Dear Editor,
Since December 2019, the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induced human disease, the coronavirus disease 2019 (COVID-19), has been reported in more than 200 countries and areas around the world (Wu et al. 2020). By May 13, there are 4,098,018 confirmed cases with 283,271 deaths according to the World Health Organization (WHO) report, and the numbers are keeping growing fast. Rapid and accurate diagnosis of suspected cases, effective isolation of infected patients and active treatments are the most important procedures for epidemic prevention and control.
At present, the conventional methods used in virological diagnosis include: (1) Virus isolation and culture; (2) detection of viral-specific antibodies; (3) detection of viral nucleic acid by PCR, sequencing, etc. (Zu et al. 2020). However, there are some obvious disadvantages to each of those diagnosis methods. For example, virus isolation and culture are difficult to implement in routine clinical laboratories. Antibodies can usually be detected on 7–21 days post-infection, which is not suitable for early diagnosis. For biosafety reason, the heat inactivation of the virus is required and this will reduce the antibody levels by an average of 50%. Furthermore, cross-reaction cannot be entirely avoided because of the conserved sequence and structure between human coronaviruses. Detection of viral nucleic acid by qRT-PCR is the most popular method till now (Wang et al. 2020), but a high rate of false-negative results is reported during the COVID-19 tests (Chan et al. 2020; Zu et al. 2020). The most possible reason for the failure of detection is that the swab samples from the nasal cavity or throat may not always contain enough genetic material to provide an accurate test. Therefore, developing a novel method to supplement the current diagnosis of COVID-19 has become imperative.
During the outbreak of SARS-CoV in 2003, scientists have reported that the nucleocapsid (N) protein of SARS-CoV is a good diagnostic biomarker (Che et al. 2004; Lau et al. 2004; Li et al. 2005). In the early stage of SARS infection (day 1–10), the detection of viral N protein is much more sensitive (90% positive) than that of viral nucleic acid (42.9%) or antibodies (21.4%) in patients’ sera (Li et al. 2005). Another study suggested that N protein in the blood samples of SARS patients can be detected as early as day 1 after disease onset. The sensitivity of detection was up to 94% with serum samples taken during the first 5 days after onset (Che et al. 2004). Compared with the detection of N antibody, catching the N antigen shows no cross-reaction with other related human and animal coronaviruses. N protein can also be detected in various patient samples including nasopharyngeal aspirate, urine, and fecal (Lau et al. 2004). The ELISA based viral antigen detection may have greater sensitivity, specificity, and reliability than the nucleic acid amplification assay.
Aptamers, in vitro selected by systematic evolution of ligands by exponential enrichment (SELEX) technology (Tuerk and Gold 1990), are DNA or RNA molecules that are capable of binding a wide range of molecules with high affinity and specificity. Aptamers that recognize viral proteins have been employed in the rapid detection of viruses and as antiviral agents in treating infection (Zou et al. 2019). For example, aptamers for the hemagglutinin (HA) proteins have been used in the detection of various strains of influenza virus including H1N1, H3N2, H5N1, and H9N2 (Zou et al. 2019). Zika NS1-binding ssDNA aptamers can be used for medical diagnosis of Zika virus infection (Lee and Zeng 2017). DNA aptamers against the dengue virus (DENV) envelop protein can neutralize the infections by all four serotypes of DENVs (Chen et al. 2015). Aptamers may also suppress viral infection by interfering with the enzymatic activities of viral encoded enzymes (Jang et al. 2008; Shum and Tanner 2008).
The purpose of this study was to identify aptamers that may be applied to the early diagnosis of COVID-19. In a previous study, Korean scientists have shown that a single strand DNA aptamer can specifically bind to SARS-CoV N protein (Cho et al. 2011). Since the N protein (YP_009724397.2) of SARS-CoV-2 shares 91% sequence homology with the N protein (NP_828858.1) of SARS-CoV (Supplementary Figure S1), we wondered whether the reported DNA aptamer against SARS-CoV N protein binds to SARS-CoV-2 N protein. Furthermore, we modified the reported DNA aptamer (Aptamer 1) into different variants and examined their binding capacities to SARS-CoV-2 N protein.
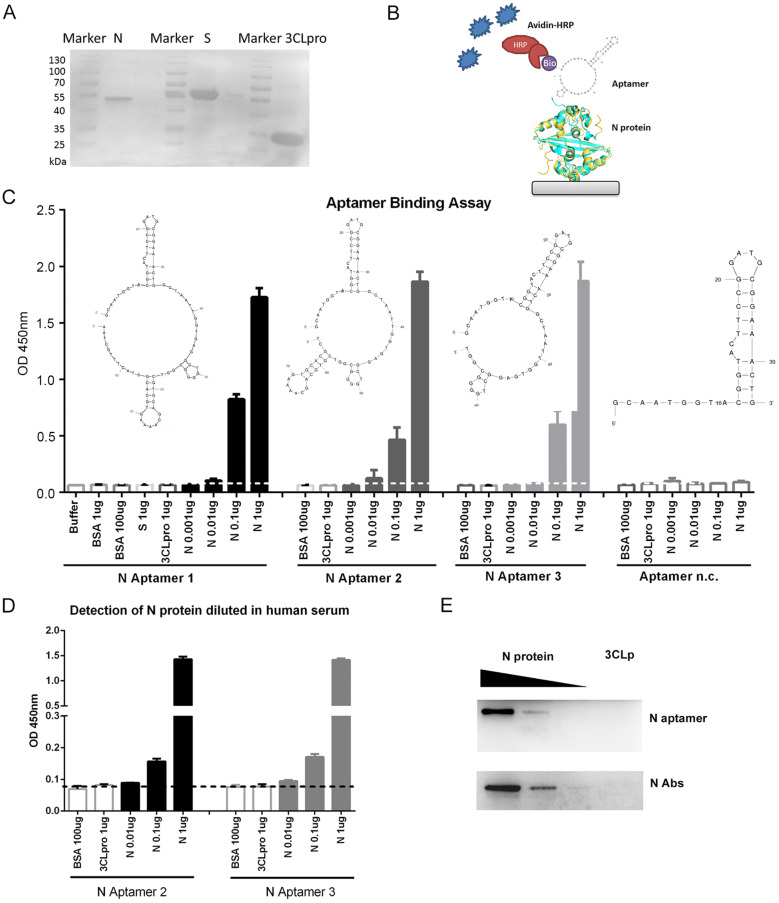
The 5′ biotinylated Aptamer 1 and its derivatives, Aptamer 2, 3, and negative control (Aptamer n.c.) (Supplementary Table S1) were synthesized by Sangon Biotech (Sangon, Shanghai, China). The secondary structures of the aptamers were predicted by the MFOLD program (http://unafold.rna.albany.edu) (Zuker 2003). The 6 × Histidine tagged N, Spike protein receptor-binding domain (S-RBD) (fused with Fc) and 3C-like protease (3CLpro) proteins from SARS-CoV-2 were prokaryotic expressed in Escherichia coli BL21 (DE3) strain by the addition of 0.5 mmol/L isopropyl-b-D-thiogalactopyranoside (IPTG), respectively. The recombinant His-tag proteins were purified using cOmplete His-Tag Purification Resin (Roche, Mannheim, Germany) following the manufacturer’s instruction. The protein concentrations were analyzed using the bicinchoninic acid assay (Thermo Scientific, IL, USA) with bovine serum albumin (BSA) as a standard. Coomassie blue staining of purified proteins after sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) confirmed the purity of these proteins that could be used for the following assays (Fig. 1A).
Fig. 1.
DNA aptamers specifically bind to SARS-CoV-2 N protein. A The expression and purification of SARS-CoV-2 N, S-RBD-Fc and 3CLpro proteins with a His-Ni–NTA system. B A sketch map for the ELAA. Briefly, the N protein was serially diluted and coated on the plates. After blocking, the N protein was detected with biotinylated ssDNA aptamers followed by HRP conjugated Avidin system. C Aptamer 1, 2 and 3 binds SARS-CoV-2 N protein in an Enzyme-Linked Aptamer Binding Assay (ELAA). BSA, SARS-CoV-2 S-RBD and 3CLpro proteins served as negative controls. Aptamer n.c. binds to none of the tested proteins. D Detection of N protein in diluted human serum by ssDNA aptamers. E ssDNA aptamer 2 specifically detects SARS-CoV-2 N protein in Western Blot assay. Antibody against N protein served as controls. Serially diluted N proteins (10 μg, 1μg, and 0.1μg) and 10 μg of 3CLpro protein were probed with N Aptamer 2 and N antibody, respectively. The data shown are representative of 3 independent experiments.
To investigate the binding affinity of three aptamers to SARS-CoV-2 N protein, we performed The Enzyme-Linked Aptamer Binding Assay (ELAA) as shown in Fig. 1B. Briefly, 96-well plate (Thermo Scientific, IL, USA) was employed to immobilize the serial diluted SARS-CoV-2 N protein to the plate overnight and blocked with 5% BSA in phosphate-buffered saline with Tween-20 (PBST) at room temperature for 1 h. After washing, 100 nmol/L of the 5-biotinylated aptamers (100 μL), denatured at 90 °C for 10 min following 10 min on ice, was added into the well and incubated at room temperature for 1 h. Then, the avidin coupled with horseradish peroxidase (HRP) (1:1000) was used to detect the biotin signal. The color development was carried out using 3,3′,5,5′-tetramethylbenzidine (TMB) substrate and stop solution of 2 mol/L H2SO4, and the absorbance was measured at 450 nm using Synergy H4 Hybrid Reader (BioTek, VT, U.S.A).
Aptamer binding assay revealed that all three Aptamers (#1, 2 and 3) specifically bound to SARS-CoV-2 N protein with a similar affinity (Fig. 1C), suggesting that the first two stem-loops in the aptamer are required for binding to SARS-CoV-2 N protein. The negative control aptamer (n.c.) with only one loop cannot bind to SARS-CoV-2 N protein. With one step ELAA, the SARS-CoV-2 N protein was able to be detected with a concentration as low as 10 ng/mL (Fig. 1C). Interestingly, SARS-CoV-2 N protein diluted in human sera from three healthy donators was also detectable using aptamer 2 or aptamer 3 as probes (Fig. 1D), implicating that detection of N protein in serum samples with aptamer may be possible for the rapid diagnosis of COVID-19.
Additionally, to compare the binding efficacy of the aptamer with a commercial antibody, we performed Western blot analysis with aptamer 2 or anti-N antibody (Sino Biological, Beijing, China). 3CLpro or serial diluted SARS-CoV-2 N proteins were separated by 10% SDS-PAGE and then transferred onto polyvinylidene fluoride (PDVF) membranes. After blocking with 5% nonfat milk, the membranes were probed with 5′ biotinylated aptamer 2 or anti-N antibody, washed, and incubated with streptavidin-HRP or goat anti-mouse IgG -labeled secondary antibody, respectively. The protein bands were visualized by the enhanced chemiluminescence (ECL) kit (Tanon, Shanghai, China). The images were photographed with the Tanon Luminescent Imaging System (Tanon, Shanghai, China). When various amounts of N proteins (0.01–10 μg) were analyzed by immunoblotting with 100 nmol/L of 5′ biotinylated aptamer 2, the signal increased in a dose-dependent manner (Fig. 1E), which was similar to the results from the commercial anti-N protein antibody. This suggested that these aptamers could be used as alternative reagents to replace the anti-N antibody for the detection of SARS-CoV-2 N protein. Importantly, DNA aptamers can be synthesized easily while antibody generation requires immunization of animals. Also, aptamers are less costive and have higher stability than protein antibodies. All these features make aptamers more useful in the development of diagnostic systems.
Altogether, we have identified a novel method for the detection of SRAS-CoV-2 N protein using DNA based aptamers. Although the aptamers used in this study were designed based on an aptamer previously selected for SARS-CoV N protein, they bind to SRAS-CoV-2 N protein with a high affinity. Most importantly, SARS-CoV-2 N protein shares very low similarity (16%–38%) with N protein from other five known human coronaviruses except for SARS-CoV. Because there are no SARS-CoV cases reported since 2004, our aptamer-based method can be used as a supplementation to the current diagnosis of SARS-CoV-2. Among the three N aptamers, aptamer 3 has the smallest molecular size. It will have the minimal cross-reaction to other proteins and be more stable than other aptamers. However, with the lack of serum samples collected from SARS-CoV-2 infected patients, the diagnostic performance such as sensitivity and specificity needs further investigation. Nevertheless, we have developed a novel method based on ssDNA aptamer for the detection of SARS-CoV-2 N protein, which may have potentials for the early diagnosis of SARS-CoV-2. Given the fact that N protein is also critical for viral genome assemble and antagonism of host immune responses (Chang et al. 2014), the N protein aptamers might be further considered used in the antiviral therapy by interfering with the N protein function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, National Natural Science Foundation of China (31770933, 81971917 and 81872103), Natural Science Foundation of Colleges in Jiangsu Province (17KJA310005), Open Project Fund from State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE1903) and Key Laboratory of Reproduction Regulation of NHC (No. KF2018-01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The study was approved by the ethics committees of Soochow University. Normal human sera were obtained from healthy volunteers. All participants provided written informed consent.
Contributor Information
Xiaohua Ni, Email: xhni_sippr@163.com.
Jianfeng Dai, Email: daijianfeng@suda.edu.cn.
References
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che XY, Hao W, Wang Y, Di B, Yin K, Xu YC, Feng CS, Wan ZY, Cheng VC, Yuen KY. Nucleocapsid protein as early diagnostic marker for SARS. Emerg Infect Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Hsiao WH, Lee HC, Wu SC, Cheng JW. Selection and characterization of DNA aptamers targeting all four serotypes of dengue viruses. PLoS ONE. 2015;10:e0131240. doi: 10.1371/journal.pone.0131240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Woo HM, Kim KS, Oh JW, Jeong YJ. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J Biosci Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KJ, Lee NR, Yeo WS, Jeong YJ, Kim DE. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem Biophys Res Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Wong BH, Tsoi HW, Woo GK, Poon RW, Chan KH, Wei WI, Peiris JS, Yuen KY. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in sars patients by enzyme-linked immunosorbent assay. J Clin Microbiol. 2004;42:2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zeng H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal Chem. 2017;89:12743–12748. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- Li YH, Li J, Liu XE, Wang L, Li T, Zhou YH, Zhuang H. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J Virol Methods. 2005;130:45–50. doi: 10.1016/j.jviromet.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum KT, Tanner JA. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. ChemBioChem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kang H, Liu X, Tong Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J Med Virol. 2020 doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Wu J, Gu J, Shen L, Mao L. Application of aptamers in virus detection and antiviral therapy. Front Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus Disease 2019 (COVID-19): a perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490:200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.