Abstract
Doublecortin like kinase 1 (DCLK1) is an understudied kinase that is upregulated in a wide range of cancers, including pancreatic ductal adenocarcinoma (PDAC). However, little is known about its potential as a therapeutic target. We leveraged chemoproteomic profiling and structure-based design to develop the first selective, in vivo-compatible chemical probe of the DCLK1 kinase domain, DCLK1-IN-1. We demonstrate activity of DCLK1-IN-1 against clinically relevant patient-derived PDAC organoid models and use a combination of RNA sequencing, proteomics and phosphoproteomics analysis to reveal that DCLK1 inhibition modulates proteins and pathways associated with cell motility in this context. DCLK1-IN-1 will serve as a versatile tool to investigate DCLK1 biology and establish its role in cancer.
INTRODUCTION
Kinase target validation and drug discovery efforts are heavily biased towards proteins with well-established roles in cellular signaling.1 However, genome-wide cancer analyses have identified many understudied, highly druggable kinases as likely cancer driver genes.1 Doublecortin like kinase 1 (DCLK1) represents one of these poorly described targets.1
DCLK1 is a serine/threonine kinase that harbors two microtubule-binding domains (Supplementary Fig. 1a). DCLK1 overexpression has been reported in multiple human cancers, including colorectal cancer and gastric cancer.2 Of particular relevance is pancreatic ductal adenocarcinoma (PDAC), where its expression correlates negatively with PDAC patient lifespan.2,3 Unlike the majority of cancers, overall patient survival in PDAC has not improved over the last 40 years, underscoring a pivotal need to better understand the basic biology of PDAC and translate those findings into the development of new therapeutics.4 Genetic studies in murine models of KrasG12D-driven PDAC have shown that upregulation of a short isoform of DCLK1 which lacks the microtubule binding domains (DCLK1-S) in the pancreas occurs upon activation of oncogenic ERK pathway signaling, and indicate that the DCLK1+ cell population is required for tumor initiation and maintenance.2,5 Proteome-wide mass spectrometry analysis also recently identified significant DCLK1 upregulation during murine PDAC progression in KrasG12D driven tumors.6 Despite the potential importance of DCLK1 in PDAC and other cancers, no DCLK1-selective molecules are presently available to study the direct effects of DCLK1 kinase inhibition.7
In the absence of a bona fide chemical probe, researchers have turned to the multi-targeted kinase inhibitors LRRK2-IN-1 (1),8 XMD8–92 (2)9 and XMD8–85 (3)10 to inhibit DCLK1 (Supplementary Fig. 1b). These molecules were initially developed to target the unrelated kinases LRRK2 and ERK5, respectively, and later discovered to also inhibit BET bromodomains.11,12 LRRK2-IN-1 and XMD8–92 have been shown to reduce proliferation of human pancreatic cancer cell lines and murine KrasG12D pancreatic spheroids, and to exert cytostatic effects in murine xenograft models.13,5 However, the pleiotropic effects of BRD4 bromodomain inhibition14 and the abstruse role of ERK5 in oncogenesis15 confound interpretation of these results. This is consistent with reports indicating that mis-annotation of the targets of anti-cancer drugs is pervasive.16
Here we report the development of DCLK1-IN-1 (4) and its characterization as a potent, selective inhibitor of the DCLK1 and DCLK2 kinases. Treatment of human PDAC cell lines with DCLK1-IN-1 had minimal effects on both cell viability and gene expression. However, DCLK1-expressing patient-derived PDAC organoids were sensitive to DCLK1 inhibition, indicating a context specific role for DCLK1 in sustaining human cancer cell viability. Analysis of changes in the transcriptome and proteome that occur in these organoids upon treatment with DCLK1-IN-1 revealed enrichment of gene signatures associated with cell motility. DCLK1-IN-1 is suitable for use as a chemical probe to investigate the role of DCLK1-mediated signaling in cell-based assays and in vivo.
RESULTS
Development of a potent DCLK1 inhibitor, DCLK1-IN-1
To discover a selective DCLK1 inhibitor, we utilized a highly parallelized chemoproteomic strategy.17,18 We screened a 350 member library of 5,11-dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-one analogs against the kinome in cell lysates using KiNativ technology to generate multicomponent SAR.18 To guide our medicinal chemistry campaign, we docked the potent, multi-targeted DCLK1 inhibitor XMD8–85 into the X-ray crystal structure of the DCLK1 kinase domain (PDB:5JZN).19 Iterative rounds of structure-based design, synthesis, and testing culminated in the identification of DCLK1-IN-1 (Fig. 1a). In this study, we observed that the diazipinone amide substituent (R1) is a crucial determinant of selectivity (Supplementary Table 1). Removal of this group (R1 = H, XMD8–87, 5) was not tolerated by DCLK1, ERK5 or BRD4, and led to a modest reduction in potency for LRRK2 relative to XMD8–85. Substitution of the R1 methyl with an ethyl group (FMF-03–055-1, 6) was well-tolerated by DCLK1 and LRRK2 but resulted in greater than 10-fold reduction in affinity for ERK5. Further increase of R1 to a more rigid and bulky isopropyl group resulted in an unacceptable loss of DCLK1 activity (FMF-03–149-1, 7). Crucially, altering the polarity of the R1 group, from a hydrophobic ethyl to an electronegative trifluoroethyl group (DCLK1-IN-1) maintained DCLK1 activity, led to a dramatic improvement in selectivity against ACK, LRRK2 and BRD4 and improved selectivity against ERK5 relative to R1 = ethyl (FMF-03–055-1). In parallel, a structurally related negative control compound, DCLK1-NEG (8), was developed to aid interpretation of novel pharmacology uncovered by DCLK1-IN-1.
Fig. 1 |. DCLK1-IN-1 is a potent DCLK1/2 inhibitor.
(a) Chemical structure of DCLK1 inhibitor DCLK1-IN-1 and negative control DCLK1-NEG. (b) ITC thermograms showing titration of DCLK1-IN-1 and DCLK1-NEG into purified, recombinant DCLK1 kinase domain. Data are representative of n = 2 experiments per compound. Integrals shown with error bars depicting error estimates of peak integration, calculated according to ref. 39. (c-d) DMSO-normalized results of a 33P-labeled ATP DCLK1 kinase assay (c) and DCLK2 kinase assay (d). Assays were performed at an ATP concentration of 50 μM (DCLK1) or 100 μM (DCLK2) (Km). Data in c-d are presented as mean ± S.D. of n = 3 biologically independent samples. (e) Schematic of the DCLK1 NanoBRET assay. (f) NanoBRET evaluation of HCT116 cells expressing DCLK1-Nluc upon treatment with the indicated compounds. Data are presented as mean ± S.D. of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
The ability of DCLK1-IN-1 to inhibit DCLK1 was comprehensively assessed in orthogonal assays. DCLK1-IN-1 bound recombinant DCLK1 protein with a Kd = 109 nM in isothermal titration calorimetry (ITC) experiments (Fig. 1b, Table 1 and Supplementary Table 2). This was further confirmed using KINOMEscan binding assays20 (IC50 = 9.5 nM, Table 1) and in a kinase assay utilizing 33P-labeled ATP (IC50 = 57 nM at 50 μM ATP concentration) (Fig. 1c, Table 1, and Supplementary Fig. 2a). Inhibition of DCLK2, a close homolog of DCLK1 expressed predominantly in the brain,21 was also confirmed using both binding and kinase assays (IC50 = 31 nM, 103 nM) (Fig. 1d, Table 1 and Supplementary Fig. 2b). In addition, a biotinylated analog of DCLK1-IN-1 (FMF-04–084-1, 9) captured native DCLK1 protein from PATU-8988T cell lysates in a pulldown assay (Supplementary Fig. 3a–c). In order to assess cellular target engagement in dose-response, we synthesized a NanoBRET tracer (TAE684-NanoBRET-590, 10) and developed a DCLK1 NanoBRET assay (Fig. 1e, Supplementary Fig. 4a–e).22 This assay confirmed that DCLK1-IN-1 potently binds DCLK1 in HCT116 cells (IC50 = 279 nM, Table 1, Fig. 1f). In all assays tested, DCLK1-NEG displayed at least 100-fold lower activity against DCLK1 (Fig. 1, Table 1, and Supplementary Fig. 2–4).
Table 1 |.
DCLK1-IN-1 is an exquisitely selective inhibitor of DCLK1.
| Target | Assay (nM) | DCLK1-IN-1 (4) | DCLK1-NEG (9) | XMD8-92 (2) | LRRK2-IN-1 (1) | JQ1 | Trametinib |
|---|---|---|---|---|---|---|---|
| DCLK1 | ITC Kda | 55 | > 1000 | - | - | - | - |
| DCLK1 | binding assay IC50b | 9.5 | 2700 | 1400 | 3 | - | - |
| DCLK1 | kinase assay IC50c | 57.2 | > 10000 | 104 | 55 | - | - |
| DCLK1 | cellular assay IC50d | 279 | > 10000 | 1150 | 400 | - | - |
| DCLK2 | binding assay IC50e | 31 | 1400 | 3600 | 9.9 | - | - |
| DCLK2 | kinase assay IC50f | 103 | > 10000 | 385 | 45 | - | - |
| ERK5 | binding assay IC50g | 1800 | > 10000 | 1600 | 21 | - | - |
| ERK5 | kinase assay IC50h | 4000 | > 10000 | 364 | 160 | - | - |
| ERK5 | cellular assay IC50i | 5360 | > 10000 | 240 | 114 | - | - |
| LRRK2 | kinase assay IC50j | 6970 | > 10000 | 36 | 4.1 | - | - |
| BRD4 | binding assay IC50k | > 10000 | > 10000 | 1566 | 1347 | 37 | - |
| PATU-8988T cells | 2D monolayer culture IC50l | > 20000 | > 20000 | 2015 | 1151 | 67.4 | 60 (P.R.) |
| PATU-8988T cells | 3D spheroid culture IC50m | > 20000 | > 20000 | 6062 | 1866 | 111.2 | 1 |
| PATU-8902 cells | 2D monolayer culture IC50l | > 20000 | > 20000 | > 20000 | > 20000 | 1814 | 4.89 |
| PATU-8902 cells | 3D spheroid culture IC50m | > 20000 | > 20000 | > 20000 | > 1000 (P.R.) | > 1000 (P.R.) | < 0.63 |
| PANFR0172_T2 | 3D organoid culture IC50n | 877 | 8340 | > 20000 | 1340 | > 1000 (P.R.) | 791 |
| PANFR0172_T3 | 3D organoid culture IC50n | 2120 | > 20000 | > 20000 | 1500 | > 1000 (P.R.) | 1180 |
| PANFR0172_T4 | 3D organoid culture IC50n | > 20000 | > 20000 | > 20000 | > 20000 | > 1000 (P.R.) | > 20000 |
| PANFR0029_T2 | 3D organoid culture IC50n | > 20000 | > 20000 | 7030 | 1040 | 363 | 185 |
ITC data against purified recombinant DCLK1 protein. Data are representative of n = 2 independent experiments.
DCLK1 KINOMEscan binding assay. Data are presented as mean of n = 2 biologically independent samples
33P-ATP DCLK1 kinase assay. Assays were performed at an ATP concentration of 50 μM (Km). Data are presented as mean of n = 3 biologically independent samples.
DCLK1 nanoBRET assay. Data are presented as mean of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
DCLK2 KINOMEscan binding assay. Data are presented as mean of n = 2 biologically independent samples (f) 33P-ATP DCLK2 kinase assay. Assays were performed at an ATP concentration of 100 μM (Km). Data are presented as mean of n = 3 biologically independent samples.
ERK5 KINOMEscan binding assay. Data are presented as mean of n = 2 biologically independent samples and
ERK5 32P-ATP kinase assay. Data are presented as mean of n = 3 biologically independent samples and representative of n = 2 independent experiments.
ERK5 cellular kinase assay. Data are presented as mean of n = 2 biologically independent samples and are representative of n = 2 independent experiments.
LRRK2 ADAPTA kinase assay. Data are presented as mean of n = 2 biologically independent samples.
BRD4 AlphaScreen assay. Data are presented as the mean of n = 3 biologically independent samples and representative of n = 2 independent experiments.
Analysis of cell viability in 2D-adherent cultures (l) and ultra-low adherent 3D-spheroid suspensions (m). Data in l and m are presented as mean of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
Analysis of patient-derived pancreatic organoid viability. Data are presented as mean of n = 3 biologically independent samples and are representative of n = 2 independent experiments.
P.R., partial response.
DCLK1-IN-1 is a highly selective DCLK1/2 inhibitor
To determine the selectivity of DCLK1-IN-1 and DCLK1-NEG, we performed KINOMEscan profiling at a concentration of 1 μM to evaluate in vitro kinase binding across a panel of 489 human kinases. DCLK1-IN-1 exclusively inhibited DCLK1 and DCLK2 to below 10% of the control signal, while DCLK1-NEG did not inhibit any kinases using this cutoff (Supplementary Fig. 5 and Supplementary Dataset 1). To further evaluate the kinome profile of DCLK1-IN-1 and DCLK1-NEG in a KRASG12V-driven PDAC context, and to further verify cellular target engagement, we performed activity-based proteomic profiling against 250 kinases using KiNativ.18 DCLK1-IN-1 significantly inhibited DCLK1, and weakly inhibited ERK5, in PATU-8988T cell lysates and live cells (Fig. 2a and Supplementary Dataset 2). DCLK1-NEG did not inhibit any kinases under these conditions (Fig. 2b and Supplementary Dataset 2). To confirm that activity against known off-targets of the benzopyrimido-diazipinone scaffold had been abrogated, we further tested DCLK1-IN-1 in dose response against ERK5, ACK, LRRK2, and BRD4 (Table 1 and Supplementary Fig. 2c–h). Overall, DCLK1-IN-1 showed no significant activity against the three kinases and no measurable activity for BRD4, confirming the pronounced selectivity of DCLK1-IN-1 (Table 1 and Supplementary Fig. 2c–h).
Fig. 2 |. DCLK1-IN-1 engages DCLK1 potently and selectively in cells.
(a-b) KiNativ assay profiling in PATU-8988T cell lysates treated with 2.5 μM DCLK1-IN-1 (a) or DCLK1-NEG (b). The % inhibition represented with circles are 68.5 (DCLK1, red) and 37.9 (ERK5, pink). Data in a-b are presented as the mean of n = 3 technical replicates and associated datasets are provided in Supplementary Dataset 2. (c) Docking model of DCLK1-IN-1 into the X-ray co-crystal structure of the DCLK1 kinase domain (PDB:5JZN). DCLK1 main chain shown grey carbons, DCLK1-IN-1 shown pink carbons. Hydrogen bonds shown as black dashed lines.
We next employed docking models to evaluate modes of interaction that contribute to selectivity of DCLK1-IN-1. First, to map the binding interactions between DCLK1-IN-1 and its target, we docked DCLK1-IN-1 into the X-ray crystal structure of the DCLK1 kinase domain (PDB:5JZN, Fig. 2c). In the docking model, DCLK1-IN-1 adopts a type 1 kinase inhibitor binding mode, with the aminopyridine moiety making two hydrogen bonds to the hinge V468 residue backbone. The trifluoro-ethyl group orients towards the hydrophobic back pocket, and a putative fluorine-sulfur contact is observed with the sulfur of the gatekeeper residue (M465).23 Second, we compared our DCLK1 docking structures to the structure of ERK5 in complex with XMD8–92 (PDB:5BYY)24 to uncover the structural basis of DCLK1-IN-1 selectivity. The residues lining the pocket of the ATP-binding site of the two proteins are highly conserved, with the exception of the gatekeeper. The DCLK1 gatekeeper methionine (M465) is in direct contact with the amide substituent in DCLK1-IN-1, while the ERK5 gatekeeper is a more rigid leucine (L137), which may be unable to accommodate the larger (2,2,2)trifluoroethyl group in DCLK1-IN-1. Additionally, the hydrocarbon L137 side chain in ERK5 would be unable to make a sulfur-fluorine interaction with the CF3 group of DCLK1-IN-1. These observations are consistent with the observed SAR (Supplementary Table 1).23 Third, to rationalize the selectivity of DCLK1-IN-1 over BRD4, we examined the structure of XMD8–92 bound to BRD4 (PDB:5LRQ). The N-methyl amide group in the scaffold of XMD8–92 acts as an acetyl-lysine (Kac) mimetic, binding in the hydrophobic Kac binding site of BRD4. The base of this pocket is occupied by six tightly bound water molecules, and thus the N-(2,2,2)trifluoroethyl amide moiety in DCLK1-IN-1 is likely too bulky and polar to be accommodated by BRD4. Collectively, our structure-based analyses and SAR studies suggest that the size and polarity of the N-(2,2,2)trifluoroethyl amide moiety is a major determinant of the improved selectivity of DCLK1-IN-1.
DCLK1-IN-1 has favorable pharmacokinetic properties and low toxicity
To finalize our chemical probe characterization, we assessed the suitability of DCLK1-IN-1 for use in vivo. DCLK1-IN-1 has a favorable pharmacokinetic profile in mice, with a half-life of 2.09 h, an area under the curve (AUCinf) of 5506 hr*ng mL−1 and 81% oral bioavailability (Supplementary Fig. 6a–c). As DCLK1 has a reported role in neuronal development, DCLK1-IN-1 was tested for effects on zebrafish development and neuronal viability in rat hippocampal neurons. DCLK1-IN-1 had no discernable effect on zebrafish CNS or other organ system development from days 1 to 8 post-fertilization when added to the fish water at concentrations up to 10 μM (Supplementary Fig. 7a–b). Similarly, it had no effect on neuronal viability in rat hippocampal neurons when administered at the same concentrations (Supplementary Fig. 7c). We also evaluated the maximum tolerated dose in mice and observed that DCLK1-IN-1 was well-tolerated at doses up to 100 mg/kg with no adverse effects and no loss of body weight observed (Supplementary Fig. 7d). Taken together, these data demonstrate that DCLK1-IN-1 is an exquisitely selective inhibitor of DCLK1, suitable for use as a chemical probe when used in concert with control compound DCLK1-NEG.
Aberrant KRAS/ERK signaling controls DCLK1 expression
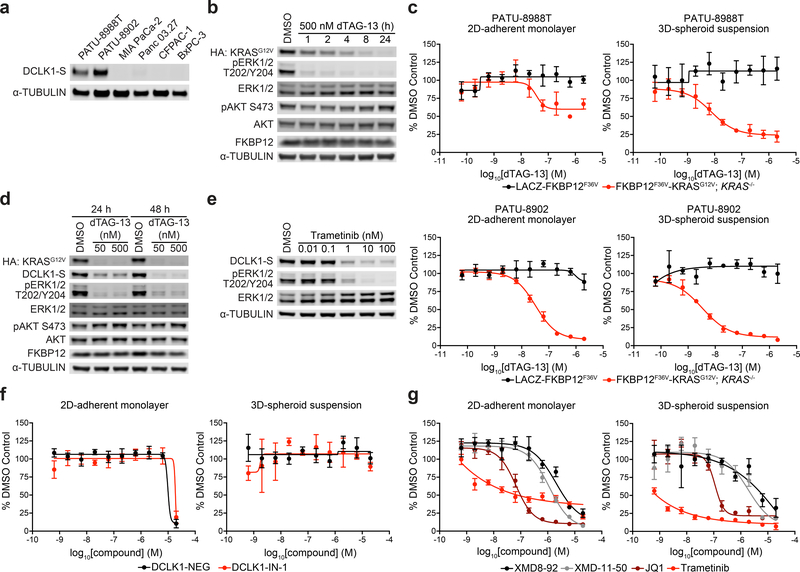
We next focused on employing DCLK1-IN-1 and DCLK1-NEG to better understand the role of this understudied kinase in human PDAC pathophysiology. We first confirmed the reported upregulation of DCLK1-S protein in KrasLSL-G12D/+ and KrasLSL-G12D/+; p53LSL-R172H/+ murine models (Supplementary Fig. 8a–b).2, 5 To identify suitable human PDAC cell lines to explore DCLK1 biology, we examined the Cancer Cell Line Encyclopedia (CCLE)25 and selected PATU-8988T and PATU-8902, which both express a short isoform of DCLK1 (uniprot isoforms O15075–3, −4) and harbor a KRASG12V mutation (Fig. 3a and Supplementary Fig. 9).25 To confirm their relevance as model systems for functional studies of DCLK1, we evaluated if the interdependence of aberrant ERK signaling pathway activation and DCLK1 expression status observed in murine models exists in PATU-8988T and PATU-8902 cell lines. We previously developed the degradation tag (dTAG) system to rapidly and selectively degrade FKBP12F36V-fusion proteins using a heterobifunctional degrader molecule, such as dTAG-13, that recruits cereblon, an E3 ubiquitin ligase.26 We employed the dTAG system to degrade mutant KRAS in isogenic PATU-8988T and PATU-8902 cell lines. We simultaneously expressed FKBP12F36V-KRASG12V, a degradable, functional KRASG12V fusion26 and Cas9/sgKRAS, which disrupts endogenous KRAS using CRISPR/Cas9 gene-editing while sparing ectopically expressed KRASG12V (referred to as FKBP12F36V-KRASG12V; KRAS−/− Supplementary Fig. 10a–d). To generate parallel control cell lines, we concurrently expressed LACZ-FKBP12F36V and Cas9/sgGFP (referred to as LACZ-FKBP12F36V, Supplementary Fig. 10a–d).
Fig. 3 |. DCLK1 is a target of aberrant KRAS-ERK signaling and is dispensable in DCLK1+ PDAC cell lines.
(a) Immunoblot analysis of the indicated cell lines. (b) Immunoblot analysis of PATU-8988T FKBP12F36V-KRASG12V; KRAS−/− clone treated with DMSO or dTAG-13 for the indicated time-course. Data in a-b are representative of n = 3 independent experiments. Uncropped immunoblots for a-b are displayed in Supplementary Fig. 18. (c) DMSO-normalized antiproliferation of PATU-8988T (top) or PATU-8902 (bottom) LACZ-FKBP12F36V or FKBP12F36V-KRASG12V; KRAS−/− clones treated with DMSO or dTAG-13 for 120 h. Cells were cultured as 2D-adherent monolayers or as ultra-low adherent 3D-spheroid suspensions. Data are presented as mean ± S.D. of n = 4 biologically independent samples and are representative of n = 3 independent experiments. (d) Immunoblot analysis of PATU-8988T FKBP12F36V-KRASG12V; KRAS−/− clone treated with DMSO or dTAG-13 for the indicated time-course. (e) Immunoblot analysis of PATU-8988T cells treated with DMSO or trametinib for 48 h. Data in d-e are representative of n = 3 independent experiments. Uncropped immunoblots for d are displayed in Supplementary Fig. 18 and uncropped immunoblots for e are displayed in Supplementary Fig. 19. (f-g) DMSO-normalized antiproliferation of PATU-8988T cells treated with the indicated compounds for 120 h. Cells were cultured as 2D-adherent monolayers or as ultra-low adherent 3D-spheroid suspensions. Data in f-g are presented as mean ± S.D. of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
Treatment of FKBP12F36V-KRASG12V; KRAS−/− cells with dTAG-13 initiated KRASG12V degradation within 1 h, with a concomitant rapid collapse in ERK signaling (Fig. 3b and Supplementary Fig. 11a–c). Proliferation was also robustly diminished in 2D-adherent monolayers and ultra-low attachment (ULA) 3D-spheroid suspensions (Fig. 3c and Supplementary Fig. 12a–b). These results are consistent with recent studies demonstrating more pronounced KRAS-dependence in 3D-spheroid suspensions27 and the increased KRAS-dependence of PATU-8902 cells compared to PATU-8988T cells.28 No toxicity or altered signaling was observed upon dTAG-13 treatment in LACZ-FKBP12F36V control cells (Fig. 3c and Supplementary Fig. 11a–b and 12a–b). Both prolonged degradation of FKBP12F36V-KRASG12V and MEK1/2 inhibition with trametinib (GSK112021)29 led to diminished DCLK1 protein levels after 48 h (Fig. 3d–e and Supplementary Fig. 13a–b). Washout of dTAG-13 led to recovery of FKBP12F36V-KRASG12V, ERK signaling and DCLK1 protein (Supplementary Fig. 13c). We note that the observed interplay between ERK signaling and DCLK1 expression may be important for only a subset of human PDAC cell lines. Although the majority of human PDAC cell lines possess KRAS mutations, only 5 out of 41 listed in the CCLE express DCLK1 transcript (Supplementary Fig. 9). Concordant with the CCLE, we detected DCLK1 protein in 2 out of 7 human PDAC cell lines tested (Fig. 3a). These data indicate that additional mechanisms may overcome aberrant activation of ERK signaling to suppress DCLK1 expression. Together, these data support the observation that aberrant ERK signaling pathway activation controls DCLK1 protein levels in a subset of human PDAC cells and provide motivation for studying DCLK1-kinase function in this population of human DCLK1+ PDAC.
Context-specific viability effects are observed upon DCLK1 inhibition in PDAC models
To evaluate the effects of DCLK1 kinase inhibition on proliferation, we treated PATU-8988T and PATU-8902 cells with a panel of inhibitors. DCLK1-IN-1 did not impact proliferation in 2D-adherent monolayers or ULA 3D-spheroid suspensions, while MEK1/2 inhibition (trametinib), BET bromodomain inhibition (JQ1)30, and pan-ERK5/LRRK2/BRD4/DCLK1 inhibition (XMD8–92 and LRRK2-IN-1) had potent effects (Fig. 3f–g, Table 1 and Supplementary Fig. 14a–b). Furthermore, treatment of PATU-8988T FKBP12F36V-KRASG12V; KRAS−/− cells with DCLK1-IN-1 after treatment and washout of dTAG-13 did not impede KRASG12V-mediated 3D-spheroid formation (Supplementary Fig. 15a–b). Taken together, this data indicates that DCLK1 kinase activity is not essential for the viability or transformation of these human PDAC cell lines. These results are consistent with data from large-scale RNAi experiments, such as Project DRIVE31 and the Cancer Dependency Map,32 which do not detect DCLK1 as a dependency for PDAC cell line proliferation, but are at odds with reported murine in vivo DCLK1 knockout studies and clinical observations.2,5
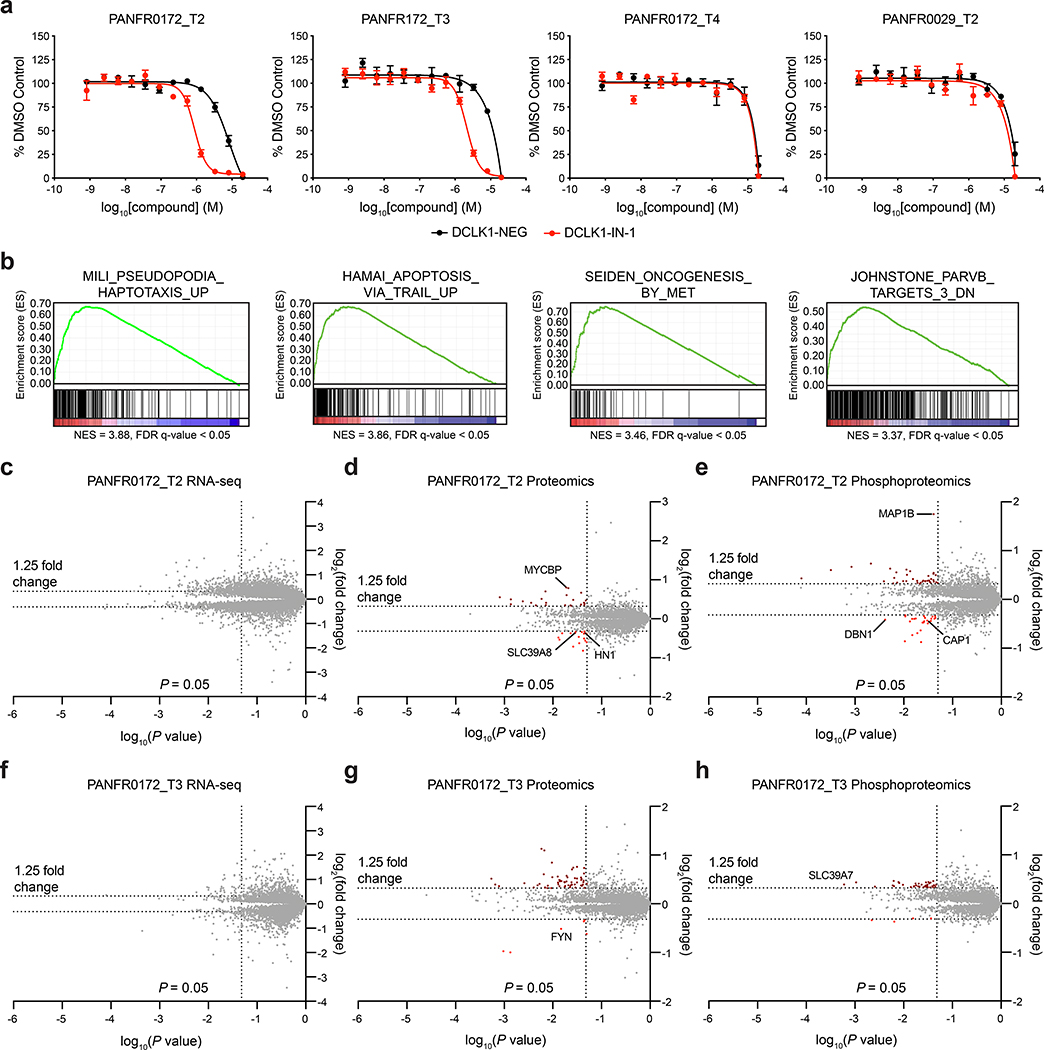
We hypothesized that the cell line cultures were not fully recapitulating PDAC disease pathology. Therefore, we tested if DCLK1 inhibition would alter the proliferation of patient-derived organoids, a culture system reported to preserve the molecular heterogeneity and patient-specific therapeutic sensitivity of human PDAC tumors.33, 34 Similar to PDAC cell lines, only a subset of human patient-derived PDAC organoids expressed detectable levels of DCLK1 (3 of 16 organoids tested, uniprot isoforms O15075–3, −4). DCLK1+ organoid cultures employed for further evaluation were derived from a chemotherapy-resistant patient with metastatic disease34 (Supplementary Fig. 16a). Three distinct specimens were taken from this patient: PANFR0172_T2 was derived from a biopsy of a metastatic liver lesion after development of resistance to FOLFIRINOX; PANFR0172_T3 was grown from a biopsy of a metastatic liver lesion obtained one month after the PANFR0172_T2 specimen and while the patient was on treatment with trametinib; and PANFR0172_T4 was derived from ascites fluid obtained approximately 6 months after the prior biopsies and after the patient had developed resistance to gemcitabine/nab-paclitaxel chemotherapy. This patient’s tumor expressed wild-type KRAS but contained an oncogenic BRAF N486_P450del mutation (Vemurafenib-resistant)35 that likely activates MAPK and ERK signaling. We tested a panel of inhibitors across this series. The organoid samples isolated earlier in disease progression (PANFR0172_T2 and PANFR0172_T3) were sensitive to DCLK1-IN-1, inducing antiproliferative effects with comparable potency to trametinib and JQ1 (Fig. 4a, Table 1 and Supplementary Fig. 16b). However, the most advanced organoid specimen (PANFR0172_T4) was resistant (Fig. 4a and Table 1). The antiproliferative effects in PANFR0172_T2 and PANFR0172_T3 were abrogated with DCLK1-NEG. In addition, DCLK1-negative organoids derived from a primary resection specimen with KRASG12D mutation from an unrelated patient (PANFR0029_T2) were not differentially sensitive to DCLK1-IN-1 and DCLK1-NEG (Fig. 4a and Table 1). Taken together these data suggest that the effects of DCLK1-IN-1 are on target, and that DCLK1 is an exploitable vulnerability in a subset of human PDAC. At late stages of treatment resistance, additional mechanisms may be activated to confer broad small molecule drug resistance (Supplemental Fig. 16b).
Figure 4. |. DCLK1 is a vulnerability in DCLK1+ patient-derived organoids.
(a) DMSO-normalized antiproliferation of patient-derived organoid cultures treated with the indicated compounds for 7 d. Data are presented as mean ± S.D. of n = 3 biologically independent samples and are representative of n = 2 independent experiments. (b) GSEA signatures upon assessment of highly expressed (FPKM > 10) transcripts after treatment of PANFR0172_T2 organoids with 2.5 μM DCLK1-IN-1 for 24 h. (c-h) Transcript (c, f), protein (d, g) and phosphoprotein (e, h) abundance after treatment of PANFR0172_T2 (c-e) or PANFR0172_T3 (f-h) patient-derived organoid cultures with 2.5 μM DCLK1-IN-1 for 24 h compared to DMSO treatment. Volcano plots depict fold change abundance relative to DMSO versus P value. P value for c, f is derived from a two-tailed Welch’s t-test. P value for d, e, g and h is derived from a two-tailed Student’s t-test. Data in b-h are from n = 3 biologically independent samples and associated datasets are provided in Supplementary Datasets 3–5.
Global profiling identifies DCLK1-mediated changes in patient-derived organoids
In order to identify potential mechanisms that contribute to the observed sensitivity of the DCLK1+ organoids, we performed RNA-sequencing and multiplexed quantitative mass spectrometry-based proteomics and phosphoproteomics on sensitive organoids PANFR0172_T2 and PANFR0172_T3 treated with 2.5 μM DCLK1-IN-1 for 24 h. To identify potential discrepancies between organoids and cell line sensitivity, we also performed these assessments in the insensitive PATU-8988T cell line (Fig. 4b–h, Supplementary Fig. 17a–d and Supplementary Datasets 3–5). Modest changes in the transcriptomes were observed in all cell types upon DCLK1-IN-1 treatment, indicating that DCLK1 does not play a direct role in modulating transcription (Fig. 4c,f and Supplementary Fig. 17b). Gene Set Enrichment Analysis (GSEA)36 of the downregulated genes in both sensitive organoid cultures identified haptotaxis and PARVB signatures, indicating that DCLK1 phosphosignaling may regulate cell motility in this context (Fig. 4b and Supplementary Fig. 17a). These signatures were not identified in the PATU-8988T cell line data. Consistent with the observed effects on cell viability, GSEA also identified apoptosis and MET-driven oncogenesis signatures in the most sensitive organoid sample PANFR0172_T2, but not in PANFR0172_T3 or PATU-8988T cells (Fig. 4b).
In line with the RNA-sequencing data, mild effects on the total protein levels were observed by global proteomics analysis upon treatment with DCLK1-IN-1 (Fig. 4d,g and Supplementary Fig. 17c). However, a few proteins associated with cell motility, including HN1, ZIP8 and FYN, were significantly downregulated in the organoid cultures (Fig. 4d,g). Moreover, phosphorylation of the actin binding proteins CAP1 and DBN1 were also significantly reduced by DCLK1-IN-1 treatment in the PANFR0172_T2 organoid in line with the RNA-sequencing results of the potential modulation of motility pathways upon DCLK1-IN1 treatment (Fig. 4e).
DISCUSSION
We report the discovery of DCLK1-IN-1, a new potent, selective chemical probe for the DCLK1 kinase using chemoproteomic profiling and structure-guided inhibitor design. Through development of target engagement assays including nanoBRET and pulldown probes and a negative control compound, DCLK1-NEG, we demonstrate that DCLK1-IN-1 displays pronounced on-target DCLK1 activity in vitro. Toxicity and pharmacokinetic studies further highlight that DCLK1 is suitable probe for in vivo evaluation. These assays and chemical tools enable comprehensive investigation of the precise roles of DCLK1 kinase function.
In this study, we used DCLK1-IN-1 to investigate the roles of DCLK1 in PDAC. We validated that DCLK1 expression in human PDAC is downstream of aberrant ERK signaling, and establish that DCLK1 is an exploitable vulnerability in a subset of patient-derived chemorefractory DCLK1+ PDAC organoids. The observed organoid sensitivity to DCLK1 inhibition is in agreement with the reported role of DCLK1 in promoting tumorigenesis in murine models of PDAC.5 Analysis of the effects of DCLK1 inhibition on phosphosignaling and the transcriptome revealed a putative role for DCLK1 in regulating cell motility in PDAC. While modest changes were evident in the global profiling measurements, further characterization is necessary to identify bona fide DCLK1 substrates and target genes. The observations in patient-derived organoids are also in contrast with the PDAC cell line results, where no sensitivity was observed. These data highlight fundamental differences between the two cell culture systems, and suggest that organoids better model the contributions of DCLK1 to tumorigenesis, potentially by capturing the reported DCLK1+ PDAC progenitor cells.5, 37 To address this discrepancy, further evaluation of the consequences of DCLK1 inhibition in in vivo models of PDAC is necessary to delineate of the effects of DCLK1 inhibition on diverse cell types, identify sensitizers to DCLK1 inhibition, and to validate DCLK1 as a therapeutic target in PDAC.
In addition to its functions in PDAC, DCLK1 in the intestinal epithelia is a marker of tuft cells, chemosensory cells that detect pathogens, regulate the type II immune response, and mediate tissue regeneration following injury.38 However, the role of DCLK1 in tuft cells is poorly described and understanding of DCLK1-mediated signaling in the contexts of the gut immunological niche and regenerative biology is lacking. The suite of assays, tools and datasets provided in this study provide an opportunity to expand the current understanding of DCLK1 function, and enable further studies into the described functions of DCLK1 in tumor biology, and further probing of its role in tuft-cell mediated type II immunity and tissue regeneration.
ONLINE METHODS
33P-ATP DCLK1 and DCLK2 kinase assay
This assay was performed by Reaction Biology Corporation according to standard methods with minor modifications to the reported protocol. Briefly, compounds were tested in 12-point dose response, at a maximum concentration of 10 μM. Assays were performed at an ATP concentration of 50 μM (Km) DCLK1 and 100 μM (Km) DCLK2.
DCLK1 Isothermal Titration Calorimetry (ITC) assay
Plasmid Information
The DNA construct consisting of N-terminally 6-His tagged human DCLK1 residues G351-H689 was obtained from Ana Clara Redondo of the Structural Genomics Consortium at the University of Oxford. The plasmid was co-transformed with lambda phosphatase under chloramphenicol selection into BL21 DE3 E.coli cells.
Protein Purification
DCLK1 protein expression was induced with 0.6 mM IPTG and expression was allowed to continue for 10 h at 18 °C. Bacteria was harvested by centrifugation and resuspended in lysis buffer (noted below) with protease inhibitors (1 mM Benzamidine and 1 mM PMSF). Lysis was performed by passing 3 times through a homogenizer. Lysate was centrifuged at 20,000 x g for 1 hour and the supernatant was filtered through a 0.2 μm membrane. Protein was captured using Nickle-NTA resin and eluted with imidazole. Eluate was concentrated to 2 mL and passed over a Superdex S200 column. Buffers used for purification: lysis buffer (50 mM Hepes pH 7.8, 350 mM NaCl, 20 mM imidazole, 5% glycerol, wash 1 (lysis buffer), wash 2 (lysis buffer with 25 mM Imidazole), elution buffer (lysis buffer with 300mM Imidazole), and S200 gel filtration buffer (10 mM Hepes pH 7.8, 700 mM NaCl, 1 mM, MgCl2, 5% glycerol).
Assay conditions
Experiments were performed using the iTC200 system (GE Healthcare) at 20 °C with titration buffer (10 mM Hepes pH 7.8, 700 mM NaCl, 1 mM MgCl2, 5% glycerol, and 1 mM DTT). 20 μM purified DCLK1 protein in 400 μL of titration buffer was placed in the sample cell, and 200 μM compound in 40 μl of titration buffer was loaded into the injection syringe. A 120 second delay at the start of the experiment was followed by 20 injections with 120 second intervals. All measured samples were stirred at 500 rpm. The data were analyzed in SEDPHAT and exported in GUSSI.
DCLK1 mobility shift assay
DCLK1 kinase activity was measured in vitro using an electrophoretic mobility shift assay. The reaction was assembled in a 384-well plate in a total volume of 20 μL. The reaction comprised 30 nM recombinant DCLK1, DMSO or inhibitors, 100 μM ATP and 1 μM FAM-labeled peptide substrate in a buffer (100 mM HEPES pH 7.5, 0.003% Brij-35, 0.004% Tween-20, 10 mM MgCl2, and 2 mM DTT). The peptide substrate employed in this assay was 5-FAM-KKLRRTLSVA-COOH. Inhibitors were dispensed using a Labcyte Echo liquid handler. The reaction was incubated at room temperature for 2 h and quenched by addition of 40 μL of termination buffer (100 mM HEPES pH 7.3, 0.015% Brij-35, 0.1% CR-3, 1 x CR-8, and 40 mM EDTA). Substrate and product peptides present in each sample were electrophoretically separated and detected using 12-channel LabChip3000 microfluidic capillary electrophoresis instrument (Caliper Life Sciences). The change in the relative fluorescence intensities of substrate and product peaks (reflecting enzyme activity) was measured. Capillary electrophoregrams were analyzed using HTS Well Analyzer software (Caliper Life Sciences). The kinase activity in each sample was determined as the product-to-sum ratio (PSR): P / (S + P), where P is the peak height of the product peptide and S is the peak height of the substrate peptide. Negative control samples (DMSO in the absence of inhibitor) and positive control samples (100% inhibition, a tested DCLK1 inhibitor) were assembled in replicates and were used to calculate percent inhibition values for each compound at each concentration. Percent inhibition (% inhibition) was determined using the following equation: % inhibition = 100 x ((PSR0% - PSRinh)/ (PSR0% - PSR100%)), where PSRinh is the product-sum ratio in the presence of inhibitor, PSR0% is the average product-sum ration in the absence of inhibitor and PSR100% is the average product-sum ratio in 100%-inhibition control samples. DCLK1 inhibitors were tested in 8-point dose-response format on each assay plate. The IC50 values were determined by fitting the inhibition curves by an eight dose-response model using GraphPad PRISM v7.
KINOMEscan binding assays
KINOMEscan competition binding assays were performed by DiscovrX, as previously described,20 with minor modifications to the reported protocol. Briefly, compounds were tested in 12-point dose response, at a maximum concentration of 10 μM.
ERK5 protein expression and purification
ERK5 protein was expressed and purified as previously described.40
ERK5 32P-ATP kinase assay
ERK5 32P-ATP kinase assay was performed as previously described,12 with minor modifications to the reported protocol, with 50 μM [γ−32P]-ATP (500 cpm/pmol) and 200 μM PIMtide (ARKKRRHPSGPPTA) used as substrates. Briefly, compounds were tested in 12-point dose response, at a maximum concentration of 10 μM.
ERK5 cellular kinase assay
ERK5 cellular kinase assay was performed in human HeLa cells as previously described,12 with minor modifications to the reported protocol. Briefly, compounds were tested in 5-point dose response, at a maximum concentration of 10 μM.
BRD4 AlphaScreen assay
This assay was performed by Reaction Biology Corporation according to standard methods, with minor modifications to the reported protocol. Briefly, compounds were tested in 5-point dose response, at a maximum concentration of 10 μM.
LRRK2 ADAPTA kinase assay
This assay was performed at Thermo Fisher as previously described, with minor modifications to the reported protocol (www.thermofisher.com/us/en/home/industrial/pharma-biopharma/drug-discovery-development/target-and-lead-identification-and-validation/kinasebiology/kinase-activity-assays/adapta-universal-kinase-assay.html). Briefly, compounds were tested in 12-point dose response, at a maximum concentration of 10 μM.
KINOMEscan kinome profiling
KINOMEscan profiling was performed by DiscovrX as previously described,20 with minor modifications to the reported protocol. Briefly, compounds were tested against 489 kinases at a concentration of 1 μM.
DCLK1 NanoBRET
DCLK1-Nluc plasmid
DCLK1-Nluc fusions were encoded in a pFN32K expression vector (Promega) and include a flexible Gly-Ser-Ser-Gly linkers between Nluc and full-length DCLK1 (NP_004755.1).
BRET DCLK1 target engagement and tracer evaluation in HEK-293 cells
HEK-293 assays were performed in 96-well format and cells were transiently transfected using FuGENE HD (Promega) according to the manufacturer’s protocol. HEK-293 cell suspensions were prepared in Assay Medium (Opti-MEM without phenol red, 1% FBS) at a density of 2 × 105 cells/mL. DNA mixtures were prepared at the following ratios in 1 mL of Opti-MEM without serum or phenol red: 9.0 μg/mL of Transfection Carrier DNA (Promega) and 1.0 μg/mL of DCLK1-Nluc plasmid. 30 μL of FuGENE HD per mL was then added to the DNA mixture to form Lipid:DNA complexes, mixed by inversion and incubated at room temperature for 20 minutes. Lipid:DNA complexes were then combined with the cell suspension at a 1:20 ratio and 85 μL of final mixture was added to white 96-well culture plates (Corning, #3917). Transfected cells were incubated in a humidified, 37°C/5% CO2 tissue culture incubator for 20 hours.
TAE684-NanoBRET-590 was prepared at a working concentration of 20X in tracer dilution buffer (12.5 mM HEPES, 31.25% PEG-400, pH 7.5). Following 20 h incubation, cells were treated with 5 μL/well of 20X TAE684-NanoBRET-590 and 10 μL/well of TAE684, followed by 2 h equilibration at 37°C. To measure BRET, NanoBRET NanoGlo Substrate (Promega) and Extracellular NanoLuc Inhibitor (Promega) were added according to the manufacturer’s recommended protocol, and filtered luminescence was measured on a GloMax Discover luminometer equipped with 450 nm BP filter (donor) and 600 nm LP filter (acceptor), using 0.5 s integration time. Milli-BRET units (mBU) are calculated by multiplying the raw BRET values by 1000. Apparent tracer affinity values (EC50) were determined using the sigmoidal dose-response (variable slope) equation available in GraphPad PRISM v7 (Equation 1): Y = Bottom + (Top-Bottom)/(1+10^((LogEC50-X)*HillSlope)). Competitive displacement data were then plotted with GraphPad PRISM v7 and data were fit to Equation 1 to determine the IC50 value.
BRET DCLK1 target engagement upon compound treatment in HCT116 cells
HCT116 assays were performed in 384-well format and cells were transiently using FuGENE HD (Promega) according to manufacturer’s instructions with the following modifications. HCT116 cell suspensions were prepared in Assay Medium (Opti-MEM without phenol red, 1% FBS) at a density of 2 × 105 cells/mL. DNA mixtures were prepared at the following ratios in 1 mL of Opti-MEM without serum or phenol red: 9.0 μg/mL of Transfection Carrier DNA (Promega) and 1.0 μg/mL of DCLK1-Nluc plasmid. 30 μL of FuGENE HD per mL was then added to the DNA mixture to form Lipid:DNA complexes, mixed by inversion and incubated at room temperature for 20 minutes. Lipid:DNA complexes were then combined with the cell suspension at a 1:20 ratio and 34 μL of final mixture was added to white 384-well culture plates (Corning, #3570). Transfected cells were incubated in a humidified, 37°C/5% CO2 tissue culture incubator for 20 hours.
Cells were treated with 100 nL of compound from compound stock plates using a Janus Workstation pin tool (PerkinElmer) and NanoBRET was performed according to manufacturer’s instructions with the following modifications. A 100X solution of TAE684-NanoBRET-590 in pure DMSO was used to prepare a Complete 20X NanoBRET Tracer Dilution Buffer (1:4 ratio of 100X TAE684-NanoBRET-590: NanoBRET Tracer Dilution Buffer (Promega)). 2 μL of Complete 20X NanoBRET Tracer Dilution Buffer and 4 μL Opti-MEM without serum phenol red were added to each well, mixed on an orbital shaker for 15 s at 700 rpm and incubated for 2 hours in a humidified, 37°C/5% CO2 tissue culture incubator. Note that a set of quadruplicate wells was assayed without the addition of DCLK1 Tracer for background correction. Plates were brought to room temperature and 20 μL of 3X Complete NanoBRET Nano-Glo Substrate (1:166 ratio of NanoBRET Nano-Glo Substrate (Promega): 1:500 Extracellular NanoLuc Inhibitor (Promega) in OptiMEM without serum or phenol red) was added to each well, incubated for 2 minutes at room temperature and donor (450 nm) and acceptor (610) emission were measured using a CLARIOstar microplate reader (BMG Labtech). Background correction was performed by subtracting average BRET ratio in the absence of tracer from BRET ratio of each sample and BRET ratios were calculated as follows: [(Acceptorsample / Donorsample) – (Acceptorno tracer control/Donorno tracer control)] x 1000. Data was normalized to DMSO-treated wells and analyzed using GraphPad PRISM v7.
Plasmid generation
dTAG plasmids
Cloning of pLEX_305-N-dTAG-KRASG12V was previously described.26 pLEX_305-C-dTAG-LACZ was generated by cloning LACZ (Addgene, #25893) into pLEX_305-C-dTAG using gateway recombination cloning technology (Invitrogen) as previously described.26 In addition to an N- or C-terminal FKBP12F36V tag, respectively, these plasmids contain tandem HA tags for monitoring of protein expression and a puromycin selectable marker.
CRISPR/Cas9 plasmids
pXPR007-sgGFP and pXPR007-sgKRAS were generated by cloning sgRNAs (sgGFP, GGCGAGGGCGATGCCACCTA and sgKRAS, AGATATTCACCATTATAGGT) using BsmbI restriction sites, as previously described.41 pXPR007 is a derivative of pXPR001 that contains a blasticidin selectable marker and Cas9. The PAM motif of sgKRAS is present in an intron, enabling cutting of wild-type or mutant KRAS alleles with concurrent cDNA rescue.
Cell line studies
Culturing
All cell lines employed in this study were purchased directly from the indicated source and cultured at 37 °C and 5% CO2 in the indicated media: PATU-8988T (source: DSMZ, media: DMEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin), PATU-8902 (source: DSMZ, media: DMEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin), MIA PaCa-2 (source: ATCC, media: DMEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin), Panc03.27 (source: ATCC, media: RPMI, 15% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 U/mL insulin), CFPAC-1 (source: ATCC, media: IMDM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin), BxPC-3 (source: ATCC, media: RPMI, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin), 293FT (source: Thermo Fisher Scientific, media: DMEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine), HEK-293 (source: ATCC, DMEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin) and HCT116 (source: ATCC, media: McCoy’s 5a medium, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin). Cells were negative for mycoplasma and were evaluated monthly using the MycoAlert Kit (Lonza). These cell lines are not among those that are commonly misidentified by ICLAC.
Lentivirus production and transduction
Lentivirus production and concentration was performed using 293FT cells as previously described.26 The only modification was that viral particles were collected 60 h after transfection. PATU-8988T and PATU-8902 cells were transduced with concentrated viral supernatants in a 1:1 ratio of LACZ-FKBP12F36V and Cas9/sgGFP or FKBP12F36V-KRASG12V and Cas9/sgKRAS in the presence of 4 μg/mL polybrene. Transduced cell lines were selected with 2 μg/mL puromycin and 10 μg/mL blasticidin.
Isolation of PATU-8988T and PATU-8902 engineered clones
Single cell clones were isolated in 96-well format from PATU-8988T and PATU-8902 cells expressing LACZ-FKBP12F36V and Cas9/sgGFP or FKBP12F36V-KRASG12V and Cas9/sgKRAS. KRAS disruption after CRISPR/Cas9 gene-editing was determined using Tracking of Indels by Decomposition (TIDE).42 Genomic DNA from parental cell lines and isolated clones was isolated as previously described.26 PCR reactions were performed using 2X Q5 hot start master mix (NEB) and the following primer pairs to amplify human KRAS for TIDE analysis: forward, 5’- AGGTAGGCAACACTGAAGTTAC-3’ and reverse, 5’-CAGACTGTGTTTCTCCCTTCTC-3’. The reverse primer was used for sanger sequencing and sequencing traces were analyzed as recommended, compared to the parental cell line (https://tide.nki.nl/). KRAS knockout was further verified using immunoblotting for KRASG12V as noted below.
Immunoblotting
Cell lines were lysed in RIPA buffer supplemented with cOmplete protease inhibitors (Roche), PhosSTOP phosphatase inhibitors (Roche), and 0.1% benzonase (Novagen) on ice for 60 minutes. Lysates were clarified by centrifugation at 20,000 x g for 10 minutes at 4 °C and immunoblotting was performed using an Odyssey CLx Imager (LI-COR) as previously described.26 The following primary antibodies were employed in this study: HA (Cell Signaling, #3724 and #2367), DCLK1 (Abcam, #ab31704), phospho-ERK1/2 T202/Y204 (Cell Signaling, #4370), ERK1/2 (Cell Signaling, #4696), phospho-AKT S473 (Cell Signaling, #4060), AKT (Cell Signaling, #2920), FKBP12 (Abcam, #ab24373), KRASG12V (Cell Signaling, #14412) and α-Tubulin (Cell Signaling, #3873). Fluorescently labelled infrared secondary antibodies (Licor, IRDye) were employed as appropriate.
Analysis of cell viability in 2D-adherent and ultra-low adherent 3D-spheroids
Cell viability was assayed in 2D-adherent conditions using 384-well culture plates (Corning, #3570) and ultra-low attachment conditions using PrimeSurface 384-well 3D culture spheroid plates (S-bio, #MS-9384WZ) as previously described.26 In brief, PATU-8988T and PATU-8902 cells were plated at a density of 100 cells per well in 50 μL media and allowed to adhere or form spheroids overnight. Cells were treated with 100 nL of compound from compound stock plates using a Janus Workstation pin tool (PerkinElmer) and incubated for 120 h. Cell viability was measured by addition of 10 μL of CellTiter-glo (Promega), followed by incubation for 15 minutes at room temperature. Luminescence was measured on an EnVision 2104 Multilabel Plate Reader (PerkinElmer). Data was normalized to DMSO-treated wells for each cell line and analyzed using GraphPad PRISM v7.
Pulldown target engagement assay
PATU-8988T cells were plated at 3,000,000 cells per 100 mm plate (Corning, #430293) and allowed to adhere overnight. Cells were washed 3 times with cold PBS and lysed with 300 μL of lysis buffer containing 50 mM TrisHCl pH 8.0, 150 mM NaCl, 1% NP-40, 5 mM EDTA, 1 mM DTT, cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Roche). After protein standardization, lysates were rotated with 5 μM of FMF-04–084-1 overnight at 4 °C. Lysates were then rotated for 2 hours at room temperature, followed by incubation with 30 μL streptavidin agarose beads for 2 hours at 4 °C. Agarose beads were pelleted at 1350 x g for 5 minutes and then washed 5 times with cold PBS. Beads were then boiled in 2X LDS buffer for 5 minutes at 95 °C and immunoblot assessment was performed.
KiNativ
KiNativ profiling was performed by ActivX as previously described18 with minor modifications to the reported protocol. In brief, PATU-8988T cells were plated at 5,000,000 cells per 150 mm plate (Corning, #430599) and allowed to adhere overnight. For live cell KiNativ assessment, cells were treated with 1 μM and 2.5 μM of DCLK1-IN-1 or 2.5 μM of DCLK1-NEG for 4 hours, washed 3 times with PBS and then harvested in cold PBS containing cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Roche). Cells were pelleted by centrifugation at 1350 x g at 4 °C for 5 minutes. Cell pellets were flash frozen in liquid N2 and KiNativ analysis was performed at ActivX. For cell lysate Kinativ assessment, untreated pellets were sent to ActivX for treatment and KiNativ analysis.
Rat primary hippocampal neuron studies
Immunofluorescence
Rat primary hippocampal neurons were isolated and maintained in culture as described previously.43 At DIV6, cells on poly-L-lysine coated coverslips were treated for 24 hours with DMSO or DCLK1-IN-1. Cells were then fixed by sequential immersions in: 1) CBS (10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 0.32 M sucrose) + 0.05% Triton X-100 + 0.25% glutaraldehyde, 60 seconds; 2) CBS + 1% glutaraldehyde; and 3) CBS + 0.1% NaBH4. Cells were then permeabilized in TBS + 0.5% Triton X-100 for 10 minutes, followed by immersion in antibody dilution buffer (AbDil) for 15 minutes. Cells were then incubated for 1 hour with Alexa 594-labeled phalloidin for 1 hour to label actin, followed by FITC-DM1α for 1 hour to label microtubules. DNA was counterstained with 5 μg/ml Hoechst 33342. Coverslips were mounted in Prolong Gold for imaging. Images were acquired on an inverted DeltaVision Elite (GE Healthcare) microscope equipped with a 60X objective (NA=1.4) (Olympus), and a CoolSnapHQ2 CCD camera (Roper). Z-sections spaced at 200 nm apart were acquired and deconvolved with SoftWorx (GE Healthcare). Images were subsequently processed with Fiji (maximum intensity Z-projections, adjusting minimum and maximum levels, rotating, and cropping).
Patient-derived organoid studies
Culturing
Patient-derived organoid cultures were initiated and maintained as previously described.44,34 In brief, biopsy or pancreatic resection tissue was obtained from patients at the Dana-Farber Cancer Institute after obtaining written informed consent. Tissue was minced and digested with digest medium (collagenase XI at 1 mg/ml, DNase, 0.5% fetal bovine serum, 10 μM Y27632, and advanced DMEM/F12) at 37 °C for 15 minutes to 1 hour. Cells were then washed and seeded in 3-dimensional Matrigel, and fed with human complete organoid medium containing advanced DMEM/F12, 10 mM HEPES, 1x Glutamax, 500 nM A83–01, 50 ng/mL mEGF, 100 ng/mL mNoggin, 100 ng/mL hFGF10, 10 nM hGastrin I, 1.25 mM N-acetylcysteine, 10 mM Nicotinamide, 1x B27 supplement, R-spondin1 conditioned media 10% final, Wnt3A conditioned media 50% final, 100 U/ml penicillin/streptomycin, and 1x Primocin. For propagation, organoids were dissociated with TrypLE before re-seeding into fresh Matrigel and culture medium.
Immunoblotting
Organoids were lysed in RIPA buffer supplemented with cOmplete protease inhibitors (Roche) on ice for 5–10 minutes. Lysates were clarified by centrifugation at 20,000 x g for 10 minutes at 4 °C and immunoblotting was performed as noted for cell lines above.
Cell viability assay
For compound testing, organoids were dissociated to single cells and 1000 viable cells per well in 20 μL of complete medium containing 10% Matrigel by volume were plated in ultra-low attachment 384-well plates (Corning, #4588). After 24 hours, cells were treated using a D300e Digital Dispenser (Tecan) and incubated for 7 days. Cell viability was measured by addition of 20 μL of CellTiter-Glo 3D (Promega) to each well followed by incubation for 1 hour at room temperature on a shaker. Luminescence was measured using an EnVision 2104 Multilabel Plate Reader (PerkinElmer). Data was normalized to DMSO-treated wells for each organoid culture and analyzed using GraphPad PRISM v7.
RNA-sequencing, proteomics and phosphoproteomics
Cell line sample preparation
For RNA-sequencing studies, PATU-8988T cells were plated at 200,000 cells in 6-well plates (Corning, #3506). After adhering overnight, cells were treated with DMSO or 2.5 μM DCLK1-IN-1, washed twice with PBS, lysed and homogenized using Qiashredder columns (Qiagen). RNA extraction was performed using the RNeasy Plus Mini Kit (Qiagen) according to manufacturer’s instructions. Prior to library preparation, ERCC RNA Spike-In Mix #1 (Life Technologies) was added to cell-count-normalized RNA samples.
For proteomics studies, PATU-8988T cells were plated at 3,500,000 per 150 mm plate (Corning, #430599) and allowed to adhere overnight. After adhering overnight, cells were treated with DMSO or 2.5 μM DCLK1-IN-1, washed 3 times with PBS and then harvested in cold PBS containing cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Roche). Cells were pelleted by centrifugation at 1350 x g at 4 °C for 5 minutes. Cell pellets were flash frozen in liquid N2.
Patient-derived organoid sample preparation
For RNA-sequencing and proteomics studies, patient-derived organoids were plated at 3,000,000 cells in T25 ultra-low attachment flasks (Corning, #3815) containing complete organoid medium with 10% Matrigel by volume. After 24 hours in culture, organoids were treated with DMSO or 2.5 μM DCLK1-IN-1. Treated organoids were collected using Cell Recovery Solution (Corning) and split into samples for RNA-sequencing and proteomics. For RNA sequencing, cells were lysed with 1 mL of TRIzol reagent (Thermo Fisher Scientific) and total RNA was extracted using the AllPrep DNA/RNA/miRNA Kit (Qiagen) according to the manufacturer’s instructions. For proteomics analysis, cell pellets were collected and flash frozen in liquid N2.
RNA-sequencing library preparation and analysis
Illumina sequencing libraries were prepared using the KAPA mRNA HyperPrep kit (Roche). Equimolar multiplexed libraries were sequenced on a NextSeq 500 (Illumina, 75 bp single-end reads) by the Molecular Biology Core Facility at the Dana-Farber Cancer Institute. Fastq files were aligned to human genome build hg19 using HiSat with default parameters. Transcripts were assembled and cuffquant and cuffnorm from the cufflinks pipeline were used to generate FPKM values as previously described.45 For cell line RNA-sequencing studies, FPKM values were then normalized to synthetic ERCC spike-in RNAs as previously described previously.46 A transcript was considered to be expressed in each data set if in at least one experimental condition the normalized FPKM > 1. GSEA was performed using curated gene sets (C2)36 as indicated in the figure legends.
Proteomics and phosphoproteomics preparation and analysis
Materials used in proteomics and phosphoproteomics assessments: Isobaric TMT reagents (Thermo Fisher Scientific), BCA protein concentration assay kit (Thermo Fisher Scientific), Empore-C18 material for in-house made StageTips (3 M), Sep-Pak cartridges (Waters), Mass spectrometry (MS)-grade trypsin (Thermo Fisher Scientific), Lys-C protease (Wako), High-Select Fe-NTA phospohpeptide enrichment kit (Thermo Fisher Scientific), and cOmplete protease and phosphatase inhibitors (Millipore Sigma). Unless otherwise noted, all solvents used for liquid chromatography (LC) were purchased from J.T. Baker all other chemicals were purchased from Thermo Fisher Scientific.
MS sample processing
Frozen cell pellets from PATU-8988T and patient-derived organoids were lysed using 8 M urea, 200 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) at pH 8.5 with protease and phosphatase inhibitors. Samples were further homogenized, and DNA was sheered via sonication using a probe sonicator (20 × 0.5 second pulses; level 3). Total protein was determined using a BCA assay and cell lysates were used immediately or stored at −80 oC until future use. A total of 100 μg of protein was aliquoted for each condition and TMT channel for further downstream processing. Protein extracts were reduced using 5 mM dithiothreitol (DTT) for 30 minutes at room temperature. For large scale phosphopeptide analyses, 1 mg per condition and TMT channel were used. Next, samples were alkylated with 20 mM iodoacetamide for 45 minutes in the dark at room temperature. To facilitate the removal of incompatible reagents, proteins were precipitated using chloroform methanol. Briefly, to 100 μL of each sample, 400 μL of methanol was added, followed by 100 μL of chloroform with thorough vortexing. Next, 300 μL of HPLC grade water was added and samples were vortexed thoroughly. Each sample was centrifuged at 14000 x g for 5 minutes at room temperature. The upper aqueous layer was removed, and the protein pellet was washed twice with methanol and centrifuged at 14000 x g for 5 minutes at room temperature. Protein pellets were re-solubilized in 200 mM EPPS buffer and digested overnight with Lys-C (1:100, enzyme: protein ratio) at room temperature. The next day, trypsin (1:100 ratio) was added and incubated at 37 oC for an additional 6 hours in a ThermoMixer set to 1000 rpm.
TMT labeling
To each digested sample, 30% anhydrous acetonitrile was added and 100 μg of peptides were labeled using 200 μg of TMT reagent (TMT1-TMT11). For large scale phosphopeptide enrichment, peptides post enrichment were quantified using BCA and labeled using a 1:2 ratio of peptide to TMT as described above. To equalized protein loading, a ratio check was performed by pooling 2 μg of each TMT-labeled sample. Samples were pooled and desalted using in-house packed C18 StageTips and analyzed by LC-MS/MS. Normalization factors were calculated from this label check, samples were mixed 1:1 across all TMT channels and desalted using a 100 mg Sep-Pak solid phase extraction cartridge. Eluted pooled peptides were stored at subjected to phospohpeptide enrichment or stored at −80 oC until future use.
Phosphopeptide enrichment
The High-Select FE-NTA Phosphopeptides Enrichment kit (Pierce) was used to enrich phosphopeptides from TMT labeled and digested samples following manufacturer’s instructions. Briefly, peptide samples were dissolved using 200 μL of Binding/Washing buffer. For small scale phosphopeptide enrichment, one enrichment column was used per TMT11-plex experiment. For large scale phosphopeptide enrichment one column was used per TMT channel. Columns were equilibrated by passing 200 μL of Binding/Washing buffer twice for 30 seconds at 1000 x g using a centrifuge. Next, the enrichment columns were plugged using the provided rubber seal and the dissolved peptides were added to the column. Peptides were incubated for 30 minutes at room temperature, with gentle tapping every 10 minutes. Unbound peptides were collected in a separate tube for proteomic analysis by spinning the column for 30 sesconds at 1000 x g using a centrifuge. Nonspecific peptides were washed away using 200 μL of Binding/Washing buffer twice and once with 200 μL of HPLC grade water for 30 seconds at 1000 x g using a centrifuge. These wash steps were saved and combined with the unbound fraction for proteomic analyses. Phosphopeptides were eluted into a new tube containing 100 μL of 10% formic acid. Peptides were eluted twice with 100 μL of Elution Buffer for 1 minutes at 1000 x g using a centrifuge. Both the phosphopeptides and pooled proteome sample were dried using a Speedvac and stored at −80 oC until future use.
Basic PH reversed-phase fractionation
Pooled peptide samples from the phosphopeptide flow-through were resuspended in 10 mM ammonium bicarbonate, 5% acetonitrile, pH 8.0 buffer and were fractionated with basic-pH reverse-phase (bRP) HPLC using an Agilent 300 extend C18 column and collected into a 96 deep-well plate. Peptides were subjected to a 50 minute linear gradient from 13 to 43% buffer B (10 mM ammonium bicarbonate, 90% acetonitrile, pH 8.0) at a flow rate of 0.6 mL/min. Samples were consolidated into 24 fractions as previously described, and 12 nonadjacent fractions were desalted using StageTips prior to analyses using LC-MS/MS47–49. For large scale phosphopeptide were separated using a 50 minute linear gradient from 5 to 32% buffer B, and 24 fractions desalted using StageTips prior to analyses using LC-MSMS.
MS data acquisition
All mass spectrometry data was acquired using an Orbitrap Fusion Lumos mass spectrometer in-line with a Proxeon NanoLC-1000 UHPLC system. Peptides were separated using an in-house 100 μm capillary column packed with 40 cm of Accucore 150 resin (2.6 um, 150 Å) (ThermoFisher Scientific) using a 180 min LC gradient from 4 to 25% acetonitrile in 0.125% formic acid per fraction. Eluted peptides were acquired using synchronous precursor selection (SPS-MS3) method for TMT quantification as previously described.50 Briefly, MS1 spectra were acquired at 120K resolving power with a maximum of 50 ms in the Orbitrap. MS2 spectra were acquired by selecting the top 10 most abundant features via collisional induced dissociation (CID) in the ion trap using an automatic gain control (AGC) of 15K, quadrupole isolation width of 0.5 m/z and a maximum ion time of 100 ms. For MS3 acquisition, a synchronous precursor selection of 10 fragment ions was acquired with an AGC of 150K for 150 ms and a normalized collision energy of 55. For small scale phosphopeptide analyses, two methods were used on the same sample. For both methods, full scan MS spectra were acquired using the orbitrap at a resolution of 120000. In the first method, collision induced dissociation (CID) with MultiStage Activation (MSA) was used, while the second method was acquired using higher energy collision-induced dissociation (HCD). In addition, phosphopeptide were separated using a 180 minute LC gradient from 1 to 20% acetonitrile. Large scale phosphopeptides were acquired using the CID plus MSA method, on a 90 minute gradient.
MS data analysis
All acquired data was processed using SEQUEST51 and a previously described in-house informatics pipeline52–54. Briefly, peptide spectral libraries were first filtered to a peptide false discovery rate (FDR) of less than 1% using linear discriminant analysis employing a target decoy strategy. Spectral searches were done using a custom fasta formatted database which included common contaminants, reversed sequences (Uniprot Human, 2014) and the following parameters: 50 ppm precursor tolerance, fully tryptic peptides, fragment ion tolerance of 0.9 Da and a static modification of TMT (+229.163 Da) on lysine and peptide N-termini, carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. For phosphopeptide analyses, phosphorylation (+79.966 Da) on serine, threonine and tyrosine was included as a variable mode. Resulting peptides were further filtered to obtain a 1% protein FDR and proteins were collapsed into groups. Phosphopeptides were further filtered using an Ascore cutoff of 13 as previously described.54 Reporter ion intensities were adjusted to correct for impurities during synthesis of different TMT reagents according to the manufacturer’s specifications. For quantitation, a total sum signal-to-noise of all report ion ions of 200 was required for analysis, and 100 for phosphopeptide analyses. Lastly, protein quantitative values were normalized so that the sum of the signal for all protein in each channel was equal to account for sample loading.
Animal studies
Zebrafish studies
Zebrafish were all of the AB background strain. All zebrafish studies (Supplementary Fig. 7a–b) were approved and performed in accordance with Institute Animal Care and Use Committee (IACUC) at Dana-Farber Cancer Institute.
Toxicity evaluation during embryonic development
1-day-old zebrafish embryos were placed in 48-well plates with 5 embryos per well, and treated with DMSO, DCLK1-IN-1 or DCLK1-NEG in standard egg water. The treated embryos were checked every day for 7 days and dead embryos were removed on daily basis. A Leica MZ10F fluorescence microscope equipped with a Leica EC3 camera was used for capturing bright field images of treated fish.
Mouse studies
Immunohistochemistry
Pancreases were harvested from mice at indicated ages and fixed in 10% formalin overnight at room temperature. Fixed tissue was processed into paraffin blocks and cut into 5 μM sections using a Leica RM2235 microtome. Tissue sections were deparaffinized in a xylene and ethanol series and then washed with water. Antigen retrieval was performed by incubating slides in Target Retrieval Buffer (DAKO, #S1699) in a pressure cooker. Immunohistochemical analysis was performed using the EnVision+ HRP Kit (DAKO, #401111) and DCLK1 (Cell Signaling, #62257) according the manufacturer’s instructions. Slides were counterstained with hematoxylin, dehydrated through an ethanol and xylene series, and coverslips were mounted using Permount (Fisher, #SP15–100). Images were acquired using an Olympus BX-UCB slide scanner.
Immunoblotting
Pancreases were harvested from 8-week-old mice and flash frozen in liquid N2 immediately upon dissection. Frozen tissue was lysed in Bio-Plex Lysis Buffer (Bio-Rad, #171304011) supplemented with Factor I (Bio-Rad, #171304011), Factor II (Bio-Rad, #171304011), and cOmplete protease inhibitor cocktail (Sigma-Aldrich) according to the manufacturer’s instructions. Lysates were clarified by centrifugation and immunoblotting was performed as previously described.55 The following primary antibodies were employed in this study: DCLK1 (Cell Signaling, #62257) and GAPDH (Cell Signaling, #D16H11). Immunoglobulin G horseradish peroxidase linked secondary antibodies were employed as appropriate.
Animal studies for immunohistochemistry and immunoblotting (Supplementary Fig. 8a–b) were approved and performed in accordance with the IACUC at Beth Israel Deaconess Medical Center. All experiments were adherent to institutional standards and were performed in pathogen-free animal facilities. Pdx1-Cre mice were obtained from The Jackson Laboratory (#014647); K-RasLSL-G12D (#01XJ6) and Tp53LSL-R172H (#01XM2) mice were obtained from the NCI Mouse Repository.
Pharmacokinetics
A group of eighteen 8- to 12-week-old male swiss albino mice were divided into two groups (Group 1: 2 mg/kg/IV; Group 2: 10 mg/kg/PO) with each group comprising of nine mice. Animals in Group 1 and Group 2 were administered with DCLK1-IN-1 solution formulation in 5% NMP, 5% Solutol in normal saline through intravenous, and oral route at a dose of 2 mg/kg and 10 mg/kg, respectively. Blood samples (approximately 60 μL) were collected under light isoflurane anesthesia from retro orbital plexus at pre-dose, 0.08, 0.25, 0.5, 1, 2, 4, 8 and 24 hr (IV) and Pre-dose, 0.25, 0.5, 1, 2, 4, 6, 8 and 24 hr (PO). Plasma samples were separated by centrifugation of whole blood and stored below −70 °C until bioanalysis. All samples were processed for analysis by protein precipitation using acetonitrile and analyzed with fit-for-purpose LC/MS/MS method (LLOQ – 9.83 ng/mL). Pharmacokinetic parameters were calculated using the non-compartmental analysis tool of Phoenix WinNonlin (Version 6.3).
Animal studies for pharmacokinetics (Supplementary Fig. 6a–c) were in accordance with the guidelines provided by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) as published in The Gazette of India, December 15, 1998. Prior approval of the Institutional Animal Ethics Committee (IAEC) was obtained before initiation of the study. Swiss albino mice were obtained from Global Bioresearch Solution.
Maximum dose tolerability evaluation
Three 8- to 12-week-old C57B6/6J mice were enrolled into each of the four dosing groups (0, 10 mg/kg, 30 mg/kg and 100 mg/kg), and all mice were dosed by oral gavage daily for two weeks. Weight was measured twice a week and at the end of two weeks, mouse tissues including brain, liver and kidney were harvested for histology analysis.
Animal studies for maximum dose tolerability (Supplementary Fig. 7d) were approved by the IACUC at Dana-Farber Cancer Institute. All experiments were adherent to institutional standards and were performed in pathogen-free animal facilities. C57B6/6J mice were obtained from The Jackson Laboratory (#000058).
Statistical analysis
Information regarding center values, error bars, number of replicates or samples, number of independent experiments, and statistical analyses are described in the corresponding figure and table legends. Experiments were not blinded nor randomized, and sample sizes were not predetermined using statistical analyses.
Reagent availability
Reagents are freely available at: http://graylab.dana-farber.org/probes.html.
Data availability statement
KINOMEscan and KiNativ data are provided in Supplementary Datasets 1–2. Cell line RNA-sequencing data has been deposited to the NCBI GEO (accession number: GSE140490). Cell line and deidentified patient-derived organoid RNA-sequencing analyzed data files are provided in Supplementary Dataset 3. Cell line and deidentified patient-derived organoid mass spectrometry-based proteomics and phosphoproteomics analyzed data files are provided in Supplementary Datasets 4–5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Kostic for critical reading of the manuscript and S. Nabet and members of the Gray laboratory for helpful discussions. We gratefully acknowledge S. Gygi for use of CORE for mass spectrometry data analysis software. This work was supported by an American Cancer Society Postdoctoral Fellowship PF-17-010-01-CDD (B.N.), Claudia Adams Barr Program in Innovative Basic Cancer Research Award (B.N.), Katherine L. and Steven C. Pinard Research Fund (N.S.G. and B.N.), Hope Funds for Cancer Research Postdoctoral Fellowship (S.R.), Harvard Catalyst KL2/CMeRIT Fellowship (S.R.), Perry Levy Fellowship (S.R.), the Lustgarten Foundation (S.R., B.M.W., A.J.A., W.C.H.), NCI HCMI program (A.J.A., S.R.), American Cancer Society 129089-PF-16-088-01-TBG (E.J.P.), KU-KIST Graduate School of Converging Science and Technology Program (T.S.), Spanish Ministerio de Economia y Competitividad (MINECO) grant SAF2015-60268R, co-funded by Fondo Europeo de Desarrollo Regional (FEDER) funds (J.M.L.), Pancreatic Cancer Action Network Catalyst Award (A.J.A.), Doris Duke Charitable Foundation Clinician Scientist Development Award (A.J.A.), NCI K08 CA218420 (A.J.A.), NCI U01 CA176058 (W.C.H.), NCI U01 CA199253 (W.C.H.) and NCI U01 CA224146 (W.C.H.), American Cancer Society Award 132205-RSG-18-039-01-DMC (K.D.W.), Welch Foundation Grant I1829 (K.D.W.), 2017 AACR-Bayer Innovation and Discovery Grant 17-80-44-GRAY (N.S.G.), DF/HCC GI SPORE Developmental Research Project Award P50CA127003 (N.S.G. and K.M.H.) and Hale Center for Pancreatic Research (J.D.M., B.M.W., A.J.A., W.C.H., and N.S.G.).
COMPETING FINANCIAL INTERESTS
F.M.F. and N.S.G. are inventors on a patent application related to the DCLK1 inhibitors described in this manuscript (WO/2018/075608). B.N. is an inventor on patent applications related to the dTAG system described in this manuscript (WO/2017/024318, WO/2017/024319, WO/2018/148443, WO/2018/148440). Z.Z., C.R.C., J.D.V., and M.B.R. are employees of Promega Corporation. B.M.W. receives research funding from Celgene, Inc, and is a consultant for G1 Therapeutics, BioLineRx, and GRAIL. A.J.A. has consulted for Oncorus, Inc. W.C.H. is a consultant for ThermoFisher, AjuIB, MPM Capital, iTeos and Paraxel and is a Scientific Founder and serves on the Scientific Advisory Board (SAB) for KSQ Therapeutics. K.D.W. is a member of the SAB for Vibliome Therapeutics. N.S.G. is a Scientific Founder, member of the SAB and equity holder in C4 Therapeutics, Syros, Soltego, B2S, Gatekeeper and Petra Pharmaceuticals. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voroni, Her2llc, Deerfield and Sanofi.
REFERENCES
- 1.Fedorov O, Muller S & Knapp S The (un)targeted cancer kinome. Nat. Chem. Biol 6, 166–169 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Westphalen CB, Quante M & Wang TC Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases 8, 164–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishio K et al. Doublecortin and CaM kinase-like-1 as an independent prognostic factor in patients with resected pancreatic carcinoma. World J. Gastroenterol 23, 5764–5772 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamska A, Domenichini A & Falasca M Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. International Journal of Molecular Sciences 18, E1338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westphalen CB et al. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell stem cell 18, 441–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker DK et al. Proteogenomic Network Analysis of Context-Specific KRAS Signaling in Mouse-to-Human Cross-Species Translation. Cell Syst 9, 258–270 e256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antolin AA et al. Objective, Quantitative, Data-Driven Assessment of Chemical Probes. Cell Chem Biol 25, 194–205 e195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat. Chem. Biol 7, 203–205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q et al. Pharmacological Inhibition of BMK1 Suppresses Tumor Growth Through PML. Cancer Cell 18, 258–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X et al. Discovery of a benzo[e]pyrimido-[5,4-b][1,4]diazepin-6(11H)-one as a Potent and Selective Inhibitor of Big MAP Kinase 1. ACS Med. Chem. Lett 2, 195–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin EC et al. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl. Acad. Sci. U. S. A 113, 11865–11870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J et al. Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains. ACS Chem Biol 13, 2438–2448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weygant N et al. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol. Cancer 13, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y & Vakoc CR Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb. Perspect. Med 7, a026674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang VT et al. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 392, 51–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin A et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Science Translational Medicine 11, eaaw8412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miduturu CV et al. High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors. Chem. Biol 18, 868–879 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patricelli MP et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 18, 699–710 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel O et al. Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1. Structure 24, 1550–1561 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Davis MI et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol 29, 1046–1051 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Vasta JD et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem Biol 25, 206–214 e211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beno BR, Yeung KS, Bartberger MD, Pennington LD & Meanwell NA A Survey of the Role of Noncovalent Sulfur Interactions in Drug Design. J. Med. Chem 58, 4383–4438 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Chen H et al. Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site. Acta crystallographica. Section D, Structural biology 72, 682–693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabet B et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janes MR et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589 e517 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Muzumdar MD et al. Survival of pancreatic cancer cells lacking KRAS function. Nat Commun 8, 1090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilmartin AG et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 17, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald ER 3rd et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592 e510 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gendoo DMA et al. Whole genomes define concordance of matched primary, xenograft, and organoid models of pancreas cancer. PLoS Comput. Biol 15, e1006596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiriac H et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 8, 1112–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster SA et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer Cell 29, 477–493 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drost J & Clevers H Organoids in cancer research. Nat. Rev. Cancer 18, 407–418 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Ting HA & von Moltke J The Immune Function of Tuft Cells at Gut Mucosal Surfaces and Beyond. J. Immunol 202, 1321–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller S et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem 84, 5066–5073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erazo T et al. Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol. Cell. Biol 33, 1671–1686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B et al. ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Rep 18, 1543–1557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banker G & Goslin K Culturing nerve cells. (The MIT Press, Cambridge; 1998). [Google Scholar]
- 44.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loven J et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulo JA Nicotine alters the proteome of two human pancreatic duct cell lines. JOP 15, 465–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulo JA & Gygi SP Nicotine-induced protein expression profiling reveals mutually altered proteins across four human cell lines. Proteomics 17, 1–2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarrete-Perea J, Yu Q, Gygi SP & Paulo JA Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J Proteome Res 17, 2226–2236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAlister GC et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eng JK, McCormack AL & Yates JR An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 52.Elias JE & Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Huttlin EL et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beausoleil SA, Villen J, Gerber SA, Rush J & Gygi SP A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Lyons J et al. Integrated in vivo multiomics analysis identifies p21-activated kinase signaling as a driver of colitis. Sci Signal 11, eaan3580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
KINOMEscan and KiNativ data are provided in Supplementary Datasets 1–2. Cell line RNA-sequencing data has been deposited to the NCBI GEO (accession number: GSE140490). Cell line and deidentified patient-derived organoid RNA-sequencing analyzed data files are provided in Supplementary Dataset 3. Cell line and deidentified patient-derived organoid mass spectrometry-based proteomics and phosphoproteomics analyzed data files are provided in Supplementary Datasets 4–5.