Abstract
Background
The axillary bud is an important index of cotton plant-type traits, and the molecular mechanism of axillary bud development in upland cotton has not yet been reported. We obtained a mutant (designated mZ571) with a high-budding phenotype in axillary bud development from the low-budding phenotype variety G. hirsutum Z571 (CCRI 9A02), which provided ideal materials for the study of complex regulatory networks of axillary bud development. In this study, RNA sequencing was carried out to detect gene expression levels during three stages of axillary buds in Z571 (LB, low budding) and mZ571 mutant (HB, high budding).
Results
A total of 7162 DEGs were identified in the three groups (HB-E vs. LB-E, HB-G1 vs. LB-G1, HB-G2 vs. LB-G2), including 4014 downregulated and 3184 upregulated DEGs. Additionally, 221 DEGs were commonly identified in all three groups, accounting for approximately 3.09% of the total DEGs. These DEGs were identified, annotated and classified. A significant number of DEGs were related to hormone metabolism, hormone signal transduction, and starch and sucrose metabolism. In addition, 45, 22 and 9 DEGs involved in hormone metabolic pathways and 67, 22 and 19 DEGs involved in hormone signal transduction pathwayspathway were identified in HB-E vs. LB-E, HB-G1 vs. LB-G1, and HB-G2 vs. LB-G2, respectively, suggesting that endogenous hormones are the primary factors influencing cotton axillary bud growth. Hormone and soluble sugar content measurements revealed that mZ571 exhibited higher concentrations of zeatin, gibberellins and soluble sugar in all three stages, which confirmed that these hormone metabolism-, hormone signal transduction- and starch metabolism-related genes showed interaction effects contributing to the divergence of axillary bud growth between mZ571 and Z571.
Conclusions
Our results confirmed the importance of endogenous hormones and sugars in the development of axillary buds, and we found that mZ571 plants, with a high-budding phenotype of axillary buds, exhibited higher endogenous hormone and sugar concentrations. Overall, we present a model for the emergence and development of cotton axillary buds that provides insights into the complexity and dynamic nature of the regulatory network during axillary bud emergence and development.
Keywords: Upland cotton, Axillary bud, Transcriptome, DEGs
Background
The axillary buds are an important agronomic trait that shapes plant structure, biomass and yield. In most higher plants, the branches originate from axillary meristems, while the main stem is derived from the apical meristems [1–3]. In different species, axillary buds show different degrees of development and growth during the vegetative period [4]. In the vegetative reproductive growth of Arabidopsis thaliana, the formation of axillary buds is inhibited until the start of flowering [4–6]. Similarly, in pea (Pisum sativum), the outgrowth of axillary buds is usually inhibited after the transformation of axillary meristems to buds with immature leaves at most nodes [7]. In rice (Oryza sativa), the growth of axillary buds is released to form tillers because of its characteristic growth habit [3], while in maize (Zea mays), another monocot species, the outgrowth of the axillary buds remains relatively restricted [8]. In sorghum (Sorghum bicolor), the Nondormant Axillary Bud 1 gene was identified from a mutant plant and functionally characterized, and the mutant showed an increase in tillers and reduced plant height [1].
Plant hormones are the primary factors involved in the regulation of axillary bud growth. Three types of hormones, auxin, cytokinin and gibberellins (GAs), have long been studied. Among these hormones, auxin supplied by the terminal bud indirectly inhibits the growth of the axillary bud, while cytokinin directly induces the outgrowth of the axillary bud [9]. Studies on broad bean (Vicia faba) showed that removing the shoot tip of broad bean can stimulate the outgrowth of axillary buds, while this outgrowth could be largely suppressed by spraying auxin on the head of the plant residues [10]. However, since auxin is produced by young leaves and transported to the stem without entering the bud, it is believed that auxin has an indirect effect on bud growth [11–13]. Cytokinin has long been regarded as a bud growth-promoting factor [14, 15]. Recent studies have suggested that cytokinin regulates bud growth by controlling auxin transport [16]. The outgrowth of axillary buds can be promoted by the exogenous supply of an appropriate dose of cytokinin [17–19]. A study on cytokinin synthesis and signalling mutants of Arabidopsis indicated that cytokinin is related to the branches of intact plants but not to the branches of headless plants [20]. GAs are important endogenous hormones and play important roles in the overall growth of crops [21]. The promotion of axillary bud outgrowth by GAs has been reported in many plants, such as rose [22], sweet cherry [23] and Jatropha curcas [18].
Cotton provides excellent natural fibre and is one of the most important economic crops in the world. In cotton production, simplified cultivation and mechanical harvesting are emerging trends that depend on whether the cotton plant type meets production requirements. Cotton varieties with ideal plant types not only accelerate mechanized cotton production but also exhibit better ventilation, light permeability and leaf falling effects. The overgrowth of axillary buds in cotton increases the workload of artificial pruning and is not conducive to mechanized management and harvesting. In addition, the ideal cotton plant type shows good ventilation and light transmittance and is not vulnerable to pests, thus reducing dependence on pesticides. In cotton plants with few axillary buds, it is less likely that fallen leaves will hang on the plants, thereby reducing impurities during the cotton harvest, which is conducive to improving the quality of fibre. Therefore, branch types with axillary buds are an important index of plant type traits that have been well studied in many different species [3, 5, 24–28]. The mechanisms of axillary bud growth and development have not been reported in cotton.
In this study, the upland cotton variety Z571 (CCRI 9A02) and an axillary bud mutant thereof (mZ571) were selected as materials for identifying the regulatory networks involved in the development of axillary buds in upland cotton. Z571 has an ideal plant type for simplified cultivation and machine mining because of its fewer axillary buds and vegetative shoot characteristics. The low-budding phenotype of the axillary buds in Z571 is helpful for the mechanization of cotton harvesting. mZ571 is a phenotypic mutant inbred line with a high-budding phenotype in the development of axillary buds compared with Z571. Z571 and mZ571 provide ideal materials with the same genetic background subject to little interference from the environment for the study of complex regulatory networks of axillary bud development. Similarly, the contents of three kinds of phytohormones (cytokinin, gibberellins and auxin) associated with axillary buds were detected to analyse the underlying regulatory mechanism in upland cotton. Additionally, transcriptome sequencing of the first stalk nodes of the two plant types was performed at four, five and six weeks to study the regulatory network in the early stage of axillary bud development. This research not only provides data for studying the molecular mechanism of cotton axillary bud development but also lays a solid foundation for ideal plant type breeding in cotton.
Results
Phenotype, endogenous hormones and soluble sugar content of G. hirsutum Z571 and its mutant mZ571
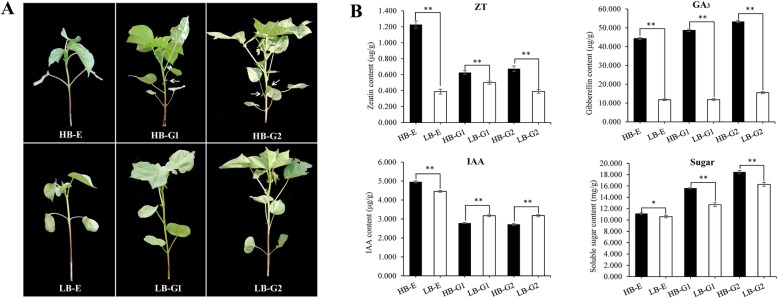
The stalk nodes used in the present study were collected from three stages of axillary bud development at the first branch position. The development of axillary buds in Z571 is limited, while that in mZ571 is very rapid (Fig. 1a). To monitor changes in cytokinin, gibberellin and auxin during axillary bud development, we measured the concentrations of zeatin (ZT), gibberellins (Gas) and indole acetic acid (IAA) in six samples by HPLC [29]. ZT showed the highest concentration in HB-E and slightly decreased concentrations in HB-G1 and HB-G2, and the concentrations of ZT in LB-E and LB-G2 were significantly lower than those in HB-E and HB-G2. Both types of plant materials showed an increasing tendency of the GA concentration during all stages. In contrast, IAA showed a decreasing pattern during the first two stages and then remained stable in the third stage. However, the IAA concentrations of HB-G1 and HB-G2 were significantly lower than those of LB-G1 and LB-G2, respectively. In addition, with the development of the cotton plants, the soluble sugar content in the axillary buds of the two plant types gradually increased, and the soluble sugar contents of HB-E, HB-G1 and HB-G2 were higher than those of LB-E, LBL-G1 and LB-G2, respectively (Fig. 1b). These results suggested that endogenous hormones and sugars are critical regulators during cotton axillary bud development.
Fig. 1.
Phenotypic analysis and identification of DEGs in axillary buds at different stages in G. hirsutum. a The phenotypes of Z571 and mZ571 in three stages. b The changes in ZT, GA, IAA and soluble sugar concentrations at different stages of the two axillary bud types. “HB-E” and “LB-G” represent the “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively. “*” and “**” indicate significant differences at P < 0.05 and P < 0.01, by the LSD test
RNA sequencing and transcriptome analysis
In total, 845,061,558 raw reads were generated from the HB-E, HB-G1, HB-G2, LB-E, LB-G1 and LB-G2 libraries (three replicates for each sample). The percentage of nucleotides with a quality score above 20 (Q20) was approximately 97.60% (Table S2). After the filtering of reads with adapters at the 3′ ends and quality scores below Q20, over 93.30% of the reads were confirmed as clean reads. As a result, 93.98–95.46% of the total clean reads were mapped to the upland cotton genome by using the TopHat2 upgrade version HISAT2 [30]. Multiple mapped reads and uniquely mapped reads accounted for 9.66–10.94% and 89.14–90.34% of these reads, respectively (Table S3). The mapped reads were assembled using StringTie software based on the reference genome, and compared with known transcripts, a total of 74,324 unigenes were generated.
Functional annotation of unigenes
The functional annotation of the unigenes was carried out by comparison with five different databases. In total, 10,197 unigenes (approximately 13.72%) were successfully matched in all five databases (Table 1). These unigenes were categorized into 26 descriptions according to the eggNOG database functional classification among which unigenes involved in signal transduction mechanisms and transcription were highly represented (Fig. S1).
Table 1.
Annotation of unigenes in five different databases
| Database | Number | Percentage |
|---|---|---|
| Annotation in NR | 70,889 | 95.38 |
| Annotation in GO | 46,000 | 61.89 |
| Annotation in KEGG | 11,968 | 16.1 |
| Annotation in eggNOG | 66,635 | 89.66 |
| Annotation in Swissprot | 55,926 | 75.25 |
| In all database | 10,197 | 13.72 |
| Total unigenes | 74,324 | 100.00 |
Identification and analysis of DEGs
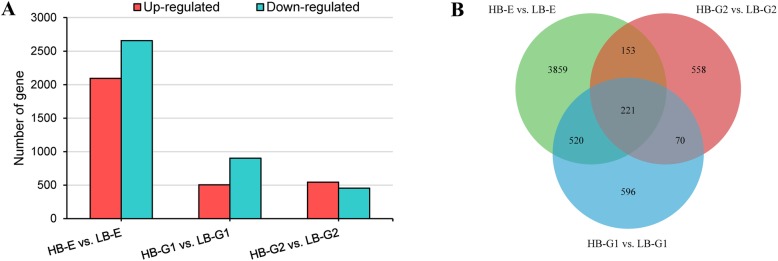
FPKM values were used to represent the abundance of each gene. A cutoff of a log2 (fold change) > 1 and a p-value < 0.05 were used to define DEGs. As a result, 7162 unigenes were found to show significantly differential expression in all three groups, accounting for 9.64% of the total unigenes. Among the DEGs, 4014 were downregulated and 3148 were upregulated (Fig. 2a). Additionally, 221 DEGs were commonly identified in the two axillary bud types in the three comparison groups (HB-E vs. LB-E, HB-G1 vs. LB-G1, HB-G2 vs. LB-G2), representing approximately 3.09% of the total DEGs. (Fig. 2b).
Fig. 2.
The identification of DEGs. a Numbers of DEGs in HB-E vs. LB-E, HB-G1 vs. LB-G1 and HB-G2 vs. LB-G2. b Venn diagram of the DEGs in the three groups. “HB-E” and “LB-G” represent the “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively
GO functional classification of DEGs
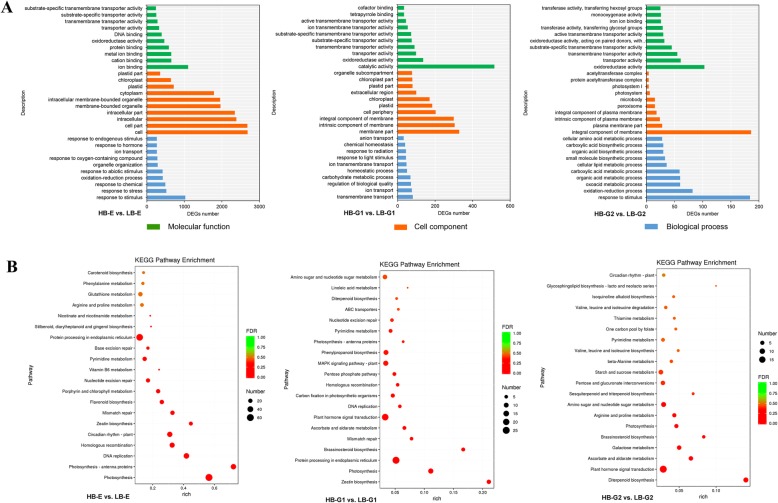
Based on GO categories, the functions of all DEGs were assessed. The results showed that 3783 (79.59%), 1068 (75.91%) and 738 (73.65%) DEGs in HB-E vs. LB-E, HB-G1 vs. LB-G1 and HB-G2 vs. LB-G2, respectively were enriched in the three main categories of cellular components (CC), biological processes (BP) and molecular functions (MF) under the criterion of a p-value< 0.05 (Fig. 3a). The DEGs annotated in the CC category were related to cell and membrane parts. Under the BP category, the DEGs were involved in the response hormones and stimuli. The DEGs annotated in the MF category were mainly involved in ion binding, catalytic activity and oxidoreductase activity. In addition, 45 DEGs (7 upregulated, 38 downregulated) identified in HB-E vs. LB-E, 22 DEGs (3 upregulated, 19 downregulated) in HB-G1 vs. LB-G1, and 9 DEGs (5 upregulated, 4 downregulated) in HB-G2 vs. LB-G2 were involved in hormone metabolic processes, which indicates that endogenous hormones play a significant role in the development of axillary buds.
Fig. 3.
GO function and KEGG pathway classification of DEGs. a GO functional classification and numbers of DEGs in three groups. b KEGG pathways and numbers of DEGs in the three groups
Pathway analysis of DEGs based on KEGG
In total, 6893 DEGs were assigned to 123 pathways based on KEGG annotation, and 57 pathways were found to be significantly enriched under the criterion of p<0.05. The top 30 pathways with the most abundant DEGs involved in axillary bud development are shown in Fig. 3b, which included the photosynthesis, DNA replication, plant hormone signal transduction, photosynthesis-antenna proteins, and protein processing in the endoplasmic reticulum pathways. Among these pathways, plant hormone-associated, photosynthesis-associated, DNA replication and starch and sucrose metabolism pathways accounted for most of the DEGs. The top five pathways with the most abundant DEGs are shown in Table 2.
Table 2.
Pathways of differentially expressed unigenes annotated in KEGG
| NO. | Pathway annotated in KEGG | DEG number | Total number | P-value | Pathway ID | Level 1 | Level 2 |
|---|---|---|---|---|---|---|---|
| HB-E vs. LB-E | |||||||
| 1 | Plant hormone signal transduction | 67 | 666 | 0.138944617 | ko04075 | Environmental Information Processing | Signal transduction |
| 2 | Photosynthesis | 61 | 108 | 2.63445E-36 | ko00195 | Metabolism | Energy metabolism |
| 3 | Protein processing in endoplasmic reticulum | 56 | 485 | 0.022169926 | ko04141 | Genetic Information Processing | Folding, sorting and degradation |
| 4 | DNA replication | 36 | 86 | 1.97547E-16 | ko03030 | Genetic Information Processing | Replication and repair |
| 5 | Photosynthesis - antenna proteins | 34 | 47 | 4.46141E-26 | ko00196 | Metabolism | Energy metabolism |
| HB-G1 vs. LB-G1 | |||||||
| 1 | Protein processing in endoplasmic reticulum | 25 | 485 | 2.21325E-05 | ko04141 | Genetic Information Processing | Folding, sorting and degradation |
| 2 | Plant hormone signal transduction | 22 | 666 | 0.018159619 | ko04075 | Environmental Information Processing | Signal transduction |
| 3 | Photosynthesis | 12 | 108 | 2.04012E-06 | ko00195 | Metabolism | Energy metabolism |
| 4 | MAPK signaling pathway - plant | 12 | 353 | 0.058286698 | ko04016 | Environmental Information Processing | Signal transduction |
| 5 | Phenylpropanoid biosynthesis | 11 | 320 | 0.063782317 | ko00940 | Metabolism | Biosynthesis of other secondary metabolites |
| HB-G2 vs. LB-G2 | |||||||
| 1 | Plant hormone signal transduction | 19 | 666 | 0.000772591 | ko04075 | Environmental Information Processing | Signal transduction |
| 2 | Diterpenoid biosynthesis | 8 | 57 | 5.31481E-07 | ko00904 | Metabolism | Metabolism of terpenoids and polyketides |
| 3 | Amino sugar and nucleotide sugar metabolism | 8 | 276 | 0.023594718 | ko00520 | Metabolism | Carbohydrate metabolism |
| 4 | Starch and sucrose metabolism | 7 | 277 | 0.060857028 | ko00500 | Metabolism | Carbohydrate metabolism |
| 5 | Ascorbate and aldarate metabolism | 6 | 91 | 0.001004811 | ko00053 | Metabolism | Carbohydrate metabolism |
“HB-E” and “LB-G” for “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively
qRT-PCR validation
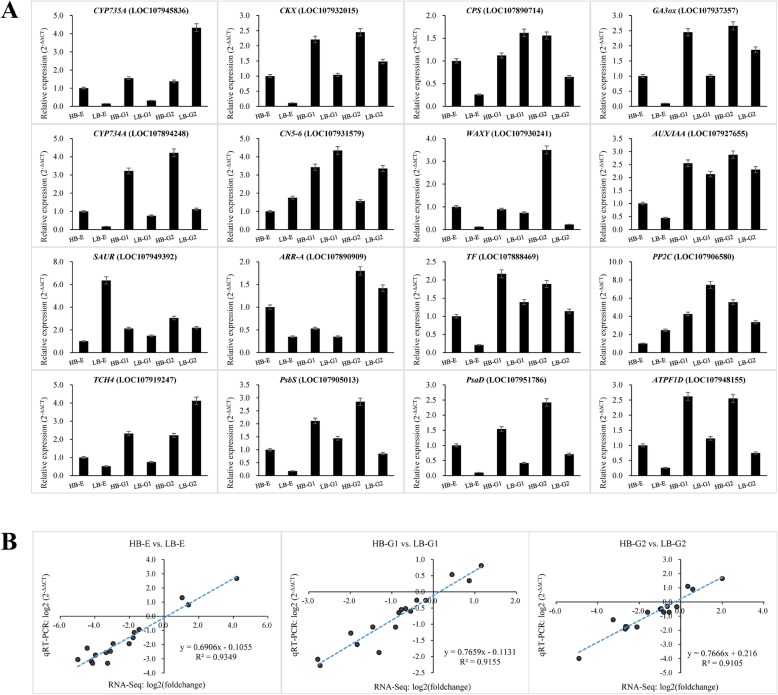
To verify the reliability of the RNA-Seq DEG data, sixteen DEGs (three photosynthesis-related genes, two zeatin biosynthesis genes, two starch and sucrose metabolism genes, two gibberellin biosynthesis genes, one brassinosteroid biosynthesis gene and six plant hormone signal transduction genes) were used for qRT-PCR analysis. The results showed that the expression patterns of these DEGs were mainly in agreement with the RNA-Seq results (Fig. 4a), with correlation coefficients (R2) > 0.91 (Fig. 4b). The results therefore confirmed the reliability of the RNA-Seq data.
Fig. 4.
Validation of RNA-Seq data by qRT-PCR analysis. a Expression of representative genes in the different developmental stages of axillary buds validated by qRT-PCR. b Correlations of the fold changes of sixteen differentially expressed genes determined by RNA-Seq and qRT-PCR analysis
Analysis of DEGs involved in hormone biosynthesis and signal transduction
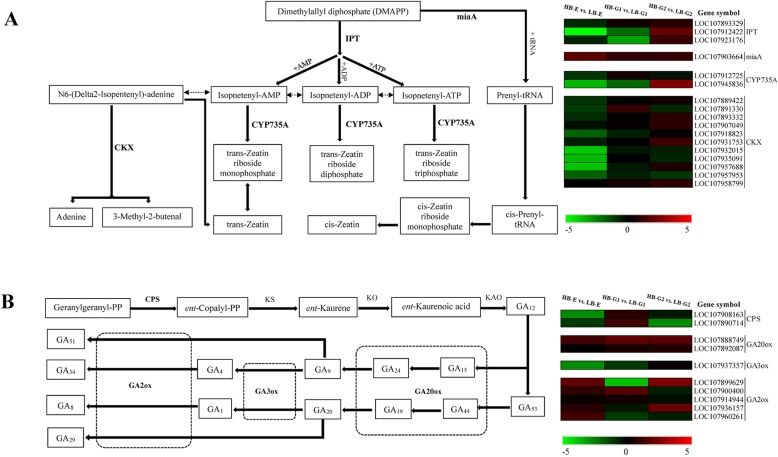
Previous studies have shown that endogenous hormones are crucial to plant growth and development. Here, several genes involved in the plant hormone signal transduction, zeatin, and gibberellin pathways were found to be differentially expressed between HB-E and LB-E, HB-G1 and LB-G1, and HB-G2 and LB-G2. For example, 14 and 10 DEGs were differentially expressed in the pathways of cytokinin (zeatin) and GA biosynthesis, respectively, which are widely believed to be involved in regulating cotton plant growth [31, 32].
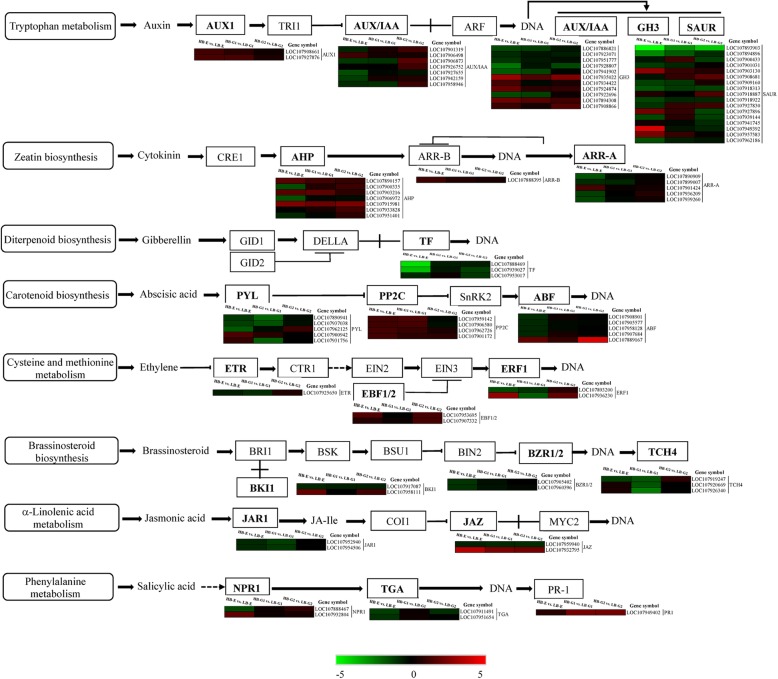
In the zeatin biosynthesis pathway, the expression level of miaA was upregulated in all three stages of mZ571. One IPT (LOC107912422) and one CYP735A (LOC107945836) were upregulated, but most CKX genes were still downregulated in HB-G2 (Fig. 5a). The expression level of the first gene encoding carbumoyl pyrophosphate synthetase that enters the GA synthesis pathway, CPS, was slightly downregulated in HB-E and HB-G2, while there was no significant change in the HB-G1 stage. One GA20ox gene (LOC107888749) was significantly upregulated in HB-G1 and HB-G2. Three GA2ox genes (LOC107899629, LOC107936157, LOC107960261) were significantly downregulated in HB-G1. In addition, GA3ox (LOC107937357) was downregulated in HB-E, HB-G1 and HB-G2, but the rate of reduction gradually decreased (Fig. 5b). For AUX1, which is involved in auxin signal transduction, the expression level was significantly upregulated in HB-E and HB-G1 cells. One AUX/IAA (LOC107906498), four CH3 (LOC107935022, LOC107934422, LOC107924874, LOC107894308), and four SAUR (LOC107903130, LOC107927896, LOC107949392, LOC107957583) genes were significantly upregulated in HB-E. Three AUX/IAA (LOC107901319, LOC107906873, LOC107926752), three CH3 (LOC107935022, LOC107894308, LOC107908866) and two SAUR (LOC107908681, LOC107918887) genes were significantly upregulated in HB-G2 (Fig. 6). For AHP, ARR-B and ARR-A, genes involved in cytokinin (zeatin) signal transduction, two AHP (LOC107890157, LOC107903216) genes in HB-G1, one ARR-B (LOC107888395) and one ARR-A (LOC107901424) gene in HB-E, and one AHP (LOC107915981) gene in all three stages of mZ571 were significantly upregulated. The BZR1/2 genes. Genes involved in brassinosteroid signal transduction, were downregulated in HB-E, after which their expression increased gradually in HB-G1 and HB-G2 but was still lower than that of LB. One BKI1 (LOC107958111) gene was significantly upregulated in HB-E and HB-G2. There was no significant upregulation of TCH4 genes. In addition, other genes involved in signal transduction, such as PYL, PP2C, and ABF, associated with abscisic acid; ETR, EBF1/2, and ERF1, associated with ethylene; JAR1 and JAZ, associated with jasmonic acid; and NPR1, TGA, and PR1, associated with salicylic acid, also showed differential expression. One ABF (LOC107889167), one ERF1 (LOC107936230) and one JAZ (LOC107932795) gene were significantly upregulated in HB-E (Fig. 6).
Fig. 5.
Transcript profiling of genes in the HB-E vs. LB-E, HB-G1 vs. LB-G1 and HB-G2 vs. LB-G2 groups. a The zeatin biosynthetic pathway. b The gibberellin biosynthetic pathway. “HB-E” and “LB-G” represent the “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively. The colour scale from green to red represents relative expression levels in the heatmap
Fig. 6.
The plant hormone signal transduction pathways. “HB-E” and “LB-G” represent the “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively. The colour scale from green to red represents the relative expression levels in the heatmap
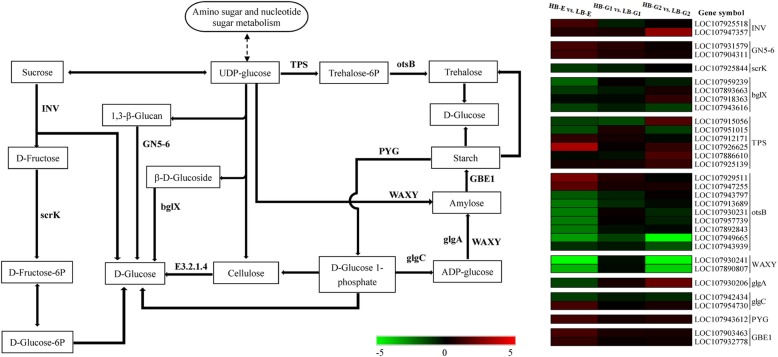
Analysis of DEGs involved in starch and sucrose metabolism
Sugar plays an important role in the growth and development of plants. Recent studies have shown that sugar is specifically an important signalling regulator of bud growth [33]. In the starch and sucrose metabolism pathway, INV was upregulated in HB-E and HB-G2 compared to LB-E and LB-G2. The expression levels of GN5–6, PYG, and GBE1 were upregulated in HB-E, HB-G1 and HB-G2 cells. Among these genes, GN5–6 presented the highest expression level in HB-E, which then gradually decreased in the HB-G1 and HB-G2 stages. The high expression of GN5–6 guarantees the transformation of 1,3-β-glucan to D-glucose. scrK, bglX, and most TPS and otsB genes were downregulated in HB-E and HB-G1. One TPS (LOC107926625) and two otsB (LOC107929511, LOC107947255) genes were upregulated in all three stages of HB.HB In addition, WAXY exhibited a higher expression level in HB-E, then decreased in HB-G1 and significantly increased in HB-G2 (Fig. 7).
Fig. 7.
The starch and sucrose metabolism pathway. “HB-E” and “LB-G” represent the “high-budding phenotype, early stage” and “low-budding, growth stage”, respectively. The colour scale from green to red represents the relative expression levels in the heatmap
Discussion
The plant type of a crop is the comprehensive expression of the morphological structure and unique physiological and ecological functions of a plant and is closely related to crop production and breeding; a favourable plant type can improve the leaf area coefficient, photosynthetic efficiency and fertility tolerance, thus increasing the harvest index [34–36]. The characteristics that affect the formation of different upland cotton plant types include the fruit branch type, number of shoot branches, number of fruiting branches, and plant height. Among these characteristics, the shoot branch, which is formed from the axillary bud, is one of the main factors affecting mechanical harvesting and the establishment of cultivation measures. Transcriptome research can provide new insight into the gene expression network of axillary bud development [37, 38].
Cytokinin and gibberellin are important regulators of axillary bud development
In-depth research on the hormonal control of axillary buds has been performed over time. A series of branching mutants in pea, morning glory, rice, and Arabidopsis plants have been analyzed to further demonstrate that the activation of buds can be controlled by auxin transporter-based systems [27, 39–43]. Cytokines have the function of positively regulating meristems and are necessary to maintain them. In general, endogenous cytokinins are transported acropetally in the xylem sap to promote axillary bud outgrowth [14]. Gibberellin belongs to the diterpene family and is essential for the growth and development of plants [18]. Previous studies have also shown that a series of changes in endogenous cytokinin and auxin levels are key factors regulating axillary bud development [14, 44–46].
In this study, to elucidate the differential regulatory networks of genes involved in axillary bud emergence and development in upland cotton, a transcriptome analysis was performed in Z571 and its mZ571 mutant involving 41 genes categorized as being involved in the zeatin biosynthesis, gibberellin biosynthesis, starch and sucrose metabolism and plant hormone signal transduction pathways. Many of the differentially expressed genes between Z571 and mZ571 related to the axillary bud type coincided with recent reports. In particular, an miaA gene (LOC107903664) and one IPT gene (LOC107912422) that were upregulated in apple [18, 47] were also found to be upregulated in mZ571. In fact, recent studies have explored the functions of IPT and CKX genes in various plant species. A study on Pisum sativum showed that the expression of the PsIPT1 and PsIPT2 genes, related to zeatin biosynthesis, in the stem nodule region differed before and after decapitation. The expression of PsIPT was maintained when the decapitated plants were maintained in buffer without IAA solution, whereas when IAA was added to the buffer, the expression of PsIPT was repressed again [14, 48]. Transgenic tobacco plants overexpressing the IPT gene showed higher trans-zeatin and zeatin riboside concentrations in the roots and stems [49]. In Arabidopsis, IPT single or multiple mutants exhibited significant reductions in branching [50]. According to the researchers, it is possible that the primary endogenous cytokinin synthesized mainly by adenylate IPTs plays a greater functional role in plant development and is more highly regulated [47, 51]. In contrast, most of the CKX genes were downregulated in HB-E, HB-G1 and HB-G2 (Fig. 5a), which indicates that CKX family genes are involved in the negative regulation of the growth and development of cotton axillary buds. This result is consistent with studies in Arabidopsis showing, for example, that the overexpression of the CKX7 gene lead to decreased contents of cytokinin-related metabolites, such as N-glucoside cytokinins and cis-zeatin [47]. The ARR-A gene belongs to the cytokinin response gene family, which is involved in the negative regulation of cytokinin signal transduction [52]. A study on yucca mutants showed that the expression of the ARR-A gene was upregulated in association with reduced auxin levels [53]. Our determination of the concentration of zeatin in the axillary buds of Z571 and mZ571 confirmed the above conclusion.
Gibberellins are key hormones that regulate a series of activities during plant growth and development [54]. The function of the GA20ox1 and GA2ox1 genes has been studied in several species; GA20ox1 positively regulates GA synthesis, while GA2ox1 negatively regulates GA synthesis [54]. According to In our results, one GA20ox family gene (GA20ox1; LOC107888749) that was shown to be upregulated in Jatropha curcas [18] was also upregulated in HB-G1 and HB-G2. Most of the GA2ox genes were downregulated in all three stages in HB, especially the GA2ox family gene GA2ox1 (LOC107899629), which showed the downregulation corresponding to a log2FoldChange of − 4.09 in HB-G1 (Fig. 5b). The GAs content of mZ571 was higher than that of Z571 in all three stages, and the concentration of GAs increased in the HB-G1 and HB-G2 stages. In fact, the zeatin and GA contents of the Z571 plant samples were lower than those of the mZ571 plant samples in the three stages, consistent with the gene expression pattern related to zeatin and gibberellin biosynthesis. For instance, in the zeatin biosynthesis pathway, the expression levels of most IPT and CKX genes in the low-budding phenotype plants were downregulated compared with those in the high-budding phenotype plants (Fig. 5a). The expression levels of GA3ox and GA20ox were downregulated in the low-budding phenotype plants (Fig. 5b). These dynamics of cytokinin and gibberellin play important roles during upland cotton axillary bud development [12, 15, 21, 55].
Possible roles of other hormones in axillary bud development
In addition to cytokinin and gibberellin, genes related to the signal transduction of other hormones, including auxin, abscisic acid, ethylene, brassinosteroid, jasmonic acid and salicylic acid, were also differentially expressed in HB-E, HB-G1 and HB-G2 (Fig. 6). The auxin signal transduction gene AUX1 was significantly upregulated in HB-E and HB-G1. Endogenous auxin (IAA) showed higher levels in mZ571 than in Z571, but the concentration of IAA decreased from HB-E to HB-G1 and remained stable in HB-G2. These auxin dynamics play a very important role in the development of axillary bud embryogenesis [56]. Previous studies have also shown a direct correlation between bud activity and auxin transport from the bud [14]. Our results regarding auxin signal transduction and the concentration of IAA were consistent with previous studies. Among abscisic acid signal transduction-related genes, the PP2C family was upregulated in HB-E and HB-G1; the PYL family was significantly downregulated in mZ571, with the exception of the abscisic acid receptors PYL4 and PYL9 in HB-E; and the ABF family except for ABF5 was downregulated in mZ571. According to the investigation of the roles of abscisic acid in Arabidopsis, abscisic acid regulates bud outgrowth in response to the red light/far-red light ratio, thus extending the known hormonal pathways associated with the regulation of branching [57]. Our KEGG annotation results showed that light-related pathways such as photosynthesis were also significantly enriched (Table 2, Fig. 3b).
Ethylene mediates several abiotic stresses in plants [58]. Among genes related to ethylene signal transduction, ETR was downregulated in HB-G1, EBF1/2 were upregulated in HB-E and HB-G2, one ERF1B (LOC107893200) gene was downregulated in HB-E and HB-G1, and one ERF1B (LOC107936230) gene was upregulated in HB-E and HB-G2. Brassinosteroid signal transduction-related genes, such as BZR1/2, TCH4 and BKI1 (LOC107917087), were downregulated or not significantly upregulated in mZ571, with the exception of one BKI1 (LOC107958111) gene that was upregulated in HB-E and HB-G2. Among jasmonic acid signal transduction-related genes, JAR1 was downregulated in mZ571, but JAZ was significantly upregulated in mZ571. Genes related to salicylic acid signal transduction and the regulatory proteins NPR1 and PR1 were upregulated in mZ571, and NPR3 and TGA were downregulated in mZ571. These hormones may play different roles in axillary bud emergence and development in upland cotton, and the exact regulatory mechanisms involved will be discussed in our future work.
The role of sugars in axillary bud development
In addition to plant hormones, sugars are important for plant growth and development because they provide energy and are a source of carbon for protein and cell wall synthesis [33]. Recent research has shown that sugars are one kind of important regulatory factor for bud growth and act as an early signal triggering bud activity [59]. According to our measurements, with cotton plant growth and development, the soluble sugar content of the axillary buds in the two types plants gradually increased, and the soluble sugar contents in HB-E, HB-G1 and HB-G2 were higher than those in LB-E, LB-G1 and LB-G2, respectively. These findings supported the traditional opinion that the bud is a site of the nutrient accumulation and storage and that plants with more buds show more nutrient accumulation [60]. A previous study also indicated that tubers are storage organs that accumulate starch during the development of potato [61]. Starch degradation results in an increase in the soluble sugar concentration; thus, starch may play an important role in the accumulation of endogenous soluble sugars in cells [62]. According to our starch and sucrose metabolism analysis results, INV, GN5–6, PYG, GBE1 and most TPS genes were upregulated in the three stages of mZ571, and WAXY showed significant downregulation in HB-E and HB-G2 (Fig. 7). In fact, the upregulation of PYG promotes the degradation of starch to D-glucose 1-phosphate, which will be further converted to D-glucose. TPS catalyses the transformation of UDP-glucose into trehalose-6P, which is then converted into trehalose under in a reaction catalysed by otsB, and trehalose if finally converted into D-glucose. These gene expression patterns are consistent with the dynamic change in the soluble sugar concentration and suggest that sugars are an energy source or nutrient aiding in bud growth in high-budding phenotype cotton plants.
Conclusion
In summary, we performed a comparative analysis of mRNA expression in the axillary buds of mZ571 and Z571 by transcriptomics. The analysis results indicated that some hormone metabolism-, hormone signal transduction- and starch metabolism-related genes showing interaction effects contribute to the divergence in axillary bud growth between mZ571 and Z571. Based on our results and previous reports, we propose a hypothetical interaction network related to axillary bud emergence and development in cotton (Fig. 8). In this model, we propose that sugars are the carbon energy source, which is essential for protein and cell wall synthesis during the emergence and development of axillary buds, hormone biosynthesis and signal transduction are the top-layer players required for cell differentiation and elongation regulating the development of axillary buds. In addition, according to KEGG analysis, we believe that photosynthesis is an important factor in the differences that arise during the development of axillary buds, which will be discussed in future work. These results further confirm the major findings previously reported for axillary buds, and important roles of genes involved in hormone biosynthesis, hormone signal transduction, and starch and sucrose metabolism were revealed. It provides us a better understanding of the complexity of the gene network involved in axillary bud emergence and development.
Fig. 8.
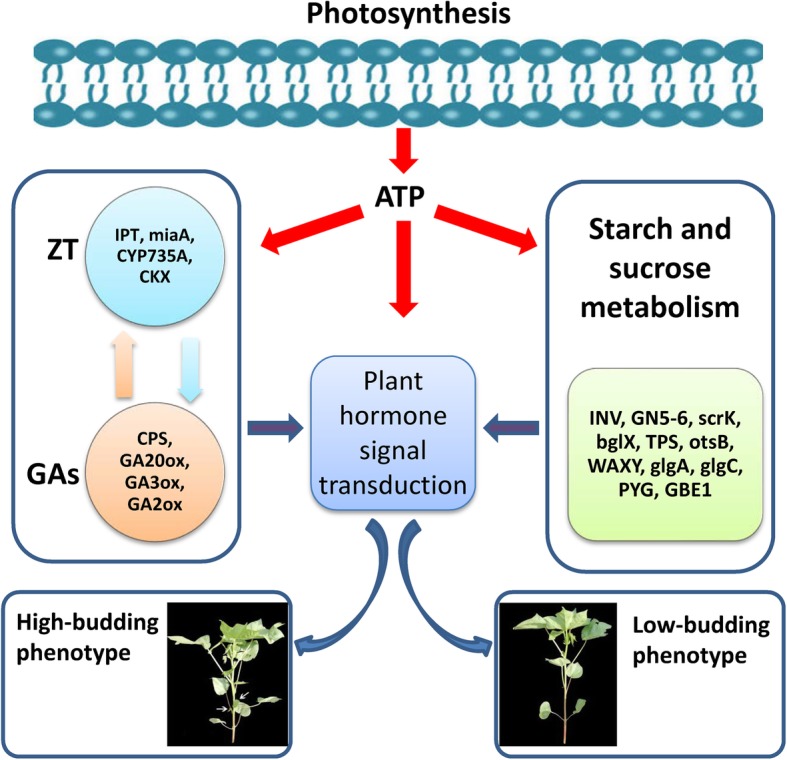
Hypothetical interaction network model of axillary bud emergence and development in upland cotton
Methods
Plant materials
Upland cotton plants of two different bud types, Z571 (CCRI 9A02, low-budding phenotype) and its mZ571 mutant (high-budding phenotype), were used for this study. Z571 shows an ideal plant type for simplified cultivation and machine harvesting because of its fewer axillary buds and vegetative shoot characteristics. mZ571 shows a high-budding phenotype in the development of axillary buds compared with Z571. The plants were grown in a growth chamber at the Chinese Academy of Agricultural Sciences Cotton Research Institute (Anyang, Henan, China). The sterilized seeds were grown in a light incubator under conditions of 28 °C/25 °C and a 12/12-h light/dark cycle, 70% relative humidity and light intensity of 11,000 Lx. The seedlings were randomly divided into three groups, and each group included nine pots. The first stalk nodes of the plants were collected in the fourth week (early stage), fifth week (growth stage 1) and sixth week (growth stage 2). Three biological replicates were performed. After collection, all samples were divided into three parts: one to be used for RNA-Seq, one for endogenous hormone determination, and one for qRT-PCR. All samples were collected and stored at − 80 °C for further use.
RNA isolation and sequencing
Total RNA was isolated from each cotton sample according to the improved CTAB extraction protocol [54]. The total RNA concentration was evaluated using a nucleic acid analyser. The construction of the cDNA library refers to the following methods. Briefly, using RNA as a template, the first strand of cDNA was synthesized with 6-base random primers and reverse transcriptase. The double-stranded cDNA library was enriched by PCR amplification and evaluated with an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), and the total and effective concentrations were determined by qRT-PCR. After RNA extraction, purification and library concentration, next-generation sequencing (NGS) based on the Illumina HiSeq sequencing platform was used for the paired-end (PE) sequencing of the library.
Mapping and sequence annotation
Raw reads were pre-processed for quality filtering, in which low-quality reads, such as reads with adapters at the 3′ end and reads with an average quality score below Q20, were removed. Eventually, the clean reads were mapped to the whole genome of Gossypium hirsutum (GCF_000987745.1) by using the TopHat2 upgrade version HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml) [30]. The expression levels of the unigenes were normalized by the FPKM (fragments per kilobase per million fragments) method so that the expression levels of different genes and samples could be compared.
Identification and analysis of DEGs
DESeq was employed to analyse the differences in gene expression between samples, and only genes with an absolute log2 (fold change) > 1 and p-value < 0.05 were identified as DEGs. Gene Ontology (GO) annotation was performed by using the Blast2GO program. The gene list and the numbers of each term were calculated for the annotated DEGs. The hypergeometric distribution method was selected to calculate the p-values (the criterion for significant enrichment was a p-value < 0.05), and the GO terms with significantly enriched DEGs were identified. Kyoto Encyclopedia of Gene and Genomes (KEGG) annotations were obtained from the KEGG automatic annotation server [63].
Determination of endogenous hormones and soluble sugars
According to previous studies, ZT is an important cytokinin used to promote plant regeneration [64, 65]. ZT induces the formation of a greater number of shoot primordia, many of which are converted into individual shoots in upland cotton tissue culture [31]. IAA and GAs are commonly applied in plant growth regulation research [21, 33]. Therefore, the concentrations of ZT, IAA and GAs in axillary buds were tested using high-performance liquid chromatography (HPLC), and the soluble sugar contents were determined using a colorimetric method. Three biological repeats were performed.
Validation of RNA-Seq by qRT-PCR
Here, to verify the validity of the transcriptome data, sixteen genes were chosen for qRT-PCR. The specific primers were designed by using Primer Premier 5 software (Table S1), and a Fast Quant RT Kit (Tiangen Biotech) was used to conduct reverse transcription. qRT-PCR was performed in an IQ5 fluorescent quantitative PCR system (Bio-Rad, USA) by using an Ultra SYBR Mixture (with ROX 1) kit (CWBIO, Beijing, China). The results were analysed by the 2−ΔΔCT method with normalization against the UBQ7 gene [54]. Each amplification reaction was performed in a total volume of 10 μL, containing 5 μL of 2× Ultra SYBR Mixture, 0.4 μL of each gene-specific primer (10 μmol/L), 0.7 μL of diluted cDNA, and 3.5 μL of sterile distilled water. The program for each reaction was 95 °C for 10 min, followed by 40 cycles of 95 °C for 20 s and 65 °C for 30 s. At the end of the reaction, the specificity of the amplified product was determined by the melting curve method. Three biological and technical replicates were performed to ensure reliability.
Supplementary information
Additional file 1: Figure S1. The eggNOG classification of assembled unigenes. X-axis: the name of 26 categories in eggNOG, Y-axis: the number of unigene annotated in the category.
Additional file 2: Table S1. Primer sequences of genes used for quantitative RT-PCR verification.
Additional file 3: Table S2. Summary of raw data analysis.
Additional file 4: Table S3. Summary of RNA-Seq Map.
Acknowledgements
Not applicable.
Abbreviations
- PAT
Polar auxin transport
- IPT
Adenosine phosphate-isopentenyl transferase
- TCP
TEOSINTE BRANCHED1, CYCLOIDEA, PCF
- BRC1
BRANCHED1
- FC1
Fine culm 1
- RBOH1
Respiratory burst oxidase homolog 1
- GO
Gene ontology
- WFI1
Whitefly induced 1
- PIN
PIN-FORMED
- qRT-PCR
Quantitative real-time PCR
- NGS
Next-Generation Sequencing
- DEGs
Differentially expressed genes
- ZT
Zeatin
- GAs
Gibberellins
- IAA
Indole acetic acid
- HPLC
High-performance liquid chromatography
- CKX
Cytokinin dehydrogenase
- GN5–6
Glucan endo-1,3-beta-glucosidase 5/6
- TPS
Trehalose 6-phosphate synthase/phosphatase
- WAXY
Granule-bound starch synthase
Authors’ contributions
GY and JS designed this experiment. NW and JS prepared this experiment. HZ and QX carried out data analysis. JS and NW wrote and revised this manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the program of National Key R&D Program of China (2018YFD0100300), the Modern Agricultural Industry Technology System of China (CARS-18-05) and the basal research fund of central public-interest scientific institution (1610162019010104). The funding bodies were not involved in the design of the study, collection, analysis, interpretation of data, and in writing the manuscript.
Availability of data and materials
The RNA-Seq data of all samples have been submitted to GenBank of the National Center for Biotechnology Information (NCBI: http://www.ncbi.nlm.nih.gov/sra), and the Sequence Read Archive (SRA) accession number is PRJNA598982.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jianbin Shi and Ning Wang contributed equally to this work.
Contributor Information
Jianbin Shi, Email: 15837252017@163.com.
Ning Wang, Email: 731667219@163.com.
Hong Zhou, Email: 593782186@qq.com.
Qinghua Xu, Email: xqh158@qq.com.
Gentu Yan, Email: yangentu@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02436-x.
References
- 1.Chen J, Zhang LM, Zhu MJ, Han LJ, Lv Y, Liu YS, Li P, Jing HC, Cai HW. Non-dormant axillary bud 1 regulates axillary bud outgrowth in sorghum. J Integr Plant Biol. 2018;60(10):938–955. doi: 10.1111/jipb.12665. [DOI] [PubMed] [Google Scholar]
- 2.McSteen P, Leyser O. Shoot branching. Annu Rev Plant Biol. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- 3.Wai AH, An G. Axillary meristem initiation and bud growth in rice. J Plant Biol. 2017;60(5):440–451. [Google Scholar]
- 4.Waldie T, Hayward A, Beveridge CA. Axillary bud outgrowth in herbaceous shoots: how do strigolactones fit into the picture? Plant Mol Biol. 2010;73(1–2):27–36. doi: 10.1007/s11103-010-9599-2. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Grandio E, Poza-Carrion C, Sorzano COS, Cubas P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell. 2013;25(3):834–850. doi: 10.1105/tpc.112.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gou JQ, Fu CX, Liu SJ, Tang CR, Debnath S, Flanagan A, Ge YX, Tang YH, Jiang QZ, Larson PR, et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017;216(3):829–840. doi: 10.1111/nph.14758. [DOI] [PubMed] [Google Scholar]
- 7.Dun EA, Hanan J, Beveridge CA. Computational modeling and molecular physiology experiments reveal new insights into shoot branching in pea. Plant Cell. 2009;21(11):3459–3472. doi: 10.1105/tpc.109.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McSteen P. Hormonal regulation of branching in grasses. Plant Physiol. 2009;149(1):46–55. doi: 10.1104/pp.108.129056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S, et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010;51(7):1127–1135. doi: 10.1093/pcp/pcq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett T, Hines G, van Rongen M, Waldie T, Sawchuk MG, Scarpella E, Ljung K, Leyser O. Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol. 2016;14(4):e1002446. doi: 10.1371/journal.pbio.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booker J, Chatfield S, Leyser O. Auxin acts in xylem-associated or medullary cells to mediate apical dominance. Plant Cell. 2003;15(2):495–507. doi: 10.1105/tpc.007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad TK, Li X, Abdel-Rahman AM, Hosokawa Z, Cloud NP, Lamotte CE, Cline MG. Does auxin play a role in the release of apical dominance by shoot inversion in Ipomoea nil ? Ann Bot-London. 1993;71(3):223–229. [Google Scholar]
- 13.Hall SM, Hillman JR. Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. timing of bud growth following decapitation. Planta. 1975;123(2):137–143. doi: 10.1007/BF00383862. [DOI] [PubMed] [Google Scholar]
- 14.Mueller D, Leyser O. Auxin, cytokinin and the control of shoot branching. Ann Bot-London. 2011;107(7):1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris SE, Cox MCH, Ross JJ, Krisantini S, Beveridge CA. Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 2005;138(3):1665–1672. doi: 10.1104/pp.104.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldie T, Leyser O. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 2018;177(2):803–818. doi: 10.1104/pp.17.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni J, Zhao ML, Chen MS, Pan BZ, Tao YB, Xu ZF. Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6-benzyladenine in Jatropha curcas. Sci Rep-Uk. 2017;7:11417. doi: 10.1038/s41598-017-11588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni J, Gao CC, Chen MS, Pan BZ, Ye KQ, Xu ZF. Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2015;56(8):1655–1666. doi: 10.1093/pcp/pcv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman H, Girault T, Barbier F, Peron T, Brouard N, Pencik A, Novak O, Vian A, Sakr S, Lothier J, et al. Cytokinins are initial targets of light in the control of bud outgrowth. Plant Physiol. 2016;172(1):489–509. doi: 10.1104/pp.16.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva-Navas J, Moreno-Risueno MA, Manzano C, Tellez-Robledo B, Navarro-Neila S, Carrasco V, Pollmann S, Gallego FJ, del Pozo JC. Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell. 2016;28(6):1372–1387. doi: 10.1105/tpc.15.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan M, Li GF, Liu XJ, Cheng F, Ma JJ, Zhao CP, Zhang D, Han MY. Exogenous application of GA3 inactively regulates axillary bud outgrowth by influencing of branching-inhibitors and bud-regulating hormones in apple (Malus domestica Borkh.) Mol Gen Genomics. 2018;293(6):1547–1563. doi: 10.1007/s00438-018-1481-y. [DOI] [PubMed] [Google Scholar]
- 22.Choubane D, Rabot A, Mortreau E, Legourrierec J, Peron T, Foucher F, Ahcene Y, Pelleschi-Travier S, Leduc N, Hamama L, et al. Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J Plant Physiol. 2012;169(13):1271–1280. doi: 10.1016/j.jplph.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Elfving DC, Visser DB. Improving the efficacy of cytokinin applications for stimulation of lateral branch development in young sweet cherry trees in the orchard. Hortscience. 2007;42(2):251–256. [Google Scholar]
- 24.Kotov AA, Kotova LM. Interaction of phytohormones in regulating the axillary bud growth in pea. Russ J Plant Physl+ 2018;65(5):628–641. [Google Scholar]
- 25.Braun N, de Saint GA, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012;158(1):225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Cell Bio. 2011;12(4):211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- 27.Leyser O. The control of shoot branching: an example of plant information processing. Plant Cell Environ. 2009;32(6):694–703. doi: 10.1111/j.1365-3040.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–U22. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki J, Tan BH, Errington SG. Eradication of endophytic bacteria via treatment for axillary buds of Petunia hybrida using plant preservative mixture (PPMTM) Plant Cell Tiss Org. 2010;102(3):365–372. [Google Scholar]
- 30.Sirén J, Välimäki N, Mäkinen V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans Comput Biol Bioinform. 2014;11(2):375–388. doi: 10.1109/TCBB.2013.2297101. [DOI] [PubMed] [Google Scholar]
- 31.Tafvizi F, Farahanei F, Sheidai M, Nejadsattari T. Effects of zeatin and activated charcoal in proliferation of shoots and direct regeneration in cotton (Gossypium hirsutum L.) Afr J Biotechnol. 2009;8(22):6220–6227. [Google Scholar]
- 32.Chen YA, Ye GY, Zhang L, Wang YH, Zhang X, Chen DH. Effect of trans-bacillus thuringiensis gene on gibberellic acid and zeatin contents and boll development in cotton. Field Crop Res. 2007;103(1):5–10. [Google Scholar]
- 33.Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA. An update on the signals controlling shoot branching. Trends Plant Sci. 2019;24(3):220–236. doi: 10.1016/j.tplants.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Pan QC, Xu YC, Li K, Peng Y, Zhan W, Li WQ, Li L, Yan JB. The genetic basis of plant architecture in 10 maize recombinant inbred line populations. Plant Physiol. 2017;175(2):858–873. doi: 10.1104/pp.17.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo YF, McCarty JC, Jenkins JN, Saha S. QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica. 2008;163(1):113–122. [Google Scholar]
- 36.Qi HK, Wang N, Qiao WQ, Xu QH, Zhou H, Shi JB, Yan GT, Huang Q. Construction of a high-density genetic map using genotyping by sequencing (GBS) for quantitative trait loci (QTL) analysis of three plant morphological traits in upland cotton (Gossypium hirsutum L.) Euphytica. 2017;213(4):83. [Google Scholar]
- 37.Wang ZR, Cui YY, Vainstein A, Chen SW, Ma HQ. Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front. Plant Sci. 2017;8:1990. [DOI] [PMC free article] [PubMed]
- 38.Tuttle JR, Nah G, Duke MV, Alexander DC, Guan XY, Song QX, Chen ZJ, Scheffler BE, Haigler CH. Metabolomic and transcriptomic insights into how cotton fiber transitions to secondary wall synthesis, represses lignification, and prolongs elongation. BMC Genomics. 2015;16:477. doi: 10.1186/s12864-015-1708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17(12):1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol. 2004;14(14):1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 41.Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51(6):1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 42.Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell. 2005;17(3):746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006;48(5):687–698. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- 44.Chatfield SP, Stirnberg P, Forde BG, Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000;24(2):159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- 45.Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. P Natl Acad Sci USA. 2004;101(21):8039–8044. doi: 10.1073/pnas.0402504101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386(6624):485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 47.Tan M, Li GF, Qi SY, Liu XJ, Chen XL, Ma JJ, Zhang D, Han MY. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Males domestica Borkh.) Gene. 2018;651:106–117. doi: 10.1016/j.gene.2018.01.101. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006;45(6):1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- 49.Faiss M, Zalubìlová J, Strnad M, Schmülling T. Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 1997;12(2):401–415. doi: 10.1046/j.1365-313x.1997.12020401.x. [DOI] [PubMed] [Google Scholar]
- 50.Müller D, Waldie T, Miyawaki K, To JPC. Melnyk CW, Kieber JJ, Kakimoto T, Leyser O. Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J. 2015;82(5):874–886. doi: 10.1111/tpj.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haberer G, Kieber JJ. Cytokinins. New insights into a classic phytohormone. Plant Physiol. 2002;128(2):354–362. doi: 10.1104/pp.010773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.To JP. Haberer G, Ferreira FJ. Type-a Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. Hormonal control of the shoot stem-cell niche. Nature. 2010;465(7301):1089–U154. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
- 54.Shi JB, Wang N, Zhou H, Xu QH, Yan GT. The role of gibberellin synthase gene GhGA2ox1 in upland cotton (Gossypium hirsutum L.) responses to drought and salt stress. Biotechnol Appl Bioc. 2019;66(3):298–308. doi: 10.1002/bab.1725. [DOI] [PubMed] [Google Scholar]
- 55.Smigocki AC. Cytokinin content and tissue distribution in plants transformed by a reconstructed isopentenyl transferase gene. Plant Mol Biol. 1991;16(1):105–115. doi: 10.1007/BF00017921. [DOI] [PubMed] [Google Scholar]
- 56.Cheng WH, Zhu HG, Tian WG, Zhu SH, Xiong XP, Sun YQ, Zhu QH, Sun J. De novo transcriptome analysis reveals insights into dynamic homeostasis regulation of somatic embryogenesis in upland cotton (G-hirsutum L.) Plant Mol Biol. 2016;92(3):279–292. doi: 10.1007/s11103-016-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy SK, Holalu SV, Casal JJ, Finlayson SA. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 2013;163(2):1047–1058. doi: 10.1104/pp.113.221895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues MA, Freschi L, Purgatto E, Lima VFGAP, Kerbauy GB. Ethylene modulates the developmental plasticity and the growth balance between shoot and root systems in the in vitro grown epiphytic orchid Catasetum fimbriatum. J Plant Growth Regul. 2014;33(3):513–525. [Google Scholar]
- 59.Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA. Sugar demand, not auxin, is the initial regulator of apical dominance. P Natl Acad Sci USA. 2014;111(16):6092–6097. doi: 10.1073/pnas.1322045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang SG, Pei JL, Li J, Tang GJ, Zhao JW, Peng XP, Nie SX, Ding YL, Wang CM. Sucrose and starch metabolism during Fargesia yunnanensis shoot growth. Physiol Plantarum. 2020;168(1):188–204. doi: 10.1111/ppl.12934. [DOI] [PubMed] [Google Scholar]
- 61.Sergeeva L, Claassens M, Jamar D, Plas L, Vreugdenhil D. Starch-related enzymes during potato tuber dormancy and sprouting. Russ J Plant Physl+ 2012;59(4):556. [Google Scholar]
- 62.Mohammadkhani N, Heidari R. Effects of drought stress on soluble proteins in two maize varieties. Turk J Biol. 2008;32(1):23–30. [Google Scholar]
- 63.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–WW85. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ikram-ul-Haq Callus proliferation and somatic embryogenesis in cotton (Gossypium hirsutum L.) Afr J Biotechnol. 2005;4(2):206–209. [Google Scholar]
- 65.Zhang BH, Feng R, Liu F, Wang QL. High frequency somatic embryogenesis and plant regeneration of an elite Chinese cotton variety. Bot Bull Acad Sinica. 2001;42(1):9–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The eggNOG classification of assembled unigenes. X-axis: the name of 26 categories in eggNOG, Y-axis: the number of unigene annotated in the category.
Additional file 2: Table S1. Primer sequences of genes used for quantitative RT-PCR verification.
Additional file 3: Table S2. Summary of raw data analysis.
Additional file 4: Table S3. Summary of RNA-Seq Map.
Data Availability Statement
The RNA-Seq data of all samples have been submitted to GenBank of the National Center for Biotechnology Information (NCBI: http://www.ncbi.nlm.nih.gov/sra), and the Sequence Read Archive (SRA) accession number is PRJNA598982.