Abstract
The unknown immune stimulation by nucleic acid nanoparticles (NANPs) has become one of the major impediments to a broad spectrum of clinical developments of this novel technology. Having evolved to defend against bacterial and viral nucleic acids, mammalian cells have established patterns of recognition that are also the pathways through which NANPs can be processed. Explorations into the immune stimulation brought about by a vast diversity of known NANPs have shown that variations in design correlate with variations in immune response. Therefore, as the mechanisms of stimulation are further elucidated, these trends are now being taken into account in the design phase to allow for development of NANPs that are tailored for controlled immune activation or quiescence.
Keywords: nucleic acids, innate immune response, toll-like receptors, RNA nanotechnology, drug delivery
Introduction
Nucleic acid biopolymers (RNA and DNA) have evolved to preserve and regulate the flow of genetic information across all forms of life. Drawing from the variety of available structures of naturally occurring or experimentally selected nucleic acid motifs, mostly manifested in RNAs, a vast library of nucleic acid nanoparticles (NANPs) has been demonstrated (Figure 1) and further investigated for the delivery of therapeutic moieties1, 2, material organization3, 4, or conditional operations in mammalian cells5–9.
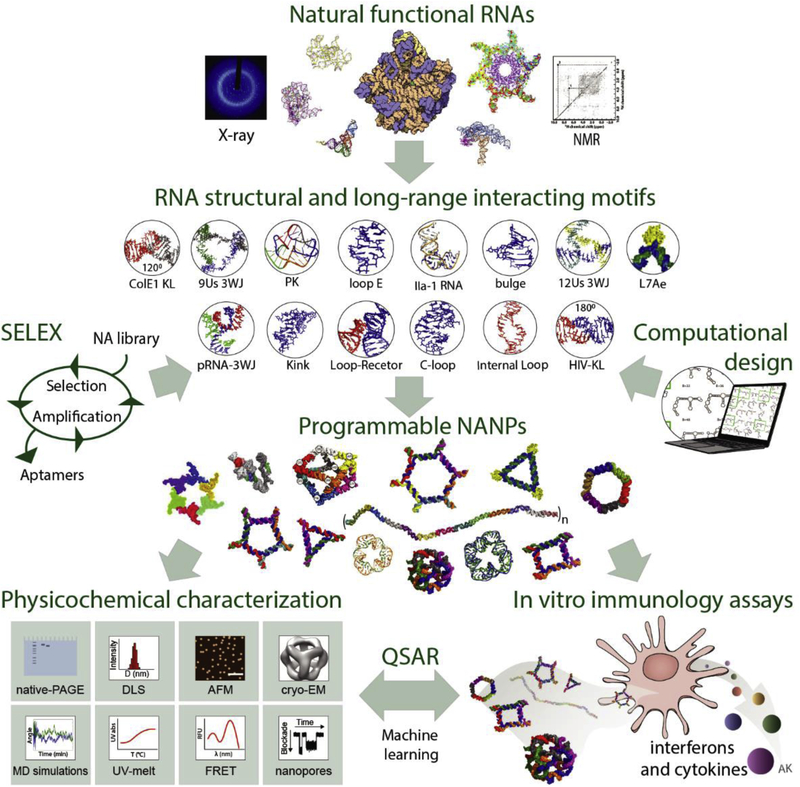
Figure 1.
The flow of NANP design and characterization. Structural and long-range interacting motifs that can be either mined from natural NAs, selected via systematic evolution of ligands by exponential enrichment (SELEX), or designed computationally are combined for the rational design of programmable NANPs. All new NANPs are then extensively characterized and their immunostimulation is assessed. Machine learning approaches such as quantitative structure-activity relationship (QSAR) modeling which relates the physicochemical parameters to relative immune response can be utilized to predict and optimize future NANP designs suitable for specific biomedical tasks. Some parts of lower right panel are adapted with permission from Nano Letters 2018, 18 (7), 4309–4321. Copyright 2018 American Chemical Society.
However, as a result of evolutionary ubiquity, nucleic acids (NAs) have well-established patterns of recognition and thus, the manner in which mammalian cells can interpret NANPs is built upon the pre-existing machinery for bacterial and viral immune recognition. While the recognition of exogenous NAs serves to defend against pathogen invasion, one key challenge for cells remains to avoid an innate immune response to their own endogenous NAs10. Four main determinants have been identified to balance the recognition of self from non-self NAs: patterns (foreign NAs are recognized based on the structure, sequence, or composition), location (occurrence of NAs in compartments unusual for their presence), quantity (changes in relative amounts of NAs compared to physiological conditions), and threshold (regulation of expression of components for NA sensing and downstream signaling)11. Accordingly, cells express pattern recognition receptors (PRRs) that precisely identify signature motifs termed pathogen-associated or danger-associated molecular patterns (PAMPs and DAMPs) in main cellular locations12. Numerous key PRRs specializing in NA recognition will be discussed in this review with the emphasis on NANP recognition pathways (Figure 2).
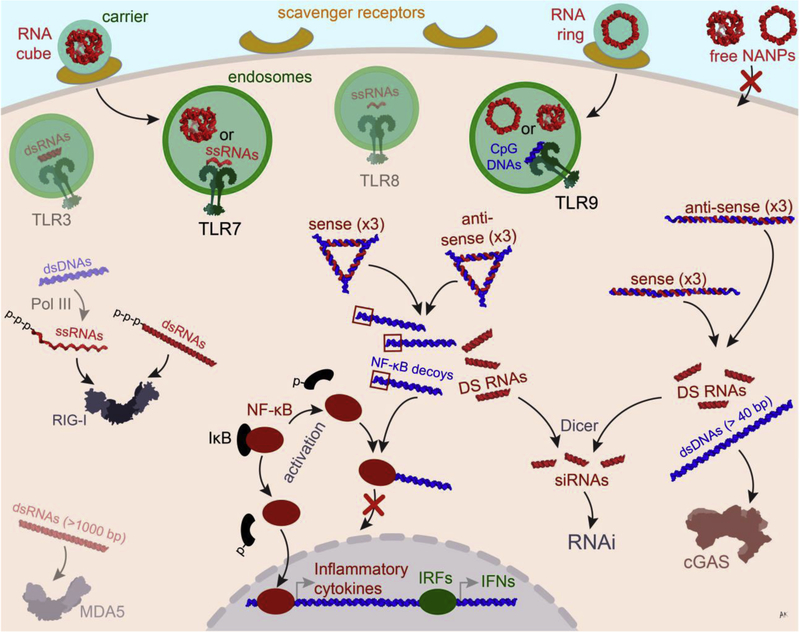
Figure 2.
Possible ways of NANP processing in the cellular environment. NANPs complexed with a polycationic carrier enter the cell via scavenger receptor-mediated endocytosis and get recognized by TLRs (e.g., TLR7 for RNA cubes and TLR9 for both RNA cubes and rings). In the cytoplasm, non-functional RNA/DNA hybrid NANPs can dynamically interact with each other to activate pre-programmed functionalities such as the release of Dicer Substrate (DS) RNAs, later processed into siRNAs, and NF-κB decoy containing dsDNAs which prevent NF-κB translocation into the nucleus and the subsequent production of inflammatory cytokines. The use of longer byproduct dsDNAs helps to activate the cGAS-STING pathway leading to the expression of inflammatory genes.
As the first line of cellular defense, four endosomal membrane-located Toll-like receptors (TLRs 3, 7, and 8 sensing RNAs and TLR9 sensing DNA) compose the group of PRRs that recognize extracellularly invading bacterial and viral NAs13. Another group of PRRs that resides in the cytoplasm and nucleus include RIG-I-like (RLRs) and MDA5 receptors sensing non-self RNAs along with cyclic GMP-AMP synthase (cGAS) and interferon-γ-inducible protein 16 (IFI16) sensing cytoplasmic DNA14, 15. The abundance of individual PRRs differs among various tissues and cell types16. While TLRs are mostly specific for cells of the immune system, intracellular PRRs are broadly expressed. Detection of NA-based PAMPs triggers intricate signaling cascades that pass through the pathway’s specific adaptor proteins defined by the type of NA trigger and finally merge to the transcription factors NF-κB, IRF3, and IRF714, 17. PRR activation culminates in expression of host defense genes and translation of water-soluble proteins (e.g., interferons, proinflammatory cytokines, and chemokines) essential for defense against pathogens. However, the same immune responses that diligently defend against pathogens can create an immunological hurdle for broad applications of therapeutic nucleic acids (TNAs) and NANPs. While some recent TNA formulations have successfully overcome immunological toxicities using chemical modifications and carriers, unwanted immunostimulation remains the major challenge for further clinical translation18. Establishing trends in NANP recognition based on design and composition introduces new possibilities to tailor NANPs for emulating the activations of specific immune pathways. Additionally, viral and bacterial pathogens have evolved methods to circumvent, avoid, or enhance the innate surveillance system. Therefore, critically examining the mechanisms of pathogen recognition and manipulation of NA immune responses will provide valuable insight into the development of NANP technology for clinical applications.
Recognition Receptors
As the field moves towards the development of rationally engineered NANPs with controlled immunostimulation8, 19–22, it becomes crucial to consider PRRs’ expression which is often cell type-specific. For example, TLRs 7 and 8 can both recognize ssRNA; however, plasmacytoid dendritic cells (pDC) and B cells can only express TLR7 while monocytes, macrophages, and myeloid DCs preferentially express TLR8 with minimal expression of TLR723. Also, TLR7 can detect short stretches of dsRNAs24–26 and as a result, pDCs have been shown to be the primary source of interferon production in response to cubic RNA NANPs via the TLR7 pathway22, 27. Recent studies have also suggested the possible involvement of TLR9 in the recognition of RNA cubes and rings in human peripheral blood mononuclear cells (PBMCs)27. However, the mechanism of recognition has yet to be determined.
It is important to note that the expression of PRRs often changes during a diseased state and, for example, the upregulation of TLR9 has been observed in patients with autoimmune thyroid disease28. As such, when choosing the designing principles of therapeutic NANPs, it is vital to consider any disease-specific changes in expression patterns of relevant PRRs.
Expression of PRRs is also tightly regulated on the subcellular level to strategically encounter pathogenic NAs while avoiding any recognition of self-NAs. The endosomal TLRs 3, 7, and 8 are synthesized in the ER and traffic from the Golgi to either endosomes or lysosomes, while TLR9 traffics from the ER directly to the endosomal compartment17, 29. Additionally, proteolytic processing of TLR ectodomains is required for receptor signaling and, therefore, is limited to the endosomal compartment30. This tightly regulated trafficking of TLRs provides an important means for avoiding the recognition of self-NAs. However, some pathogens evolved several mechanisms aimed at escaping TLR-mediated detection. For example, some bacteria utilize the endosomal compartment to create an intracellular replication niche31, 32, whereas other microbes use effector proteins to decrease phagosomal calcium concentration, increase phagosome pH, and avoid or reduce fusion with lysosomes33–36. Manipulating phagosome maturation prevents bacterial degradation and reduces the presence of NA ligands for endosomal TLRs. Alternating phagosome maturation may also alter the presence or function of these TLRs in the endocytic compartment. Therefore, pathogen recognition within the endosome is dependent on the presence of functional TLRs.
While mechanisms of endosomal and TLR escape by pathogens may have deleterious consequences for the host, they could be employed for designing SMART (Specific, Manageable, Adjustable, Reproducible, and Targeted) NANPs for biomedical applications. Since NANPs are made of nucleic acids which, when delivered into the endosomal compartment, could elicit TLR-driven interferon responses, this property is beneficial for applications in which activation of the immune system is desirable (e.g., vaccines and immunotherapies). In contrast, the mechanisms analogous to those utilized by microbes escaping immune recognition could potentially be implemented into the NANP design to diminish the immunorecognition of therapeutic cargo in conditions for which immunostimulation is undesirable (e.g., drug delivery).
Similarly, the delivery and intracellular trafficking of NANPs are key determinants in immune receptor recognition or avoidance. Due to their negative charge, free NANPs are unable to enter the cell without the use of a carrier and are immunoquiescent22. Activation of the NANPs’ immune recognition can, therefore, be controlled by selecting a delivery carrier with specificity to certain routes of uptake, and, consequently, to various intracellular compartments. For example, delivery via receptor-mediated endocytosis allows for the targeting of endosomal TLRs, while delivery to the cytosol would introduce NANPs directly to cytosolic NA sensors such as RIG-I and MDA5. Since TLR and RIG-I/MDA5 pathways have different threshold concentrations for activation by NA ligands, such flexibility in delivery would allow for dose-control over beneficial type I interferon (IFN) responses. For example, when a robust type I IFN response is wanted, delivery into the endosomal TLR-rich compartment is the optimal solution. In contrast, when it is desired to activate a type I IFN by a higher concentration of NANPs, then delivery into cytosol is the most optimal route.
An alternative strategy is the use of dynamic hybrid DNA-RNA NANPs which can surpass recognition until they intracellularly re-associate with one another to release functional RNA interference inducers (Dicer Substrate (DS) RNAs) and double-stranded (ds) DNA byproducts (Figure 2)37, 38. Longer dsDNA, however, can activate the cGAS-cGAMP-STING pathway and trigger the expression of inflammatory genes38. To avoid this, the designed dsDNA byproducts can be shortened and programmed to carry additional functions such as binding NF-κB and lowering the subsequent production of proinflammatory cytokines19. Tightly controlling NANP intracellular trafficking via the use of specific carriers or development of tools that emulate bacterial phagosome manipulation are essential next steps in the therapeutic application of NANPs.
Signature Motifs
Mirroring pathogenic strategies, NANPs can be designed to either avoid PRRs entirely by mimicking host NAs or to elicit specific signaling brought about by selective binding or inhibition39, 40. While PRRs may have evolved to detect and disallow foreign NAs from entering the cell, the use of NANPs has the potential to take advantage of well-established and predictable NA processing. Using this strategy, an additional layer of programmability—tailored processing—can be embedded into NANP structures.
Vaccine adjuvants which serve to enhance the immune response against an antigen are an especially promising route for NANP technology. In addition to incorporating the most immunostimulatory design principles into a NANP, there are also motifs which can direct immune stimulation. Unmethylated CpG oligodeoxynucleotides which are common in bacterial genomes are processed by TLR941. Many nanoformulations have utilized CpG motifs to consistently induce strong immune responses42. For NANPs, sequence-specific activation can be incorporated directly as part of a multi-stranded assembly21.
In order to evade detection and be seen as “self,” pathogens can mimic host mRNA by protecting their own RNA with a 5’ end cap. Since many viral RNAs lack RNA cap modification, its absence is sensed by interferon-stimulated genes that regulate protein synthesis43, 44. Utilizing these approaches, RNA strands in NANP assemblies can be modified with a 5’ end cap if detection is not desirable. Besides alternative routes to obtain cap structures or use cap-independent translation, alphaviruses or filoviruses can use secondary structural motifs in the 5′ UTR to alter IFIT (interferon-induced proteins with tetratricopeptide repeats) binding and function43. Almost each virus is generating noncoding RNAs (ncRNAs) with diverse roles in the virus life cycle. These ncRNAs modulating immune responses in favor of viral infection have medical potential as targets for the development of novel antiviral therapeutics. Furthermore, RNA motifs or RNA modifications involved in subverting cellular immunity can enrich the field of NA nanotechnology. Embedding such motifs in NANPs would allow for designing assemblies with attenuated immunogenicity and enhanced stability for transfected or in vivo co-transcriptionally assembled nanoparticles.
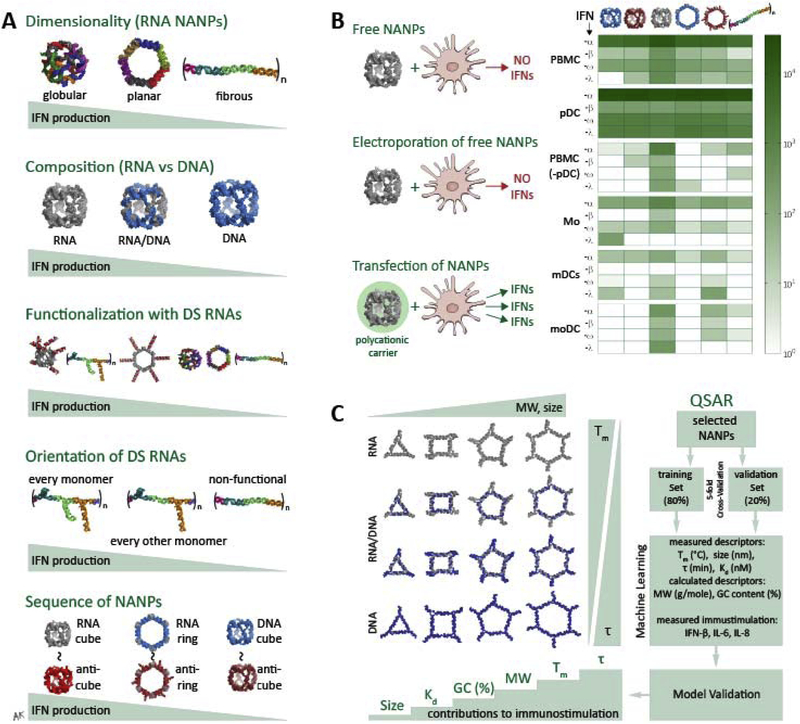
Taking the programmability of NANPs into consideration at the very initial stages of design, the composition and dimensionality of NANPs have been shown to greatly dictate their processing and subsequent initiation of immune responses22. Globular NANPs have been shown to be more immunostimulatory than planar NANPs, which in turn are more immunostimulatory than fibrous NANP structures (Figure 3A). Within the same dimensions, the composition (RNA, DNA, or an RNA/DNA hybrid) also influences the extent of immune response22, 45. NANPs made of DNA have been shown to be less immunostimulatory than the RNA counterparts46 and thus can act as an immunoquiescent carrier for the delivery of therapeutic cargos. However, DNA NANPs can also be advantageous for strategic activation, as DNA constructs have been utilized to bind to TLR9 in the endosome, causing downstream production of type I IFNs for immune modulation47–49. With increasing numbers of RNAs in their composition, NANPs become more immunostimulatory46. RNA NANP interactions with endosomal TLRs 7 and 922 as well as with RLRs27 induce the production of pro-inflammatory cytokines and type I IFNs downstream. Upon functionalization with DS RNAs, each NANP becomes relatively more immunostimulatory. However, the orientation of functional moieties has been shown to regulate the magnitude of stimulation42, 50. Additionally, despite the basic sequence of the NANP, the structural trends prevail, with reverse complement “anti” NANPs producing the same relative stimulation. In order to stimulate any immune response, NANPs must be taken up by cells utilizing a carrier and the greatest IFN production comes from pDCs (Figure 3B). Other trends in NANP design parameters have been investigated using QSAR modeling, during which a library of polygonal NANPs was designed based upon the minimal changes in their sequences, but varying between their composition, relative blood stability, melting temperature, molecular weight, GC content, Kd, and size45. Analyzing these descriptors in addition to their relative levels of immune stimulation allowed for correlations between physicochemical and immunostimulatory properties of NANPs, suggesting that molecular weight, melting temperature, and relative blood stability might be the most closely linked descriptors to immune response (Figure 3C).
Figure 3.
Trends in immune stimulation by NANPs. (A) The dimensionality, composition, functionalization, orientation, and sequence of NANPs have been evaluated relative to contributions to immunostimulation. Globular NANPs are more immunostimulatory than planar NANPs, which are in turn more immunostimulatory than fiborous. For composition, an increasing number of RNA strands in an assembly over DNA strands yields a greater subsequent immune response. Increased functionalization of NANPs with DS RNAs increases relative IFN production, while the orientation of DS RNAs within a single fibrous structure can decrease the effect. Finally, the sequences between variations of the structure have no effect on immune stimulation, while the structure itself is what dictates the response. (B) Neither free NANPs without a carrier nor electroporated free NANPs induce any IFN response. Instead, transfection using a polycationic carrier is necessary to trigger the IFN production. Across multiple immune cell types, pDCs show the greatest production of types I and III IFNs in response to various NANPs. (C) A library of RNA, RNA/DNA, and DNA NANP polygons composed of the same set of sequences but varying in relative blood stability, melting temperature, molecular weight, GC content, Kd, and size revealed that those descriptors had the respective impact on NANP-induced immune stimulation. Some parts of (b) are adapted with permission from Nano Letters 2018, 18 (7), 4309–4321. Copyright 2018 American Chemical Society. Some parts of (c) are used with permission from © 2017 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Safety Considerations and Future Directions
The immune stimulation by TNAs and NANPs has been a significant challenge to the transition of these biotechnologies into the clinical setting51. Since overwhelming immunostimulation may have deleterious consequences to the host, understanding the mechanisms by which NANPs activate the immune cells while monitoring the biomarkers of inflammation in the context of NANPs’ physicochemical properties constitute a framework for responsible and safe use of these materials. Infusion reactions resulting from cytokine storms or complement activation-related pseudoallergies (CARPA) have adverse systemic effects which can surpass the efficacy of therapeutics and deprecate their biocompatibility. Therefore, recent studies have investigated patterns of immune recognition between different designing strategies of NANPs in order to more accurately predict their immune responses22, 27, 45.
Preestablished and well-evolved immunorecognition pathways by bacterial and viral pathogens present a direct means for NANP recognition, but also offer a great advantage to the field of TNAs by offering a known road map around which therapeutic strategies can be planned. With this in mind, predictable immune activation can be incorporated into the design of NANPs which could be a boon for immunotherapy and the use of vaccine adjuvants in one direction, as well as for immunoquiescent drug delivery in another.
Acknowledgements
Research reported in this publication was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R01GM120487 (to K.A.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Martin Panigaj’s work on the subject was supported by the MediPark, Košice - Phase II ITMS2014+: 313011D103 supported by the Operational Programme Research and Development Programme, funded by the ERDF; and NEXO II (Network of Excellence in Oncology) under project ITMS 26220120039. The authors thank Dr. Marina Dobrovolskaia (NCL) for helpful feedback and discussion.
References
- 1.Jasinski D; Haque F; Binzel DW; Guo P, Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11 (2), 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA, In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc. Chem. Res 2014, 47 (6), 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz-Schilling M; Dupin A; Chizzolini F; Krishnan S; Mansy SS; Simmel FC, Optimized Assembly of a Multifunctional RNA-Protein Nanostructure in a Cell-Free Gene Expression System. Nano letters 2018, 18 (4), 2650–2657. [DOI] [PubMed] [Google Scholar]
- 4.Jepsen MDE; Sparvath SM; Nielsen TB; Langvad AH; Grossi G; Gothelf KV; Andersen ES, Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat Commun 2018, 9 (1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata T; Fujita Y; Ohno H; Suzuki Y; Hayashi K; Komatsu KR; Kawasaki S; Hidaka K; Yonehara S; Sugiyama H; Endo M; Saito H, Protein-driven RNA nanostructured devices that function in vitro and control mammalian cell fate. Nat Commun 2017, 8 (1), 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.●.Chandler M; Afonin KA, Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior. Nanomaterials (Basel) 2019, 9 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]; This latest review on the tailorable immune stimulation of NANPs describes additional directions for controlled conditional activation by NANPs of preprogrammed therapeutic functions in cells.
- 7.Bindewald E; Afonin KA; Viard M; Zakrevsky P; Kim T; Shapiro BA, Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano letters 2016, 16 (3), 1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN; Shapiro BA, Triggering of RNA Interference with RNA-RNA, RNA-DNA, and DNA-RNA Nanoparticles. ACS Nano 2015, 9 (1), 251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohno H; Akamine S; Saito H, RNA nanostructures and scaffolds for biotechnology applications. Current Opinion in Biotechnology 2019, 58, 53–61. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki T; Kawai T, Discrimination Between Self and Non-Self-Nucleic Acids by the Innate Immune System. International review of cell and molecular biology 2019, 344, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roers A; Hiller B; Hornung V, Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 2016, 44 (4), 739–54. [DOI] [PubMed] [Google Scholar]
- 12.Patel S, Danger-Associated Molecular Patterns (DAMPs): the Derivatives and Triggers of Inflammation. Current allergy and asthma reports 2018, 18 (11), 63. [DOI] [PubMed] [Google Scholar]
- 13.Tan X; Sun L; Chen J; Chen ZJ, Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annual review of microbiology 2018, 72, 447–478. [DOI] [PubMed] [Google Scholar]
- 14.Luecke S; Paludan SR, Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine 2017, 98, 4–14. [DOI] [PubMed] [Google Scholar]
- 15.Lee J-H; Chiang C; Gack MU, Endogenous Nucleic Acid Recognition by RIG-I-Like Receptors and cGAS. Journal of Interferon & Cytokine Research 2019, 39 (8), 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broz P; Monack DM, Newly described pattern recognition receptors team up against intracellular pathogens. Nature reviews. Immunology 2013, 13 (8), 551–65. [DOI] [PubMed] [Google Scholar]
- 17.Miyake K; Shibata T; Ohto U; Shimizu T; Saitoh S-I; Fukui R; Murakami Y, Mechanisms controlling nucleic acid-sensing Toll-like receptors. International Immunology 2018, 30 (2), 43–51. [DOI] [PubMed] [Google Scholar]
- 18.Dobrovolskaia MA; McNeil SE, Strategy for selecting nanotechnology carriers to overcome immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics. Expert Opinion on Drug Delivery 2015, 12 (7), 1163–1175. [DOI] [PubMed] [Google Scholar]
- 19.●●.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic acids research 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; The nucleic acid nanostructures from this approach demonstrate controlled immune stimulation by using different shapes of RNA-DNA hybrid assemblies which, when combined with their cognates, release therapeutic RNAi inducers as well as DNA decoys capable of binding NF-κB to prevent pro-inflammatory cytokine production.
- 20.Bui MN; Brittany Johnson M; Viard M; Satterwhite E; Martins AN; Li Z; Marriott I; Afonin KA; Khisamutdinov EF, Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine 2017, 13 (3), 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P, Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res 2014, 42 (15), 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.●●.Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Letters 2018, 18 (7), 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]; This research paper describes the very first systematic study of the effects of design variables (size, shape, sequence, dimensionality, composition, connectivity, etc) in a comprehensive library of NANPs on immune stimulation, establishing that NANPs can be used as a powerful tool for controlled immune stimulation.
- 23.Eng HL; Hsu YY; Lin TM, Differences in TLR7/8 activation between monocytes and macrophages. Biochemical and biophysical research communications 2018, 497 (1), 319–325. [DOI] [PubMed] [Google Scholar]
- 24.Hornung V; Guenthner-Biller M; Bourquin C; Ablasser A; Schlee M; Uematsu S; Noronha A; Manoharan M; Akira S; de Fougerolles A; Endres S; Hartmann G, Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature medicine 2005, 11 (3), 263–70. [DOI] [PubMed] [Google Scholar]
- 25.Judge AD; Sood V; Shaw JR; Fang D; McClintock K; MacLachlan I, Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature biotechnology 2005, 23 (4), 457–62. [DOI] [PubMed] [Google Scholar]
- 26.Sioud M, Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol 2005, 348 (5), 1079–90. [DOI] [PubMed] [Google Scholar]
- 27.Hong E; Halman JR; Shah A; Cedrone E; Truong N; Afonin KA; Dobrovolskaia MA, Toll-Like Receptor-Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells. Molecules 2019, 24 (6), 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng S; Li C; Wang X; Liu X; Han C; Jin T; Liu S; Zhang X; Zhang H; He X; Xie X; Yu X; Wang C; Shan L; Fan C; Shan Z; Teng W, Increased Toll-Like Receptors Activity and TLR Ligands in Patients with Autoimmune Thyroid Diseases. Front Immunol 2016, 7, 578–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BL; Barton GM, Trafficking of endosomal Toll-like receptors. Trends in cell biology 2014, 24 (6), 360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewald SE; Engel A; Lee J; Wang M; Bogyo M; Barton GM, Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. The Journal of experimental medicine 2011, 208 (4), 643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebreton A; Stavru F; Cossart P, Organelle targeting during bacterial infection: insights from Listeria. Trends in cell biology 2015, 25 (6), 330–8. [DOI] [PubMed] [Google Scholar]
- 32.Omotade TO; Roy CR, Manipulation of Host Cell Organelles by Intracellular Pathogens. Microbiology spectrum 2019, 7 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaughnessy LM; Hoppe AD; Christensen KA; Swanson JA, Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell Microbiol 2006, 8 (5), 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergne I; Chua J; Lee HH; Lucas M; Belisle J; Deretic V, Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 2005, 102 (11), 4033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vergne I; Chua J; Deretic V, Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. The Journal of experimental medicine 2003, 198 (4), 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spano S; Gao X; Hannemann S; Lara-Tejero M; Galan JE, A Bacterial Pathogen Targets a Host Rab-Family GTPase Defense Pathway with a GAP. Cell Host Microbe 2016, 19 (2), 216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA, Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat Nanotechnol 2013, 8 (4), 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; Stepler M; KewalRamani VN; Kashlev M; Blumenthal R; Shapiro BA, Co-transcriptional production of RNA–DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Research 2013, 42 (3), 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ariza-Mateos A; Gómez J, Viral tRNA Mimicry from a Biocommunicative Perspective. Front. Microbiol 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenbaum BD; Levine AJ; Bhanot G; Rabadan R, Patterns of Evolution and Host Gene Mimicry in Influenza and Other RNA Viruses. PLOS Pathogens 2008, 4 (6), e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lester SN; Li K, Toll-like receptors in antiviral innate immunity. J Mol Biol 2014, 426 (6), 1246–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo S; Li H; Ma M; Fu J; Dong Y; Guo P, Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol Ther Nucleic Acids 2017, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyde JL; Gardner CL; Kimura T; White JP; Liu G; Trobaugh DW; Huang C; Tonelli M; Paessler S; Takeda K; Klimstra WB; Amarasinghe GK; Diamond MS, A viral RNA structural element alters host recognition of nonself RNA. Science (New York, N.Y.) 2014, 343 (6172), 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi YJ; Bowman JW; Jung JU, A Talented Duo: IFIT1 and IFIT3 Patrol Viral RNA Caps. Immunity 2018, 48 (3), 474–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA, Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13 (42), 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.●.Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA, Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Research 2017, 45 (4), 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]; The described novel approach introduces cognate NANPs designed for intracellular re-association to activate pre-programmed functions in cells and showing that the composition (DNA vs RNA) of cognate NANPs also alters their physicochemical and immunomodulatory properties.
- 47.Tursi SA; Lee EY; Medeiros NJ; Lee MH; Nicastro LK; Buttaro B; Gallucci S; Wilson RP; Wong GCL; Tükel Ç, Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLOS Pathogens 2017, 13 (4), e1006315–e1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lande R; Lee EY; Palazzo R; Marinari B; Pietraforte I; Santos GS; Mattenberger Y; Spadaro F; Stefanantoni K; Iannace N; Dufour AM; Falchi M; Bianco M; Botti E; Bianchi L; Alvarez M; Riccieri V; Truchetet M-E; Wong G. CL; Chizzolini C; Frasca L, CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis. Nature Communications 2019, 10 (1), 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee EY; Zhang C; Di Domizio J; Jin F; Connell W; Hung M; Malkoff N; Veksler V; Gilliet M; Ren P; Wong GCL, Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nature Communications 2019, 10 (1), 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.●.Rackley L; Stewart JM; Salotti J; Krokhotin A; Shah A; Halman JR; Juneja R; Smollett J; Lee L; Roark K; Viard M; Tarannum M; Vivero-Escoto J; Johnson PF; Dobrovolskaia MA; Dokholyan NV; Franco E; Afonin KA, RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition. Advanced Functional Materials 2018, 28 (48), 1805959. [DOI] [PMC free article] [PubMed] [Google Scholar]; The immune stimulation of functional fibrous NANPs was explored and compared to planar and globular NANPs, demonstrating that the orientation of functional moieties plays a crucial role in their immunorecognition.
- 51.Dobrovolskaia MA; McNeil SE, Immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics. Expert opinion on biological therapy 2015, 15 (7), 1023–48. [DOI] [PubMed] [Google Scholar]