Abstract
Background
Avian pathogenic Escherichia coli (APEC), an important extraintestinal pathogenic E. coli, causes colibacillosis, an acute and mostly systemic disease involving multiple organ lesions such as meningitis. Meningitis-causing APEC can invade the host central nervous system by crossing the blood–brain barrier (BBB), which is a critical step in the development of meningitis. However, the bacteria-host interaction mechanism in this process remains unclear.
Methods
In this study, we examined E. coli and bEnd.3 cells transcriptomes during infection and mock infection to investigate the global transcriptional changes in both organisms using RNA sequencing approach.
Results
When APEC infected the bEnd.3 cells, several significant changes in the expression of genes related to cell junctional complexes, extracellular matrix degradation, actin cytoskeleton rearrangement, immune activation and the inflammatory response in bEnd.3 cells were observed as compared to the mock infection group. Thus, the immune activation of bEnd.3 cells indicated that APEC infection activated host defenses. Furthermore, APEC may exploit cell junction degradation to invade the BBB. In addition, amino acid metabolism and energy metabolism related genes were downregulated and the protein export pathway related genes were upregulated in APEC cultured with bEnd.3 cells, compared to that in control. Thus, APEC may encounter starvation and express virulence factors during incubation with bEnd.3 cells.
Conclusion
This study provides a comprehensive overview of transcriptomic changes that occur during APEC infection of bEnd.3 cells, and offers insights into the bacterial invasion strategies and the subsequent host defense mechanism.
Keywords: Dual RNA-seq, Host–pathogen interactions, Meningitis, APEC, bEnd.3 cells
Introduction
Avian pathogenic Escherichia coli (APEC), an important extraintestinal pathogenic E. coli (ExPEC), causes colibacillosis, an acute systemic disease that involves multiple organ lesions including respiratory, digestive, vascular and nervous system diseases (Dziva & Stevens, 2008; Ewers et al., 2004). Previous studies have demonstrated that APEC strains with different serotypes (O18, O2, O1) can induce meningitis in newborn mammals, such as mice, with varying degrees of septicemia via pathogenic mechanisms that are similar to those of neonatal meningitis-causing E. coli (NMEC) strains (Krishnan et al., 2015; Mellata, Johnson & Curtiss, 2018; Zhu Ge et al., 2014). The APEC XM strain (O2:K1), isolated from the brain of a duck with septicemia and meningitis, was shown to be involved in the systemic infection of 7-day-old ducks and 5-week-old Institute of Cancer Research mice, causing severe meningitis in a neonatal mouse model (Hejair et al., 2017; Ma et al., 2014).
Various bacterial factors have been recognized as potent or putative virulence factors of APEC strains, including adhesins (Fim, Pap and Mat), iron acquisition systems (siderophores), two-component regulatory systems (RstAB system, ArcA/B system), vacuolating autotransporter toxin located in the chromosome, and the ColV plasmid encoded virulence genes (Breland, Eberly & Hadjifrangiskou, 2017; Gao et al., 2012; Johnson et al., 2006; Zhao et al., 2015). In particular, it has been reported that the virulence genes ibeA and gimB, which contribute to the invasion of host cells, are shared between APEC and NMEC (Barbieri et al., 2013; Peigne et al., 2009). Moreover, the host cell cytosolic phospholipase A2 and E. coli ibeA gene have been proved to be involved in the invasion of brain microvascular endothelial cells (BMECs) (Das et al., 2001; Maruvada & Kim, 2012; Zhu, Pearce & Kim, 2010). Since APEC is a potential reservoir of ExPEC virulence genes and pathogenic to humans (Rodriguez-Siek et al., 2005), APEC infection may pose a potential risk for zoonotic transfer (Mitchell et al., 2015). E. coli usually causes meningitis via several steps involving bacteria–host interactions: entry into the gastrointestinal tract mucosa (Birchenough et al., 2017), invasion of the intravascular space, survival and multiplication in the serum to a particular order of magnitude (Sullivan, Lascolea & Neter, 1982), traversing through the blood–brain barrier (BBB), and ultimately contributing to central nervous system (CNS) complications and neuronal injury (Kim, 2003b; Witcomb et al., 2015). A critical step in meningitic process is the bacterial crossing of the BBB, a structural and functional barrier formed by BMECs, astrocytes and pericytes that blocks the transport of harmful substances and pathogenic microorganisms. Bacteria invade the BBB via intercellular and paracellular pathways as well as Trojan horse mechanisms (Kim, 2003a, 2008). However, the mechanisms involved in the bacteria-host interaction during this process remain unclear.
Bacteria can rapidly reprogram their gene expression networks in response to their constantly changing living environment. During in vivo infection, the bacteria compete with the host for survival or nutrition and gene expression changes are observed in both, which differ from those observed in artificial culture conditions. Dual RNA sequencing (RNA-seq) was first used to simultaneously profile host and pathogen transcriptomes in 2012 in order to better understand the host–pathogen interactions (Westermann, Gorski & Vogel, 2012). Since then, dual RNA-seq analyses have been successfully performed to assess pathogen–host interactions, including those between Pseudomonas plecoglossicida and Epinephelus coioides (Zhang et al., 2019) as well as Salmonella and HeLa cells (Westermann et al., 2016). However, the interaction between E. coli and BBB-related cells has not yet been explored by dual RNA-seq.
In this study, we investigated the potential mechanism of APEC-host cell interaction by infecting the mouse brain microvascular endothelial cell line (bEnd.3) with the APEC XM strain (O2:K1). The transcriptomes of APEC strain and bEnd.3 cells were measured by dual RNA-seq during the interaction. The findings of this study may contribute toward improving the current understanding of E. coli invasion across the BBB.
Materials and Methods
Culture conditions
The APEC XM strain (O2:K1) was isolated from the brain of a duck with symptoms of septicemia and meningitis (donated by Dr. Guoqiang Zhu, Yangzhou University), and grown aerobically on Luria-Bertani (LB) plates or in LB broth with agitation (180 rpm/min) at 37 °C. Mouse BMECs (bEnd.3; ATCC CRL-2299, American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) in 10 cm cell culture dishes at 37 °C in a 5% CO2 atmosphere.
Bacterial adherence and invasion of bEnd.3 cells
For the adherence and invasion assays, the APEC strain was grown in LB broth with agitation (180 rpm/min) until the optical density at 600 nm reached 1.0 (1 × 108 CFU/mL) in exponential phase. The bacteria were collected by centrifugation (3,500 rpm, 8 min), washed twice with phosphate-buffered saline (PBS), and resuspended in FBS-free DMEM. Then, bEnd.3 cells were infected with the APEC XM strain in 10 cm dishes at a multiplicity of infection (MOI) of 100 for 1, 2, 3, 4, 5 or 6 h at 37 °C in 5% CO2. The mock-infection cells were cultured in FBS-free DMEM as control. The bEnd.3 cells were gently washed with PBS three times to remove any non-adherent bacteria, and then lysed with 0.5% Triton X-100 for 30 min at 37 °C. The suspensions were collected, serially diluted 10-fold, and plated on LB plates. After incubation overnight at 37 °C, the number of CFUs was calculated. The time point at which the highest number of bacteria adhered to and invaded the bEnd.3 cells was selected for the sample collection and RNA-seq analysis.
Total RNA isolation
Bacteria were cultured in DMEM without FBS for 3 h and then were treated with RNAprotect Bacteria Reagent (QIAGEN, Hilden, Germany) to protect the RNA. Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions (Invitrogen Co., Ltd., San Diego, CA, USA) and genomic DNA was digested using RNase-free DNase. To sequence the bacterial transcriptome, rRNA was removed from the total RNA using a Ribo-Zero rRNA removal kit (gram-negative bacteria, Epicentre Biotechnologies, Madison, WI, USA). The total RNA of bEnd.3 cells, infected with or without APEC for 3 h, were isolated using pre-cooled TRIzol reagent (Invitrogen Co., Ltd., San Diego, CA, USA) according to the manufacturer’s instructions. RNA integrity was analyzed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA), RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA), and RNA concentration was measured using a Qubit® RNA Assay Kit with a Qubit® 2.0 Fluorometer (Invitrogen Co., Ltd., San Diego, CA, USA).
cDNA library construction and RNA-seq
Nine individual cDNA sequencing libraries (three mock infection APEC samples, three mock-infected bEnd.3 samples, and three infection samples) were prepared using a NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) according to the manufacturer’s recommendations. Index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and fragmented using divalent cations under high temperatures in NEBNext First Strand Synthesis Reaction Buffer (5×) (NEB, Ipswich, MA, USA). cDNA was synthesized using a random hexamer primer and fragments of 250–300 bp in length were preferentially selected and purified using an AMPure XP system (Beckman Coulter, Brea, CA, USA). PCR was then performed using a Phusion High-Fidelity DNA polymerase (Vazyme, Nanjing, China), universal PCR primers, and Index (X) primers. The PCR products were purified using an AMPure XP system (Beckman Coulter, Brea, CA, USA) and library quality was assessed using an Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). The index-coded samples were clustered using a cBot Cluster Generation System (Illumina, Inc., San Diego, CA, USA) with a TruSeqPE Cluster Kit v3-cBot-HS (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions. The library was then sequenced on an Illumina HiSeq platform (Illumina, Inc., San Diego, CA, USA) to generate 125/150 bp paired-end reads. The raw reads in FASTQ format were first processed using in-house Perl scripts and clean data were obtained by removing reads containing adaptor or poly-N sequences and low-quality reads. The Q20, Q30 and GC content of the clean data were then calculated. Bowtie2-2.2.3 was used to build the reference genome index and align the clean reads to the reference genome. HTSeq v0.6.1 was used to count the number of reads mapped to each gene. Gene transcription levels were estimated by calculating the fragments per kilobase of transcript per million mapped reads.
Differentially expressed genes screening and functional analysis
The DEGs in bEnd.3 cells and APEC were evaluated by comparing the transcriptome data of both cell types cultured in DMEM or during their interaction using the DESeq2 R package (1.16.1). The P values of results were adjusted using Benjamini and Hochberg’s approach to control the false discovery rate. Differential expression was determined using the following thresholds: |log2-fold change| of ≥1 or 0.5 and an adjusted P value of ≤0.05.
DEG functional annotation and enrichment were performed using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, while KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways and the GOseq R package was used to analyze the GO enrichment of DEGs. GO terms with corrected P values of less than 0.05 were considered to be significantly enriched for DEGs.
Quantitative real-time PCR
qRT-PCR was carried out using a previously described method for validating gene expression data obtained by high-throughput profiling platforms (Everaert et al., 2017). A total of 17 genes were randomly selected to analyze their relative expression level and then qRT-PCR primers were designed and validated for these genes (Table S1). qRT-PCR was carried out on a CFX CONNECT Real-time PCR machine (Bio-Rad, Louisville, KY, USA) using ChamQ SYBR qRT-PCR Master Mix (2×) (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The amplification cycles were performed as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. All qRT-PCR assays were performed in triplicate, with expression values estimated using the 2−ΔΔCt method and normalized using gapA and GADPH for APEC and bEnd.3 cells, respectively. The correlation between the fold changes obtained by qRT-PCR and RNA-seq were determined using Pearson correlation analysis.
Results
Ability of APEC to adhere to and invade bEnd.3 cells
To characterize the interactions between the APEC strain and the BBB, bEnd.3 cells were used to establish an in vitro model. The ability of adhesion and invasion at six serial time points were evaluated. MOI of 100 was identified as the appropriate infectious dose of bacteria and the maximum adhesion and invasion was achieved at 3 h (Fig. S1).
Dual RNA-seq analysis of APEC and infected bEnd.3 cells
To characterize the response of bEnd.3 cells to infection and investigate the effect of bEnd.3 cells resistance on the transcriptional response of E. coli in vitro, we used a dual RNA-seq method that enabled the simultaneous transcriptional profiling of bacteria and bEnd.3 cells. Total RNA, including cellular and bacterial RNA, was isolated from infected bEnd.3 cells at 3 h. Nine individual cDNA sequencing libraries (three mock infection APEC samples, three mock-infected bEnd.3 samples, and three infection samples) were prepared and sequenced using the Illumina paired-end method, generating more than 1 × 107 clean reads per group after removal of the low-quality reads (Table 1). The clean reads of Q20 and Q30 were above 97% and 91%, respectively, with a GC content of approximately 50%. Next, we mapped the sequenced clean reads to the Mus musculus (Ensembl release-92) and E. coli (GCF_002844685.1) genomes. In the bEnd.3 cell-APEC interaction (be_AP) group, over 65% of the clean reads were mapped to the M. musculus genome and less than 20% were mapped to the E. coli genome, providing sufficient data for further analysis. In the two control groups, more than 92% and 99% of the clean reads were mapped to the E. coli and M. musculus genomes, respectively. All further analyses were based on the uniquely mapped reads and all raw data were submitted to the ZENODO database (DOI 10.5281/zenodo.3689240, 10.5281/zenodo.3672826).
Table 1. Summary of illumina RNA-seq data.
Each row of data indicates the total reads, clean reads, total mapped clean data, Q30 (%), GC (%), and percent sequence reads mapped of every sample.
| Sample | Total reads | Clean reads | Total mapped clean data* (Gb) | Q30 (%) |
GC (%) |
Mapped rate (%) |
|---|---|---|---|---|---|---|
| APEC_1 | 11,377,278 | 11,303,464 | 1.7G | 92.84 | 51.33 | 99.43a |
| APEC_2 | 12,595,278 | 12,482,110 | 1.87G | 92.86 | 51.46 | 99.39a |
| APEC_3 | 9,850,068 | 9,786,504 | 1.47G | 94.91 | 51.45 | 99.72a |
| bEnd3_1 | 59,805,426 | 59,042,776 | 8.86G | 92.05 | 50.7 | 92.44b |
| bEnd3_2 | 49,924,482 | 49,319,300 | 7.4G | 92.1 | 51.22 | 92.46b |
| bEnd3_3 | 64,453,186 | 63,528,514 | 9.53G | 91.87 | 50.15 | 92.29b |
| bE_AP_1 | 77,516,134 | 76,516,858 | 11.48G | 92.15 | 48.91 | 19.16a |
| 79,574,798 | 78570978 | 11.79G | 92.14 | 49.1 | 67.44b | |
| bE_AP_2 | 77,558,096 | 76,635,112 | 11.5G | 92.83 | 48.42 | 16.29a |
| 80,037,182 | 79,108,450 | 11.87G | 92.81 | 48.63 | 69.38b | |
| bE_AP_3 | 75,686,054 | 74,877,152 | 11.23G | 92.91 | 48.65 | 13.96a |
| 78,470,990 | 77,655,916 | 11.65G | 92.89 | 48.85 | 71.53b |
Notes:
Clean data were obtained from raw data by removing reads containing adapter, ploy-N and low quality reads.
Clean reads were mapped to Escherichia coli genome.
Clean reads were mapped to Mus musculus genome.
Validation of dual RNA-seq data by qRT-PCR
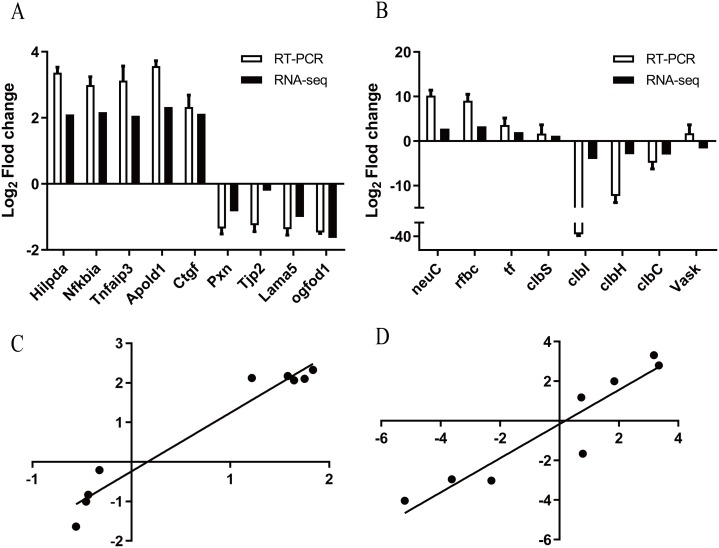
To confirm the results of the RNA-seq analysis, 17 highly expressed DEGs were randomly selected for further validation using qRT-PCR (details in Table S1). The trends of up- and down-regulation for 16 of the genes were consistent with the results of the Illumina sequencing analysis, except the Vask gene (Figs. 1A and 1B). In addition, the Pearson correlation coefficients (R2) of bEnd.3 cells and APEC strain were 0.954 and 0.876, respectively, indicating that the results of both techniques correlated strongly (Figs. 1C and 1D), thus confirming the reliability and accuracy of the transcriptome analysis.
Figure 1. Validation of dual RNA-seq analysis.
(A) qRT-PCR analysis of bEnd.3 transcriptome representative genes identified by RNA-seq. The x-axis represents individual genes and the y-axis fold change in expression determined by RNA-seq (black bars) or qRT-PCR (white bars). All data are shown as means ± SD. BEnd.3 transcriptome representative genes are Hilpda (hypoxia inducible lipid droplet associated), Nfkbia (nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha), Tnfaip3 (tumor necrosis factor, alpha-induced protein 3), Apold1 (apolipoprotein L domain containing 1), Ctgf (connective tissue growth factor), Pxn (paxillin), Tjp2 (tight junction protein 2), Lama5 (laminin, alpha 5) and Ogfod1 (2-oxoglutarate and iron-dependent oxygenase domain containing 1). (B) qRT-PCR analysis of BEnd.3 transcriptome representative genes identified by RNA-seq. APEC strain transcriptome representative genes are clbS (colibactin self-protection protein clbS), neuC (UDP-N-acetylglucosamine 2-epimerase (hydrolyzing)), tf (type 1 fimbrial protein), rfbC (dTDP-4-dehydrorhamnose 3,5-epimerase), clbI (colibactin polyketide synthase ClbI), clbH (colibactin non-ribosomal peptide synthetase clbH), VasK (type VI secretion protein VasK) and clbC (colibactin polyketide synthase ClbC). All data are shown as means ± SD. (C) The correlation coefficient (R2) between the two data sets of bEnd.3 cells. The x-axis represents the log2 fold change in expression determined by qRT-PCR and the y-axis represents the log2 fold change in expression determined by RNA-seq. (D) The correlation coefficient (R2) between the two data sets of APEC strain. The x-axis represents the log2 fold change in expression determined by qRT-PCR and the y-axis represents the log2 fold change in expression determined by RNA-seq.
Analysis of changes in the bEnd.3 cells transcriptome during infection
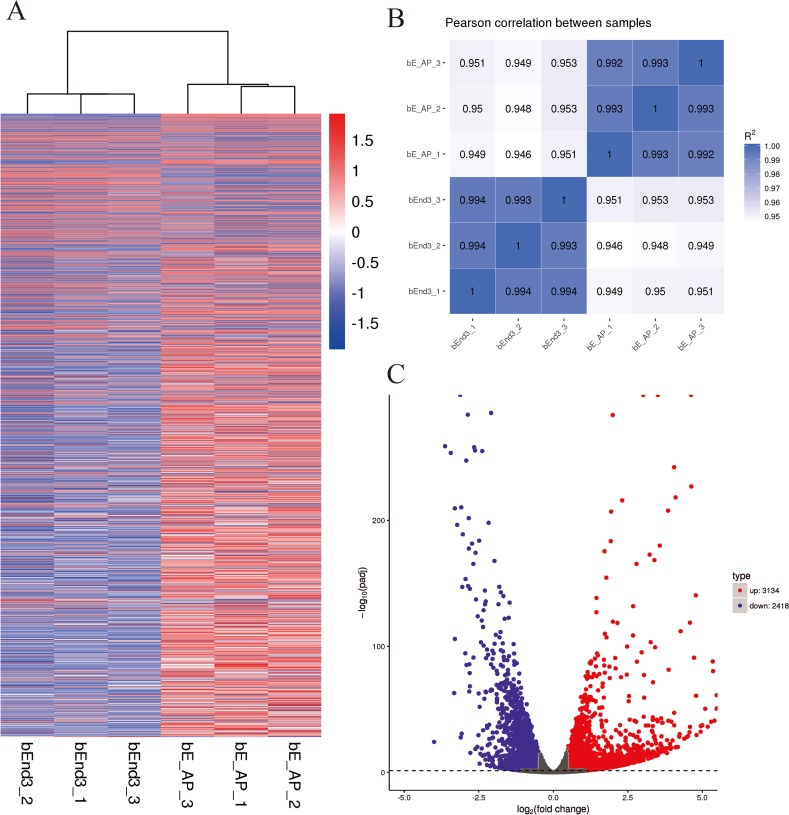
The hierarchical clustering (Fig. 2A) and RNA-seq sample Pearson correlation analysis (Fig. 2B) of gene expression datasets from the infected and uninfected bEnd.3 cells demonstrated high reproducibility within group. The DESeq2 R package identified 5,552 DEGs between the uninfected and infected bEnd.3 cells, among which 3,134 were upregulated and 2,418 were downregulated in response to infection (Fig. 2C; |log2-fold change| ≥ 0.5 and adjusted P value < 0.05).
Figure 2. Differential expression overview profiles of bEnd.3 cells transcriptome data.
(A) Heat maps of bEnd.3 gene expression during infection or mock infection samples. The read counts of each cellular mRNA were normalized by the sum of the total reads. Colors from white to red indicate upregulated cellular genes; colors from white to blue indicate downregulated cellular genes. (B) Pearson correlation between infection and mock infection samples. (C) Volcano plot of P values as a function of weighted fold change for mRNAs in infected and control groups. The vertical dotted line delimits up- and down-regulation. Red plots represent significant upregulated and green plots represent significant downregulated. (|log2 fold change| of ≥0.5, corrected P < 0.05).
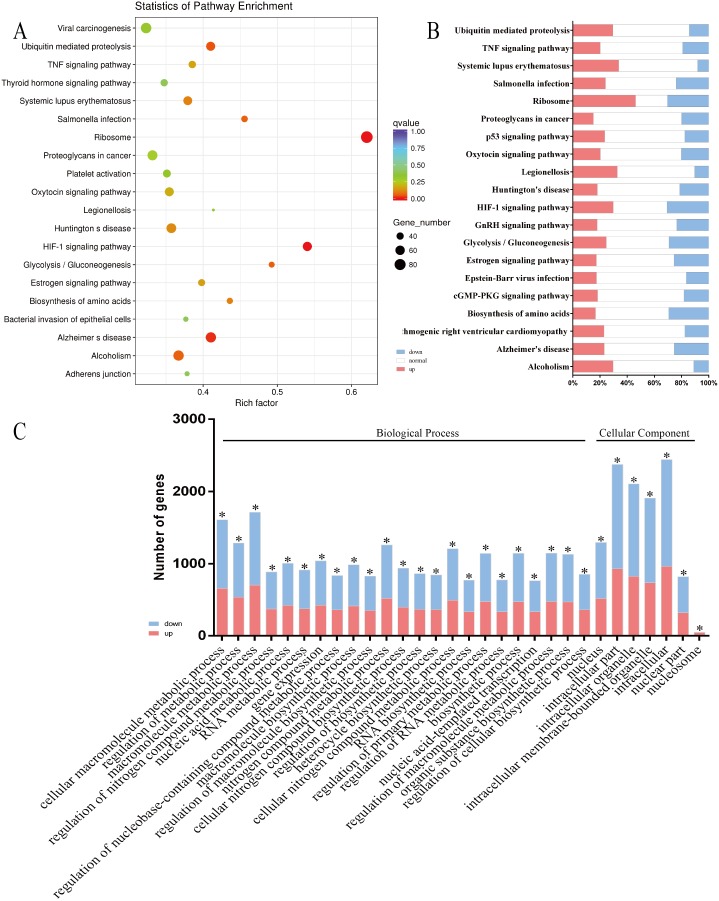
Functional classification of the DEGs using KEGG pathway enrichment analysis revealed their association with 273 pathways, indicating that many host genes, whose expression changed in response to infection, were enriched in signal transduction and immune response. The top 20 pathways are shown in Figs. 3A and 3B. The DEGs were also annotated by GO enrichment analysis using the GOseq R package, which showed that the DEGs were enriched in 12,833 GO terms, including 9,505 biological process terms, 1,158 cellular component terms and 2,152 molecular function terms. The majority of the top 30 enriched GO terms (23/30) were classified as biological processes, details of which are shown in Fig. 3C. This study focused on the interesting changes in the bEnd.3 cells, including DEGs responsible for altering the integrity of host cell junctional complexes, actin cytoskeletal rearrangements, extracellular matrix (ECM) degradation, immune activation, and inflammatory responses (Table 2; Table S2).
Figure 3. Enrichment analysis of KEGG pathways and GO terms for DEGs in bEnd.3 cells.
(A and B) The top 20 enriched KEGG pathways were classified as human disease (9/20, Salmonella infection and Legionellosis etc), endocrine system (4/20, Estrogen signaling pathway and Oxytocin signaling pathway etc), signal transduction (2/20, HIF-1 signaling pathway, TNF signaling pathway) and other pathways. The size of each circle represents the number of DEGs in each pahway (larger circles represent more DEGs) and the color represents the corrected P value of each pathway. Red bars represent significant upregulated, blue bars represent significant downregulated and white bars represent no expression or no significant expression. (C) In the top 30 enriched GO terms, most terms (23/30) were classified as biological process, seven of them were belonged to cellular component.Red bars represent significant upregulated and blue bars represent significant downregulated (|log2 fold change| ≥0.5, *corrected P < 0.05).
Table 2. DEGs of bEnd.3 cells between the two groups.
Different expression genes of bEnd.3 cells between the infection and mock-infection group.
| Components | Genes | log2 | padj | Description | |
|---|---|---|---|---|---|
| Fold change | |||||
| DEGs related to cell junctional complexes | |||||
| Tight junctions (TJs) | Claudins | Cldn5 | −0.51506 | 0.000166 | claudin 5 |
| Adherens junctions (AJs) | Cadherin | Cdh5 | −0.73355 | 3.27E−19 | cadherin 5 |
| Cdh24 | −0.78667 | 0.041049 | cadherin-like 24 | ||
| nectin | Nectin1 | −1.7466 | 1.22E−50 | nectin cell adhesion molecule 1 | |
| Nectin2 | −0.53373 | 0.00102 | nectin cell adhesion molecule 2 | ||
| Nectin3 | 0.59157 | 3.14E−07 | nectin cell adhesion molecule 3 | ||
| DEGs related to actin cytoskeletal rearrangements | |||||
| Regulation of actin cytoskeleton | Cfl1 | −0.54554 | 3.27E−10 | cofilin 1, non-muscle | |
| Actn1 | −0.60337 | 2.06E−09 | actinin, alpha 1 | ||
| Limk1 | −0.6284 | 0.001253 | LIM-domain containing, protein kinase | ||
| Pxn | −0.83324 | 3.61E−10 | paxillin | ||
| Actn4 | −1.6743 | 1.54E−27 | actinin alpha 4 | ||
| Actb | 2.0004 | 1.10E−136 | actin, beta | ||
| Itgav | 0.78575 | 1.76E−21 | integrin alpha V | ||
| Rock1 | 0.61447 | 2.59E−12 | Rho-associated coiled-coil containing protein kinase 1 | ||
| Rock2 | 0.50308 | 2.05E−09 | Rho-associated coiled-coil containing protein kinase 2 | ||
| DEGs of immune activation and inflammatory response | |||||
| Pattern recognition receptors (PRRs) | Tlr13 | 1.2499 | 6.48E−16 | toll-like receptor 13 | |
| Tlr4 | 0.82039 | 2.48E−17 | toll-like receptor 4 | ||
| Complement system | C3 | −0.58759 | 0.005068 | complement component 3 | |
| C3ar1 | 0.74408 | 2.90E−05 | complement component 3a receptor 1 | ||
| Cfp | −0.99477 | 3.44E−05 | complement factor properdin | ||
| Masp1 | 3.9332 | 0.00077 | mannan-binding lectin serine peptidase 1 | ||
| Chemokines | C subfamiliy | Xcr1 | 1.749 | 6.96E−06 | chemokine (C motif) receptor 1 |
| C–C subfamiliy | Ccl2 | 0.61885 | 0.000328 | chemokine (C–C motif) ligand 2 | |
| Ccrl2 | 0.72992 | 0.03855 | chemokine (C–C motif) receptor-like 2 | ||
| C–X3–C subfamiliy | Cx3cl1 | −0.54859 | 0.000456 | chemokine (C–X3–C motif) ligand 1 | |
| C–X–C subfamiliy | Cxcl1 | 1.7688 | 2.22E−06 | chemokine (C–X–C motif) ligand 1 | |
| Cxcl16 | 1.4227 | 0.000779 | chemokine (C–X–C motif) ligand 16 | ||
| Cxcl2 | 1.7977 | 0.00055 | chemokine (C–X–C motif) ligand 2 | ||
The BBB is a highly specialized structural and biochemical barrier; its properties are primarily determined by junctional complexes between the endothelial cells (ECs), comprising of adherens junctions (AJs) and tight junctions (TJs). In this study, genes encoding AJs (e.g., Cdh5, Cdh24, Pcdh1, Pcdhgc3, Nectin1 and Nectin2), which were involved in supporting cadherin association and regulating out-in signaling processes, were downregulated, but Nectin3 was upregulated in E. coli-infected bEnd.3 cells. Moreover, genes encoding TJs (e.g., Cldn5, Tjap1, Actn1 and Actn4), which are involved in sealing the interendothelial cleft, were downregulated in the infected cells. These findings indicated that the structural integrity, permeability and paracellular barriers of the BBB were destroyed during infection.
Previously, it has been shown that the actin cytoskeleton was rearranged and the ECM, with related receptors, were closely regulated when NMEC traversed the BBB (Kim, Kang & Kim, 2005; Kim, 2002). In the present study, some DEGs (Rock1, Rock2, Vav3, Itgav, Lamc2, Sdc4, Gp1ba, and Thbs1) related to actin arrangement and the ECM were upregulated, while other DEGs with similar functions (Actn4, Actn1, Itgb4, Pxn, Cfl1, Wasf1, Wasl, Dag1, Col5a3, Col5a1, Itga5, Col27a1, Lama5, Itgb4, Fn1, Agrn, Comp, Col1a1 and Hspg2) were downregulated. These findings suggested an increase in Rho/ROCK pathway activation, F-actin cytoskeleton rearrangement, and BBB permeability.
Inflammation is a hallmark of bacterial meningitis and is mediated mainly by cytokines and chemokines, which occurs in response to bacteria or their products. Additionally, Tlr4, Tlr13, Cxcl2, Cxcl1, Xcr1, Cxcl16, Nfkbia, Tnfaip3, Il6, Casp12, Nod2, Ccl2, Vcam1, Ncf2, Cfd, Cd46 and C3ar1 were upregulated and C3 was downregulated in the infection group, which suggested that the permeability of the BBB increased and the recruitment of monocytes, neutrophils, T cells, and natural killer cells was enhanced during the infection process.
E. coli transcriptome changes during bEnd.3 cells infection
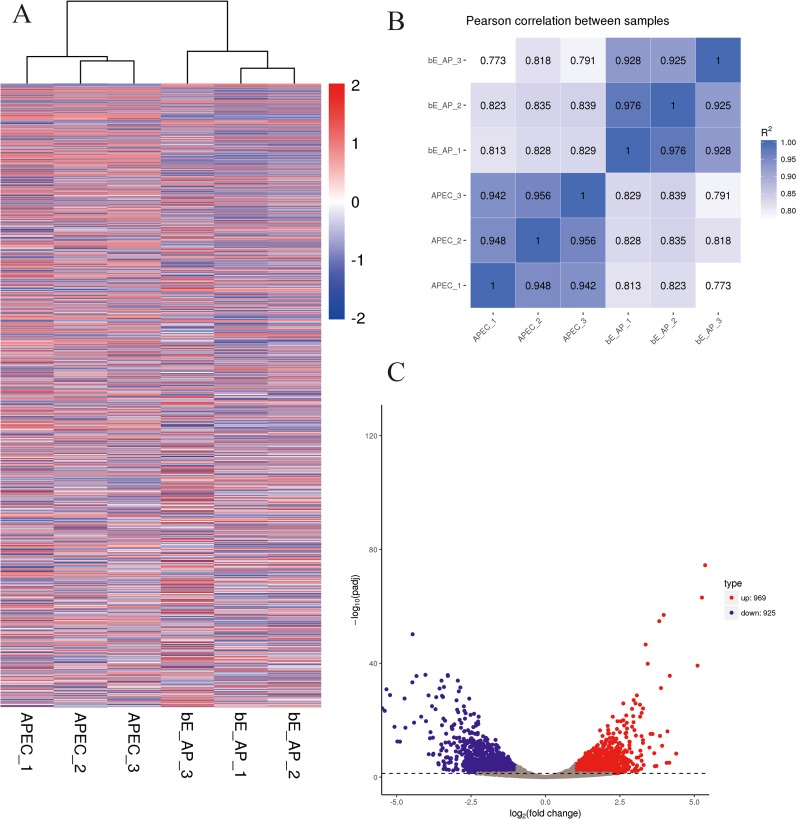
The hierarchical clustering (Fig. 4A) and RNA-seq sample Pearson correlation analysis (Fig. 4B) of gene expression datasets demonstrated high reproducibility within group from the infected and mock-infected bEnd.3 cells. The DESeq2 R package identified 1,894 DEGs between the two infection conditions, including 969 upregulated and 925 downregulated genes (Fig. 4C, |log2 fold change| ≥ 1 and adjusted P-value < 0.05).
Figure 4. Differential expression overview profiles of APEC strain transcriptome data.
(A) Heat maps of APEC strain gene expression during infection or control samples. The read counts of each cellular mRNA were normalized by the sum of the total reads. Colors from white to red indicate upregulated cellular genes; colors from white to blue indicate downregulated cellular genes. (B) Pearson correlation between infection and control samples. (C) Volcano plot of P-values as a function of weighted fold change for mRNAs in infection and control groups. The vertical dotted line delimits up- and down-regulation. Red plots represent significant upregulated and green plots represent significant downregulated (|log2 fold change| ≥1, corrected P < 0.05).
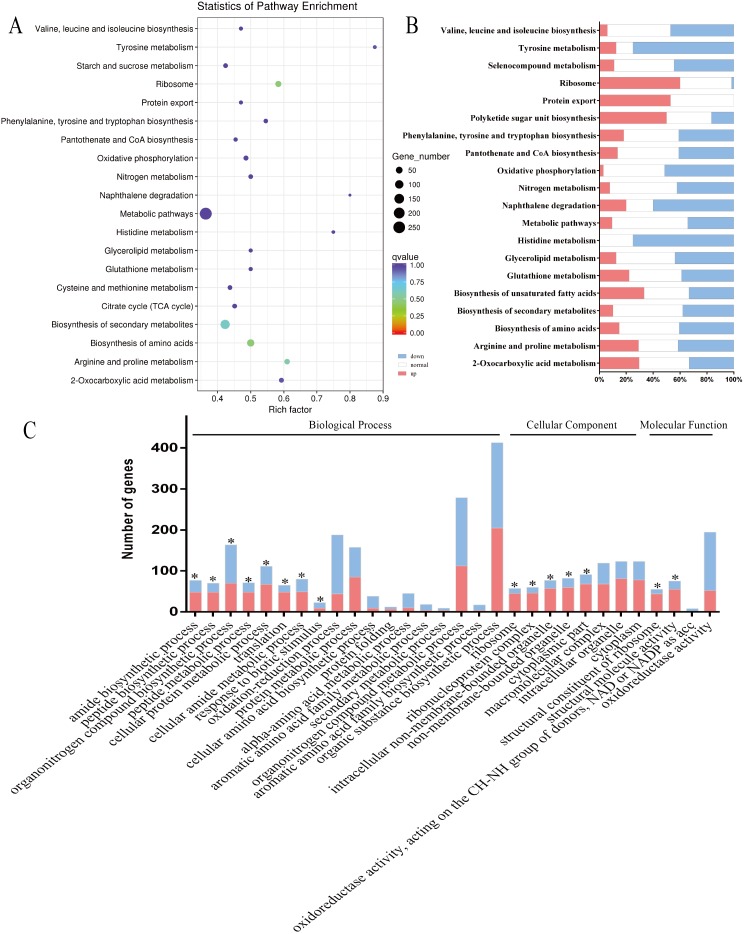
KEGG pathway enrichment analysis was used to functionally classify the DEGs for 88 pathways, revealing that many E. coli genes were enriched in amino acid and energy metabolism when APEC was cultured with the cells. The top 20 pathways are shown in Figs. 5A and 5B. The DEGs were also annotated by GO enrichment analysis using the GOseq R package, and enriched for 2,482 GO terms, including 1,376 biological process terms, 311 cellular component terms and 795 molecular function terms. The majority of the top 30 enriched GO terms (18/30) were classified as biological processes, which are shown in detail in Fig. 5C. In this study, we also identified several interesting changes in E. coli, particularly in the DEGs related to virulence factors, protein export systems and amino acid metabolism (Table 3; Table S3).
Figure 5. Enrichment analysis of KEGG pathways and GO terms for DEGs in APEC strain.
(A and B) The top 20 enriched KEGG pathways were classified as amino acid metabolism (7/20, Valine, leucine and isoleucine biosynthesis and Selenocompound metabolism, etc.), lipid metabolism (2/20, Biosynthesis of unsaturated fatty acids and Glycerolipid metabolism), energy metabolism (2/20, Nitrogen metabolism and Oxidative phosphorylation) and othe pathways. The size of each circle represents the number of DEGs in each pahway (larger circles represent more DEGs) and the color represents the corrected P value of each pathway. Red bars represent significant upregulated, blue bars represent significant downregulated and white bars represent no expression or no significant expression. (C) In the top 30 enriched GO terms, most terms (18/30) were classified as biological process, eight of them belonged to cellular component and four of them were classified as molecular function. Red bars represent significant upregulated and blue bars represent significant downregulated (|log2 fold change| ≥1, *corrected P < 0.05).
Table 3. DEGs of APEC strain between two groups.
Different expression genes of APEC strain between the infection and mock-infection group.
| Components | Gene name |
log2 Fold Change |
padj | Description | |
|---|---|---|---|---|---|
| Virulence factors related to meningitis | |||||
| Outer membrane protein | CXG97_RS20855 | – | −2.3638 | 3.43E−06 | membrane protein |
| CXG97_RS13750 | – | −1.8361 | 0.0083262 | membrane protein | |
| CXG97_RS15480 | – | −1.7665 | 0.014095 | membrane protein | |
| CXG97_RS10635 | – | −1.1948 | 0.039525 | membrane protein | |
| CXG97_RS12085 | – | 3.121 | 3.42E−09 | membrane protein | |
| CXG97_RS18795 | – | 1.5797 | 0.003432 | membrane protein | |
| CXG97_RS10035 | – | 1.1784 | 0.032157 | membrane protein | |
| CXG97_RS08525 | – | 1.3149 | 0.015745 | autotransported outer membrane protein involved in cell adhesion | |
| CXG97_RS01930 | – | 1.7816 | 0.0014647 | autotransporter outer membrane beta-barrel domain-containing protein | |
| CXG97_RS01635 | – | 1.4776 | 0.0098823 | autotransporter outer membrane beta-barrel domain-containing protein | |
| CXG97_RS09010 | – | 1.6293 | 0.049761 | fimbrial biogenesis outer membrane usher protein | |
| CXG97_RS18505 | – | 1.3315 | 0.045966 | fimbrial biogenesis outer membrane usher protein | |
| CXG97_RS09580 | slyB | 1.3411 | 0.0099186 | outer membrane lipoprotein SlyB | |
| CXG97_RS15430 | – | 1.5911 | 0.0023044 | outer membrane protein assembly factor BamD | |
| CXG97_RS15540 | – | 1.2863 | 0.015568 | outer membrane protein assembly factor BamE | |
| fimbrial | CXG97_RS01490 | – | −3.3352 | 2.07E−05 | fimbrial chaperone EcpB |
| CXG97_RS17665 | – | −1.2869 | 0.0229 | fimbrial protein SteB | |
| CXG97_RS25945 | fimA | −1.1275 | 0.041784 | type 1 fimbriae major subunit | |
| CXG97_RS13610 | – | −1.7529 | 0.002143 | flagella biosynthesis regulator Flk | |
| CXG97_RS25950 | – | −2.2246 | 0.00011843 | fimbrin fimI | |
| CXG97_RS09010 | – | 1.6293 | 0.049761 | fimbrial biogenesis outer membrane usher protein | |
| CXG97_RS09025 | – | 2.7839 | 2.36E−07 | fimbrial chaperone protein FimC | |
| fimbrial | CXG97_RS09015 | – | 1.7675 | 0.0015729 | fimbrial chaperone protein FimC |
| CXG97_RS17675 | – | 1.996 | 8.94E−05 | type 1 fimbrial protein | |
| CXG97_RS20510 | – | 1.2399 | 0.02421 | type 1 fimbrial protein | |
| CXG97_RS09020 | – | 2.3148 | 5.86E−06 | Fml fimbriae subunit | |
| flagellin | CXG97_RS11050 | – | 1.2589 | 0.039065 | flagellin FliC |
| LPS biosynthesis | CXG97_RS21640 | – | 2.4423 | 3.2651E−06 | ligase |
| CXG97_RS21660 | – | 1.9943 | 0.00014874 | LPS 1%2C2-glucosyltransferase | |
| CXG97_RS21655 | – | 2.0378 | 0.00015131 | LPS core heptose(II) kinase RfaY | |
| CXG97_RS21665 | – | 1.8557 | 0.00042876 | lipopolysaccharide 3-alpha-galactosyltransferase | |
| CXG97_RS06255 | – | −1.3255 | 0.017077 | lipid A biosynthesis lauroyl acyltransferase | |
| CXG97_RS21670 | – | 1.2497 | 0.019185 | lipopolysaccharide core heptose(I) kinase RfaP | |
| CXG97_RS21630 | – | −1.7497 | 0.038733 | ADP-heptose–LPS heptosyltransferase | |
| Proteins export and amino acid metabolism | |||||
| Protein export | CXG97_RS19815 | SecY | 2.0511 | 4.25E−05 | protein translocase subunit SecY |
| CXG97_RS23005 | tatC | 1.9541 | 0.00021961 | twin-arginine translocase subunit TatC | |
| CXG97_RS02115 | YajC | 1.7879 | 0.00069815 | preprotein translocase subunit YajC | |
| CXG97_RS22995 | TatA | 1.3288 | 0.012952 | twin-arginine translocase subunit TatA | |
| CXG97_RS23000 | TatB | 1.2944 | 0.014643 | twin-arginine translocase subunit TatB | |
| CXG97_RS02125 | SecF | 2.2484 | 0.020017 | protein translocase subunit SecF | |
| CXG97_RS21575 | SecB | 1.237 | 0.025291 | protein-export protein SecB | |
| CXG97_RS19155 | SecG | 1.1816 | 0.027567 | protein-export membrane protein SecG | |
| Arginine and proline metabolism | CXG97_RS23620 | argB | 3.3547 | 1.35E−05 | acetylglutamate kinase |
| CXG97_RS17080 | arcC | 1.6337 | 0.008478 | carbamate kinase | |
| CXG97_RS08705 | patD | 1.3149 | 0.037641 | gamma-aminobutyraldehyde dehydrogenase | |
| CXG97_RS25795 | argF | 8.588 | 1.37E−24 | ornithine carbamoyltransferase | |
| CXG97_RS03780 | speF | 1.5823 | 0.034956 | ornithine decarboxylase SpeF | |
| CXG97_RS06060 | putA | −3.5858 | 0.005959 | bifunctional proline dehydrogenase/L-glutamate gamma-semialdehyde dehydrogenase PutA | |
| CXG97_RS10130 | astB | −4.4096 | 0.006318 | succinylarginine dihydrolase | |
| CXG97_RS10135 | astD | −5.5193 | 0.000153 | N-succinylglutamate 5-semialdehyde dehydrogenase | |
| CXG97_RS17430 | speB | −1.8935 | 0.000611 | agmatinase | |
| CXG97_RS23160 | glnA | −1.458 | 0.009095 | glutamate–ammonia ligase | |
A critical step in the development of meningitis is the adhesion and invasion of ECs. Therefore, virulence factors related to fimbriae, flagella, outer membrane proteins, and lipoproteins are highly important for allowing pathogenic E. coli to resist blood flow and cross the BBB.
The RNA-seq data showed that the expression of seven genes related to outer membrane proteins (e.g., CXG97_RS09580, CXG97_RS01930), eight fimbrial genes (e.g., CXG97_RS09010, CXG97_RS09025), one flagellin gene (CXG97_RS11050), two pilus genes (CXG97_RS17695 and CXG97_RS17680), and one lipoprotein gene (CXG97_RS19095) increased significantly during infection, whereas the expression of nine fimbrial genes (e.g., CXG97_RS01490, CXG97_RS25950) decreased during infection (Table 3; Table S3).
Lipopolysaccharide (LPS) is produced by most Gram-negative bacteria and can activate the host immune system via TLR4. RNA-seq data revealed that eight DEGs were enriched in the LPS biosynthesis pathway, five of which (e.g., CXG97_RS21640, CXG97_RS21660) were upregulated in the infection group. Colicins are class III bacteriocins, which are produced during nutrient or oxygen stress and regulated by the SOS response (Smarda & Smajs, 1998). The expression of six genes related to colicin biosynthesis and transport (e.g., CXG97_RS26815, CXG97_RS27750) increased significantly during infection. In addition, thirteen colibactin genes (e.g., CXG97_RS11550, CXG97_RS11545), which induce chromosomal instability and DNA damage in eukaryotic cells and lead to EC senescence and immune cell apoptosis, were downregulated during infection, while only one colibactin gene (CXG97_RS11495) was upregulated. The type VI secretion system (T6SS) contributes to the pathogenicity of bacteria (Zhou et al., 2012) and bacteria–bacteria interactions (Basler, Ho & Mekalanos, 2013). Four T6SS genes (e.g., CXG97_RS01210, CXG97_RS01220) were downregulated during infection in the present study. ABC transporters are essential bacterial virulence factors, which play roles in the secretion of toxins and antimicrobial agents, and are associated with physiological processes (Davidson & Chen, 2004). In RNA-seq data, numerous DEGs were enriched in the ABC transporter pathway, with CXG97_RS18120, CXG97_RS13545, CXG97_RS23390 and CXG97_RS05305 being the most significantly upregulated genes in the APEC strain cultured with cells. These results suggested that the occurrence of meningitis was related to synergistic effects of many virulence factors.
Compared to the negative control, the prokaryotic protein export pathway was one of the most enriched E. coli KEGG pathways, which is composed of the general secretory system (Sec system), twin-arginine translocase (Tat) system, and a single peptide. The Sec and Tat systems are responsible for the transport of unstable or unfolded bacterial structural proteins to the periplasm or cytoplasmic membrane. Some genes (secYFBG, tatABC, yajc and lepB) from three parts of this pathway were upregulated in the infected groups, which suggested that protein secretion may be increased in the infected group. Moreover, several DEGs related to amino acid metabolism in APEC changed in response to bEnd.3 cells, as compared to those cultured in DMEM. The majority of these genes, which were involved in arginine and proline metabolism (argB, astDB, speBF and patD), histidine metabolism (hisHDFAF), and valine, leucine and isoleucine biosynthesis (leuCB and ilvA), were downregulated in the infected samples, suggesting that APEC may encounter a difficult and complex nutritional environment during the infection process.
Discussion
Bacterial meningitis is an inflammatory disease of the CNS, which not only causes high morbidity and mortality but also leaves survivors with long-term neurological sequelae. To infect the CNS, bacteria must interact with and cross the BBB via a critical step involving the adherence and invasion of BMECs, an important component of the BBB. In the present study, we used RNA-seq to measure genome-wide transcriptional changes in both APEC and bEnd.3 cells, including cell junctional complexes, cell signaling, inflammatory responses, bacterial adhesion and invasion factors and metabolic competition for similar nutritional substrates. The findings of the present study may contribute toward an improved understanding of the microbe-cell interaction during the invasion process.
TJ and AJ proteins can form junctional complexes and thus play important roles in maintaining the integrity of the BBB (Tietz & Engelhardt, 2015). We found that major components of TJs and AJs, such as Cldn5 and Cdh5, were downregulated in infected cells. Cdh5 is involved in neuroinflammation development, BBB dysregulation (Gijbels et al., 1990), and leukocyte transmigration in vitro (Orsenigo et al., 2012), and affects the expression of other TJ and AJ proteins (Dejana & Vestweber, 2013; Orsenigo et al., 2012). Conversely, Cldn5 highly expressed in ECs in the CNS (Daneman et al., 2010; Ohtsuki et al., 2008), which plays a key role in the paracellular barrier and forms mechanical links to maintain the structural integrity and high electrical resistance of vasculature (Abbott et al., 2010). The changes in Cdh5 and Cldn5 expression observed in the present study are consistent with the previous findings reported in a Staphylococcus aureus and group B Streptococcus model of meningitis (Kim et al., 2015; McLoughlin et al., 2017). In addition, Cdh5 controls Cldn5 by triggering its transcriptional repression via FoxO1 and β-catenin (Taddei et al., 2008). Transcriptome data in the present study revealed a significant decrease in FoxO1 expression during infection but only a slight increase in β-catenin expression, which may be related to the up-regulation of β-catenin protein levels and the activation of Wnt/β-catenin signaling by LPS (Xing et al., 2019). Moreover, Wnt/β-catenin signaling was shown to be involved in BBB development, where its blockade decreased Cdh5 expression in primary ECs of newborn mouse brain but not in that of the adult (Hubner et al., 2018). The results of the present study as well as previous studies (Kim et al., 2015; McLoughlin et al., 2017) suggest that Cdh5 and Cldn5 are major determinants of BBB deterioration during infection, while Wnt/β-catenin signaling may contribute to the maintenance of BBB integrity. However, further studies are required to investigate the complex relationship between Cdh5, Cldn5 and Wnt/β-catenin signaling during the development of E. coli meningitis.
As a dual (physical and immunological) barrier, the BBB is also a central determinant of protective homeostatic surveillance during CNS infections (Klein & Hunter, 2017). CNS ECs are semiprofessional antigen-presenting cells that present antigens to T cells and regulate the multistep cascade for immune cell trafficking into the CNS (Meyer, Martin-Blondel & Liblau, 2017). In the present study, Tlr4 and Tlr13 were upregulated in infected cells. However, a previous study on mouse meningitis induced by E. coli showed that TLRs were activated in brain tissues, with elevated Tlr2, Tlr4 and Tlr7 expression (Bottcher et al., 2003). TLR activation has also been shown to modulate microvascular EC permeability and the expression of coagulation pathway intermediaries (Khakpour, Wilhelmsen & Hellman, 2015). Tlr13 was highly expressed in almost all mouse CNS cell types and specifically detected 23S ribosomal RNA from E. coli (Li & Chen, 2012). Recent studies have identified that the recognition of E. coli mRNA stimulated helper T cell differentiation, promoted vaccine responses, and helped to distinguish between live and dead microbes (Sander et al., 2011; Ugolini et al., 2018). To our knowledge, this is the first study to report the high expression of Tlr13 in E. coli meningitis model in vitro. However, further investigations are necessary to elucidate the specific microbial components that activate Tlr13 in E. coli meningitis and reveal the details of the related pathways involved in infection. Moreover, future studies should investigate TLR inhibitors as potential targets to prevent serious meningitis-related complications.
Following inflammatory activation, host cells release cytokines and chemokines to maintain immune surveillance, facilitate leukocyte traffic, and recruit other inflammatory factors (Takeshita & Ransohoff, 2012). In the present study, Cxcl1, Cxcl2 and Cxcl16 were significantly upregulated in infected cells. A similar innate immune response was previously observed in different bacterial meningitis as well. Cxcl1, Cxcl2 and CXCL16 were associated with the migration of immune cells to sites of inflammation, matrix metalloproteinase activity, increased cell–cell adhesion, NF-кB-dependent cell proliferation, and proinflammatory gene transcription (Chandrasekar, Bysani & Mummidi, 2004; Girbl et al., 2018; Griffith, Sokol & Luster, 2014; Hofnagel et al., 2011; Semple, Kossmann & Morganti-Kossmann, 2010; Van Der Voort et al., 2005). In addition, Cxcl1 and Cxcl2 have been shown to alter human BMEC permeability and disrupt EC junctions during the migration of neutrophils and monocytes (Girbl et al., 2018; Zhang et al., 2013).
The mechanism of E. coli pathogenesis involves complex patterns of adhesion, protein export into host cells, changes in signaling mechanisms, impaired immune responses with colonization, disrupted membrane potential, and cytoskeletal manipulation (Bhavsar, Guttman & Finlay, 2007; Hornef et al., 2002; Kim et al., 2010). In the present study, many bacterial DEGs related to fimbrial and flagellin components were upregulated in the infection group. Type 1 fimbriae are mainly formed by FimAGHF proteins and mediate the mannose-sensitive adhesion of E. coli to various eukaryotic cells (Hanson & Brinton, 1988; Klemm, 1984; Teng et al., 2005). Conversely, the expression of S fimbriae in E. coli promoted adhesion to cow, human, and rat BMECs but not the systemic vascular endothelium (Prasadarao, Wass & Kim, 1997). Moreover, it has been shown that flagella, the locomotive organelles of bacteria, are an association factor rather than an invasion factor in human BMECs (Parthasarathy, Yao & Kim, 2007). The results of the present study also indicated that fimbrial and flagellin components were highly important virulence factors of the APEC XM strain for BBB attachment and invasion.
E. coli exerts physiological or pathogenic functions by exporting proteins via eight different systems (Crane & Randall, 2017). In the present study, many DEGs (secYFBG and LepB) related to the Sec system were upregulated in the infection group. SecB plays a crucial role as a chaperone during protein secretion by binding to precursors and delivering them to the membrane for translocation. Previous studies have shown that many virulence factors, such as P pilus, type 1 pilus, curli, OmpT and OmpA, were secreted into the extracellular environment or localized in the outer membrane by the SecYEG complex or SecB chaperone (Baars et al., 2006; Stathopoulos et al., 2000; Stones & Krachler, 2015). Recently, LepB has been proved to be a potential target for an attractive new antibacterial agent due to its crucial role in the Sec pathway; LepB inhibition leads to preprotein accumulation at the phospholipid bilayer and thus cell death (De Rosa et al., 2017; Ferrandez & Condemine, 2008). On the basis of these results, Sec pathway-mediated secretion may play an important role in bacterial pathogenesis. Thus, Sec pathway-associated proteins could be potential antibiotic drug targets for the prevention and treatment of meningitis. Another major component of the protein export pathway is the Tat system. In the present study, the three primary components of the Tat system (tatABC) were upregulated in the infected group. The Tat system was shown to take part in the development of bacteremia as well as the production of Shiga toxin 1 (Stx1) and H7 flagellin (Siddiqui, Beattie & Khan, 2012). Therefore, these results suggest that the Tat system may have a potential role in virulence during meningitis.
In addition to the deterioration of physical and immunological barrier functions, host cells and pathogens fiercely competed for nutrition. Indeed, the metabolic competition between the host and bacteria could influence both bacterial virulence and host responses, which determine the outcome of infection (Olive & Sassetti, 2016). In the present study, we identified a series of DEGs related to arginine and proline metabolism, which are particularly important nutrients for the host–pathogen interaction. Arginine is the unique substrate for nitric oxide synthase (NOS) to generate nitric oxide (NO) (Palmer, Ashton & Moncada, 1988), which has a variety of physiological functions, including vasodilation, leukocyte activation, and killing of virus and bacteria in diseases (Chicoine et al., 2004). In the present study, four host genes (Gm15587, Nos2, Pycr1, Arg2) were enriched in this metabolic pathway, two of which (Nos2, Pycr1) were downregulated in the infection group. Nos2 encoded a NOS and contributed to BBB breakdown and thus early mortality in murine Streptococcus pneumoniae meningitis (Yau et al., 2016). Previous studies showed that E. coli utilized arginine via the arginine decarboxylase and the arginine succinyltransferase pathway to produce polyamines (putrescine, spermidine, and spermine) for proliferation or survival in an acidic environment (Schneider, Kiupakis & Reitzer, 1998; Stancik et al., 2002). Since arginase and iNOS have the same substrate, E. coli may exploit their competition to block NO production and thus avoid being killed by NO in a similar manner to other pathogens (Bronte & Zanovello, 2005; Eckmann et al., 2000; Talaue et al., 2006). In our RNA-seq data set, some DEGs related to arginine and proline metabolism (argBF, arcC, patD, and speF) were upregulated in the infection group, while others (putA, astBD, speB and glnA) were downregulated. On the basis of these results, arginine/ornithine ABC transporters may be an effective target for preventing meningitis. Furthermore, the simultaneous changes in arginine and proline metabolism in the host and microbe may provide novel insights into nutrient and metabolite competition that occurs during meningitis development. Further experiments are required to explore this field and discover the molecular mechanisms underlying the host–pathogen relationship during infection.
Conclusion
This study provided a comprehensive overview of the transcriptomic changes that occurred when APEC infected the bEnd.3 cells. APEC may exploit the degradation of cell junctional connections to invade the BBB and secrete virulence factors to promote bacterial infection. Meanwhile, bEnd.3 cells resisted the bacterial infection via immune activation and inflammatory response. Therefore, this study provides insights into the process of bacterial invasion and the subsequent host defense mechanism, which can be used as reference for further investigations in this field.
Supplemental Information
Adherence to and invasion of bEnd.3 cells by APEC strain (at MOI of 100). Data are means + standard errors of three independent experiments, each performed in triplicate. An asterisk indicates that the adherence and invasion values of others’ time points were significantly higher than the values of the first one hour at P-value < 0.01.
Funding Statement
This study was supported by grants from the National Key R&D Program (2017YFD0500203), National Natural Science Foundation of China (No. 31672614, 31802253), the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS[2019]4610), Graduate Research Innovation Program of Yangzhou University (XKYCX19_127), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (TAPP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Peili Wang conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Xia Meng conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Jianji Li conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Yanfei Chen performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Dong Zhang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Haoran Zhong performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Pengpeng Xia conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Luying Cui analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Guoqiang Zhu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Heng Wang conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The APEC RNA-sequences and bEnd.3 cells RNA-sequences are available at SRA: (PRJNA612631) SRR11307869, SRR11307868, SRR11307867, SRR11307872, SRR11307871 and SRR11307870; (PRJNA612931) SRR11318238, SRR11318235, SRR11318236, SRR11318234, SRR11318233 and SRR11318237.
The data is also available at Zenodo: wang peili. (2019). Raw data of RNA-seq and IGV -2 [Data set]. Zenodo. DOI 10.5281/zenodo.3689240.
and
Wang peili. (2019). Raw data of RNA-seq and IGV -1 [Data set]. Zenodo. DOI 10.5281/zenodo.3672826.
References
- Abbott et al. (2010).Abbott NJ, Patabendige AA, Dolman DE, Yusof SR, Begley DJ. Structure and function of the blood–brain barrier. Neurobiology of Disease. 2010;37(1):13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Baars et al. (2006).Baars L, Ytterberg AJ, Drew D, Wagner S, Thilo C, van Wijk KJ, De Gier JW. Defining the role of the Escherichia coli chaperone SecB using comparative proteomics. Journal of Biological Chemistry. 2006;281(15):10024–10034. doi: 10.1074/jbc.M509929200. [DOI] [PubMed] [Google Scholar]
- Barbieri et al. (2013).Barbieri NL, De Oliveira AL, Tejkowski TM, Pavanelo DB, Rocha DA, Matter LB, Callegari-Jacques SM, De Brito BG, Horn F. Genotypes and pathogenicity of cellulitis isolates reveal traits that modulate APEC virulence. PLOS ONE. 2013;8(8):e72322. doi: 10.1371/journal.pone.0072322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler, Ho & Mekalanos (2013).Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell–cell interactions. Cell. 2013;152(4):884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar, Guttman & Finlay (2007).Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449(7164):827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- Birchenough et al. (2017).Birchenough GM, Dalgakiran F, Witcomb LA, Johansson ME, McCarthy AJ, Hansson GC, Taylor PW. Postnatal development of the small intestinal mucosa drives age-dependent, regio-selective susceptibility to Escherichia coli K1 infection. Scientific Reports. 2017;7(1):83. doi: 10.1038/s41598-017-00123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher et al. (2003).Bottcher T, Von Mering M, Ebert S, Meyding-Lamade U, Kuhnt U, Gerber J, Nau R. Differential regulation of Toll-like receptor mRNAs in experimental murine central nervous system infections. Neuroscience Letters. 2003;344(1):17–20. doi: 10.1016/S0304-3940(03)00404-X. [DOI] [PubMed] [Google Scholar]
- Breland, Eberly & Hadjifrangiskou (2017).Breland EJ, Eberly AR, Hadjifrangiskou M. An overview of two-component signal transduction systems implicated in extra-intestinal pathogenic E. coli infections. Frontiers in Cellular and Infection Microbiology. 2017;7:162. doi: 10.3389/fcimb.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte & Zanovello (2005).Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nature Reviews Immunology. 2005;5(8):641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Chandrasekar, Bysani & Mummidi (2004).Chandrasekar B, Bysani S, Mummidi S. CXCL16 signals via Gi, phosphatidylinositol 3-kinase, Akt, IκB kinase, and nuclear factor-κB and induces cell–cell adhesion and aortic smooth muscle cell proliferation. Journal of Biological Chemistry. 2004;279(5):3188–3196. doi: 10.1074/jbc.M311660200. [DOI] [PubMed] [Google Scholar]
- Chicoine et al. (2004).Chicoine LG, Paffett ML, Young TL, Nelin LD. Arginase inhibition increases nitric oxide production in bovine pulmonary arterial endothelial cells. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2004;287(1):L60–68. doi: 10.1152/ajplung.00194.2003. [DOI] [PubMed] [Google Scholar]
- Crane & Randall (2017).Crane JM, Randall LL. The sec system: protein export in Escherichia coli. EcoSal Plus. 2017;7 doi: 10.1128/ecosalplus.ESP-0002-2017. Epub ahead of print 21 November 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman et al. (2010).Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood–brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLOS ONE. 2010;5(10):e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das et al. (2001).Das A, Asatryan L, Reddy MA, Wass CA, Stins MF, Joshi S, Bonventre JV, Kim KS. Differential role of cytosolic phospholipase A2 in the invasion of brain microvascular endothelial cells by Escherichia coli and listeria monocytogenes. Journal of Infectious Diseases. 2001;184(6):732–737. doi: 10.1086/322986. [DOI] [PubMed] [Google Scholar]
- Davidson & Chen (2004).Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annual Review of Biochemistry. 2004;73(1):241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- De Rosa et al. (2017).De Rosa M, Lu L, Zamaratski E, Szalaj N, Cao S, Wadensten H, Lenhammar L, Gising J, Roos AK, Huseby DL, Larsson R, Andren PE, Hughes D, Brandt P, Mowbray SL, Karlen A. Design, synthesis and in vitro biological evaluation of oligopeptides targeting E. coli type I signal peptidase (LepB) Bioorganic & Medicinal Chemistry. 2017;25(3):897–911. doi: 10.1016/j.bmc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Dejana & Vestweber (2013).Dejana E, Vestweber D. The role of VE-cadherin in vascular morphogenesis and permeability control. Progress in Molecular Biology and Translational Science. 2013;116:119–144. doi: 10.1016/b978-0-12-394311-8.00006-6. [DOI] [PubMed] [Google Scholar]
- Dziva & Stevens (2008).Dziva F, Stevens MP. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathology. 2008;37(4):355–366. doi: 10.1080/03079450802216652. [DOI] [PubMed] [Google Scholar]
- Eckmann et al. (2000).Eckmann L, Laurent F, Langford TD, Hetsko ML, Smith JR, Kagnoff MF, Gillin FD. Nitric oxide production by human intestinal epithelial cells and competition for arginine as potential determinants of host defense against the lumen-dwelling pathogen Giardia lamblia. Journal of Immunology. 2000;164(3):1478–1487. doi: 10.4049/jimmunol.164.3.1478. [DOI] [PubMed] [Google Scholar]
- Everaert et al. (2017).Everaert C, Luypaert M, Maag JLV, Cheng QX, Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Scientific Reports. 2017;7(1):621. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers et al. (2004).Ewers C, Janssen T, Kiessling S, Philipp H-C, Wieler LH. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Veterinary Microbiology. 2004;104(1–2):91–101. doi: 10.1016/j.vetmic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Ferrandez & Condemine (2008).Ferrandez Y, Condemine G. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Molecular Microbiology. 2008;69(6):1349–1357. doi: 10.1111/j.1365-2958.2008.06366.x. [DOI] [PubMed] [Google Scholar]
- Gao et al. (2012).Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, Gao S, Liu X. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiology. 2012;12(1):143. doi: 10.1186/1471-2180-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijbels et al. (1990).Gijbels K, Van Damme J, Proost P, Put W, Carton H, Billiau A. Interleukin 6 production in the central nervous system during experimental autoimmune encephalomyelitis. European Journal of Immunology. 1990;20(1):233–235. doi: 10.1002/eji.1830200134. [DOI] [PubMed] [Google Scholar]
- Girbl et al. (2018).Girbl T, Lenn T, Perez L, Rolas L, Barkaway A, Thiriot A, Del Fresno C, Lynam E, Hub E, Thelen M, Graham G, Alon R, Sancho D, Von Andrian UH, Voisin MB, Rot A, Nourshargh S. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity. 2018;49(6):1062–1076.e6. doi: 10.1016/j.immuni.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, Sokol & Luster (2014).Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annual Review of Immunology. 2014;32(1):659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- Hanson & Brinton (1988).Hanson MS, Brinton CC., Jr Identification and characterization of E. coli type-1 pilus tip adhesion protein. Nature. 1988;332(6161):265–268. doi: 10.1038/332265a0. [DOI] [PubMed] [Google Scholar]
- Hejair et al. (2017).Hejair HMA, Zhu Y, Ma J, Zhang Y, Pan Z, Zhang W, Yao H. Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. Microbial Pathogenesis. 2017;107:29–37. doi: 10.1016/j.micpath.2017.02.033. [DOI] [PubMed] [Google Scholar]
- Hofnagel et al. (2011).Hofnagel O, Engel T, Severs NJ, Robenek H, Buers I. SR-PSOX at sites predisposed to atherosclerotic lesion formation mediates monocyte-endothelial cell adhesion. Atherosclerosis. 2011;217(2):371–378. doi: 10.1016/j.atherosclerosis.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Hornef et al. (2002).Hornef MW, Wick MJ, Rhen M, Normark S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nature Immunology. 2002;3(11):1033–1040. doi: 10.1038/ni1102-1033. [DOI] [PubMed] [Google Scholar]
- Hubner et al. (2018).Hubner K, Cabochette P, Dieguez-Hurtado R, Wiesner C, Wakayama Y, Grassme KS, Hubert M, Guenther S, Belting HG, Affolter M, Adams RH, Vanhollebeke B, Herzog W. Wnt/β-catenin signaling regulates VE-cadherin-mediated anastomosis of brain capillaries by counteracting S1pr1 signaling. Nature Communications. 2018;9(1):4860. doi: 10.1038/s41467-018-07302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson et al. (2006).Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. Journal of Bacteriology. 2006;188(2):745–758. doi: 10.1128/JB.188.2.745-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakpour, Wilhelmsen & Hellman (2015).Khakpour S, Wilhelmsen K, Hellman J. Vascular endothelial cell Toll-like receptor pathways in sepsis. Innate Immunity. 2015;21(8):827–846. doi: 10.1177/1753425915606525. [DOI] [PubMed] [Google Scholar]
- Kim (2002).Kim KS. Strategy of Escherichia coli for crossing the blood–brain barrier. Journal of Infectious Diseases. 2002;186(Suppl. 2):S220–S224. doi: 10.1086/344284. [DOI] [PubMed] [Google Scholar]
- Kim (2003a).Kim KS. Emerging molecular targets in the treatment of bacterial meningitis. Expert Opinion on Therapeutic Targets. 2003a;7(2):141–152. doi: 10.1517/14728222.7.2.141. [DOI] [PubMed] [Google Scholar]
- Kim (2003b).Kim KS. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nature Reviews Neuroscience. 2003b;4(5):376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- Kim (2008).Kim KS. Mechanisms of microbial traversal of the blood–brain barrier. Nature Reviews Microbiology. 2008;6(8):625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim et al. (2010).Kim M, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Sasakawa C. Bacterial interactions with the host epithelium. Cell Host & Microbe. 2010;8(1):20–35. doi: 10.1016/j.chom.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2015).Kim BJ, Hancock BM, Bermudez A, Del Cid N, Reyes E, Van Sorge NM, Lauth X, Smurthwaite CA, Hilton BJ, Stotland A, Banerjee A, Buchanan J, Wolkowicz R, Traver D, Doran KS. Bacterial induction of snail1 contributes to blood–brain barrier disruption. Journal of Clinical Investigation. 2015;125(6):2473–2483. doi: 10.1172/JCI74159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Kang & Kim (2005).Kim BY, Kang J, Kim KS. Invasion processes of pathogenic Escherichia coli. International Journal of Medical Microbiology. 2005;295(6–7):463–470. doi: 10.1016/j.ijmm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Klein & Hunter (2017).Klein RS, Hunter CA. Protective and pathological immunity during central nervous system infections. Immunity. 2017;46(6):891–909. doi: 10.1016/j.immuni.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm (1984).Klemm P. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli: nucleotide sequence and primary structure of the protein. European Journal of Biochemistry. 1984;143(2):395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- Krishnan et al. (2015).Krishnan S, Chang AC, Hodges J, Couraud PO, Romero IA, Weksler B, Nicholson BA, Nolan LK, Prasadarao NV. Serotype O18 avian pathogenic and neonatal meningitis Escherichia coli strains employ similar pathogenic strategies for the onset of meningitis. Virulence. 2015;6(8):777–786. doi: 10.1080/21505594.2015.1091914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Chen (2012).Li X-D, Chen ZJ. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife. 2012;1:e00102. doi: 10.7554/eLife.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2014).Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infection and Immunity. 2014;82(9):3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruvada & Kim (2012).Maruvada R, Kim KS. IbeA and OmpA of Escherichia coli K1 exploit Rac1 activation for invasion of human brain microvascular endothelial cells. Infection and Immunity. 2012;80(6):2035–2041. doi: 10.1128/IAI.06320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin et al. (2017).McLoughlin A, Rochfort KD, McDonnell CJ, Kerrigan SW, Cummins PM. Staphylococcus aureus-mediated blood–brain barrier injury: an in vitro human brain microvascular endothelial cell model. Cellular Microbiology. 2017;19(3):e12664. doi: 10.1111/cmi.12664. [DOI] [PubMed] [Google Scholar]
- Mellata, Johnson & Curtiss (2018).Mellata M, Johnson JR, Curtiss R., III Escherichia coli isolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses and Public Health. 2018;65(1):103–113. doi: 10.1111/zph.12376. [DOI] [PubMed] [Google Scholar]
- Meyer, Martin-Blondel & Liblau (2017).Meyer C, Martin-Blondel G, Liblau RS. Endothelial cells and lymphatics at the interface between the immune and central nervous systems: implications for multiple sclerosis. Current Opinion in Neurology. 2017;30(3):222–230. doi: 10.1097/WCO.0000000000000454. [DOI] [PubMed] [Google Scholar]
- Mitchell et al. (2015).Mitchell NM, Johnson JR, Johnston B, Curtiss R, III, Mellata M, Bjorkroth J. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Applied and Environmental Microbiology. 2015;81(3):1177–1187. doi: 10.1128/AEM.03524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki et al. (2008).Ohtsuki S, Yamaguchi H, Katsukura Y, Asashima T, Terasaki T. mRNA expression levels of tight junction protein genes in mouse brain capillary endothelial cells highly purified by magnetic cell sorting. Journal of Neurochemistry. 2008;104:147–154. doi: 10.1111/j.1471-4159.2007.05008.x. [DOI] [PubMed] [Google Scholar]
- Olive & Sassetti (2016).Olive AJ, Sassetti CM. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nature Reviews Microbiology. 2016;14(4):221–234. doi: 10.1038/nrmicro.2016.12. [DOI] [PubMed] [Google Scholar]
- Orsenigo et al. (2012).Orsenigo F, Giampietro C, Ferrari A, Corada M, Galaup A, Sigismund S, Ristagno G, Maddaluno L, Koh GY, Franco D, Kurtcuoglu V, Poulikakos D, Baluk P, McDonald D, Grazia Lampugnani M, Dejana E. Phosphorylation of VE-cadherin is modulated by haemodynamic forces and contributes to the regulation of vascular permeability in vivo. Nature Communications. 2012;3(1):1208. doi: 10.1038/ncomms2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, Ashton & Moncada (1988).Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333(6174):664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- Parthasarathy, Yao & Kim (2007).Parthasarathy G, Yao Y, Kim KS. Flagella promote Escherichia coli K1 association with and invasion of human brain microvascular endothelial cells. Infection and Immunity. 2007;75(6):2937–2945. doi: 10.1128/IAI.01543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peigne et al. (2009).Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Medigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. The plasmid of Escherichia coli strain S88 (O45: K1: H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infection and Immunity. 2009;77(6):2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasadarao, Wass & Kim (1997).Prasadarao NV, Wass CA, Kim KS. Identification and characterization of S fimbria-binding sialoglycoproteins on brain microvascular endothelial cells. Infection and Immunity. 1997;65(7):2852–2860. doi: 10.1128/IAI.65.7.2852-2860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Siek et al. (2005).Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, Nolan LK. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151(6):2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- Sander et al. (2011).Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474(7351):385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, Kiupakis & Reitzer (1998).Schneider BL, Kiupakis AK, Reitzer LJ. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. Journal of Bacteriology. 1998;180(16):4278–4286. doi: 10.1128/JB.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, Kossmann & Morganti-Kossmann (2010).Semple BD, Kossmann T, Morganti-Kossmann MC. Role of chemokines in CNS health and pathology: a focus on the CCL2/CCR2 and CXCL8/CXCR2 networks. Journal of Cerebral Blood Flow & Metabolism. 2010;30(3):459–473. doi: 10.1038/jcbfm.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, Beattie & Khan (2012).Siddiqui R, Beattie R, Khan NA. The role of the twin-arginine translocation pathway in Escherichia coli K1 pathogenicity in the African migratory locust, Locusta migratoria. FEMS Immunology & Medical Microbiology. 2012;64(2):162–168. doi: 10.1111/j.1574-695X.2011.00870.x. [DOI] [PubMed] [Google Scholar]
- Smarda & Smajs (1998).Smarda J, Smajs D. Colicins–exocellular lethal proteins of Escherichia coli. Folia Microbiologica. 1998;43(6):563–582. doi: 10.1007/BF02816372. [DOI] [PubMed] [Google Scholar]
- Stancik et al. (2002).Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. Journal of Bacteriology. 2002;184(15):4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos et al. (2000).Stathopoulos C, Hendrixson DR, Thanassi DG, Hultgren SJ, St. Geme III J W, Curtiss III R. Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: an evolving story. Microbes and Infection. 2000;2(9):1061–1072. doi: 10.1016/S1286-4579(00)01260-0. [DOI] [PubMed] [Google Scholar]
- Stones & Krachler (2015).Stones DH, Krachler A-M. Fatal attraction: how bacterial adhesins affect host signaling and what we can learn from them. International Journal of Molecular Sciences. 2015;16(2):2626–2640. doi: 10.3390/ijms16022626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, Lascolea & Neter (1982).Sullivan TD, Lascolea LJ, Neter E. Relationship between the magnitude of bacteremia in children and the clinical disease. Pediatrics. 1982;69:699. [PubMed] [Google Scholar]
- Taddei et al. (2008).Taddei A, Giampietro C, Conti A, Orsenigo F, Breviario F, Pirazzoli V, Potente M, Daly C, Dimmeler S, Dejana E. Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nature Cell Biology. 2008;10(8):923–934. doi: 10.1038/ncb1752. [DOI] [PubMed] [Google Scholar]
- Takeshita & Ransohoff (2012).Takeshita Y, Ransohoff RM. Inflammatory cell trafficking across the blood-brain barrier: chemokine regulation and in vitro models. Immunological Reviews. 2012;248(1):228–239. doi: 10.1111/j.1600-065X.2012.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaue et al. (2006).Talaue MT, Venketaraman V, Hazbon MH, Peteroy-Kelly M, Seth A, Colangeli R, Alland D, Connell ND. Arginine homeostasis in J774.1 macrophages in the context of Mycobacterium bovis BCG infection. Journal of Bacteriology. 2006;188(13):4830–4840. doi: 10.1128/JB.01687-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng et al. (2005).Teng C-H, Cai M, Shin S, Xie Y, Kim K-J, Khan N-A, Di Cello F, Kim K-S. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infection and Immunity. 2005;73(5):2923–2931. doi: 10.1128/IAI.73.5.2923-2931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz & Engelhardt (2015).Tietz S, Engelhardt B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. Journal of Cell Biology. 2015;209:493–506. doi: 10.1083/jcb.201412147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini et al. (2018).Ugolini M, Gerhard J, Burkert S, Jensen KJ, Georg P, Ebner F, Volkers SM, Thada S, Dietert K, Bauer L, Schafer A, Helbig ET, Opitz B, Kurth F, Sur S, Dittrich N, Gaddam S, Conrad ML, Benn CS, Blohm U, Gruber AD, Hutloff A, Hartmann S, Boekschoten MV, Muller M, Jungersen G, Schumann RR, Suttorp N, Sander LE. Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. Nature Immunology. 2018;19(4):386–396. doi: 10.1038/s41590-018-0068-4. [DOI] [PubMed] [Google Scholar]
- Van Der Voort et al. (2005).Van Der Voort R, Van Lieshout AW, Toonen LW, Sloetjes AW, Van Den Berg WB, Figdor CG, Radstake TR, Adema GJ. Elevated CXCL16 expression by synovial macrophages recruits memory T cells into rheumatoid joints. Arthritis and Rheumatism. 2005;52(5):1381–1391. doi: 10.1002/art.21004. [DOI] [PubMed] [Google Scholar]
- Westermann et al. (2016).Westermann AJ, Forstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Muller L, Reinhardt R, Stadler PF, Vogel J. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature. 2016;529(7587):496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- Westermann, Gorski & Vogel (2012).Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nature Reviews Microbiology. 2012;10(9):618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- Witcomb et al. (2015).Witcomb LA, Collins JW, McCarthy AJ, Frankel G, Taylor PW. Bioluminescent imaging reveals novel patterns of colonization and invasion in systemic Escherichia coli K1 experimental infection in the neonatal rat. Infection and Immunity. 2015;83(12):4528–4540. doi: 10.1128/IAI.00953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing et al. (2019).Xing Y, Zhang Y, Jia L, Xu X. Lipopolysaccharide from Escherichia coli stimulates osteogenic differentiation of human periodontal ligament stem cells through Wnt/β‐catenin–induced TAZ elevation. Molecular Oral Microbiology. 2019;34(1):699. doi: 10.1111/omi.12249. [DOI] [PubMed] [Google Scholar]
- Yau et al. (2016).Yau B, Mitchell AJ, Too LK, Ball HJ, Hunt NH. Interferon-γ-induced nitric oxide synthase-2 contributes to blood/brain barrier dysfunction and acute mortality in experimental Streptococcus pneumoniae meningitis. Journal of Interferon & Cytokine Research. 2016;36(2):86–99. doi: 10.1089/jir.2015.0078. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2013).Zhang K, Tian L, Liu L, Feng Y, Dong YB, Li B, Shang DS, Fang WG, Cao YP, Chen YH. CXCL1 contributes to beta-amyloid-induced transendothelial migration of monocytes in Alzheimer’s disease. PLOS ONE. 2013;8(8):e72744. doi: 10.1371/journal.pone.0072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2019).Zhang B, Zhuang Z, Wang X, Huang H, Fu Q, Yan Q. Dual RNA-Seq reveals the role of a transcriptional regulator gene in pathogen–host interactions between Pseudomonas plecoglossicida and Epinephelus coioides. Fish & Shellfish Immunol. 2019;87:778–787. doi: 10.1016/j.fsi.2019.02.025. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2015).Zhao Y, Wang S, Yang D, Liu X, Han X, Tian M, Ding C, Liu Z, Yu S. Vacuolating autotransporter toxin affects biological characteristics and pathogenicity of avian pathogenic Escherichia coli. Wei Sheng Wu Xue Bao. 2015;55:1208–1214. [PubMed] [Google Scholar]
- Zhou et al. (2012).Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, Kim KS, Zhao GP, Guo X, Yao Y. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infection and Immunity. 2012;80(3):1243–1251. doi: 10.1128/IAI.05994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Ge et al. (2014).Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, Wang H, Leung FC, Dai J, Fan H. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2: K1: H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1: K1 and human ExPEC O18: K1 strains. PLOS ONE. 2014;9(11):e112048. doi: 10.1371/journal.pone.0112048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Pearce & Kim (2010).Zhu L, Pearce D, Kim KS. Prevention of Escherichia coli K1 penetration of the blood–brain barrier by counteracting the host cell receptor and signaling molecule involved in E. coli invasion of human brain microvascular endothelial cells. Infection and Immunity. 2010;78(8):3554–3559. doi: 10.1128/IAI.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Adherence to and invasion of bEnd.3 cells by APEC strain (at MOI of 100). Data are means + standard errors of three independent experiments, each performed in triplicate. An asterisk indicates that the adherence and invasion values of others’ time points were significantly higher than the values of the first one hour at P-value < 0.01.
Data Availability Statement
The following information was supplied regarding data availability:
The APEC RNA-sequences and bEnd.3 cells RNA-sequences are available at SRA: (PRJNA612631) SRR11307869, SRR11307868, SRR11307867, SRR11307872, SRR11307871 and SRR11307870; (PRJNA612931) SRR11318238, SRR11318235, SRR11318236, SRR11318234, SRR11318233 and SRR11318237.
The data is also available at Zenodo: wang peili. (2019). Raw data of RNA-seq and IGV -2 [Data set]. Zenodo. DOI 10.5281/zenodo.3689240.
and
Wang peili. (2019). Raw data of RNA-seq and IGV -1 [Data set]. Zenodo. DOI 10.5281/zenodo.3672826.