1AH coding region, Cmr | This study |
| pMMB-tli2AH | pMMB207 carrying the tli2AH coding region, Cmr | This study |
| pMMB-tli1tli2AH | pMMB207 carrying the tli1tli2AH coding region, Cmr | This study |
| pMMB- tle1AH(6*His) | pMMB207 carrying the tle1AH+ 6*His coding region, Cmr | This study |
| pMMB-Kan | pMMB207 carrying the Kan coding region, Cmr, Kanr | This study |
| pMMB-Gen | pMMB207 carrying the Gen coding region, Cmr, Ger | This study |
| pBAD/HisA | Expression vector, Ampr | Invitrogen |
| pBAD-tle1AH | pBAD/HisA carrying tle1AH sequence | This study |
| pBAD-peri-tle1AH | pBAD/HisA carrying peri + tle1AH sequence, peri is the PelB leader sequence | This study |
| pBAD-peri-tle1AHS303A | pBAD/HisA carrying peri + tle1AHS303A sequence, peri is the PelB leader sequence | This study |
| pGEX-4T-1 | Expression vector, Ampr | Invitrogen |
| pGEX-tle1AH | pGEX-4T-1 carrying tle1AH sequence | This study |
| pET-28a | Expression vector, Kanr | Invitrogen |
| His-vgrG | pET-28a carrying vgrG sequence | This study |
| His-hcp | pET-28a carrying hcp sequence | This study |
All reagents used in this study were supplied by Sigma (St. Louis, MO, USA) unless otherwise indicated.
Construction of gene deletion mutants and A. hydrophila complementation strains
Single or double gene mutants were constructed by homologous recombination using the suicide plasmid pYAK1 [28]. All oligonucleotide primers are listed in Additional file 1. The left and right flanking regions of the target gene were PCR-amplified and ligated in-frame using fusion PCR. Then, the fusion fragments were cloned into pYAK1 with the restriction enzyme BamHI and then chemically transformed into E. coli SM10 competent cells [29]. For conjugation, the recombinant E. coli SM10 (Cm resistant, Cmr) and A. hydrophila NJ-35 (Amp resistant, Ampr) grown to log phase were mixed at a ratio of 3:1 (vol/vol) and then spotted on a 0.22-µm nylon filter (OKLABS, Tianjin, China) on LB plates. After incubation at 28 °C for 20 h, the bacteria were washed from the filter and grown on LB plates containing Amp and Cm. The positive colonies were verified by PCR and cultured in LB medium without NaCl for three generations. Then, the double crossover mutants were selected on LB agar plates containing 20% sucrose. The final mutants were confirmed by PCR amplification for the deleted region and flanking DNA, followed by DNA sequencing. In the same way, a triple-deletion mutant was also constructed.
The corresponding complemented strains of the mutants were constructed with the shuttle plasmid pMMB207 [30]. The target genes were amplified from the chromosomal DNA of A. hydrophila NJ-35 and then ligated into the pMMB207 vector. The recombinant plasmids were transformed into E. coli SM10 to serve as a donor strain, and then transferred into the mutant strains by conjugation as above. The transconjugants were selected on LB agar plates containing Amp and Cm and further confirmed by PCR.
Growth curve determination
Aeromonas hydrophila strains were cultured in LB broth at 28 °C until OD600 values were up to 0.5 (2.5 × 108 CFU/mL, as determined by counting the number of colony forming units (CFU) on agar plates). Then, the bacteria in the early logarithmic stage of growth were inoculated into flasks (OKLABS) containing 25 mL of fresh LB medium at 1:100. The flasks were placed in a shaker at 28 °C and incubated for 16 h. Every 1 h, the OD600 was measured using a SmartSpec Plus spectrophotometer (BIO-RAD, USA). The experiment was repeated three times independently.
Biofilm formation assay
Biofilm formation was assayed as previously described [31] with some modification. In brief, A. hydrophila strains were cultured in LB broth at 28 °C to an OD600 of 0.6–0.8 and normalized to an OD600 of 0.1. Then, 200 µL aliquots of bacterial suspensions (1:100 dilution in LB) were dispensed into 96-well polystyrene plates (Beyotime, Shanghai, China), and an equal volume of fresh medium was used as the blank control. Each strain was replicated in eight wells. To avoid edge effect resulting from evaporation and temperature fluctuation, no samples were added in the outermost two rows and two columns of the 96-well plates. After the plates were incubated at 28 °C for 24 h without shaking, the culture supernatant was discarded, and the wells were washed three times with sterile phosphate buffered saline (PBS) by removing the fluid with pipettors. After air-drying for 15 min, 200 µL methanol per well was added, and adherent bacterial cells were fixed for 15 min. Then, methanol was removed, and the wells were dried for 15 min. The attached bacteria were stained with 200 μL 1% crystal violet solution for 10 min. The plates were then washed with double deionized water (ddH2O) five times as above, followed by air-drying for 10 min. Subsequently, the bound crystal violet was solubilized using absolute ethanol, and the OD595 value of each well was measured. The assay was performed in three independent experiments.
Antibacterial competition assay
Competition experiments were performed as previously described [32] with some modifications. To construct a T6SS− strain, we inactivated the clpV gene, which encodes a putative ATPase required for T6SS function [33]. A. hydrophila NJ-35 and its derived strains were used as the predator strains, while E. coli BL21, V. parahaemolyticus RIMD 2210633, and other Aeromonas strains (Table 1) served as the preys. The predator and prey strains were cultured for 5 h, respectively; the cultures were adjusted to an OD600 of 1.0 and concentrated 10 times. Cells were mixed together at a ratio of 5:1 (predator to prey); 25 μL of the mixture was spotted onto a 0.22-μm nylon filter on LB plates, and the plates were incubated for 3 h at 28 °C. Prey cells that were mixed with an equal volume of LB media were used as a control. Then, the cultures on the spots were suspended in 1 mL of LB broth. The CFU of surviving prey cells were enumerated by serial dilution and plating onto the correspondingly selective medium. The experiment was repeated three times independently.
Protein secretion assay
A protein secretion assay was performed to explore the secretion of Tle1AH in A. hydrophila as described elsewhere [26]. The tle1AH gene was expressed in A. hydrophila through the recombinant plasmid pMMB207-tle1AH (fused with 6*His-tag). Then, the strains were grown in 500-mL glass culture flasks with 200 mL LB medium for 18 h, and bacterial cultures were collected by centrifugation at 10 000 × g for 10 min. The cell pellets were resuspended using 5 mL PBS and 50 µL 5 × SDS-PAGE (sodium dodecyl sulfate/polyacrylamide gel electrophoresis; KeyGEN BioTECH, Nanjing, China) sample loading buffer. The culture supernatants were filtered using a 0.22-µm membrane filter (OKLABS), and 100% ice-cold trichloroacetic acid (TCA) solution (to 10%) was added to precipitate proteins on ice for 1 h. The proteins were then centrifuged at 15 000 × g for 15 min at 4 °C, and the supernatant was discarded. The concentrated protein precipitate was washed twice with 100% acetone and centrifuged at 15 000 × g for 10 min. After air-drying in a sterile laminar flow hood for 20 min, the proteins were collected and treated with 5× SDS-PAGE buffer. Then, the protein samples from cell pellets and culture supernatants were analyzed by SDS-PAGE and Western blot with anti-His mouse monoclonal antibody (mAb; Abmart, Shanghai, China) or anti-GroEL (heat shock protein Hsp60) polyclonal antiserum [34]. Here GroEL (a cytoplasmic protein) serves as a loading control.
Growth curves for Tle1AH toxicity assays
The Tle1AH toxicity assay was carried out as described previously [35]. pBAD/HisA (Invitrogen) was used for construction of the expression vectors for tle1AH and its point mutant tle1AHS303A (the catalysis site of Tle1AH at position 303 mutated from serine to alanine). Point mutation S303A was generated by fusion PCR and further verified by sequencing [34]. To achieve periplasmic localization, the PelB leader sequence [36] was fused in front of the tle1AH and tle1AHS303A. E. coli TOP10 containing pBAD/HisA–tle1AH or pBAD/HisA–tle1AHS303A were grown overnight at 37 °C in a 5-mL eppendorf tube (GeGene Tech, Shanghai, China) with LB medium containing Amp; the OD600 was adjusted to 0.5, and the cultures were inoculated into a 50-mL glass flask with 20 mL LB broth at a ratio of 1:100. Cultures were induced with 0.25% l-arabinose after 1.5 h of growth. A growth curve was drawn by measuring the OD600 every 30 min. The experiment was repeated three times independently.
Expression and purification of proteins
Primers were designed according to the sequences of the tle1AH and vgrG genes of A. hydrophila NJ-35 in GenBank (accession number NZ_CP006870). Tle1AH was cloned into the pGEX-4T-1 vector (Invitrogen) for expression with an N-terminal glutathione-S-transferase (GST) tag. VgrG/hcp was cloned into the pET-28a vector (Invitrogen) with a His tag. The GST-Tle1AH and His-VgrG/Hcp proteins were expressed in BL21 (DE3) cells (CWBIO, Beijing, China). The transformed cells were cultured in LB medium at 37 °C to an OD600 of 0.8, at which time the fusion protein expression was at the highest level and most was expressed as soluble protein not as an inclusion body based on our preliminary experiment. Protein expression was induced with 100 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 16 °C for 20 h. The cultures were harvested by centrifugation at 8000 × g, resuspended in 1 × PBS and lysed by sonication. The lysate was centrifuged at 12 000 × g for 15 min at 4 °C to remove precipitate, and then the supernatant was loaded on a Ni2+-NTA column (GE Healthcare, Shanghai, China) to purify the proteins. The eluted proteins were collected and dialyzed for pull-down assays.
GST pull-down assay
A GST pull-down assay was employed to identify the interactions between TleAH and VgrG/Hcp. Briefly, GST-Tle1AH proteins were incubated with prepared glutathione Sepharose beads (25 µm; Enriching Biotechnology, Shanghai, China) on a rotating incubator for 3 h at 4 °C, and then the beads were collected and washed three times with PBS (pH 7.4). Then, His-VgrG/Hcp proteins were added to the beads and incubated for 3 h at 4 °C. The beads were washed with PBS buffer five times, and the bound proteins were washed off the beads with elution buffer (50 mM Tris–HCl, 10 mM GSH, pH 8.0). The elution was analyzed and detected by SDS-PAGE and Western blotting with anti-His or anti-GST mouse mAb (Abmart).
Western blotting analysis
Protein samples with SDS loading buffer were boiled for 10 min; 10 μL of each sample was loaded on an SDS-PAGE gel, which was run at 80-120 V by Bio-Rad PowerPac Basic for 1 h and transferred to a 0.22-µm NC (nitrocellulose) filter membrane (Solarbio, Beijing, China) by electroblotting apparatus (Bio-Rad, USA). The membrane was blocked with 5% (wt/vol) skimmed milk in TBST (20 mM Tris–HCl, 150 mM NaCl, 0.05% (V/V) Tween 20) buffer for 2 h at 37 °C, incubated with anti-His, anti-GST or anti-GroEL antibody (1:5000) for 1.5 h at room temperature and washed three times with TBST buffer, incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or goat anti-rabbit IgG antibody (1:5000; Linc-Bio Science, Shanghai, China) for 1.5 h, and washed three times in TBST buffer. The blots were then detected using the Enhanced Chemiluminescence (ECL) Detection Kit (CMCTAG, USA) and ChemiDocTM Touch imaging system (Bio-Rad, USA).
Determination of the bacterial median lethal dose (LD50)
A LD50 challenge assay was performed as previously reported [33]. Briefly, the mid-logarithmic bacterial cultures were washed three times with sterile PBS and serially diluted tenfold from 5 × 106 to 5 × 101 CFU/mL. For each A. hydrophila strain, six groups of 10 zebrafish per group were intraperitoneally (i.p.) injected with 20 µL of the bacterial suspension in PBS. Additionally, 10 zebrafish that were injected only with PBS served as a negative control. Mortality was recorded twice per day for 7 days, and the LD50 values were calculated following the method of Reed and Muench [37].
Tissue colonization by wild-type and tle1AH mutant strains
A competitive colonization assay was performed as previously reported [35]. To assay the competitive colonization of NJ-35 and its mutant derivatives in heart, hepatopancreas, spleen and kidney of crucian carp, WT and mutant bacteria were suspended in PBS to achieve a final concentration of 6.5 × 107 CFU/mL, respectively and mixed at a 1:1 ratio. Then, 100 µL of the mixture was intraperitoneally injected into five fish. After 24 h, the organ samples were collected in a sterile environment, homogenized by vortexing in 900 µL of PBS, and diluted tenfold in PBS; aliquots were plated onto LB agar with the corresponding antibiotics. The bacteria used here carried either a Gm or Kan resistance vector, allowing them to be easily screened. For graphical and statistical purposes, the viable plate counts (CFU per gram of sample) were log10 transformed.
Bioinformatics analysis
Nucleotide and protein sequences were acquired from the National Center for Biotechnology Information database [38]. Phylogenetic tree reconstruction was performed using MEGA 7.0 [39], and sequence logos were generated using Geneious Prime 2019.
Statistical analyses
Data were analyzed and plotted using GraphPad Prism version 7 software. Multiple comparisons were performed by analysis of variance (ANOVA) followed by the Turkey multiple-comparison test. P-values < 0.05 were considered to be statistically significant. The error bars presented in the figures represent the standard deviations of the means of multiple replicate experiments.
Results
A potential T6SS effector is predicted in A. hydrophila NJ-35
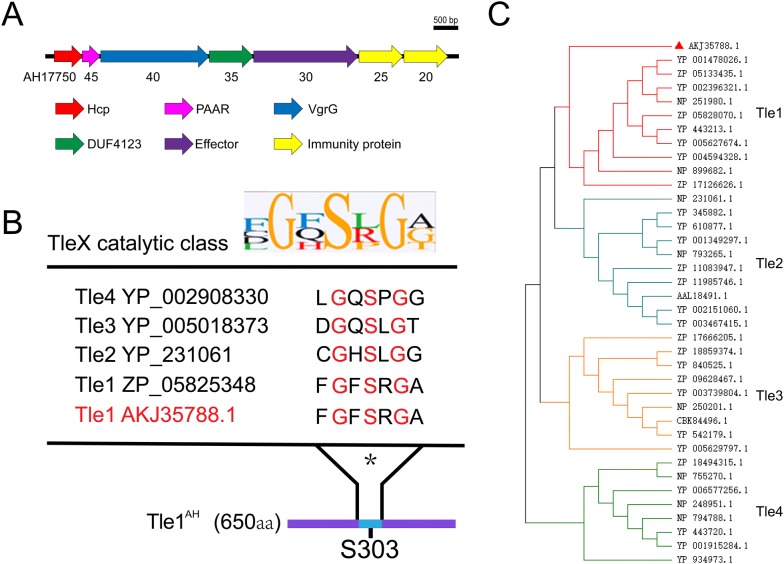
Based on the conserved domain DUF4123 [26], we searched potential T6SS effectors in A. hydrophila NJ-35. A putative effector-immunity (AH17720-17730) pair encoded downstream of the DUF4123 (AH17735) domain was predicted (Figure 1A). Bioinformatics analysis indicated that the AH17730 protein possesses a highly conserved DUF2235 domain, with a catalytic motif Gly-X-Ser-X-Gly (X is for any amino acid), which is common in esterases and lipases (Figure 1B). Further phylogenetic analysis suggested that AH17730 belongs to the Tle1 family (Figure 1C); therefore, AH17730 was named Tle1AH.
Figure 1.
Tle1AHis a potential T6SS effector inA. hydrophilaNJ-35. A Genetic organization of T6SS-related proteins containing the DUF4123 domain in A. hydrophila NJ-35. The numbers below refer to the gene locus tag (U876-XXXXX). Sequencing data for NJ-35 can be obtained from the National Center for Biotechnology Information (accession number: CP006870). B Sequence alignment of conserved catalytic motifs (labeled in red) compared between Tle families. Sequence logos were generated from alignments of the catalytic motifs from the families Tle1-4 (Gly-X-Ser-X-Gly, X is for any amino acid). * represents the catalytic residues. C Phylogenetic analyses of Tle1AH (AKJ35788.1) with representative members of the families Tle1-4. Figure was prepared using MEGA7.0.
Tle1AH mediates interbacterial antagonism
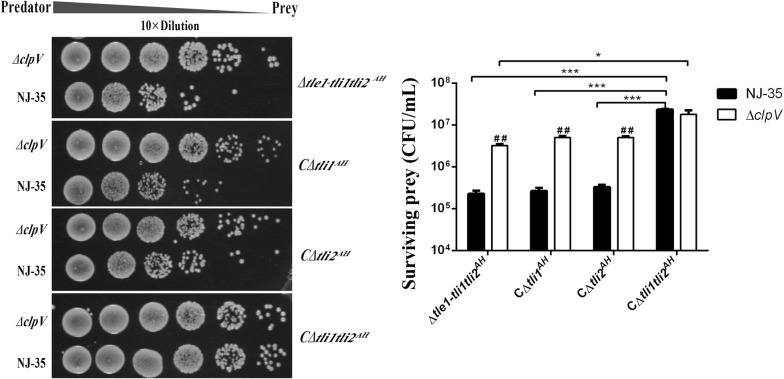
To determine whether tle1AH participates in inter-generic antagonism of A. hydrophila NJ-35, we performed a quantitative bacterial competition assay using E. coli BL21 and V. parahaemolyticus RIMD 2210633 as prey strains. The CFU of surviving prey cells were enumerated on LB agar containing Kan. As shown in Figure 2A, compared with the LB group (without the predator), the wild-type NJ-35 and its derived mutants ∆tle1AH and ∆clpV (T6SS−) caused a considerable reduction in survival of E. coli (P < 0.001). Mutant strains ∆tle1AH (P < 0.01) and ∆clpV (P < 0.001) exhibited significantly decreased killing ability compared to the wild-type strain, while the capacity of the former to kill E. coli was stronger than the latter (P < 0.01). The killing ability of the complement strain C∆tle1AH was restored to the wild-type level. Similar results to E. coli were obtained for V. parahaemolyticus (Figure 2B).
Figure 2.
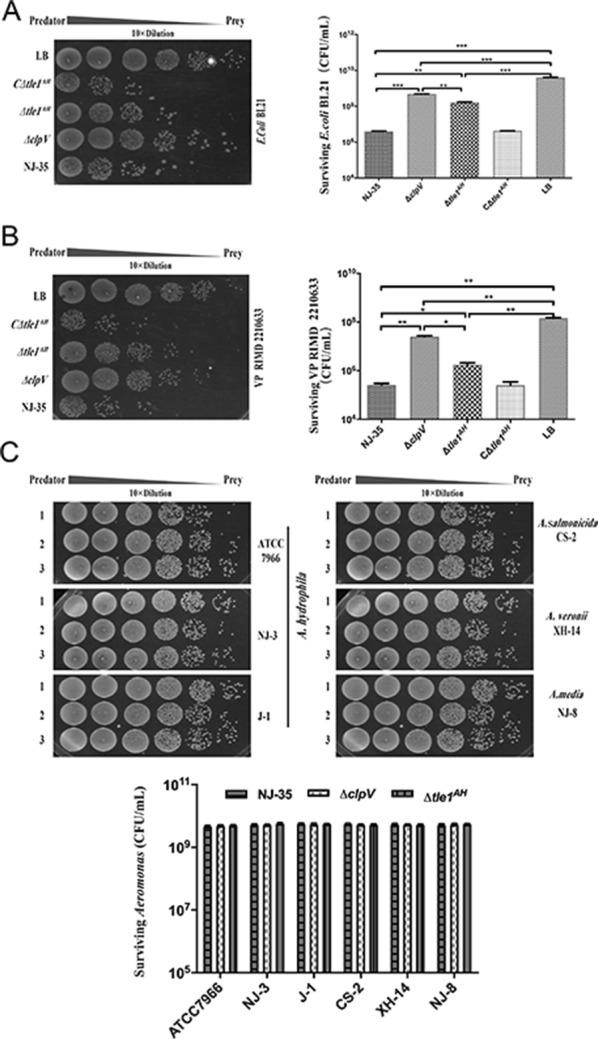
Tle1AHis required for the interbacterial antagonism ofA. hydrophilaNJ-35. Predator and prey cells at a ratio of 5:1 were cocultured to assay the recovery of surviving prey cells by determining colony forming unit (CFU). A. hydrophila NJ-35 and its mutant derivatives ∆clpV, ∆tle1AH or C∆tle1AH were used as the predator strains. ClpV, which encodes a putative ATPase required for T6SS function, was deleted to construct the T6SS− strain (∆clpV). “LB” indicates incubation of E. coli with sterile LB medium alone and serves as the control. AE. coli BL21 as the prey strain. BV. parahaemolyticus RIMD 2210633 as the prey strain. CAeromonas strains as the preys, including A. hydrophila strains ATCC 7966, J-1 and NJ-3, A. sobria CS-2, A. media NJ-8 and A. veronii XH-14. Lane 1, the wild-type A. hydrophila NJ-35; Lane 2, ∆clpV (T6SS−); Lane 3, ∆tle1AH. Data are presented as the mean ± standard deviation (error bars) of three independent experiments. ***P < 0.001, **P < 0.01. *P < 0.05.
Further, we evaluated the intra-generic role of tle1AH by examining the abilities of A. hydrophila NJ-35 and its derived deletion mutant ∆tle1AH to outcompete other A. hydrophila strains (ATCC 7966, J-I and NJ-3) or related species including A. salmonicida CS-2, A. media NJ-8 and A. veronii XH-14. Notably, no significant difference was observed in CFU of surviving Aeromonas when the wild-type NJ-35, ∆tle1AH or ∆clpV were used as the predator strains (Figure 2C). These data suggest that Tle1AH may be involved in inter-generic but not intra-generic bacterial competition of A. hydrophila NJ-35.
Tle1AH is a phospholipase effector secreted by T6SS
To determine if the secretion of Tle1AH is accomplished via T6SS in A. hydrophila NJ-35, we constructed a pMMB-tle1AH overexpression vector labeled with a 6*His-tag. Then, the recombinant vector from the donor strain E. coli SM10 was conjugated into the recipient strains NJ-35 and ∆clpV. Western blot analysis showed that Tle1 protein could be detected in the supernatant of the wild-type strain NJ-35 but not in that of the ∆clpV mutant strain. For reference, Tle1AH was detected in the whole cells of both strains (Figure 3A). The results demonstrate that the secretion of Tle1 protein depends on T6SS.
Figure 3.
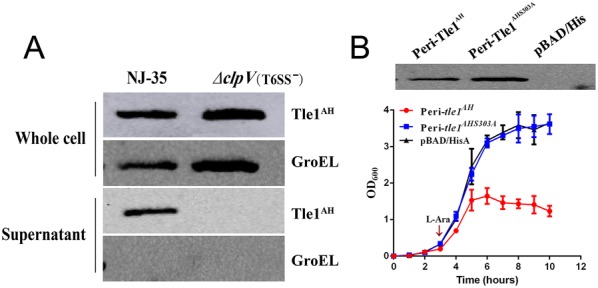
Tle1AHis a phospholipase effector secreted by T6SS ofA. hydrophilaNJ-35. A T6SS-dependent secretion of Tle1AH was confirmed by Western blot on whole cells and supernatants of A. hydrophila NJ-35 and the ∆clpV strain. ClpV, which encodes a putative ATPase required for T6SS function, was deleted to construct the T6SS− strain (∆clpV). The anti-His antibody was used to measure the production of Tle1AH and anti-GroEL antibody served as an internal reference. GroEL: heat shock protein Hsp60. B Growth of E. coli TOP10 producing peri-Tle1AH and peri-Tle1AHS303A in LB broth. pBAD/His was used for construction of the expression vectors for tle1AH and its point mutant tle1AHS303A (the catalysis site of Tle1AH at position 303 mutated from serine to alanine). To achieve periplasmic localization, the PelB leader sequence was fused in front of the tle1AH and tle1AHS303A. Cultures were induced by l-arabinose (l-Ara) at the indicated time by the arrow. A growth curve was drawn by measuring the OD600 every 30 min. Data are presented as the mean ± standard deviation (error bars) of three independent experiments. The expression of peri-Tle1AH and peri-Tle1AHS303A was detected in E. coli TOP10 by Western blot using anti-His antibody.
We reasoned that if Tle1AH is a phospholipase with the Gly-X-Ser-X-Gly sequence between amino acids 303 and 307 as the catalytic motif and is secreted by T6SS, a point mutation of the catalytic motif will abolish its enzymatic activity. To prove its catalytic activity, we mutated the serine catalysis site (S303A) and constructed expression vectors for tle1AH and its point mutant tle1AHS303A. Western blot analysis indicates that peri-tle1AH and peri-tle1AHS303A recombinant vectors were well expressed in E. coli TOP10 cells. The growth curve indicates an obvious decrease in the survival of E. coli containing the peri-tle1AH recombinant plasmid, while the E. coli containing peri-tle1AHS303A plasmid shows a similar growth rate as the negative control bacteria (Figure 3B). The results proved that Tle1AH is a T6SS phospholipase effector and toxic to E. coli when artificially localized to the periplasm via a sec-dependent leader sequence.
Tle1AH and Tli1Tli2AH are an effector-immunity pair
In T6SS, immunity genes are generally tightly linked next to their isogenous effector genes, and the immunity proteins are produced to specifically bind and neutralize their cognate toxins [40]. Unexpectedly, downstream of tle1AH, there exists two open reading frames (ORF) with unknown function, AH17725 (tli1AH) and AH 17720 (tli2AH), both of which belong to the DUF2931 family. To determine which ORF plays an important role in protection against Tle1AH, we constructed three complementary plasmids that harbored the tli1AH or tli2AH gene alone or together and introduced these plasmids separately into the Δtle1-tli1tli2AH mutant (deletion of tle1AH and its two cognate immunity genes). Using these mutants as preys and the wild-type NJ-35 and ∆clpV as predators, a competitive experiment was carried out. As shown in Figure 4, when ∆clpV was used as the predator strain, there exist no statistically significant differences in surviving prey cells between Δtle1-tli1tli2AH and its single restoration strain of tli1AH or tli2AH, or between the single and double restoration strains, except for a significant difference between Δtle1-tli1tli2AH and the double restoration strain (P < 0.05). However, using the wild-type NJ-35 as the predator strain, the double restoration strain exhibits a significant surviving advantage over Δtle1-tli1tli2AH or the single restoration strains (P < 0.001), while no significant differences were observed between Δtle1-tli1tli2AH and its single restoration strains of tli1AH or tli2AH. Compared with the ∆clpV, NJ-35 could cause substantial decreases in the number of surviving cells of Δtle1-tli1tli2AH and its single gene restoration strain of tli1AH or tli2AH (P < 0.01), but not make a significant difference in the surviving populations of the tli1tli2AH double restoration strain. These data indicate that single gene restoration of tli1AH or tli2AH could not protect the ∆tle1-tli1tli2AH mutant from antagonism by Tle1AH, and only when the two immunity proteins were coexpressed could the effective protection be conferred.
Figure 4.
Tli1Tli2AHare the cognate immunity proteins to Tle1AH. A. hydrophila NJ-35 and the ∆clpV strain were used as the predator strains. ClpV, which encodes a putative ATPase required for T6SS function, was deleted to construct the T6SS− strain (∆clpV). The prey strains included the gene deletion mutant ∆tle1-tli1tli2AH and its single or double restoration strains of immunity genes, they are, C∆tli1AH (∆tle1-tli1tli2AH/pMMB-tli1AH), C∆tli2AH (∆tle1-tli1tli2AH/pMMB-tli2AH) and C∆tli1tli2AH (∆tle1-tli1tli2AH/pMMB-tli1tli2AH). The predator and prey strains were cultured at a ratio of 5:1, and surviving prey cells were serially diluted and determined on the LB plate containing antibiotics. Data are presented as the mean ± standard deviation (error bars) of three independent experiments. ##P < 0.01 indicates a significant difference between this group and the NJ-35 group. ***P < 0.001 or *P < 0.05 indicate significant differences between the two specified groups.
Tle1AH interacts with the VgrG protein
The VgrG and Hcp proteins have been proposed as carriers of T6SS effectors [19]. To gain insights into the secretion mechanism of Tle1AH, we investigated whether the VgrG or Hcp proteins encoded upstream of Tle1AH were involved in Tle1AH delivery. We tested the interactions of Tle1AH with either VgrG or Hcp using a pull-down assay. Our results indicate that VgrG was able to bind to Tle1AH (Figure 5), whereas Hcp failed to do so (data not shown), suggesting that VgrG is essential for Tle1AH export into target cells.
Figure 5.
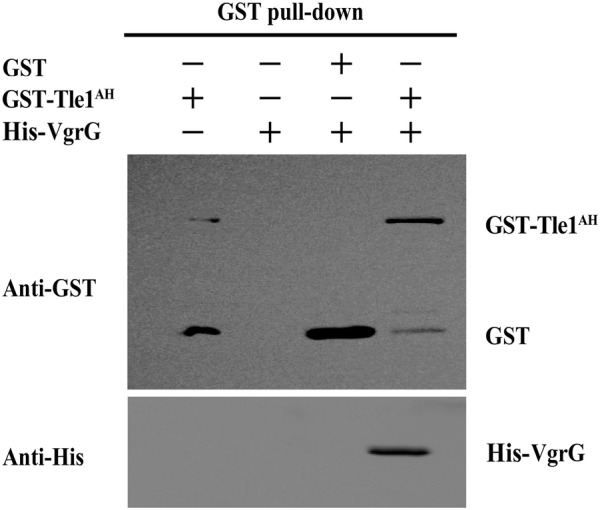
The interaction of Tle1AHwith VgrG was verified by an in vitro pull-down assay. Purified GST, GST-Tle1AH and His-VgrG were incubated with magnetic beads for the GST pull-down assay. Controls included the incubation with GST-Tle1AH alone, His-VgrG alone, or GST with His-VgrG. Bound proteins were washed off the beads with the elution buffer and detected by immunoblotting using anti-GST or anti-His antibodies.
Tle1AH is required for virulence and colonization of A. hydrophila NJ-35
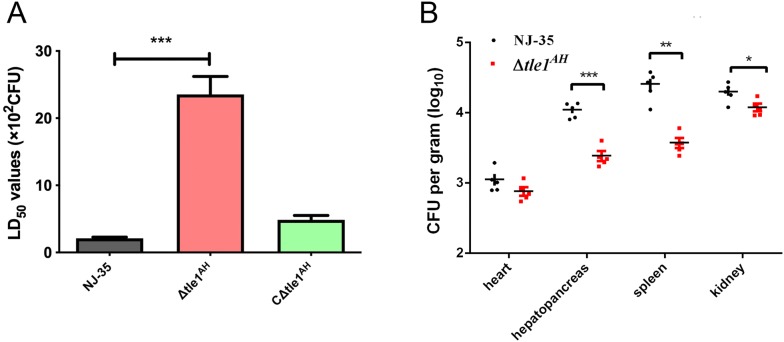
Zebrafish is a well-established animal model to evaluate Aeromonas virulence [41]. To determine whether the tle1AH gene affected bacterial virulence, the LD50 values of the wild-type and tle1AH mutant strains were investigated using a zebrafish model. The LD50 value of A. hydrophila NJ-35 was 2.11 × 102 CFU, while the tle1AH mutant strain had an approximately 11-fold higher LD50 value than the wild-type strain (P < 0.001) (Figure 6A). Most dead fish showed typical clinical features of hemorrhagic septicemia. The results indicate that Tle1AH affects the virulence of A. hydrophila NJ-35. Furthermore, we explored whether Tle1AH was required for the colonization of the NJ-35 strain in crucian carp. As expected, the colonization ability of the ∆tle1AH strain was significantly lower than that of the wild-type strain in the hepatopancreas (P < 0.001), spleen (P < 0.01) and kidney (P < 0.05) (Figure 6B). These data indicate that Tle1AH facilitates the survival and colonization of A. hydrophila within the host, which is a common characteristic of antibacterial effectors.
Figure 6.
Tle1AHis required for the virulence and colonization ofA. hydrophilaNJ-35. A Determination of the LD50 values of the wild-type and tle1AH mutant strains in zebrafish. Zebrafish were intraperitoneally (i.p.) injected with tenfold serially diluted bacterial suspensions. The control group was i.p. injected with sterile PBS only. B Competitive assays of NJ-35 and Δtle1AH in crucian carp. Strains were mixed at a ratio of 1:1 and inoculated to fish by intraperitoneal injection. After 24 h, heart, hepatopancreas, spleen and kidney were harvested for counting of the number of colony-forming units (CFU) per gram of sample. Data are presented as the mean ± standard deviation (error bars) of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05.
Tle1AH is associated with biofilm formation of A. hydrophila
To examine whether tle1AH is involved in some biological characteristics of A. hydrophila, like the classical effector proteins Hcp or VgrG, we further assayed bacterial growth, motility and biofilm formation. Our results indicate that there were no significant changes in the growth rate (Additional file 2) and bacterial motility (Additional file 3) in the tle1AH mutant compared to the wild-type strain. However, the biofilm formation phenotype of the ∆tle1AH strain was weaker than that of the NJ-35 strain, and the decreased biofilm formation ability could be restored to the wild-type level in the C∆tle1AH strain (Figure 7). This result suggests that in addition to antimicrobial activity, the tle1AH gene may directly or indirectly influence the biofilm formation of A. hydrophila.
Figure 7.
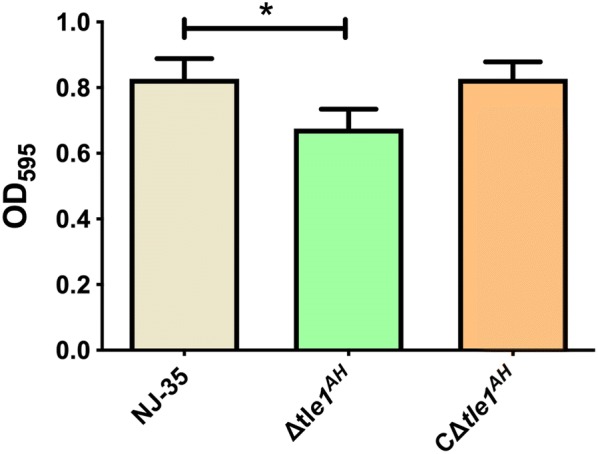
Biofilm formation of the wild-type andtle1AHmutant strains. Biofilm formation was determined by crystal violet staining using 96-well plates, and the values were measured at OD595. Data are presented as the mean ± standard deviation (error bars) of three independent experiments. *P < 0.05.
Search for DUF4123 and downstream genes in A. hydrophila strains of the known T6SS components
Given that the DUF4123 domain has been used to predict unknown T6SS effectors, we expanded our analyses for the large-scale search of effectors in A. hydrophila strains (Table 2). Many potential effectors were identified using DUF4123 as a signal (Figure 8). Most of the effectors encode the DUF2235 domain at the C-terminus and have a Gly-X-Ser-X-Gly motif, indicating that they are members of the Tle family. And genes encoding putative immunity proteins, which belong to the DUF2931 family, could be found downstream of their cognate effector genes. The number of immunity genes varies from none to three, indicating that E-I pairs exist in a variety of forms. In addition, there were also a few strains without the DUF2235 domain but with other colicin or unknown effector proteins.
Table 2.
The A. hydrophila strains used to search for putative effectors
| Strains | Accession number | Sequence range of DUF4123-immunity | Conserved domain of putative protein | Protein ID of putative protein |
|---|---|---|---|---|
| NJ-35 | NZ_CP006870 | 3 862 649–3 866 785 | DUF2235 | WP_047234910 |
| J-1 | NZ_CP006883 | 3 759 517–3 763 012 | DUF2235 | WP_016349832 |
| 4AK4 | NZ_CP006579 | 3 417 209–3 422 000 | Colicin | WP_025327967 |
| AH10 | NZ_CP011100 | 3 826 164–3 830 669 | Lipase | WP_045790457 |
| JBN2301 | NZ_CP013178 | 3 873 138–3 876 633 | DUF2235 | WP_016349832 |
| 112 134–117 054 | Unknown | WP_139118716 | ||
| D4 | NZ_CP013965 | 3 835 001–3 838 496 | DUF2235 | WP_016349832 |
| 112 134–117 054 | Unknown | WP_139118716 | ||
| GYK1 | NZ_CP016392 | 3 750 771–3 754 266 | DUF2235 | WP_016349832 |
| ML09_119 | NC_021290 | 1 315 162–1 318 657 | DUF2235 | WP_016349832 |
| YL17 | NZ_CP007518 | 2 331 025–2 336 071 | DUF2235 | WP_016349832 |
| AL09_71 | NZ_CP007566 | 1 314 833–1 318 328 | DUF2235 | WP_016349832 |
| pc104A | NZ_CP007576 | 1 314 832–1 318 327 | DUF2235 | WP_016349832 |
| AL06_06 | CP010947 | 1 271 104–1 276 922 | Colicin | WP_016349832 |
| 4 374 167–4 376 145 | HNH endonuclease | WP_016349832 | ||
| AHNIH1 | NZ_CP016380 | 1 185 505–1 190 948 | Unknown | WP_016349832 |
| 2 892 361–2 897 678 | Tox-HNH-EHHH | WP_016349832 | ||
| ATCC 7966T | NC_008570 | 1 215 532–1 219 858 | DUF2235 | WP_016349832 |
Figure 8.
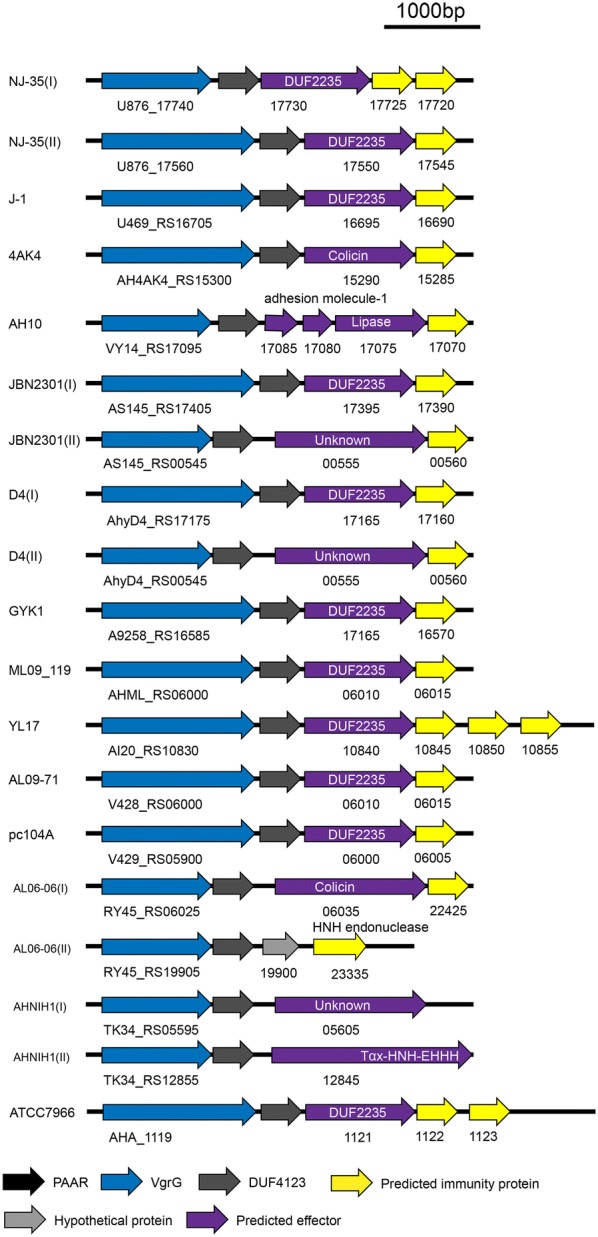
Predicted DUF4123-associated T6SS effector-immunity (EI) gene modules in 14A. hydrophilastrains. All the strains used to search for putative effectors are listed in Table 2. The gray arrows indicate DUF4123, the purple arrows indicate the putative effectors, and yellow arrows indicate immunity genes. The arrows indicate the direction of transcription.
Discussion
To adapt to the complexity of the living environment, microbes have evolved many mechanisms to compete with other species for limited nutrition [42–44]. Some Gram-negative bacteria encode T6SS weapons that deliver toxins to either prokaryotic or eukaryotic cells to mediate different signals [45]. Many studies have successfully identified effector proteins using diverse methods [21, 24, 46]. In this study, we predicted a T6SS effector protein, AH17730, in the genome of A. hydrophila NJ-35 based on the conserved domain DUF4123. By analyzing the amino acid sequence of AH17730, we identified a catalytic motif Gly-X-Ser-X-Gly, which can be found in the Tle lipase family. Tle proteins have been reported as a superfamily of T6SS phospholipase effectors that directly target the cell membrane by hydrolyzing its lipid components [47]. The known Tle proteins are classified into five groups according to their protein sequences and phylogenetic distribution. In this study, we demonstrate that AH17730 has phospholipase activity and belongs to the Tle1 family; therefore, AH17730 was named Tle1AH.
Previously, Tle1BT was shown to be required for the antibacterial activity of Burkholderia thailandensis [47]. Similarly, our study demonstrates that Tle1AH is essential for A. hydrophila to kill E. coli or V. parahaemolyticus in a T6SS-dependent manner. Notably, the antibacterial competition experiment shows that Tle1AH could not act on closely related strains or species, such as A. hydrophila strains ATCC 7966, J-1 and NJ-3, A. salmonicida CS-2, A. media NJ-8 and A. veronii XH-14. For this reason, we speculated that it might be related to cognate immunity proteins. In our work, we identified two putative immunity proteins, AH17725 and AH17720, downstream of Tle1AH, and bioinformatics analysis shows that the two proteins belong to the DUF2931 superfamily. Some members of this superfamily have been annotated as outer membrane lipoproteins, which is consistent with the localization of Tle1AH. Functional analysis indicates that the two immunity proteins were required to work together to protect A. hydrophila NJ-35 against toxicity of Tle1AH. To further determine whether a correlation exists between the toxicity of Tle1AH and the distribution of immune genes, we searched for the known genomes of two A. hydrophila strains, including ATCC 7966 (accession number NC_008570), a well-characterized type strain for the species, originally isolated from “a tin of milk with a fishy odor” [48], and J-1 (accession number CP006883), an epidemic piscine strain of China [49]. As expected, the genome of ATCC 7966 contains the same E-I module (AHA_1121-1123) as AH17720-17730 of NJ-35. In the J-1 genome, a gene that has 100% identity with tle1AH, V469_RS16695, was found, and a DUF2931 family protein is encoded downstream of this gene. Therefore, it is possible that the same or similar immune proteins in the above Aeromonas strains play a role in resisting the toxicity of Tle1AH from NJ-35. Unfortunately, no complete genome sequences are available for strains NJ-3, CS-2, NJ-8 or XH-14. A recent study indicated that an additional non-native T6SS auxiliary cluster can be acquired and used by a V. cholerae strain to kill kin cells lacking the immunity protein [50]. Whether such evolutionary dynamics of the V. cholerae T6SS actually exist in Aeromonas will be the subject of our future study.
Interestingly, our results show that only when two immunity proteins worked together could A. hydrophila NJ-35 assure self-protection against toxicity of Tle1AH. Similar findings have not been previously reported. Although it is not surprising that two homologous immunity genes could be found to coexist downstream of the tle1 gene, expression of the immunity gene that is most closely adjacent to tle1 is usually sufficient to abolish Tle1 activity. A previous study on the entero-aggregative E. coli (EAEC) Sci-1 T6SS indicated that the tle1EAEC gene, which encodes an effector and is responsible for Sci-1-mediated antibacterial activity, is followed by a duplicated region encoding two putative immunity proteins, EC042_4535 and EC042_4536; the production of EC042_4535 (Tli1EAEC) in ∆EC042_4535-4536 prey cells protected themselves against EAEC killing, and the Tle1EAEC activity was completely abolished with a Tle1EAEC:Tli1EAEC molecular ratio of 1:1 [51]. We do not know much about the binding ratios of the Tle1 effector and its cognate immunity proteins in various bacteria. It is possible that future structure-based functional analysis of Tle1 and its complexes with immunity proteins will reveal the diverse mechanisms of inhibition by immunity proteins.
Different mechanisms have been proposed or identified for cargo effectors. They can directly or indirectly contact the VgrG spike, the Hcp rings or PAAR proteins [16, 52]. Genes coding for Tle superfamily proteins, which are known as cargo effectors, are commonly found in the vicinity of the vgrG gene [18, 47]. Therefore, one may hypothesize that these effectors will be transported by interaction with VgrG. To determine the Tle1AH secretion mechanism, in this work, we performed a pull-down assay and demonstrated that Tle1AH could interact with VgrG but not Hcp. We speculate that this interaction was required for proper Tle1AH delivery. In a previous study, the transport of Tle1EAEC was thought to be required for binding the C-terminal extension of VgrG1 in E. coli, and a putative protein–protein interaction module of this extension revealed a transthyretin-like (TTR) domain fold [51]. For the T6SS-mediated delivery of a given VgrG-binding Tle1, multiple binding events likely occur in a certain order that includes Tle1 binding to the cognate VgrG and to the immunity protein. In the process, we do not know whether there are other chaperone proteins involved and how the DUF4123 domain functions. In the future, structural analyses of the DUF4123, VgrG, and Tle1 proteins are required to fully understand the mechanisms of Tle1 delivery.
Although the T6SS plays a significant role in antimicrobial competition, there remains a possibility that it serves some purposes beyond competition. PA2374, an effector secreted by H3-T6SS of Pseudomonas, is important for iron uptake and functions by interacting with outer membrane vesicles and the Pseudomonas quinolone signal system [53]. It has also been reported that the VgrG and Hcp proteins are also involved in motility, protease activity, biofilm formation and virulence in addition to competitive growth in A. hydrophila SSU (now belonging to Aeromonas dhakensis species) [54]. In the present study, we demonstrate that the tle1AH gene was involved in biofilm formation and virulence, and influenced the colonization ability of A. hydrophila NJ-35 in crucian carp. The findings indicate that the effects of Tle1AH on A. hydrophila virulence are multidimensional, which also explains the complexity of the pathogenic mechanism of this bacterium.
In this study, we identified a Tle1AH effector protein of T6SS in A. hydrophila NJ-35, with two cognate immunity proteins working together to prevent sibling bacteria from intoxication. Also, we identified some putative Tle1 family effectors in A. hydrophila strains with known genome sequences, and interestingly, these strains have been determined to be virulent [55–59], implying that Tle1 may be related to A. hydrophila virulence. Hopefully, there are more diverse effectors to be identified, which will provide a deeper understanding of the T6SS strategies of A. hydrophila.
Supplementary information
Additional file 1. Primers used in this study. Underlined sequences indicate restriction sites.
Additional file 2. Growth curve of the wild-type andtle1AHmutant strains. The cells were cultured in LB broth and densities were measured every 2 h at OD600. Data are presented as the mean ± standard deviation (error bars) of three independent experiments.
Additional file 3. Motility of the wild-type andtle1AHmutant strains. Swimming ability was observed after culturing strains at 28 °C for 48 h on 0.3% LB agar plates. The migration diameters were measured to assess the motility. Data are presented as the mean ± standard deviation (error bars) of three independent experiments.
Acknowledgements
The authors thank Dr Jiale Ma for providing plasmid pBAD/HisA.
Abbreviations
- DUF
domain of unknown function
- Tle1
type VI lipase effectors
- VgrG
valine-glycine repeat protein G
- Hcp
hemolysin co-regulated protein
- PAAR
proline-alanine-alanine-arginine
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- LB
Luria–Bertani
- OD
optical density
- PBS
phosphate buffered saline
- Cm
chloramphenicol
- Amp
ampicillin
- Kan
kanamycin
- Gm
gentamicin
- PCR
polymerase chain reaction
- CFU
colony forming unit
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- GroEL
heat shock protein Hsp60
- LD50
median lethal dose
Authors’ contributions
SM and YD carried out most of the experiments described in the manuscript and wrote the article; NW, JL and CL participated in the design of the study and performed the statistical analysis. YL provided expertise and conceived the study. All authors read and approved the final manuscript.
Funding
This study was funded by the National Key R & D Program of China (2019YFC1605402), National Nature Science Foundation of China (31372454) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additonal files.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shuiyan Ma, Email: 2017107059@njau.edu.cn.
Yuhao Dong, Email: dongyuhao@njau.edu.cn.
Nannan Wang, Email: wangnn@jit.edu.cn.
Jin Liu, Email: liujin0703@163.com.
Chengping Lu, Email: lucp@njau.edu.cn.
Yongjie Liu, Email: liuyongjie@njau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13567-020-00794-w.
References
- 1.Rasmussen-Ivey CR, Figueras MJ, McGarey D, Liles MR. Virulence factors of Aeromonas hydrophila: in the wake of reclassification. Front Microbiol. 2016;7:1337. doi: 10.3389/fmicb.2016.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomas JM. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012:256–261. doi: 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu XJ, Ferguson MR, Popov VL, Houston CW, Peterson JW, Chopra AK. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect Immun. 1998;66:3501–3509. doi: 10.1128/iai.66.8.3501-3509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pemberton JM, Kidd SP, Schmidt R. Secreted enzymes of Aeromonas. FEMS Microbiol Lett. 1997;152:1–10. doi: 10.1111/j.1574-6968.1997.tb10401.x. [DOI] [PubMed] [Google Scholar]
- 5.Esteve C, Alcaide E, Canals R, Merino S, Blasco D, Figueras MJ, Tomas JM. Pathogenic Aeromonas hydrophila serogroup O:14 and O:81 strains with an S layer. Appl Environ Microbiol. 2004;70:5898–5904. doi: 10.1128/AEM.70.10.5898-5904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green ER, Mecsas J (2016) Bacterial secretion systems: an overview. Microbiol Spectr 4:VMBF-0012-2015 [DOI] [PMC free article] [PubMed]
- 7.Bhoite S, van Gerven N, Chapman MR, Remaut H (2019) Curli biogenesis: bacterial amyloid assembly by the type VIII secretion pathway. EcoSal Plus 8:ESP-0037-2018 [DOI] [PMC free article] [PubMed]
- 8.Lasica AM, Ksiazek M, Madej M, Potempa J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol. 2014;22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol. 2017;25:267–279. doi: 10.1016/j.tim.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barret M, Egan F, Fargier E, Morrissey JP, O’Gara F. Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology. 2011;157:1726–1739. doi: 10.1099/mic.0.048645-0. [DOI] [PubMed] [Google Scholar]
- 19.De Maayer P, Venter SN, Kamber T, Duffy B, Coutinho TA, Smits TH. Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics. 2011;12:576. doi: 10.1186/1471-2164-12-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 2016;12:e1005735. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lien YW, Lai EM. Type VI secretion effectors: methodologies and biology. Front Cell Infect Microbiol. 2017;7:254. doi: 10.3389/fcimb.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bondage DD, Lin JS, Ma LS, Kuo CH, Lai EM. VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc Natl Acad Sci U S A. 2016;113:E3931–E3940. doi: 10.1073/pnas.1600428113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X, Moore R, Wilton M, Wong MJ, Lam L, Dong TG. Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc Natl Acad Sci U S A. 2015;112:9106–9111. doi: 10.1073/pnas.1505317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV, Mirzaei H, Orth K. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A. 2014;111:9271–9276. doi: 10.1073/pnas.1406110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abolghait SK. Suicide plasmid-dependent IS1-element untargeted integration into Aeromonas veronii bv. sobria generates brown pigment-producing and spontaneous pelleting mutant. Curr Microbiol. 2013;67:91–99. doi: 10.1007/s00284-013-0335-4. [DOI] [PubMed] [Google Scholar]
- 29.Melton-Witt JA, McKay SL, Portnoy DA. Development of a single-gene, signature-tag-based approach in combination with alanine mutagenesis to identify listeriolysin O residues critical for the in vivo survival of Listeria monocytogenes. Infect Immun. 2012;80:2221–2230. doi: 10.1128/IAI.06196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 31.Dong Y, Liu J, Pang M, Du H, Wang N, Awan F, Lu C, Liu Y. Catecholamine-stimulated growth of Aeromonas hydrophila requires the TonB2 energy transduction system but is independent of the amonabactin siderophore. Front Cell Infect Microbiol. 2016;6:183. doi: 10.3389/fcimb.2016.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Dong Y, Wang N, Li S, Yang Y, Wang Y, Awan F, Lu C, Liu Y. Tetrahymena thermophila predation enhances environmental adaptation of the carp pathogenic strain Aeromonas hydrophila NJ-35. Front Cell Infect Microbiol. 2018;8:76. doi: 10.3389/fcimb.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Liu J, Pang M, Wu Y, Awan F, Liles MR, Lu C, Liu Y. Diverse roles of Hcp family proteins in the environmental fitness and pathogenicity of Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl Microbiol Biotechnol. 2018;102:7083–7095. doi: 10.1007/s00253-018-9116-0. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Dong Y, Wang N, Ma S, Lu C, Liu Y. Diverse effects of nitric oxide reductase NorV on Aeromonas hydrophila virulence-associated traits under aerobic and anaerobic conditions. Vet Res. 2019;50:67. doi: 10.1186/s13567-019-0683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Sun M, Pan Z, Lu C, Yao H. Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochim Biophys Acta Gen Subj. 2018;1862:1635–1643. doi: 10.1016/j.bbagen.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Lei SP, Lin HC, Wang SS, Callaway J, Wilcox G. Characterization of the Erwinia carotovora pelB gene and its product pectate lyase. J Bacteriol. 1987;169:4379–4383. doi: 10.1128/jb.169.9.4379-4383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 38.National Center for Biotechnology Information Database. Accessed 2 June 2019
- 39.Wamala SP, Mugimba KK, Dubey S, Takele A, Munang’andu HM, Evensen O, Mutoloki S, Byarugaba DK, Sorum H. Multilocus sequence analysis revealed a high genotypic diversity of Aeromonas hydrophila infecting fish in Uganda. J Fish Dis. 2018;41:1589–1600. doi: 10.1111/jfd.12873. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Long M, Shen X. Effector-immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities. Molecules. 2018;23:E1009. doi: 10.3390/molecules23051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang MD, Lin XQ, Hu M, Li J, Lu CP, Liu YJ. Tetrahymena: an alternative model host for evaluating virulence of Aeromonas strains. PLoS One. 2012;7:e48922. doi: 10.1371/journal.pone.0048922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 43.Konovalova A, Sogaard-Andersen L. Close encounters: contact-dependent interactions in bacteria. Mol Microbiol. 2011;81:297–301. doi: 10.1111/j.1365-2958.2011.07711.x. [DOI] [PubMed] [Google Scholar]
- 44.Rendueles O, Ghigo JM. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol Rev. 2012;36:972–989. doi: 10.1111/j.1574-6976.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- 45.Hachani A, Wood TE, Filloux A. Type VI secretion and anti-host effectors. Curr Opin Microbiol. 2016;29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Fitzsimons TC, Lewis JM, Wright A, Kleifeld O, Schittenhelm RB, Powell D, Harper M, Boyce JD. Identification of novel Acinetobacter baumannii type VI secretion system antibacterial effector and immunity pairs. Infect Immun. 2018;86:e00297. doi: 10.1128/IAI.00297-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang M, Jiang J, Xie X, Wu Y, Dong Y, Kwok AH, Zhang W, Yao H, Lu C, Leung FC, Liu Y. Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci Rep. 2015;5:9833. doi: 10.1038/srep09833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crisan CV, Chande AT, Williams K, Raghuram V, Rishishwar L, Steinbach G, Watve SS, Yunker P, Jordan IK, Hammer BK. Analysis of Vibrio cholerae genomes identifies new type VI secretion system gene clusters. Genome Biol. 2019;20:163. doi: 10.1186/s13059-019-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flaugnatti N, Le TT, Canaan S, Aschtgen MS, Nguyen VS, Blangy S, Kellenberger C, Roussel A, Cambillau C, Cascales E, Journet L. A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 2016;99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 52.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sha J, Rosenzweig JA, Kozlova EV, Wang S, Erova TE, Kirtley ML, van Lier CJ, Chopra AK. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology. 2013;159:1120–1135. doi: 10.1099/mic.0.063495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim YL, Roberts RJ, Ee R, Yin WF, Chan KG. Complete genome sequence and methylome analysis of Aeromonas hydrophila strain YL17, isolated from a compost pile. Genome Announc. 2016;4:e00060. doi: 10.1128/genomeA.00060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pridgeon JW, Zhang D, Zhang L. Complete genome sequence of the highly virulent Aeromonas hydrophila AL09-71 isolated from diseased channel catfish in West Alabama. Genome Announc. 2014;2:e00450. doi: 10.1128/genomeA.00450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tekedar HC, Waldbieser GC, Karsi A, Liles MR, Griffin MJ, Vamenta S, Sonstegard T, Hossain M, Schroeder SG, Khoo L, Lawrence ML. Complete genome sequence of a channel catfish epidemic isolate, Aeromonas hydrophila strain ML09-119. Genome Announc. 2013;1:e00755. doi: 10.1128/genomeA.00755-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang W, Li N, Li M, Zhang D, An G. Complete genome sequence of fish pathogen Aeromonas hydrophila JBN2301. Genome Announc. 2016;4:e01615. doi: 10.1128/genomeA.01615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu L, Zheng JS, Wang WM, Luo Y. Complete genome sequence of highly virulent Aeromonas hydrophila strain D4, isolated from a diseased blunt-snout bream in China. Microbiol Resour Announc. 2019;8:e01035. doi: 10.1128/MRA.01035-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Primers used in this study. Underlined sequences indicate restriction sites.
Additional file 2. Growth curve of the wild-type andtle1AHmutant strains. The cells were cultured in LB broth and densities were measured every 2 h at OD600. Data are presented as the mean ± standard deviation (error bars) of three independent experiments.
Additional file 3. Motility of the wild-type andtle1AHmutant strains. Swimming ability was observed after culturing strains at 28 °C for 48 h on 0.3% LB agar plates. The migration diameters were measured to assess the motility. Data are presented as the mean ± standard deviation (error bars) of three independent experiments.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additonal files.