Abstract
The recent outbreak of COVID-19 caused by SARS-CoV-2 brought a great global public health and economic concern. SARS-CoV-2 is an enveloped RNA virus, from the genus Betacoronavirus. Although few molecules have been tested and shown some efficacy against SARS-CoV-2 in humans but a safe and cost-effective attachment inhibitors are still required for the treatment of COVID-19. Natural products are gaining attention because of the large therapeutic window and potent antiviral, immunomodulatory, anti-inflammatory, and antioxidant properties. Therefore, this study was planned to screen natural products from Ayurveda that have the potential to modulate host immune system as well as block the virus entry in host cells by interfering its interaction with cellular receptor and may be used to develop an effective and broad-spectrum strategy for the management of COVID-19 as well as other coronavirus infections in coming future. To decipher the antiviral activity of the selected natural products, molecular docking was performed. Further, the drug-likeness, pharmacokinetics and toxicity parameters of the selected natural products were determined. Docking results suggest that curcumin and nimbin exhibits highest interaction with spike glycoprotein (MolDock score − 141.36 and − 148.621 kcal/mole) and ACE2 receptor (MolDock score − 142.647 and − 140.108 kcal/mole) as compared with other selected natural products/drugs and controls. Also, the pharmacokinetics data illustrated that all selected natural products have better pharmacological properties (low molecular weight; no violation of Lipinski rule of five, good absorption profiles, oral bioavailability, good blood–brain barrier penetration, and low toxicity risk). Our study exhibited that curcumin, nimbin, withaferin A, piperine, mangiferin, thebaine, berberine, and andrographolide have significant binding affinity towards spike glycoprotein of SARS-CoV-2 and ACE2 receptor and may be useful as a therapeutic and/or prophylactic agent for restricting viral attachment to the host cells. However, few other natural products like resveratrol, quercetin, luteolin, naringenin, zingiberene, and gallic acid has the significant binding affinity towards ACE2 receptor only and therefore may be used for ACE2-mediated attachment inhibition of SARS-CoV-2.
Keywords: COVID-19, SARS-CoV-2, Attachment inhibitor, Natural products, Ayurveda, CAM (Complementary and Alternative Medicine)
Introduction
A recent coronavirus pneumonia pandemic, named COVID-19 by World Health Organization, caused by a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been a major threat to global health since December 2019 [https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/novel coronavirus (2019-nCoV) is a new strain that has emerged from Wuhan City, Hubei Province of China and spread in more than 195 countries of the world including India [1]. The virus infection leads to over 3.8 million confirmed cases and 0.26 million deaths worldwide [https://www.who.int/emergencies/diseases/novel-coronavirus-2019].
SARS-CoV-2 is an enveloped RNA virus, from the genus Betacoronavirus and identified as the seventh member of the family Coronaviridae, which has infected the human population [2]. Commonly reported symptoms of a COVID-19 patient are fever, headache, vomiting, chills, dyspnea, nausea, sore throat, coughing up blood, shortness of breath, myalgia, diarrhea, and malaise. The severe infection leads to pneumonia, acute respiratory distress syndrome (ARDS) and sometimes multi-organ failures such as kidney failure, and even death [3]. SARS-CoV-2 genome showed higher sequence homology towards SARS-CoV than that of MERS-CoV through the whole genome sequence alignment analysis [4]. The coronavirus genome encodes four structural proteins: spike glycoprotein (S), a small envelope protein (E), matrix glycoprotein (M) and nucleocapsid protein (N) [5]. The spike (S) glycoprotein present in CoVs acts as a viral antigen and responsible for host-receptor binding, virus internalization and induces robust humoral and cell-mediated immune responses in humans during infection. The internalization of SARS-CoV-2 initiates via binding to its cellular receptor angiotensin-converting enzyme 2 (ACE2). The receptor binding and membrane fusion characteristic of spike glycoprotein make its an ideal target for the the attachment inhibitors for the management of COVID-19 [6].
Designing of drugs that can target the host cells or immune system as well as have direct inhibitory action against SARS-CoV-2 may be an effective approach for the treatment of COVID-19 [7]. The innate immune response is critical for controlling the replication and infection of coronavirus. With the advent of new and more efficient screening assays and prediction methods, the efficacy of natural products/drugs that can effectively treat a wide range of viral infections by interfering with different host functions need to be explored during outbreaks [8]. Therefore, the screening of natural products/drugs from Ayurveda that may have the potential to modulate the host immune system as well as block the virus entry into the host cell by interfering with its cellular receptor has been elucidated, which may be an effective and broad-spectrum approach for combating COVID-19. This study may provide the understanding of antiviral activity of natural products which may enable us to establish a potential therapy for patients diagnosed with COVID-19 in a dose-dependent manner with a reduced level of associated toxicity.
Material and methods
Ligand retrieval
A literature search was performed on PubMed and Google to identify natural products that have immunomodulatory properties, shown antiviral activity against RNA viruses, and also used for the management of similar signs and symptoms (clinical manifestations) associated with SARS-CoV-2 infection. The 3 dimensional (3D) structures of; hydroxychloroquine (CID: 3652), nafamostat (CID: 4413), captopril (CID: 44,093), nimbin (CID: 108,058), curcumin (CID: 969,516), withaferin A (CID: 265,237), piperine (CID: 638,024), mangiferin (CID: 5,281,647), thebaine (CID: 5,324,289), berberine (CID: 2353), andrographolide (CID: 5,318,517), quercetin (CID: 5,280,343), luteolin (CID: 5,280,445), resveratrol (CID: 445,154), naringenin (CID: 932), zingiberene (CID: 92,776), β-caryophyllene (CID: 5,281,515), citronellol (CID: 8842), eugenol (CID: 3314), and gallic acid (CID: 370) were retrieved from the NCBI PubChem compound database in SDF format and optimized in Discovery Studio. The chemical structures and their pharmacological properties of selected natural products were given in (Table 1).
Table 1.
List of natural products evaluated for in silico antiviral activity against SARS-CoV-2
| Ligand | Source (Common /Ayurveda or Sanskrit Name) | Structure | Pharmacological functions |
|---|---|---|---|
| Hydroxychloroquine | Synthetic drug | 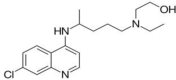 |
Antimalarial agent, Treatment of COVID-19 |
| Nafamostat | Synthetic drug | 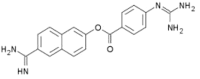 |
Antiviral agent |
| Captopril | Synthetic drug | 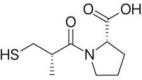 |
Antihypertensive agent |
| Nimbin | Azadirachta indica (Neem/Nimbka) | 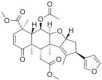 |
Antiinflammatory, antimicrobial |
| Curcumin | Curcuma longa (Turmeric/Haridra) | 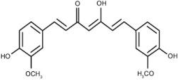 |
Antiinflammatory |
| Withaferin A | Withania somnifera (Indian ginseng/Ashwagandha) | 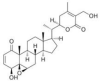 |
Immunomodulator, antiangiogenic, antitumor agent |
| Piperine | Piper nigrum (Black pepper/Maricha) | 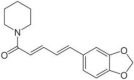 |
Antimicrobial, immunomodulator, hepatoprotective, antioxidant |
| Mangiferin | Mangifera indica (Mango/Aamra) | 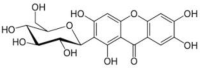 |
Antiviral, andanthelminthic, antiinflammatory, analgesic |
| Thebaine | Papaver somniferum (Poppy/Khas-khas) | 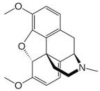 |
Analgesic, antitussive |
| Berberine | Berberis vulgaris (Barberry/Daruharidra) | 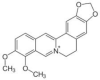 |
Antidiabetic, antihypertensive, antiinflammatory, antioxidant, antidepressant, anticancer, antimicrobial |
| Andrographolide | Andrographis paniculata (Green chireta/Kalamegha) | 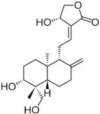 |
Antiviral, antiinflammatory, antineoplastic |
| Quercetin | Citrus aurantium (Bitter orange/Naranga) | 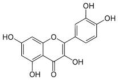 |
Antioxidant, neuroprotective, antiviral, anticancer, antimicrobial, anti-inflammatory |
| Luteolin | Capsicum annuum (Bell peppers/Mahamarichika) |  |
Antiinflammatory, antioxidant, anticarcinogenic, antihyperalgesic, anxiolytic, antidepressant |
| Resveratrol | Vaccinium angustifolium (Blueberry/Nila badari) |  |
Antiinflammatory, antioxidative |
| Naringenin | Citrus paradisi (Grapefruit/Madhukarkati) |  |
Antiinflammatory, antioxidative, antidyslipidemic, antiobesity, antidiabetic, antifibrotic, |
| Zingiberene | Zingiber officinale (Ginger/Singabera) |  |
Antiinflammatory |
| β-Caryophyllene | Syzygium aromaticum (Clove/Devakusuma) | 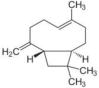 |
Antiinflammatory |
| Citronellol | Rosa damascena (Damask rose/Satapatri) | 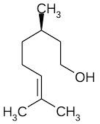 |
Antiinflammatory, antibacterial, antifungal |
| Eugenol | Ocimum sanctum (Holy basil/Tulsi) | 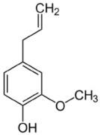 |
Antioxidant, neuroprotective, antiviral, anticancer, antimicrobial, antiinflammatory |
| Gallic acid | Emblica officinalis (Indian gooseberry/Amalaki) | 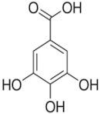 |
Antibacterial, antifungal, antiviral, antiinflammatory, antioxidant, anticancer, antidiabetic |
Protein retrieval
The 3D crystal structure of spike glycoprotein of SARS-CoV-2 (PDB ID:6VXX) and its receptor Angiotensin-converting enzyme-2 (PDB ID: 1R42) were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank [9, 10].
Molecular docking or structure-based drug design
Molegro Virtual Docker (MVD-3.0.0) has been used for the molecular docking of selected natural products/drugs with spike glycoprotein and angiotensin-converting enzyme-2 [11]. Structure-based drug design approach has been used to identify novel attachment inhibitors against SARS-CoV-2 infection. MVD offers precise predictions of ligand binding modes (87.0%) than other docking software such as (FlexX2: 57.9%, Surflex: 75.3%, GOLD: 78.2%, and Glide: 81.8%) [11]. The potential ligand binding site or cavities was predicted using MVD. Precisely 5 cavities were identified in the protein but only one cavity having maximum surface area and volume was selected as active site based on the active site residues described in the literature [11]. This cavity was utilized in further structure-based docking studies. Nafamostat (spike glycoprotein inhibitor) and captopril (angiotensin-converting enzyme-2 inhibitor) were taken as a control for selected natural products/drugs [6]. The docking score of selected natural products/drugs was compared with the docking score of nafamostat for spike glycoprotein and docking score of captopril for angiotensin-converting enzyme-2 respectively. The natural products that showed higher docking score than nafamostat and captopril was considered a potent inhibitor of spike glycoprotein and angiotensin-converting enzyme-2 respectively. Considering the suggested use in COVID-19, the hydroxychloroquine was also taken for comparison in this study.
Absorption, distribution, metabolism, excretion, toxicity, and receptor binding analysis
Physico-chemical and pharmacokinetic properties have a vital role in the discovery of novel antiviral agents. The drug-likeness properties of the selected natural products were investigated via swissADME server [12]. ADME-toxicity of the selected natural products was analyzed by the admetSAR server. The absorption, water solubility, human oral bioavailability, blood–brain barrier (BBB) transport, and plasma protein binding were calculated. Additionally, carcinogenicity, hepatotoxicity, acute oral toxicity and receptor binding properties of these compounds were also calculated [13].
Results
Inhibitory effect of natural products against spike glycoprotein of SARS-CoV-2
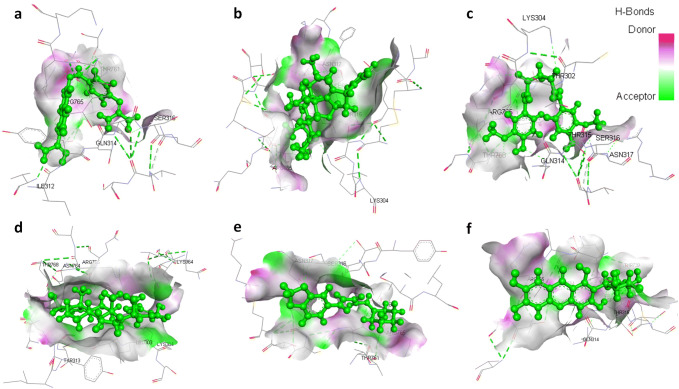
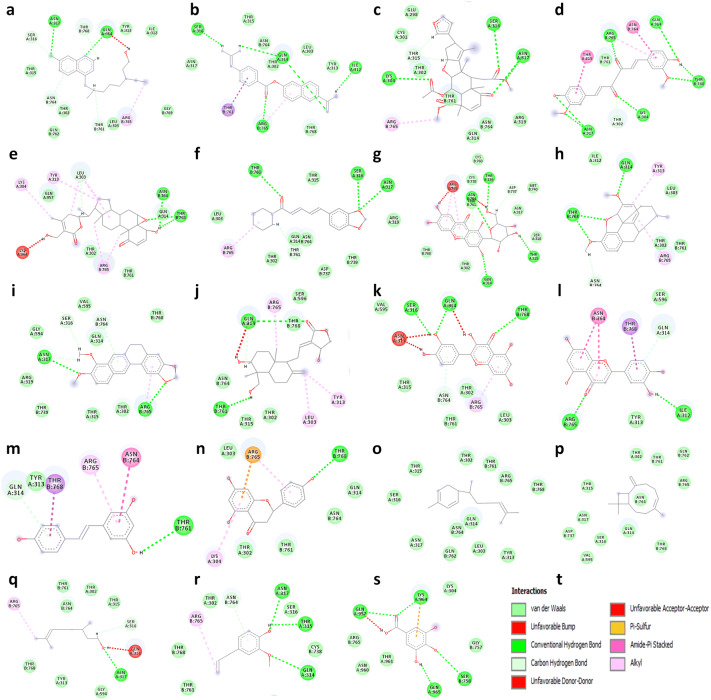
The binding energies of selected natural products (Table 1) with spike glycoprotein were studied and docking results are given in terms of MolDock score, interaction energy, H-bond energy and interacting amino acid residues present at the active site of protein (Table 2). Docking results revealed that out of 17 selected natural products/drugs; only nimbin and curcumin have shown higher score than nafamostat (MolDock score − 114.633 kcal/mole, interaction energy − 124.911 kcal/mole and forms five hydrogen bonds with Ile312, Gln314, Ser316, and Arg765 amino acid residues present at the active site of protein) and hydroxychloroquine (MolDock score − 105.28 kcal/mole, interaction energy − 116.92 kcal/mole and forms two hydrogen bond with Gln314 and Asn317) (Fig. 1). Nimbin showed the highest binding affinity toward spike glycoprotein (MolDock score − 148.621 kcal/ mole and interaction energy − 142.145 kcal/ mole) and also forms three hydrogen bond with Lys304, Ser316, and Asn317. Similarly, curcumin has been found to interact with spike glycoprotein with a high binding affinity (MolDock score − 141.36 kcal/mole and interaction energy − 143.705 kcal/mole) and forms six hydrogen bond with Lys304, Gln314, Asn317, Arg765, and Thr768. In addition, it has been found that the nimbin and curcumin have better binding affinity toward spike glycoprotein as compared to nafamostat and hydroxychloroquine. Apart from nimbin and curcumin, other selected natural products/drugs such as withaferin A, piperine, mangiferin, thebaine, berberine, and andrographolide have also demonstrated a significant binding affinity towards spike glycoprotein of SARS-CoV-2 with significant hydrogen bond interactions with various residues (Fig. 2).
Table 2.
Molecular docking results are indicated with maximum binding affinity of selected natural products against SARS-CoV-2 spike glycoprotein and its cellular receptor along with MolDock score and other parameters
| Ligand name | MolDock score (kcal/mole) | Interaction (kcal/mole) | H-bond (kcal/mole) | Interacting amino acid residues |
|---|---|---|---|---|
| Spike glycoprotein of SARS-CoV-2 (6VXX) | ||||
| Hydroxychloroquine | − 105.28 | − 116.92 | − 4.57 | Gln314, Ser316, Asn317,Thr761, Asn764, Arg765, Thr768 |
| Nafamostat | − 114.63 | − 124.91 | − 8.55 | Ile312, Gln314, Ser316, Thr761, Arg765 |
| Nimbin | − 148.62 | − 142.14 | − 15.87 | Lys304,Thr315, Ser316, Asn317, Arg765 |
| Curcumin | − 141.36 | − 143.70 | − 16.39 | Thr302, Lys304, Gln314, Thr315, Asn317, Asn764, Arg765, Thr768 |
| Withaferin A | − 108.56 | − 136.26 | − 7.69 | Lys303, Lys304, Tyr313, Gln314, Asn764, Arg765, Thr768 |
| Piperine | − 104.56 | − 110.91 | − 8.55 | Gln314, Ser316, Asn317, Arg765, Thr768 |
| Mangiferin | − 104.26 | − 136.54 | − 23.51 | Gln314,Thr315, Asp737, Thr739, Thr761, Asn764, Arg765, |
| Thebaine | − 103.58 | − 113.19 | − 10.77 | Tyr313, Gln314, Asn764, Arg765, Thr768 |
| Berberine | − 99.93 | − 117.62 | − 9.41 | Ser316, Asn317, Asn764, Arg765 |
| Andrographolide | − 98.80 | − 113.55 | − 5.21 | Lys303, Tyr313, Gln314, Thr761, Arg765 |
| Quercetin | − 86.22 | − 115.16 | − 14.48 | Gln314, Ser316, Asn317, Asn764, Arg765,Thr768 |
| Luteolin | − 85.01 | − 107.21 | − 10.32 | Ile312, Gln314, Asn764, Arg765,Thr768 |
| Resveratrol | − 83.40 | − 90.38 | − 6.06 | Gln314, Thr761, Asn764, Arg765, Thr768 |
| Naringenin | − 82.10 | − 103.13 | − 11.73 | Lys304, Arg765, Thr768 |
| Zingiberene | − 75.49 | − 81.61 | 0 | - |
| β-caryophyllene | − 72.37 | − 71.15 | 0 | - |
| Citronellol | − 69.44 | − 68.55 | − 5.59 | Gln314, Ser316, Asn317, Arg765 |
| Eugenol | − 66.55 | − 73.74 | − 6.19 | Gln314, Thr315, Asn317, Asn764, Arg765, |
| Gallic acid | − 63.54 | − 72.46 | − 16.36 | Ser758, Gln957, Lys964, Gln965 |
| Angiotensin-converting enzyme 2 (1R42) | ||||
| Hydroxychloroquine | − 105.05 | − 113.20 | − 3.25 | Ala348, Trp349, Asp350, Glu375, His378, Asp382, His401 |
| Captopril | − 73.69 | − 81.15 | − 13.07 | His378, Asn394, Glu398, His401, Glu402 |
| Curcumin | − 142.64 | − 139.52 | − 9.49 | Ala348, His378, Asn394, Tyr385, His401, Glu402 |
| Nimbin | − 140.10 | − 125.30 | − 12.76 | His378, Asn394, His401, Tyr510, Tyr515, Arg518 |
| Piperine | − 112.83 | − 120.05 | − 1.10 | Gly399, His401, Glu402, Arg514, Arg518 |
| Withaferin A | − 112.38 | − 139.67 | − 11.16 | Thr347, Ala348, Glu375, His378, Phe390, Arg393, Phe400, His401 |
| Mangiferin | − 104.15 | − 137.74 | − 23.11 | Asp206, Ala348, Asp382, Asn394, Gly395, Glu398, His401, Arg514 |
| Thebaine | − 100.77 | − 108.58 | − 6.23 | His378, Asn394, His401 |
| Andrographolide | − 99.354 | − 116.74 | − 14.38 | Ala348, His378, Asp382, Tyr385, Asn397, Phe400, His401, Glu402, Arg514 |
| Berberine | − 97.54 | − 116.92 | − 0.429 | Ala348, Glu375, His378, Phe390, His401 |
| Resveratrol | − 96.81 | − 103.7 | − 5.26 | Asp382, Phe390, Arg393, His401 |
| Quercetin | − 92.05 | − 112.59 | − 7.86 | Asp350, Tyr385, Phe390, Arg393 |
| Luteolin | − 89.95 | − 108.26 | − 22.58 | His378, Tyr385, Glu398, His401, Glu402, Arg514 |
| Naringenin | − 83.42 | − 102.46 | − 5 | Ala348, Asp350, His378, Asp382, Tyr385, Arg393, Asn394, His401 |
| Zingiberene | − 82.85 | − 88.32 | 0 | His378, His401 |
| Gallic acid | − 75.64 | − 84.60 | − 16.88 | Asp206, Asn394, Gly395, Asn397, His401, Arg514 |
| β-caryophyllene | − 73.42 | − 72.20 | 0 | Phe390, Leu391, Arg393, Phe400 |
| Citronellol | − 69.61 | − 71.37 | − 5 | Phe390, Arg393, Phe400, His401 |
| Eugenol | − 67.46 | − 77.25 | − 2.5 | His401, Glu402, Arg514 |
Fig. 1.
3D structural views of ligand-binding site. The 3D structures display H-bond interactions (green dashed lines) of ligands a nafamostat, b nimbin, c curcumin, d withaferin A, e piperine, and f mangiferin with the Spike glycoprotein of SARS-CoV-2 (6VXX) (color figure online)
Fig. 2.
2D visualization of molecular interaction of Spike glycoprotein of SARS-CoV-2 (6VXX) with a hydroxychloroquine, b nafamostat, c nimbin, d curcumin, e withaferin A, f piperine, g mangiferin, h thebaine, i berberine, j andrographolide, k quercetin, l luteolin, m resveratrol, n naringenin, o zingiberene, p β-caryophyllene, q citronellol, r eugenol, s gallic acid and t displays the color indication for interactions. H-bonding (dark green circles associated with the green dashed lines); van der Waals forces (medium light green circles); carbon–oxygen dipole–dipole interaction (light green circles with dashed lines); alkyl-pi interactions (light pink circles with dashed lines); T-shaped pi–pi stacking and (parallel) pi–pi stacking (both indicated with dark violet circles); cation-pi interaction (orange circle). The blue halo surrounding the interacting residues represents the solvent accessible surface that is proportional to its diameter. Images were generated using discovery studio visualizer 4 (color figure online)
Inhibitory effect of natural products against Angiotensin-converting enzyme 2
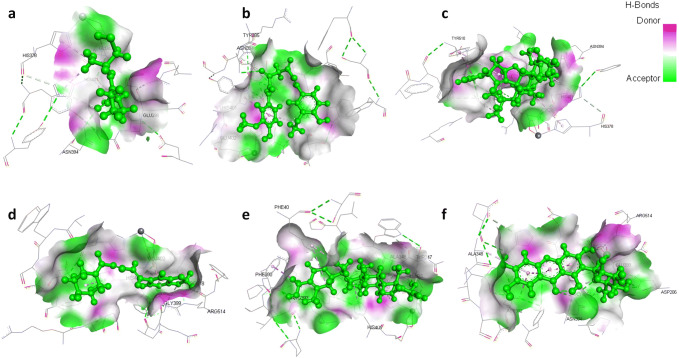
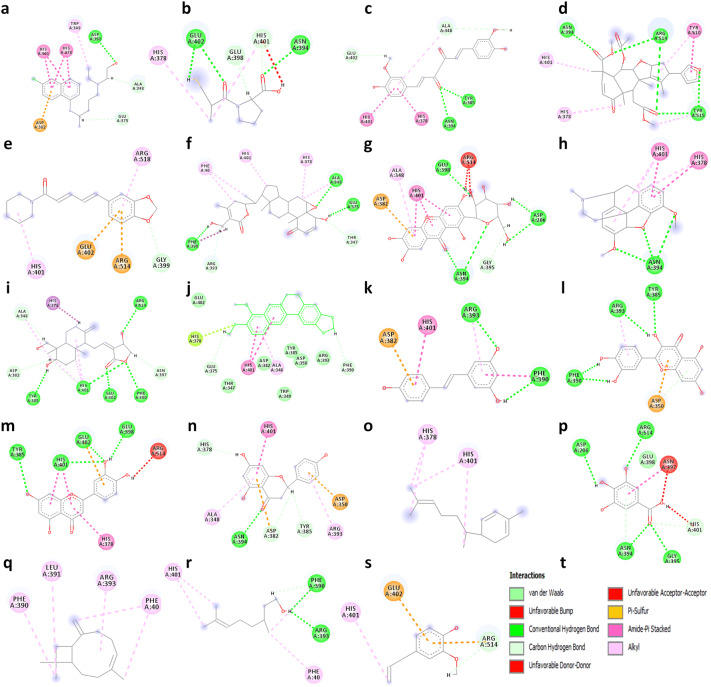
Many studies have shown that host ACE2 acts as a specific receptor for the Spike receptor binding domain (RBD) of SARS-CoV-2. Therefore, targeting ACE2 may be considered as host-specific therapy to block SARS-CoV-2 from entering into the host cells and may facilitate the design of the novel attachment inhibitors for the treatment of COVID-19. The docking results revealed that all the selected natural drugs showed higher binding affinity towards ACE2 except β-caryophyllene, citronellol, and eugenol as compared to captopril which is a known inhibitor of the enzyme. We have found that captopril interacts with amino acid residues His378, Asn394, Glu398, His401, and Glu402 of ACE2 to generate a binding score (MolDock score of − 73.6975 kcal/mole and interaction energy − 81.1588 kcal/mole) and forms three hydrogen bonds with Glu402 and Asn394 (Fig. 3). In addition, hydroxychloroquine is also evaluated for ACE2 binding property, which exhibited significant interaction with the protein (MolDock score − 105.05 kcal/mole and interaction energy − 113.20 kcal/mole) and form hydrogen bond with Asp350 present at the active site of protein. Among the selected natural products, curcumin has the highest binding affinity for ACE2 followed by nimbin, piperine, withaferin A, mangiferin, thebaine, andrographolide, berberine, resveratrol, quercetin, luteolin, naringenin, zingiberene, and gallic acid. Curcumin interacts with Ala348, His378, Asn394, Tyr385, His401, and Glu402 at the active site of the protein and generated MolDock score and interaction energy (− 142.647 kcal/mole and − 139.525 kcal/mole) and forms two hydrogen bonds with Tyr385 and Asn394. Similarly, nimbin also interacts with His378, Asn394, His401, Tyr510, Arg514, and Tyr515 at the active site and generated moldock score and interaction energy (− 140.108 kcal/mole and − 125.304 kcal/mole) and forms four hydrogen bonds with Asn394, Arg514, and Tyr515. Unlike spike glycoprotein, curcumin and nimbin again showed better binding affinity toward ACE2 compared to selected natural drugs, controls, hydroxychloroquine and ranked top two molecules that have inhibitory effect for SARS-CoV-2 spike glycoprotein as well as its cellular receptor ACE2 (Fig. 4).
Fig. 3.
3D structural views of ligand-binding site. The 3D structures display H-bond interactions (green dashed lines) of ligands a captopril, b curcumin, c nimbin, d piperine, e withaferin A, and f mangiferin with the Angiotensin-converting enzyme 2 (1R42) (color figure online)
Fig. 4.
2D visualization of molecular interaction of Angiotensin-converting enzyme 2 (1R42) with a hydroxychloroquine, b captopril, c curcumin, d nimbin, e piperine, f withaferin A, g mangiferin, h thebaine, i andrographolide, j berberine, k resveratrol, l quercetin, m luteolin, n naringenin, o zingiberene, p gallic acid q β-caryophyllene r citronellol, s eugenol and t displays the color indication for interactions. H-bonding (dark green circles associated with the green dashed lines); van der Waals forces (medium light green circles); carbon–oxygen dipole–dipole interaction (light green circles with dashed lines); alkyl-pi interactions (light pink circles with dashed lines); T-shaped pi–pi stacking and (parallel) pi–pi stacking (both indicated with dark violet circles); cation–pi interaction (orange circle). The blue halo surrounding the interacting residues represents the solvent accessible surface that is proportional to its diameter. Images were generated using Discovery Studio Visualizer 4.5 (color figure online)
Absorption, distribution, metabolism, excretion, toxicity and receptor binding analysis:
Drug-likeness properties play a crucial role in antiviral drug development. The calculated drug-likeness parameters of selected natural products using swissADME are given in (Table 3). The absorbtion profile is directly related to the availability of drug in systemic circulation. The admetSAR results revealed that all the selected natural drugs exhibited good intestinal absorption after the oral uptake in the gastrointestinal tract. In addition, all the selected natural drugs also showed good water solubility and oral bioavailability whereas molecules such as nimbin, piperine, mangiferin, thebaine, berberine, andrographolide, quercetin, luteolin, resveratrol, naringenin, zingiberene, citronellol, and eugenol have shown poor human oral bioavailability. Distribution of drugs to the various tissue and organs guaranteed its better treatment efficiency. After the drug distribution analysis we have found that all the natural drugs have good blood–brain barrier penetration except curcumin, mangiferin, quercetin, luteolin, resveratrol, naringenin, and gallic acid. Furthermore, based on distribution profile, non-substrates or non-inhibitors of P-glycoprotein (ABCB1) and renal organic cationic transporter (OCT2) of the drugs were predicted which facilitate efflux of xenobiotics from the cells. The drug metabolism analysis suggested that all the selected natural drugs are predicted as either non-substrates or non-inhibitors of CYP1A2, CYP2D6, CYP2C9, CYP2C19, and CYP3A4 enzymes. Further, the drugs were evaluated for carcinogenicity and hepatotoxicity and no drugs have shown carcinogenic effect whereas nimbin, curcumin, mangiferin, berberine, quercetin, luteolin, resveratrol, and naringenin were found to be associated with hepatotoxicity. The receptor binding analysis suggested that except the zingiberene, β-caryophyllene, citronellol, eugenol, and gallic acid, all the selected natural drugs have shown significant binding with cellular receptors such as estrogen receptor, androgen receptor, thyroid receptor, glucocorticoid receptor, aromatase and PPAR gamma receptor (Tables 4, 5 and 6).
Table 3.
Drug likeness properties of selected natural products
| Ligand | Molecular formula | Molecular weight (g/mol) | LogP | Hydrogen bond donor | Hydrogen bond acceptor | Topological polar surface area (Å2) |
|---|---|---|---|---|---|---|
| Hydroxychloroquine | C18H26ClN3O | 335.9 | 3.6 | 2 | 4 | 48.4 |
| Nafamostat | C19H17N5O2 | 347.4 | 2.0 | 4 | 4 | 141 |
| Captopril | C9H15NO3S | 217.29 | 0.3 | 2 | 4 | 58.6 |
| Nimbin | C30H36O9 | 540.6 | 2.3 | 0 | 9 | 118 |
| Curcumin | C21H20O6 | 368.4 | 3.2 | 2 | 6 | 93.1 |
| Withaferin A | C28H38O6 | 470.6 | 3.8 | 2 | 6 | 96.4 |
| Piperine | C17H19NO3 | 285.34 | 3.5 | 0 | 3 | 38.8 |
| Mangiferin | C19H18O11 | 422.3 | − 0.4 | 8 | 11 | 197 |
| Thebaine | C19H21NO3 | 311.4 | 2.2 | 0 | 4 | 30.9 |
| Berberine | C20H18NO4+ | 336.4 | 3.6 | 0 | 4 | 40.8 |
| Andrographolide | C20H30O5 | 350.4 | 2.2 | 3 | 5 | 87 |
| Quercetin | C15H10O7 | 302.23 | 1.5 | 5 | 7 | 127 |
| Luteolin | C15H10O6 | 286.24 | 1.4 | 4 | 6 | 107 |
| Resveratrol | C14H12O3 | 228.24 | 3.1 | 3 | 3 | 60.7 |
| Naringenin | C15H12O5 | 272.25 | 2.4 | 3 | 5 | 87 |
| Zingiberene | C15H24 | 204.35 | 5.2 | 0 | 0 | 0 |
| β-caryophyllene | C15H24 | 204.35 | 4.4 | 0 | 0 | 0 |
| Citronellol | C10H20O | 156.26 | 3.2 | 1 | 1 | 20.2 |
| Eugenol | C10H12O2 | 164.2 | 2.0 | 1 | 2 | 29.5 |
| Gallic acid | C7H6O5 | 170.12 | 0.7 | 4 | 5 | 98 |
Table 4.
Pharmacokinetics, toxicities and receptor binding properties of hydroxychloroquine, nafamostat, captopril, nimbin, curcumin withaferin A and piperine
| Parameters | Hydroxychloroquine | Nafamostat | Captopril | Nimbin | Curcumin | Withaferin A | Piperine |
|---|---|---|---|---|---|---|---|
| Absorption | |||||||
| Human intestinal absorption | + | + | + | + | + | + | + |
| Caco-2 | + | – | + | – | – | – | + |
| Human oral bioavailability | + | – | + | – | + | + | - |
| Water solubility | − 3.566 | − 4.074 | − 1.798 | − 4.455 | − 3.364 | − 4.203 | − 3.398 |
| Distribution | |||||||
| Subcellular localization | Lysosomes | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria |
| Blood–brain barrier penetration | + | + | + | + | – | + | + |
| Plasma protein binding | 0.756 | 0.739 | 0.367 | 0.788 | 0.832 | 0.944 | 1.058 |
| P-glycoprotein inhibitior | – | – | – | + | + | + | – |
| P-glycoprotein substrate | + | – | – | + | – | + | – |
| Metabolism | |||||||
| OATP2B1 inhibitior | – | – | – | – | – | – | – |
| OATP1B1 inhibitior | + | + | + | - | + | + | + |
| OATP1B3 inhibitior | + | + | + | - | + | + | + |
| MATE1 inhibitior | – | + | – | – | – | – | – |
| OCT2 inhibitior | – | – | – | – | – | – | – |
| BSEP inhibitior | + | + | – | + | + | + | + |
| CYP3A4 substrate | – | – | – | + | – | + | – |
| CYP2C9 substrate | + | – | – | – | – | – | – |
| CYP2D6 substrate | – | – | – | – | – | – | – |
| CYP3A4 inhibition | + | – | – | + | – | – | + |
| CYP2C9 inhibition | – | – | – | – | + | – | – |
| CYP2C19 inhibition | – | – | – | – | + | – | – |
| CYP2D6 inhibition | – | – | – | – | + | – | + |
| CYP1A2 inhibition | – | + | – | – | + | – | + |
| CYP inhibitory promiscuity | – | – | – | + | + | – | + |
| Toxicity | |||||||
| Carcinogenicity | – | – | – | – | – | – | – |
| Human either-a-go-go inhibition | + | – | – | + | – | + | + |
| Hepatotoxicity | – | + | – | + | + | – | – |
| Acute oral toxicity(kg/mol) | 2.665 | 2.33 | 1.853 | 3.955 | 1.992 | 3.276 | 2.201 |
| Receptor binding | |||||||
| Estrogen receptor binding | – | + | – | + | + | + | + |
| Androgen receptor binding | – | + | – | + | + | + | + |
| Thyroid receptor binding | + | + | – | + | + | + | + |
| Glucocorticoid receptor binding | – | + | – | + | + | + | – |
| Aromatase binding | + | + | – | + | + | + | + |
| PPAR gamma | + | + | – | + | + | + | + |
OATP2: organic anion-transporting polypeptide; OCT2: organic cation transporter 2; BSEP: bile salt export pump; CYP: cytochrome P450; Human ether-a-go-go-related Gene (hERG); (–): absence of activity; ( +): presence of activity
Table 5.
Pharmacokinetics, toxicities and receptor binding properties of mangiferin, thebaine, berberine, andrographolide, quercetin and luteolin
| Parameters | Mangiferin | Thebaine | Berberine | Andrographolide | Quercetin | Luteolin |
|---|---|---|---|---|---|---|
| Absorption | ||||||
| Human intestinal absorption | + | + | + | + | + | + |
| Caco-2 | – | + | + | + | – | + |
| Human oral bioavailability | – | – | – | – | – | – |
| Water solubility | − 2.398 | − 2.979 | − 2.974 | − 2.853 | − 2.999 | − 2.999 |
| Distribution | ||||||
| Subcellular localization | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Mitochondria |
| Blood–brain barrier penetration | – | + | + | + | – | – |
| Plasma protein binding | 0.97 | 0.718 | 0.834 | 0.536 | 1.175 | 1.043 |
| P-glycoprotein inhibitior | - | - | + | - | - | - |
| P-glycoprotein substrate | – | + | – | – | – | – |
| Metabolism | ||||||
| OATP2B1 inhibitior | + | - | – | – | + | + |
| OATP1B1 inhibitior | + | + | + | + | + | + |
| OATP1B3 inhibitior | + | + | + | + | + | + |
| MATE1 inhibitior | – | – | – | – | + | + |
| OCT2 inhibitior | – | – | – | – | – | – |
| BSEP inhibitior | – | – | + | + | – | – |
| CYP3A4 substrate | + | + | + | + | + | - |
| CYP2C9 substrate | – | – | – | – | – | – |
| CYP2D6 substrate | – | + | – | – | – | – |
| CYP3A4 inhibition | – | – | – | – | + | + |
| CYP2C9 inhibition | – | - | - | - | - | - |
| CYP2C19 inhibition | – | – | – | – | – | – |
| CYP2D6 inhibition | – | + | + | – | – | – |
| CYP1A2 inhibition | – | – | + | – | + | + |
| CYP inhibitory promiscuity | – | – | + | – | + | + |
| Toxicity | ||||||
| Carcinogenicity | – | – | – | – | – | – |
| Human either-a-go-go inhibition | – | – | – | – | – | – |
| Hepatotoxicity | + | – | + | – | + | + |
| Acute oral toxicity (kg/mol) | 2.979 | 3.597 | 1.545 | 2.795 | 2.559 | 2.525 |
| Receptor binding | ||||||
| Estrogen receptor binding | + | + | + | + | + | + |
| Androgen receptor binding | + | - | + | + | + | + |
| Thyroid receptor binding | - | + | + | + | + | + |
| Glucocorticoid receptor binding | + | + | + | + | + | + |
| Aromatase binding | - | - | - | + | + | + |
| PPAR gamma | + | + | + | - | + | + |
OATP2: organic anion-transporting polypeptide; OCT2: organic cation transporter 2; BSEP: bile salt export pump; CYP: cytochrome P450; Human ether-a-go-go-related Gene (hERG); (–): absence of activity; ( +): presence of activity
Table 6.
Pharmacokinetics, toxicities and receptor binding properties of resveratrol, naringenin, zingiberene, β-caryophyllene, citronellol, eugenol, and gallic acid
| Parameters | Resveratrol | Naringenin | Zingiberene | β-caryophyllene | Citronellol | Eugenol | Gallic acid |
|---|---|---|---|---|---|---|---|
| Absorption | |||||||
| Human intestinal absorption | + | + | + | + | + | + | + |
| Caco-2 | + | + | + | + | + | + | – |
| Human oral bioavailability | – | – | – | + | – | – | + |
| Water solubility | − 2.778 | − 3.19 | − 4.937 | − 4.687 | − 2.78 | − 1.918 | − 1.097 |
| Distribution | |||||||
| Subcellular localization | Mitochondria | Mitochondria | Lysosomes | Lysosomes | Lysosomes | Mitochondria | Mitochondria |
| Blood Brain Barrier penetration | – | – | + | + | + | + | – |
| Plasma protein binding | 0.677 | 1.001 | 1.038 | 0.832 | 0.769 | 0.94 | 0.668 |
| P-glycoprotein inhibitior | – | – | – | – | – | – | – |
| P-glycoprotein substrate | – | – | – | – | – | – | – |
| Metabolism | |||||||
| OATP2B1 inhibitior | – | – | – | – | – | – | – |
| OATP1B1 inhibitior | + | + | + | + | + | + | + |
| OATP1B3 inhibitior | + | + | + | + | + | + | + |
| MATE1 inhibitior | – | – | – | – | – | – | – |
| OCT2 inhibitior | – | – | – | – | – | – | – |
| BSEP inhibitior | – | – | – | – | – | – | – |
| CYP3A4 substrate | – | – | – | + | – | – | – |
| CYP2C9 substrate | – | – | – | – | – | – | – |
| CYP2D6 substrate | – | – | – | – | – | + | – |
| CYP3A4 inhibition | + | + | – | – | – | – | – |
| CYP2C9 inhibition | + | + | – | – | – | – | – |
| CYP2C19 inhibition | + | + | – | – | – | – | – |
| CYP2D6 inhibition | – | – | – | – | – | – | – |
| CYP1A2 inhibition | + | + | – | – | – | – | – |
| CYP inhibitory promiscuity | + | + | – | – | – | – | – |
| Toxicity | |||||||
| Carcinogenicity | – | – | – | – | – | – | – |
| Human either-à-go-go inhibition | – | – | + | + | – | – | – |
| Hepatotoxicity | + | + | – | – | – | – | – |
| Acute Oral Toxicity (kg/mol) | 2.644 | 1.87 | 1.827 | 2.366 | 2.077 | 1.645 | 1.552 |
| Receptor binding | |||||||
| Estrogen receptor binding | + | + | – | – | – | – | – |
| Androgen receptor binding | + | + | – | – | – | – | + |
| Thyroid receptor binding | + | + | – | – | – | – | – |
| Glucocorticoid receptor binding | + | + | – | – | – | – | – |
| Aromatase binding | + | + | – | – | – | – | – |
| PPAR gamma | + | + | – | – | – | – | – |
OATP2: organic anion-transporting polypeptide; OCT2: organic cation transporter 2; BSEP: bile salt export pump; CYP: cytochrome P450; Human ether-a-go-go-related Gene (hERG); (–): absence of activity; ( +): presence of activity
Discussion
Coronavirus is an enveloped RNA virus, from the genus Betacoronavirus that is transmitted among birds, humans, and other mammals. The recent outbreak of COVID-19 caused by SARS-CoV-2 is a serious global public health and economic concern that emphasizes the significance of restricting infectious agents at international borders [14]. In the absence of approved antiviral agent for SARS-CoV-2 infection, more than 80 clinical trials of 20 drugs such as traditional Chinese medicines (TCM), human immunoglobulin, interferons, methylprednisolone, chloroquine, arbidol, remdesivir, favipiravir, lopinavir, ritonavir, oseltamivir, bevacizumab, and hydroxychloroquine have been launched for the treatment of COVID-19 [15]. Natural products are gaining interest because of the large therapeutic window and potent antiinflammatory and antioxidant properties. Recently, various drugs from natural origin have shown antiviral activity against the number of viruses such as Influenza virus, Zika virus, Dengue virus, Japanese encephalitis virus, Herpes simplex virus, Hepatitis B and C virus, Respiratory syncytial virus, and Human immunodeficiency virus [16]. The importance of natural products for health and disease treatment have immense importance throughout human evolution. Several natural products derived from plants for thousands of years have been traditionally used to treat various types of human illnesses including viral infections. Natural plants such as Azadirachta indica, Curcuma longa, Withania somnifera, Ocimum sanctum, Emblica officinalis, Piper nigrum, Mangifera indica, Andrographis paniculata, and Zingiber officinale etc. have been listed in AYUSH system of medicine and extensively used as complementary and alternative medicine for the management of chronic fever, inflammation and auto-immune disorders. In addition, these natural products/drugs from Ayurveda have significant pharmacological activities that regulate various vital cell signaling pathways that cause cytotoxic, genotoxic, and mitogenic reactions leading to various disease pathologies [17].
The development of drugs with broad-spectrum antiviral activities is a long pursued goal in the drug discovery. This research is primarily focused to develop therapeutics targeting the spike glycoprotein of SARS-CoV-2 and its host receptor ACE2 to find potential attachment inhibitors for the treatment of COVID-19. Recently various studies have shown that hydroxychloroquine may have potential while combating the SARS-CoV-2 infection [18–20]. However, we have found that natural drugs such as nimbin, curcumin, withaferin A, piperine and nafamostat (known spike glycoprotein inhibitor) have better binding affinity toward spike glycoprotein and its cellular receptor ACE2 compared to hydroxychloroquine. It has been reported that patients receiving hydroxychloroquine may experience several severe adverse effects such as visual disturbances, ventricular tachycardia, cardiovascular collapse, convulsions, hypokalemia, rhythm and conduction disorders including QT prolongation, and ventricular fibrillation. Therefore, development of potential attachment inhibitor from natural products is a safe and effective strategy for the treatment of COVID-19. Our docking results demonstrated that curcumin and nimbin are the best-ranked drugs among the selected natural products showing an inhibitory effect for spike glycoprotein and ACE2. Both the drugs have been traditionally used in AYUSH system of medicine for the management of various disease conditions such as inflammation, fever, and treatment of infections, autoimmune disorders like arthritis and neurological disorders [21, 22]. Most of these natural products/drugs have been recognized as safe for human usage. Besides, these natural products/drugs are also well-known regulator of various interleukins, chemokines, and cytokines. Recent studies have shown that the level of IL-6, TNF-, and IFNs were found to be elevated in patients with SARS-CoV-2 [23]. Therefore, these natural drugs can be used as alternative therapy for the management of various sign and symptoms associated SARS-CoV-2 infection as well as other coronaviruses in the absence of specific antiviral drug.
We further analyzed the drug-likeness and ADME toxicity and receptor binding properties of selected natural products by using the swissADME and admetSAR server. According to In Lipinski rule for drug-likeness, any drug molecule having MW > 500 g/mol, hydrogen-bond-donating atoms > 5, Hydrogen-bond-accepting atoms > 10, or ClogP > 5 is not considered as a good pharmaceutical agent in terms of oral activity. Even though they may have strong docking energies, compounds violating more than 2 Lipinski’s rule of 5 were not considered for drug-likeness. Molecules violating more than one of these rules may have problems with oral bioavailability [24]. Similarly, the knowledge of pharmacokinetics and toxicity profile of drugs is essential in order to measure the safety and efficacy of in biological system. Therefore, designing an attachment inhibitor from natural products may provide a safe, effective and broad-spectrum approach while combating viral infections. The findings of the present study would facilitate us to understand the antiviral mechanism of natural products which enables us to establish a potential therapy for patients diagnosed with COVID-19 in a dose-dependent manner with a reduced level of associated toxicity.
Collectively, for the first time our data demonstrated the efficacy of selected natural products against spike glycoprotein of SARS-CoV-2 and its cellular receptor ACE2. We have found that curcumin, nimbin, withaferin A, piperine, mangiferin, thebaine, berberine, and andrographolide may effectively inhibit the interaction of spike glycoprotein with its receptor compared to other selected natural drugs. Also, the pharmacokinetics data illustrated that all selected natural products have better pharmacological properties (low molecular weight; no violation of Lipinski rule of five, good absorption profiles, oral bioavailability, good blood–brain barrier penetration, and low toxicity risk). Therefore, based on our results it is suggested that these natural products/drugs from Ayurveda may be useful as a therapeutic or prophylactic agent during COVID-19 for restricting viral attachment to the host cells via inhibition of spike glycoprotein or its cellular receptor. Consequently, resveratrol, quercetin, luteolin, naringenin, zingiberene, and gallic acid could be a useful starting material for the development of potent antiviral agents for the prevention and treatment of SARS-CoV-2 and other coronaviruses if tested further.
Acknowledgements
The authors are grateful to the Vice Chancellor, King George’s Medical University (KGMU), Lucknow, India for the encouragement for this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Funding
None.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Vimal K. Maurya and Shailendra K. Saxena contributed equally to this work.
References
- 1.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) Virusdisease. 2020;31(1):13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, thrusting Coronaviruses into the spotlight. Viruses. 2019;11(1):E59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G, De Clercq E. Therapeutic options for the 2019 novel Coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim KJ, Liu X, Komabayashi T, Jeong SI, Selli S. Natural products for infectious diseases. Evid Based Compl Alternat Med. 2016;2016:9459047. doi: 10.1155/2016/9459047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020 Mar 6. doi: 10.1016/j.cell.2020.02.058. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 10.Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA, Patane MA, Pantoliano MW. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 12.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, et al. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 14.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID-19 treatment. Rev Panam Salud Publica. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akram M, Tahir IM, Shah SMA, Mahmood Z, Altaf A, Ahmad K, et al. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother Res. 2018;32(5):811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- 17.Lin YJ, Xu Y, Xu J. Complementary and alternative therapies for inflammatory diseases 2018. Evid Based Complement Alternat Med. 2018;2018:2068724. doi: 10.1155/2018/2068724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yazdany J, Kim AHJ. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: What every clinician should know. Ann Intern Med. 2020:M20–1334. [DOI] [PMC free article] [PubMed]
- 19.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mégarbane B. Chloroquine and hydroxychloroquine to treat COVID-19: between hope and caution. Clin Toxicol (Phila). 2020 doi: 10.1080/15563650.2020.1748194. [DOI] [PubMed] [Google Scholar]
- 21.Praditya D, Kirchhoff L, Brüning J, Rachmawati H, Steinmann J, Steinmann E. Antiinfective properties of the golden spice Curcumin. Front Microbiol. 2019;10:912. doi: 10.3389/fmicb.2019.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azab A, Nassar A, Azab AN. AntiInflammatory activity of natural products. Molecules. 2016;21(10):1321. doi: 10.3390/molecules21101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conti P, Ronconi G, Caraffa A, Gallenga CE, Ross R, Frydas I, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): antiinflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2):1. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]