T lymphocytes are crucial for antiviral responses and provide a promising repertoire for potential therapies of viral diseases such as cytomegalovirus (CMV) infection1 and the ongoing COVID-19 pandemic caused by SARS-CoV-2.2 CMV-related diseases occur once the host immune system is impaired or lacks a protective repertoire of virus-specific T lymphocytes.3 Adoptive transfer of T-cell receptor (TCR)-engineered T cells (TCR-Ts) provides an encouraging alternative treatment option for patients with CMV reactivation.4 However, generating TCR-Ts requires the identification of epitope-specific and functional TCR pairs. Modern single-cell sequencing techniques open up the ability to unravel TCR repertoires,5 which offers a potential opportunity to screen functional TCR pairs for TCR-T therapy. Here, we report an efficient approach that combines ex vivo CD8+ T-cell stimulation with single-cell RNA and TCR V(D)J sequencing to identify CMV-specific TCRs for generating TCR-Ts.
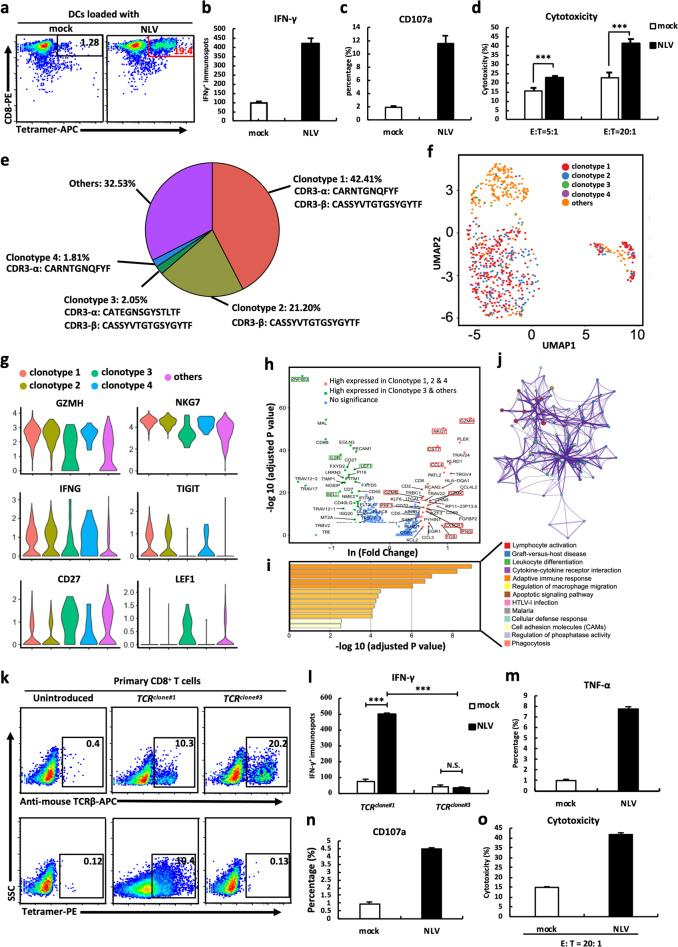
To identify functional CMV-specific TCRs, autologous dendritic cells (DCs) were loaded with synthetic CMV-NLV peptides (pp65495-503, an immunodominant CMV epitope). Purified CD8+ T lymphocytes isolated from healthy HLA-A*02:01+ donors were stimulated with CMV-NLV-loaded DCs. Following two consecutive stimulations (14 days), an NLV-MHC tetramer assay showed that 19.4% of stimulated CD8+ T lymphocytes reacted specifically to NLV, compared with 1.28% in the controls (Fig. 1a). To investigate the functionality of NLV-specific T cells, we evaluated various immune responses against NLV-loaded T2 cells. Fourfold higher amounts of IFN-γ were produced by NLV-specific T cells when stimulated with NLV-loaded T2 cells than by NLV-specific T cells when stimulated with mock-loaded T2 cells (Fig. 1b). FACS-based analysis of CD107a degranulation validated that the NLV-specific T cells responded actively to NLV-loaded T2 cells (Fig. 1c). CFSE/PI cell-mediated cytotoxicity assays further showed that NLV-specific T cells could efficiently lyse NLV-loaded T2 cells at E:T ratios of 5:1 to 20:1 (Fig. 1d). These results collectively suggested the successful ex vivo generation of functionally competent CD8+ NLV-specific T cells.
Fig. 1.
TCRs were identified by single-cell sequencing of engineered CD8+ T cells. a NLV-specific CD8+ T cells stained by NLV-MHC tetramer. IFN-γ secretion (b), CD107a degranulation (c), and cytotoxic activity (d) of NLV-stimulated T cells in response to T2 cells loaded with mock or NLV peptide. e The percentage and CDR3 amino acid sequences of distinct clonotypes. f Cells colored by clonotypes on the UMAP projection. g Violin plots of differentially expressed genes. h Differential gene expression analysis of functional gene sets in clusters 1 and 2. Each dot represents a gene. i GO and KEGG pathway enrichment analyses using genes upregulated in cluster 1. j Network of enriched terms in Fig. 2h colored by clonotype ID, in which nodes that share the same clonotype ID are typically close to each other. k Expression of introduced TCRclone#1 and TCRclone#3 in CD8+ T cells measured by NLV-MHC tetramer and anti-mouse TCRβ mAb. l IFN-γ ELISPOT results of TCRclone#1- and TCRclone#3-transduced CD8+ T cells in response to T2 cells loaded with mock or NLV peptide. TNF-α secretion (m), CD107a degranulation (n), and cytotoxic activity (o) of TCRclone#1-transduced CD8+ T cells in response to T2 cells loaded with mock or NLV peptide. Data are the mean ± SEM. Paired two-tailed Student’s t tests were conducted. P < 0.05 was considered significant. ***p < 0.001; N.S. not significant
To identify sequences of NLV-specific TCRs, NLV-MHC tetramer+ and CD8+T cells were sorted by flow cytometry and analyzed with single-cell RNA and TCR V(D)J sequencing. A total of 781 single cells with high sequencing quality were captured by both single-cell RNA and TCR sequencing. T cells with identical TCR sequences were defined as one TCR clonotype and ranked according to frequencies (Fig. 1e). Unsupervised clustering showed that clonotypes 1, 2, and 4 were located in one cluster (cluster 1), while clonotype 3 and other low-frequency clonotypes had a distinct expression profile (cluster 2) (Fig. 1f). Differential gene expression (DGE) analysis revealed that cytotoxic effector molecules (GZMH and NKG7), the proinflammatory cytokine IFNG, and the exhaustion marker TIGIT were highly expressed in cluster 1, suggesting that cluster 1 cells had been activated and had differentiated. In contrast, naïve cell-related markers (CD27 and LEF1) were highly expressed in cluster 2, suggesting naïve populations (Fig. 1g). Furthermore, DGE analysis with a T-cell-specific functional gene set6 showed high expression of 40 functional genes (p value < 0.01, Wilcoxon test, and fold change ≥ 2) in cluster 1 (Fig. 1h). Gene ontology analysis and KEGG pathway enrichment analysis showed that these 40 genes are functionally involved in cytokine receptor–ligand interactions and lymphocyte activation (Fig. 1i, j). These results suggest that cluster 1 represents potentially functional NLV-specific T cells, whereas cluster 2 represents naïve CD8+ T cells.
We selected the complete TCR pair from clonotype 1 (NLV-specific) and clonotype 3 (naïve) for TCR-T generation. The reconstructed full-length TCR pairs were designated TCRclone#1 and TCRclone#3, in which the TCR constant regions were replaced with murine ones to eliminate potential mispairing and enhance the expression of TCR pairs (Fig. S1a).7 Lentiviruses expressing TCRclone#1 or TCRclone#3 were transduced into primary CD8+ T cells with high transduction efficiency (Fig. S1b, c), and the cells were evaluated 7 days after transduction with functional analyses. Approximately 10% of TCRclone#1-engineered T cells were positive for both NLV-MHC tetramer and TCRβ mAb staining. As expected, TCRclone#3-engineered T cells were positive for TCRβ mAb staining (20.2%) but negative for NLV-MHC tetramer reactivity (Fig. 1k). Using genetically engineered TCRβ-deficient Jurkat cells, we further confirmed the difference in reactivity to NLV-MHC tetramers between the two clonotypes (Fig. S1d). Analysis of IFN-γ production showed that TCRclone#1-transduced T cells significantly increased IFN-γ secretion in response to only NLV-loaded T2 target cells and not the mock control (Fig. 1l, P value < 0.001 and fold change > 6). We also evaluated other immune responses and found that TNF-α production (Fig. 1m, p value<0.001, fold change>7), CD107a expression (Fig. 1n, p value < 0.001, fold change > 4), and cytotoxicity (Fig. 1o, p value < 0.001, fold change > 2) were significantly higher in TCRclone#1-engineered T cells reacting against NLV-loaded T2 cells than in those reacting against the mock control. These results suggest that engineered CD8+ T cells with the TCR pair from clonotype 1 respond specifically and functionally to CMV-NLV.
We present a single-cell approach for the generation of TCR-engineered CD8+ T cells. This strategy can potentially be applied to other diseases in addition to CMV-associated diseases, such as the ongoing pandemic caused by SARS-CoV-2, to generate immunotherapeutic treatments for viral diseases. There are two essential steps to ensure the success of this approach: identification of viral-specific immunodominant epitopes and identification of reactive and functional TCR pairs. The first step can be achieved using computing-based large-scale screening methods.8 We show in this study that the second step can be achieved with a combination of ex vivo CD8+ T-cell stimulation followed by single-cell RNA and TCR sequencing. Although the naïve TCRs also evolved in stimulated cells (e.g., clonotype 3), we could predict naïve cell status using transcriptional information from single-cell RNA sequencing. Our findings correlate with a recent discovery that antigen-experienced CD8+ T cells can display a naïve-like phenotype.9 In summary, defined clusters of TCRs are highly associated with the activation and differentiation status of T cells. Correlative analysis of the single-cell transcriptome and TCR sequencing data provides a useful strategy to identify functional TCR pairs against viral epitopes. Our approach can enhance the development of TCR-T therapies for the treatment of infectious diseases caused by human transmissible viruses and potentially for the severe acute respiratory syndrome caused by coronaviruses.10
Supplementary information
Acknowledgements
This project is supported by the Science, Technology and Innovation Commission of Shenzhen Municipality under grant No. JCYJ20170303151334808 and grant No. JSGG20180508152912700. The first author would like to acknowledge financial support from the China Scholarship Council (CSC) (grant No. 201904910476).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Fei Wang, Qumiao Xu, Zhenkun Zhuang
Contributor Information
Xiuqing Zhang, Email: zhangxq@genomics.cn.
Linnan Zhu, Email: zhulinnan@genomics.cn.
Cheng-chi Chao, Email: chengchic@abvisioninc.com.
Supplementary information
The online version of this article (10.1038/s41423-020-0466-z) contains supplementary material.
References
- 1.Smith C, et al. T cell repertoire remodeling following post-transplant T cell therapy coincides with clinical response. J. Clin. Invest. 2019;129:5020–5032. doi: 10.1172/JCI128323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 3.Stern L, et al. Human cytomegalovirus latency and reactivation in allogeneic hematopoietic stem cell transplant recipients. Front Microbiol. 2019;10:1186. doi: 10.3389/fmicb.2019.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schub A, et al. CMV-specific TCR-transgenic T cells for immunotherapy. J. Immunol. 2009;183:6819–6830. doi: 10.4049/jimmunol.0902233. [DOI] [PubMed] [Google Scholar]
- 5.De Simone M, Rossetti G, Pagani M. Single cell T cell receptor sequencing: techniques and future challenges. Front Immunol. 2018;9:1638. doi: 10.3389/fimmu.2018.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen CJ, et al. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang GL, et al. Hotspot Hunter: a computational system for large-scale screening and selection of candidate immunological hotspots in pathogen proteomes. BMC Bioinform. 2008;9(Suppl 1):S19. doi: 10.1186/1471-2105-9-S1-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulko V, et al. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat. Immunol. 2016;17:966–975. doi: 10.1038/ni.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh HL, et al. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.