Abstract
Dengue virus (DENV) is a single-stranded RNA virus transmitted by mosquitoes in tropical and subtropical regions. It causes dengue fever, dengue hemorrhagic fever and dengue shock syndrome in patients. Each year, 390 million people are estimated to be infected by four serotypes of dengue virus, creating a great burden on global public health and local economy. So far, no antiviral drug is available for dengue disease, and the newly licensed vaccine is far from satisfactory. One large obstacle for dengue vaccine and drug development is the lack of suitable small animal models. Although some DENV infection models have been developed, only a small number of viral strains can infect immunodeficient mice. In this study, with biologically cloned viruses from a single clinical isolate, we have established two mouse models of DENV infection, one is severe lethal infection in immunocompromised mice, and the other resembles self-limited disease manifestations in Balb/c mice with transient blockage of type I IFN responses. This study not only offers new small animal models of dengue viral infection, but also provides new viral variants for further investigations on dengue viral pathogenesis.
Keywords: Dengue virus (DENV), Clinical isolate, Murine model, Interferon deficiency
Introduction
Dengue virus (DENV) is an arbovirus transmitted by Aedes aegypti and Aedes albopictus mosquitoes. It belongs to Flavivirus genus of Flaviviridae family and contains a positive single-stranded RNA genome. DENV can be antigenically classified into four serotypes (DENV-1–4), which are often co-circulating in the endemic regions with similar clinical manifestations. Though the majority of DENV infections are asymptomatic, a minority of which can result in self-limited disease including dengue fever (DF), and less than 1% may develop into dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Because of the global warming, increased air travel, and the lack of an efficacious vaccine, DENV has become the most prevalent mosquito-borne viral pathogen in recent decades (Guzman et al. 2010). Each year, DENV is estimated to infect 390 million people globally, affecting nearly half of the world population. Asia accounts for 75% of the dengue disease burden, followed by Latin America and Africa (Bhatt et al. 2013). As an imported pathogen in China, dengue virus had caused sporadic outbreaks in southern China before 2013. However, an unexpected large dengue outbreak attacked Guangzhou in 2014, resulting in 46,864 national reported cases that year (Jin et al. 2015), in which the majority was caused by dengue virus serotype 1 (DENV-1), and the others by serotype 2 (DENV-2) (Zhao et al. 2016; Li et al. 2017). Since then, 2000–5000 confirmed cases were reported to China CDC (Chinese Centers for Disease control and Prevention) each year.
In nature, non-human primates and human are the only mammalian hosts for DENV infection. Naturally acquired dengue virus does not infect any outbred or inbred mice, unless viruses are inoculated with extremely high dose (e.g. 10−9 PFU of DENV-2 16681 in 4-week old C57/BL6) (Chen et al. 2007) or through intracerebral injection. In some other models, mouse-brain adapted strains are used. But such infection of adapted strain or intracerebral infection usually results in neurological manifestations, which are not routine clinical signs of human diseases. And, after passaging in mice, characteristics of these mouse brain-adapted viral strains may differ from strains naturally acquired, attenuation in their ability to cause human disease was documented (Sabin and Schlesinger 1945; Hotta 1952; Sarathy et al. 2015). Thus, the inability of dengue virus to replicate in murine model had hampered the development of dengue vaccine and antiviral therapy. Monkey models, while more useful, are not suitable for routine preclinical testing of vaccine candidates or drug prototypes because of their high cost, limited animal availability and ethical concerns.
The species specificity of DENV infection may be partially attributed to the differences of innate immunity between primates and mice. Interferon (IFN) signaling plays an essential role in the protection against dengue disease in humans, nevertheless DENV has also evolved many mechanisms to antagonize IFN signaling and IFN production in vivo (Green et al. 2014). One of them is the binding and degradation of human STAT2 by dengue viral protein NS5 (Ashour et al. 2010). Another is the cleavage of human STING molecules by dengue viral protease NS2B3 (Aguirre et al. 2012; Yu et al. 2012). Interestingly, both of these two antagonistic mechanisms only exist in human. DENV NS5 does not bind to murine STAT2, neither does NS2B3 cleave murine STING proteins, because the target sequences on these two molecules are highly species specific (Ashour et al. 2010; Aguirre et al. 2012; Stabell et al. 2018). But deletion of mSTAT2 or mSTING could enhance the replication of DENV in mouse or mouse derived primary cells (Ashour et al. 2010; Aguirre et al. 2012). These observations also imply the necessity of IFN signaling and IFN production in restricting DENV infection in mice.
Therefore, immunocompromised mice without complete IFN responses or humanized mice are presumed more susceptible to DENV infection. As humanized mice are laborious to prepare, and often introduce mouse-to-mouse variation, they are less suitable for application in vaccine development (Akkina et al. 2011). In contrast, knockout mice are easier to manipulate and a series of IFN deficient mice have been tested in the development of dengue infection model, including those of IFNα/βR−/−, IFNγR−/−, IFNα/β/γR−/−, MAVS−/−, STAT1−/−, STAT2−/−, IRF3−/− and IRF7−/− (Shresta et al. 2006; Perry et al. 2011; Prestwood et al. 2012). Among them, type I/II IFN receptors double knock out mice usually develop lethal diseases when challenged with suitable DENV strains, thus are used more frequently. However, only a limited number of dengue virus strains are able to replicate in mice and reproduce the severe symptoms (e.g., vascular leakage) of human disease. These viruses are usually based on clinical isolates (DENV-3 C0360/94, DENV-3 703-4, DENV-4 TVP-376), obtained through alternate passage between mosquito C6/36 cells and AG129 mice (DENV-2 D2S10), or isolated after plaque-purification (DENV-2 D2Y98P-PP1, DENV-2 S221) (Sarathy et al. 2015). Such mouse models are valuable in testing vaccine efficacy and studying viral pathology or transmission.
Notably, there are an increasing number of investigations that suggest T cell immunity is an essential component for flavivirus vaccine (St John and Rathore 2019). But, type I interferon are vital for CD4 + and CD8 + T cell generation and maturation (Crouse et al. 2015). And IFN-γ is also one of the major cytokines mediating Th1/CTL function and T cell development. Therefore, IFN deficient transgenic mice are not ideal for studies of T cell immunity or T cell vaccines in an active immunization model, for the lacking of cross-talk between IFN pathway and adaptive immunity.
In this study, we have purified two dengue virus strains of serotype 2 exhibiting different plaque sizes from one clinical isolate DENV-2 GZ. Through subcutaneous injection of low doses of these viruses to type I/II IFN receptor knock-out mice, we successfully established mouse models of lethal infection. The pathogenicity of the two variants in AG6 was demonstrated to be different. Further, we compared the infectivity of these two viral variants in a self-limited infection model, in which type I IFN receptor of wild-type Balb/c mice had been transiently blocked before infection, and found only the virus strain exhibiting larger plaque size caused infectious viral particles in sera. Our research here provides two kinds of dengue virus infection mouse models, which reproduce both severe and self-limited manifestations of human diseases, expanding the current choice of dengue viral infection small animal models.
Materials and Methods
Virus and Cells
Dengue virus serotype 2 clinical strain, DENV-2 GZ, was originally isolated from a patient in Guangzhou in 2012, and was kindly provided by Dr. Xiaoyan Che of Zhujiang hospital in Guangzhou, China. The virus was propagated in mosquito C6/36 cells for no more than three passages in our lab. Virus titer was determined in Vero cells by foci forming assay.
Vero cells were passaged in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Auckland, New Zealand) and 1% penicillin/streptomycin (P/S, Gibco, NY, USA), and cultured at 37 °C in a 5% CO2 incubator.
Mosquito C6/36 cells were cultured in Minimum Essential Medium (MEM, Gibco) supplemented with 10% FBS, 1% penicillin/streptomycin and 1% non-essential amino acids (Gibco, NY, USA) in a 28 °C incubator under 5% CO2 atmosphere.
Plaque-Purification
To purify viruses of different plaque morphology from the original clinical isolate, virus stock of DENV-2 GZ was serially diluted, and used to infect C6/36 cell at 8, 4, 2, 1, and 1/2 PFU per well in a 96-well plate. Seven days post infection, supernatant of each well was harvested. The foci size of virus that produced in each well was determined subsequently in Vero cells by foci forming assay in 48-well plate. Only supernatant containing virus with uniform foci size was kept for further propagation. After three passages, two viral clones maintained different plaque sizes, herein named as DENV-2 1D4-5-SP (variant with Small Plaque size) and DENV-2 8H2-7-LP (variant with Large Plaque size), were stored in a – 80 °C freezer.
Foci Forming Assay
Titer of virus stock and infectious viral particle in serum of mice were determined using foci forming assay in Vero cells. Specifically, Vero cells were seeded at 7 × 104 cells/well in a 48-well tissue culture plate and incubated overnight at 37 °C, 5% CO2. On the second day, sera of infected mice or viral stocks were serially diluted in DMEM and added into wells containing Vero cells for a 2 h incubation period. Subsequently, the viral inoculum was removed and DMEM supplemented with 1.5% FBS, 1% P/S, 1.5% carboxymethylcellulose (CMC, Sigma, MO, USA) was added. After incubation for 96 h at 37 °C, cells were fixed with 4% paraformaldehyde (PFA). Following the removal of PFA, infected cells were stained with anti-dengue monoclonal antibody D1-11 (1:500, Santa Cruz Biotechnology, CA, USA) for 2 h, followed by addition of biotin labeled anti-mouse IgG (Santa Cruz Biotechnology, CA, USA), and then streptavidin–alkaline phosphatase (Sigma, MO, USA). Finally, the foci of dengue virus were visualized by adding BCIP/NBT substrate (Beyotime Biotechnology, Jiangsu, China). Neutralizing activity of mouse sera was evaluated using Foci Reduction Neutralization Test (FRNT) as previous described (Sun et al. 2017).
Sequencing
Genome information of two viruses was acquired through Sanger sequencing. Briefly, viral genomic RNA was extracted from the virus stocks. After synthesis of viral cDNA with specific primers, 14 fragments covering the whole coding region of genome were amplified using Phusion high-fidelity PCR polymerase (Thermofisher, Lithuania). When the PCR product concentration was high enough, they were sent directly for sequencing. Otherwise, fragments were inserted into the pEASY-blunt plasmid (Transgene, Beijing, China), which was then used to transform E. coli bacteria. Three colonies containing each of the fragments were sequenced. Full-length sequence of the single open reading frame was achieved based on the consensus sequence of each fragment, and then stringed together by overlapping them with each other. After that, sequence information of these two viruses was submitted to GenBank, and the access numbers are MN952966 (DENV-2 8H2-7-LP) and MN952967 (DENV-2 1D4-5-SP) respectively.
Elispot Assay
IFN-γ Elispot assay was performed according to manufacturer’s protocol (Mabtech, Nacka Strand, Sweden). Splenocytes of infected or control mice were isolated by homogenizing and filtering through 40 µm cell strainers. After depleting erythrocytes, cells were re-suspended in 10% FBS/PRMI-1640 and added to Elispot plates (Millipore, Co. Cork, Ireland) pre-coated with IFN-γ antibodies. Then PBS, 10 µg/mL peptide, NS1 or E80 protein or 5 µg/mL conA was added to each well to stimulate splenocytes, and the plates were incubated at 37 °C for 40–48 h. After being washed with PBS for 5 times, plates were stained with biotin conjugated antibody, and subsequently streptavidin-horse radish peroxidase (HRP). Finally, immune spots were visualized with the addition of TMB substrate. The image of spots was captured and analyzed by ImmunoSpot Analyzer (Cellular Technology Ltd. USA).
Mouse Infection and Sample Collection
Balb/c mice were purchased from Beijing Vital River Laboratory Animal Technology. Type I and Type II IFN receptor double knock-out C57BL/6 mice (AG6) were originally provided by Dr. Guangxun Meng of Institut Pasteur of Shanghai, and raised in our laboratory. All mice were bred and maintained in specific pathogen-free barrier facilities and used at 6–8 weeks of age. For virus infection experiments, all procedures were completed under BSL-2 and ABSL-2 laboratory conditions as approved by the Biosafety Committee of Institut Pasteur of Shanghai.
Female Balb/c mice of 6–8 weeks old were injected with 2 mg anti-type I IFN receptor monoclonal antibody (MAR1-5A3, BioXcell, USA) intraperitoneally (i.p.) at one day prior to challenge with 1.8 × 106 PFU parental strain, 8H2-7-LP or 1D4-5-SP through subcutaneous (s.c.) injection. Blood samples were collected though retro-orbital bleeding daily from the 1st day post infection (1 dpi) to 7th day post infection (7 dpi). Whole blood was lysed and homogenized within TRIzol-LS reagent (Invitrogen, CA, USA) for measuring viral RNA copy. Meanwhile, for further detection of infectious viral particles in serum, sera were isolated from the blood through centrifugation.
Female AG6 mice of 6–8 weeks old were infected with 1 × 103 PFU, 1 × 104 PFU or 1 × 105 PFU DENV viruses through subcutaneous (s.c.) injection on day 0. Body weight and survival rate were monitored from 1 dpi to 15 dpi, during which moribund mice with weight loss over 20% were euthanatized and recorded as death.
Measurement of DENV-induced Vascular Leakage in AG6 Mice
Vascular leakage was determined following intravascular administration of Evans blue dye (Sigma, louisiana, USA). Briefly, 200 µL Evans blue dye (1% in PBS) was intravenously injected to mice on the 7th days post-infection of the two viral variants. After 1 h dissemination, the mice were anaesthetized and perfused with 50 mL PBS. Digestive tracts and livers were collected and photographed.
Realtime-PCR Quantification of Viral RNA
To quantify viral genomic copy in blood, RNA was extracted following the user’s guide of TRIzol LS reagent. A two-step quantitative RT-PCR (qRT-PCR) was then performed. First, cDNA was synthesized using the FastQuant RT Kit (with gDNase) (Tiangen Biotech, Beijing, China). Second, amplifications were carried out using the FastFire qPCR PreMix (Probe) Kit (Tiangen Biotech, Beijing, China) with the primer pairs designed according to the conserved sequence in DENV 3′UTR: (Forward, 5′-TGAYAAGCARTCAGACAC-3′; Reverse, 5′-TCACCARRCTCCCTTTGC-3′; Fluorescence probe 5′-CCAGAGATCCTGCTGTCTC-3′).
Amplification cycles consisted of an initial incubation step of 95 °C for 1 min, 40 cycles at 95 °C for 5 s, 55 °C for 10 s, and 72 °C for 20 s. Standard RNA were in vitro transcribed from a DNA template that contains a T7 promoter and the target polynucleotide fragment for the primers indicated above. To obtain the standard curve, serially diluted standard RNA (102–109 copy) were used as templates in parallel with sample RNAs. Copy of viral RNA was calculated according to standard curves.
Statistical Analysis
All data were analyzed using GraphPad Prism software. T test or two-way ANOVA were used to analyze the significance as indicated in legends. All results were expressed as means plus and minor standard errors of mean (SEM). P values of 0.05 or less were considered statistically significant (*P < 0.05; **P < 0.01, ***P < 0.001; ****P < 0.0001).
Results
Two DENV-2 Strains from One Clinical Isolate Show Different Replication Capability in vitro
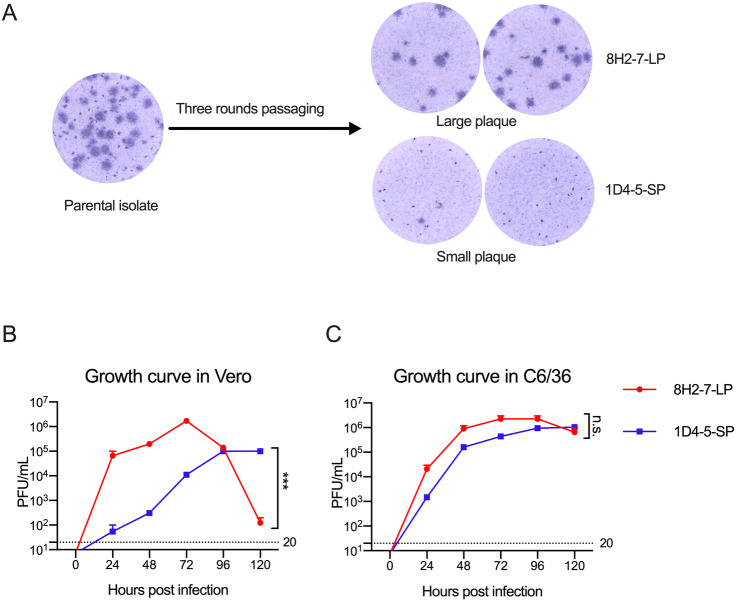
During in vitro propagation of a clinical strain of DENV-2 obtained from Guangzhou, China, heterogenous foci of different plaque sizes were observed in Vero cell cultures (Fig. 1A). Through limiting dilution and culture in mosquito C6/36 cells, two viral variants were separated. After subsequent passage for three rounds in C6/36 cells, two virus strains with stable foci morphology were obtained. One of them forms larger foci of about 1.25 mm diameter after 80 h culture, and it was termed as DENV-2 8H2-7-LP (Fig. 1A); the other forms smaller foci of about 0.25 mm diameter and was termed as DENV-2 1D4-5-SP (Fig. 1A).
Fig. 1.
Different replication capacity of two viral variants DENV-2 1D4-5-SP and DENV-2 8H2-7-LP in Vero cells. A Foci of the parental clinical isolate, and DENV-2 1D4-5-SP, DENV-2 8H2-7-LP in infected Vero cells on day 4 post infection; One-step growth curve of DENV-2 1D4-5-SP, DENV-2 8H2-7-LP in Vero cells (B), and C6/36 cells (C). Viruses were used to infect cells at MOI 2.5, supernatant of infected cells were collected at the indicated timepoints. Titers of infectious virus in supernatant were measured using foci forming assay in Vero cells. Detection limits of infectious virus titer were indicated with dashed lines. Significance of the differences was calculated with Two-way ANOVA test, ***P < 0.001, n.s., not significant.
We next compared the replication abilities of these two viruses in both Vero cells and C6/36 cells using the one-step growth curve. DENV-2 8H2-7-LP had a higher replicating efficiency in Vero cells than DENV-2 1D4-5-SP during the first 72 h after infection, followed by a decline afterwards, due to the probability of running out target cells for infection; in comparison, DENV-2 1D4-5-SP rose slowly to a peak at 96 h post infection (hpi), and kept its plateau until 120 hpi (Fig. 1B). Distinct from what was observed in Vero cells, there was no obvious growth difference between the two viruses in mosquito cells (Fig. 1C).
Taken together, these data demonstrated that two viral variants with different foci sizes are isolated from one clinical virus isolate. And these two variants have different replication abilities in Vero but not mosquito C6/36 cells.
DENV-2 8H2-7-LP and DENV-2 1D4-5-SP are Phylogenetically Linked but Show Variation in Sequence
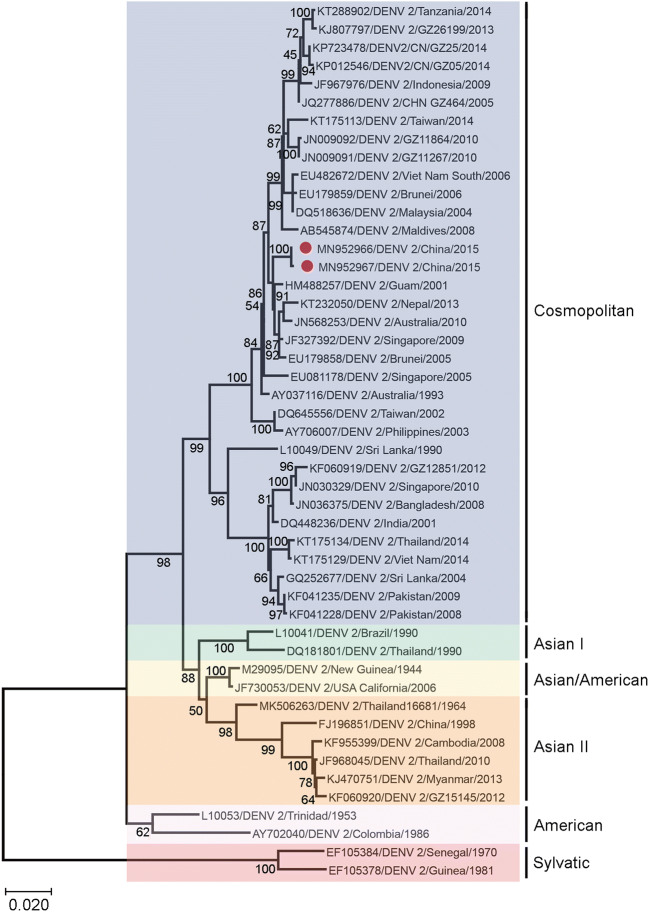
To determine whether the different replication phenotypes reflect underlying genetic divergence between the LP and SP viruses, we sequenced the whole coding sequence of DENV-2 8H2-7-LP and DENV-2 1D4-5-SP, and aligned their envelope sequence with those of representative viruses belonging to different DENV-2 genotypes, including Asian genotype, American Asian genotype, Cosmopolitan genotype and Sylvatic genotype. Additionally, DENV-2 strains isolated in Guangdong, China, from 2005 to 2014 were also included for comparison. The phylogenetic analysis showed that DENV-2 8H2-7-LP and DENV-2 1D4-5-SP belonged to the Cosmopolitan genotype and were clustered together with many strains isolated in West pacific regions, with percentages of homology ranging from 99.5% to 100% (Fig. 2).
Fig. 2.
Phylogenetic analysis of DENV-2 1D4-5-SP and DENV-2 8H2-7-LP with representative serotype-2 dengue viruses of different genotypes isolated from different geographical regions. The phylogenetic tree was obtained after analyzing envelope sequences of DENV-2 with MEGA X software, using the Maximum Likelihood method and Kimura 2-parameter model. Members of six reported genotypes of DENV-2 were included. DENV-2 1D4-5-SP (MN952967) and DENV-2 8H2-7-LP (MN952966) were denoted by red dots in the tree. The scale bar denotes an evolutionary distance of 0.02 nucleotides per position in the sequence.
Further comparison of genomic information between these two viruses also indicated that they had sequence variations in coding regions as illustrated in Table 1. Collectively, we found 14 sequence variations in E, NS3, NS4B and NS5 regions, including 7 synonymous mutations and 7 amino acid changes. Among the 7 nonsynonymous mutations, two are in domain I/II regions of E protein, two are in peptidase domain and helicase domain of NS3 respectively, another is located in a transmembrane helix of NS4B, and the rest two are in NS5 RdRp domain. All of these domains are components of viral binding, replication or processing machinery, and highly related to viral antagonisms to host immunity. The conservation of mutations was also analyzed in comparison with representative strains from different genotypes, interestingly, though most synonymous mutations were also observed in other dengue-2 viruses, the seven nonsynonymous mutations were unique, and these sites seem more conserved in dengue-2 viruses (Table 1). These genomic characteristics may contribute to the different infectivity both in vitro and in vivo.
Table 1.
Sequence variation between 8H2-7-LP and 1D4-5-SP.
| Genotype | Segment | E | NS2A | NS2B | NS3 | NS4A | NS4B | NS5 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Domain | aDI | DII | / | / | Pro | Hel | / | TM | MTase | RdRp | |||||
| / | bLocation(nt) | 1399 | 1434 | 1540 | 4080 | 4125 | 4429 | 4876 | 5065 | 5817 | 6390 | 7144 | 8182 | 9112 | 9207 |
| cLocation(aa) | 155 | 166 | 202 | 191 | 206 | 97 | 119 | 182 | 332 | 5 | 107 | 151 | 461 | 492 | |
| Cosmopolitan | 1D4-5-SP | d(A)Thr | (A)P | (A)Lys | (C)L | (G)K | (C)R | (A)Asn | (A)Ile | (G)V | (T)N | (C)Leu | (T)L | (C)Leu | (C)Asp |
| 8H2-7-LP | (G)Ala | (G)P | (G)Glu | (T)L | (A)K | (A)R | (G)Asp | (G)Val | (C)V | (C)N | (T)Phe | (C)L | (A) Ile | (A)Glu | |
| eAY037116 | – | (C)P | (G)Glu | – | – | (A)R | – | – | (C)V | – | – | – | – | – | |
| Asian I | DQ181801 | – | – | (G)Glu | – | – | (A)R | – | – | (T)V | (C)N | – | – | – | – |
| Asian II | MK506263 | – | – | (G)Glu | – | – | (A)R | – | – | (C)V | (C)N | – | – | – | – |
| Asian/American | M29095 | – | – | (G)Glu | – | – | (A)R | – | – | (T)V | (C)N | – | – | – | – |
| American | AY702040 | – | – | (G)Glu | – | – | (A)R | – | – | (T)V | (C)N | – | – | – | (T)Asp |
| Sylvatic | EF105378 | – | (C)P | (G)Glu | – | (A)K | (A)R | – | – | (T)V | – | – | (C)L | – | (T)Asp |
aDI, DII domain I, II of E protein ectodomain, Pro Protease, Hel Helicase, TM Transmembrane, MTase, methyltransferase; RdRp, RNA-dependent RNA polymerase;
bLocation of nucleotide variation in the whole viral genomes;
cLocation of both synonymous and nonsynonymous variation sites on amino acid sequences of each viral protein;
dNonsynonymous variations were presented with three-letter amino acid codes;
eRepresentative strains from each genotype were selected from sequences analyzed in Fig. 2, and listed in table with their access number for comparison. “–” means the nucleotide and residue are identical to the first strain 1D4-5-SP.
DENV-2 8H2-7-LP is More Pathogenic than DENV-2 1D4-5-SP in Type I and Type II IFN Receptors Double Knock-out Mice
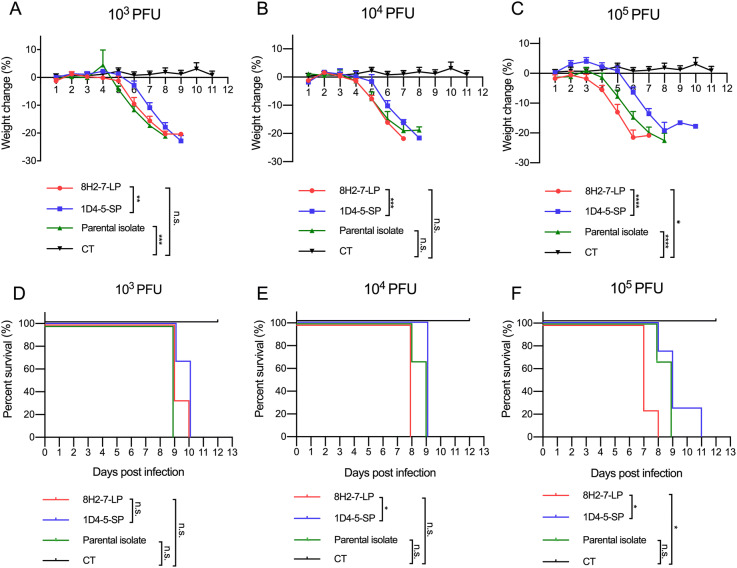
As type I/II interferon deficient mice are more susceptible to dengue virus infection, we determined the in vivo infectivity of these two viruses in type I and type II IFN receptors double knock-out AG6 mice. AG6 mice between 6 and 8 weeks old were infected through s.c. injection with 103 PFU, 104 PFU and 105 PFU DENV-2 8H2-7-LP or DENV-2 1D4-5-SP (Fig. 3). It is observed that infection of either virus led to 100% mortality within 7–11 days, even at a dose as low as 103 PFU, though DENV-2 8H2-7-LP caused more weight loss and faster death than DENV-2 1D4-5-SP at the same dosage. Specifically, weight loss began on 3 dpi, 4 dpi and 5 dpi in mice infected with 105, 104, and 103 PFU DENV-2 8H2-7-LP, respectively (Fig. 3A, 3B, 3C), and death began on 7 dpi, 8 dpi and 9 dpi correspondingly (Fig. 3D, 3E, 3F). In contrast, weight loss was first observed 1–3 days later in mice infected with DENV-2 1D4-5-SP, on 5 dpi or 6 dpi with inoculation of 104–105 PFU or 103 PFU, respectively. The death was also delayed, began on 8 dpi or 9 dpi correspondingly. As a mixture of two types of viral variants, the parental strain was also able to cause weight loss and lethal diseases in AG6 mice. Comparing to the two purified variants, the parental quasispecies presented intermediate level of pathology, which was most obvious in the dose groups of 105 PFU. Through Evans blue staining, we also detected plasma leakage in sick mice on day 7 post infection by either DENV-2 8H2-7-LP or DENV-2 1D4-5-SP viruses (Fig. 4). Consistent with the progress of weight loss and survival curves, leakage of Evans blue in liver or digestive tract was more severe in mice infected with 8H2-7-LP on 7 dpi.
Fig. 3.
Weight loss and survival curve of AG6 mice after infection with DENV-2 8H2-7-LP and DENV-2 1D4-5-SP. AG6 mice between 6-8 weeks old were infected with two viral variants and the parental isolate through s.c. injection. Three different dose of viruses, 103 PFU (A, D; n = 3), 104 PFU (B, E; n = 3), and 105 PFU (C, F; LP and SP n = 4, Parental n = 3), were inoculated into mice. Mice injected with PBS were included within each panel for control (CT, n = 6). Weight change was recorded and presented as a ratio to the initial weight on day 0. Moribund mice with weight loss over 20% were euthanatized and recorded as death. Two-way ANOVA was used in statistical analysis of differences among groups, *P < 0.05, **P < 0.01, ***P < 0.001,**** P < 0.0001, n.s., not significant.
Fig. 4.
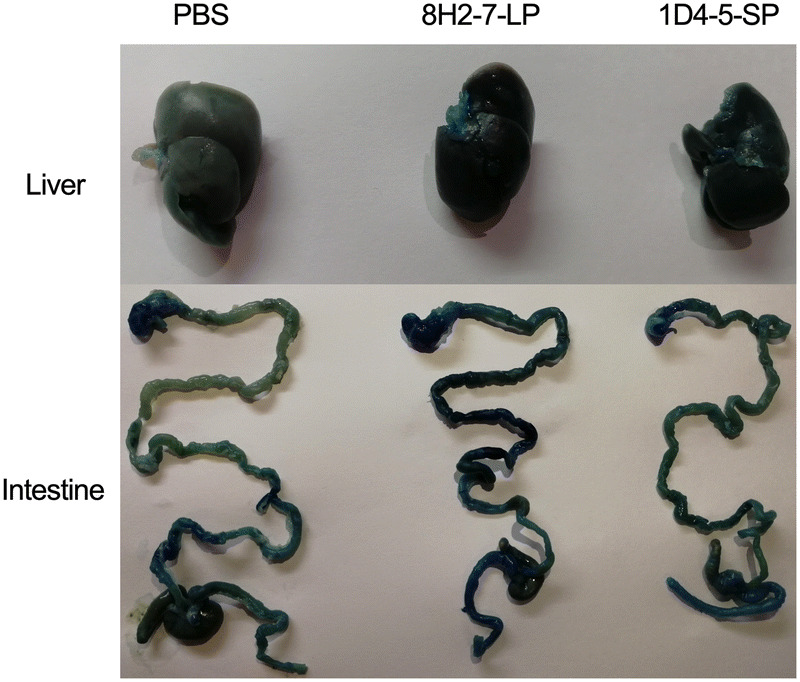
Vascular leakage in AG6 mice induced by infection of DENV-2 8H2-7-LP and DENV-2 1D4-5-SP. AG6 mice of 8 weeks old were infected with 105 PFU DENV-2 8H2-7-LP or 1D4-5-SP through s.c. injection. Seven days post infection, mice were injected intravenously with Evans blue and DENV-induced vascular leakage was visualized in different tissues. PBS infected mice were used as negative controls.
The evidence above demonstrated that the two viral isolates show differences on the pathology and disease kinetics, though they both have capabilities for causing lethal infection in IFN deficient mice.
DENV-2 8H2-7-LP Replicates More Efficiently than DENV-2 1D4-5-SP in WT Balb/c Mice with Transient Blockage of Type I IFN Receptor
Although both viruses can cause lethal infection in type I and type II double knock-out mice, whether the LP and SP viruses behave differently in type II IFN competent host is unknown. We have recently developed a ZIKV infection model in Balb/c mice with transient blockage of type I IFN receptor (IFNAR) (Liang et al. 2018), we thus used the same strategy to examine these two dengue viruses for further comparison.
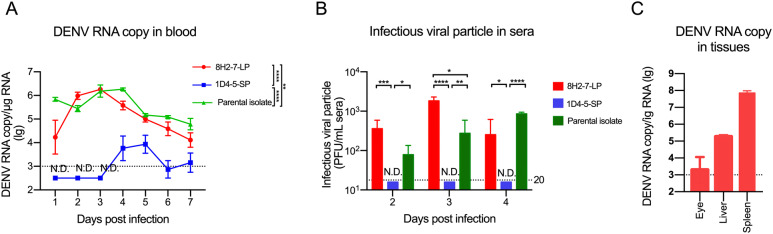
WT Balb/c mice were injected with antibody specific for IFNAR at one day prior to s.c. infection with 106 PFU of either viral strain, and monitored for seven days. The viral RNA of DENV-2 8H2-7-LP could be detected throughout the week after infection, and the peak RNA level of about 106 genomic copy per microgram total RNA was reached on day 3 post infection, followed by gradually dropping to 104 copy on the 7th day (Fig. 5A). In contrast, viral RNA of DENV-2 1D4-5-SP was not detected until day 4 post infection, when merely 104 copy per microgram of total RNA was transiently detected for 2 days. Consistently, infectious virus was detected in sera of DENV-2 8H2-7-LP infected mice from 2 to 4 dpi, during which peak viremia of 103 PFU/mL appeared on day 3 post infection. Meanwhile, infectious virus was not detectable in mice infected by DENV-2 1D4-5-SP (lower than detection limit of 20 PFU/mL) (Fig. 5B). Moreover, 8H2-7-LP viral RNA was also detected in liver, spleen and eyes on 3 dpi (Fig. 5C), further confirming replication of 8H2-7-LP in susceptible organs. The parental strain also replicated in Balb/c mice (Fig. 5A and 5B), and the kinetics of viremia were closer to that of 8H2-7-LP, except for the delay of peak viremia to the 4th day post infection in both assays.
Fig. 5.
Viral replication of DENV-2 8H2-7-LP and DENV-2 1D4-5-SP in Balb/c mice pretreated with IFNAR blocking antibodies. Female Balb/c mice of 6–8 weeks old were injected through i.p. with 2 mg antibody to type I IFN receptor (MAR1-5A3) at one day prior to infection. On the next day, 106 PFU of viruses were inoculated into mice through s.c. route. Then, blood samples were collected daily after infection. A Viral RNA copies in blood were measured using qRT-PCR, Two-way ANOVA was used in statistical analysis of the differences among groups, **P < 0.01, ****P < 0.0001, and B Titers of infectious virus in serum samples were determined through foci forming assay in Vero cells; n = 5. T test was used in statistical analysis of the differences among groups, *P < 0.05, **P < 0.01, *** P < 0.001, ****P < 0.0001. C Eye, liver and spleen were collected from 8H2-7-LP virus-infected mice, and viral RNA loads were determined, n = 4. Detection limits of RNA copy and infectious virus titer were indicated with dotted lines. N.D. not detected.
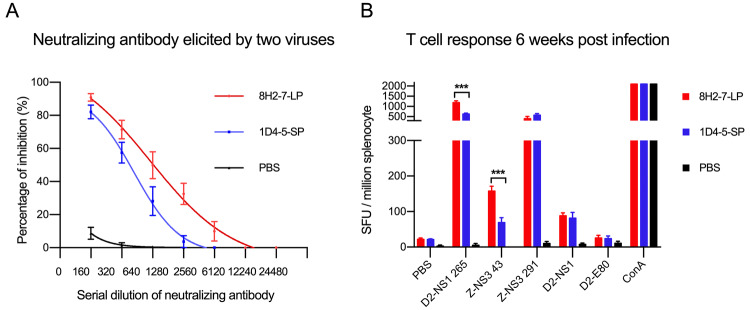
To examine whether the above observed different virological properties of the two DENV variants affect adaptive immunity in host, we evaluated adaptive immune responses induced by their infection in Balb/c mice. DENV-2 8H2-7-LP infection elicited antibodies cross-neutralizing the reference strain DENV-2 16681 (Fig. 6A) and it also stimulated T cell responses specific to peptides of DENV-2 NS1 and ZIKV NS3 (Fig. 6B). Similarly, neutralizing antibody and T cell responses were also detected in mice which were previously infected by DENV-2 1D4-5-SP, though both responses were weaker than those elicited by DENV-2 8H2-7-LP. Considering genome similarity between the two variants, such different magnitudes of adaptive immune responses are mainly related to the different viral replication of two variants and different expression level of immunogens in vivo.
Fig. 6.
DENV-2 specific adaptive immune responses elicited by DENV-2 8H2-7-LP and DENV-2 1D4-5-SP infection. At 40 days post infection, FRNT (Foci Reduction Neutralization Test) A and IFN-γ Elispot assay (B) were performed with mouse sera and splenocytes, respectively. Reference strain DENV-2 16681 was used in FRNT assay, peptides and proteins derived from DENV-2 16681 (D2-NS1 265, D2-NS1, D2-E80) and Zika virus (Z-NS3 43, Z-NS3 291) were included in Elispot assay to measure specific and cross-reactive responses; n = 4. Significance of the differences was calculated with t test, ***P < 0.001.
In summary, we isolated from the same viral quasispecies two DENV variants that showed distinct in vitro replication phenotype in Vero cells, and different in vivo infectivity and pathogenicity in mice. Using either of these two viral variants, we have established a lethal infection model in immune deficient AG6 mice. With the variant DENV-2 8H2-7-LP, we have also developed a self-limited infection model in WT mice pretreated with Type-I IFN receptor antibodies. These two closely-related viruses not only offer new tools for in vivo studies, but also provide resources to investigate the fundamental molecular basis that modulate viral replication.
Discussion
In this study, we have successfully separated two viral variants that show different foci sizes in Vero cells from a common serotype-2 clinical isolate of dengue virus. Using these two viral variants, we established mouse infection models that reproduce severe manifestations of dengue diseases in AG6 mice. Such lethal model with 8H2-7-LP or 1D4-5-SP viruses in AG6 mice can be used in pathology study, for example, to explore the viral and host factors that mediate hemorrhagic manifestation of dengue viruses. On the other hand, DENV-2 8H2-7-LP was used in another infection model which resembles self-limited infection of dengue virus in Balb/c mice. And this transient infection model is more suitable for study of T cell responses, since the immune cell lineages are intact and the IFN-γ responses are normal during the development of adaptive immunity. Additionally, this self-limited infection model is also suitable for investigating how murine host resolves dengue infection.
In previous investigations of dengue infection, only a limited number of viral strains were found to infect immune deficient mice, in which human illness without neurological disease can be studied, and lethal doses of these viruses ranged from 104 to 107 PFU (Sarathy et al. 2015). One often used virus is dengue serotype-2 strain D220, which was obtained from a neuropathic strain PL046 through alternative passage between C6/36 mosquito cells and AG129 mice. The LD50 of this virus in AG129 is 105–106 PFU (Orozco et al. 2012). Another is D2Y98P-PP1, a double-plaque purified clone from a laboratory strain D2Y98P, which had been passaged in C6/36 cell for 20 times. And the lethal dose of D2Y98P-PP1 in AG129 mice was documented higher than 105 PFU (Tan et al. 2011). In contrast, the virus strains in this study had 100% mortality within 11 days after inoculation in AG6 mice of 6–8 weeks old even when the dose was reduced to 103 PFU, i.e., approximately 100 times more lethal than the D220 and D2Y98P-PP1 viral strains, and thus making DENV-2 8H2-7-LP virus a unique tool for studying the pathogenesis of dengue virus. Through Evans blue staining, we also confirmed the manifestation of plasma leakage in infected mice, which is a characteristic of severe dengue diseases in human. Collectively, we show the highly virulent dengue strain 8H2-7-LP is ideal for establishing lethal infection models at low inoculum.
To increase the utility of the new dengue viral variants in the context of relatively normal immune system, we have also adapted a recently developed model that transiently blocks IFNAR with antibodies in Balb/c mice. High level of viral RNA was maintained within the first week after infection by DENV-2 8H2-7-LP, but only low level of transient viral RNA was detected at 3–4 dpi in mice administered with DENV-2 1D4-5-SP. Though no clinical symptom was observed, in mice infected with DENV-2 8H2-7-LP, we successfully detected infectious virus in sera, and viral RNA in multiple organs including liver, spleen and eyes. The viruses might have been eliminated from mice after 7 days of infection, accompanied by the maturation of DENV adaptive immunity. Thus, such characteristic and progress of infection is consistent with what was observed in patients with mild disease (WHO 2009).
In previous animal models, DENV infection of adult WT mice required extremely high inoculum, intracranially injection, or mouse adapted viruses. A portion of these models led to neurological symptoms or additional clinically irrelevant manifestations, limiting their use for pathology and therapeutic study (Plummer and Shresta 2014; Sarathy et al. 2015). To our knowledge, this is the first study which developed a DENV infection model in WT mice, using a moderate dose of a non-adapted virus strain through physiologically more relevant s.c. injection route.
It is often observed that many viruses form heterogenous plaques of different sizes in vitro, indicating that viruses exist as quasispecies during transmission or passaging (Kato et al. 2017; Moser et al. 2018), but the virological differences among various phenotypic variants have not been carefully examined. Clinically, such viral diversity may assist arbovirus to survive under different selection pressure from two distinct hosts, invertebrate and vertebrate. DENV-2 8H2-7-LP exhibits better replication capability than DENV-2 1D4-5-SP in Vero cells, whereas their growth in mosquito cells is similar, reflecting possibility that the large-plaque variants in the original isolate contribute more to viral fitness in mammalian host. Consistently, in mouse model, DENV-2 8H2-7-LP presents higher pathogenicity in AG6 mice and better replication in Balb/c mice.
Comparing the coding sequence of DENV-2 8H2-7-LP and DENV-2 1D4-5-SP, we found 7 amino acid variations on E, NS3, NS4B and NS5 proteins. E protein is responsible for virus attachment and entry; NS3, NS4B and NS5 are all the components of replication complex and essential antagonists for type I IFN responses (Green et al. 2014). Thus, further investigation of the underlying mechanism for the different phenotypes of these two viruses is valuable for understanding the molecular basis of dengue virulence and DENV-host interaction.
Acknowledgements
This work was supported in part by Science and Technology Commission of Shanghai Municipality (Grant No. 19ZR1462900) and the National Science Foundation of China (Grant No. 31300757) to JS; and the National Science Foundation of China (Grant No. 31870916 and No. 31670941) to XJ.
Author Contributions
JS and XJ designed and supervised the study. ZHZ and JS carried out the experiments. ZHZ, JS and XJ analyzed the data and wrote the paper. ZHL and ML assisted with animal experiments on AG6 mice. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This study was approved by Animal Ethics Committee of Institut Pasteur of Shanghai, Chinese Academy of Sciences (Approval number: A2018016). All mouse experiments were performed strictly according to rules of care and use of laboratory animals established by Institutional Animal Care and Use Committee of the Institut Pasteur of Shanghai, Chinese Academy of Sciences.
Contributor Information
Xia Jin, Email: jinxia@shphc.org.cn.
Jin Sun, Email: jsun01@ips.ac.cn.
References
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkina R, Berges BK, Palmer BE, Remling L, Neff CP, Kuruvilla J, Connick E, Folkvord J, Gagliardi K, Kassu A, Akkina SR. Humanized Rag1-/- gammac-/- mice support multilineage hematopoiesis and are susceptible to HIV-1 infection via systemic and vaginal routes. PLoS ONE. 2011;6:e20169. doi: 10.1371/journal.pone.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Morrison J, Laurent-Rolle M, Belicha-Villanueva A, Plumlee CR, Bernal-Rubio D, Williams KL, Harris E, Fernandez-Sesma A, Schindler C, Garcia-Sastre A. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Hofman FM, Kung JT, Lin YD, Wu-Hsieh BA. Both virus and tumor necrosis factor alpha are critical for endothelium damage in a mouse model of dengue virus-induced hemorrhage. J Virol. 2007;81:5518–5526. doi: 10.1128/JVI.02575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- Green AM, Beatty PR, Hadjilaou A, Harris E. Innate immunity to dengue virus infection and subversion of antiviral responses. J Mol Biol. 2014;426:1148–1160. doi: 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J Infect Dis. 1952;90:1–9. doi: 10.1093/infdis/90.1.1. [DOI] [PubMed] [Google Scholar]
- Jin X, Lee M, Shu J. Dengue fever in China: an emerging problem demands attention. Emerg Microbes Infect. 2015;4:e3. doi: 10.1038/emi.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato F, Tajima S, Nakayama E, Kawai Y, Taniguchi S, Shibasaki K, Taira M, Maeki T, Lim CK, Takasaki T, Saijo M. Characterization of large and small-plaque variants in the Zika virus clinical isolate ZIKV/Hu/S36/Chiba/2016. Sci Rep. 2017;7:16160. doi: 10.1038/s41598-017-16475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Pan P, He Q, Kong X, Wu K, Zhang W, Liu Y, Huang H, Liu J, Zhang Z, Wu Lai X, Liu X, Wu J. Molecular epidemiology demonstrates that imported and local strains circulated during the 2014 dengue outbreak in Guangzhou, China. Virol Sin. 2017;32:63–72. doi: 10.1007/s12250-016-3872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Yang R, Liu Z, Li M, Liu H, Jin X. Recombinant Zika virus envelope protein elicited protective immunity against Zika virus in immunocompetent mice. PLoS ONE. 2018;13:e0194860. doi: 10.1371/journal.pone.0194860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser LA, Boylan BT, Moreira FR, Myers LJ, Svenson EL, Fedorova NB, Pickett BE, Bernard KA. Growth and adaptation of Zika virus in mammalian and mosquito cells. PLoS Negl Trop Dis. 2018;12:e0006880. doi: 10.1371/journal.pntd.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco S, Schmid MA, Parameswaran P, Lachica R, Henn MR, Beatty R, Harris E. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J Gen Virol. 2012;93:2152–2157. doi: 10.1099/vir.0.045088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer EM, Shresta S. Mouse models for dengue vaccines and antivirals. J Immunol Methods. 2014;410:34–38. doi: 10.1016/j.jim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, Shresta S. Gamma interferon (IFN-gamma) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-alpha/beta receptor-deficient mice. J Virol. 2012;86:12561–12570. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin AB, Schlesinger RW. Production of immunity to dengue with virus modified by propagation in mice. Science. 1945;101:640–642. doi: 10.1126/science.101.2634.640. [DOI] [PubMed] [Google Scholar]
- Sarathy VV, Milligan GN, Bourne N, Barrett AD. Mouse models of dengue virus infection for vaccine testing. Vaccine. 2015;33:7051–7060. doi: 10.1016/j.vaccine.2015.09.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J Virol. 2006;80:10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John AL, Rathore APS. Adaptive immune responses to primary and secondary dengue virus infections. Nat Rev Immunol. 2019;19:218–230. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- Stabell AC, Meyerson NR, Gullberg RC, Gilchrist AR, Webb KJ, Old WM, Perera R, Sawyer SL. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife. 2018;7:e31919. doi: 10.7554/eLife.31919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li M, Wang Y, Hao P, Jin X. Elaboration of tetravalent antibody responses against dengue viruses using a subunit vaccine comprised of a single consensus dengue envelope sequence. Vaccine. 2017;35:6308–6320. doi: 10.1016/j.vaccine.2017.09.063. [DOI] [PubMed] [Google Scholar]
- Tan GK, Ng JK, Lim AH, Yeo KP, Angeli V, Alonso S. Subcutaneous infection with non-mouse adapted Dengue virus D2Y98P strain induces systemic vascular leakage in AG129 mice. Ann Acad Med Singapore. 2011;40:523–532. [PubMed] [Google Scholar]
- WHO . In: Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. New. Geneva: WHO Guidelines Approved by the Guidelines Review Committee; 2009. [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhang FC, Zhu Q, Wang J, Hong WX, Zhao LZ, Deng YQ, Qiu S, Zhang Y, Cai WP, Cao WC, Qin CF. Epidemiological and Virological Characterizations of the 2014 Dengue Outbreak in Guangzhou. China. PLoS ONE. 2016;11:e0156548. doi: 10.1371/journal.pone.0156548. [DOI] [PMC free article] [PubMed] [Google Scholar]