Abstract
Objective
This study aimed to investigate the role of long non‐coding RNA (lncRNA) THRIL in coronary heart disease (CHD) patients.
Methods
A total of 420 patients who underwent coronary arteriography due to suspected symptoms of CHD were enrolled, in which 220 were diagnosed as CHD and 200 were set as control subjects. LncRNA THRIL in plasma samples of CHD patients and control subjects was detected by reverse transcription‐quantitative polymerase chain reaction. Gensini score and biochemical indexes were evaluated in CHD patients and control subjects. Plasma inflammatory cytokines were detected, and major adverse cardiovascular events (MACE) were recorded in CHD patients.
Results
Both before and after adjustment by age/gender, lncRNA THRIL was increased in CHD patients compared with control subjects (both P < .001), and it well predicted enhanced CHD risk by receiver operating characteristic curves. For coronary artery stenosis, it was positively correlated with Gensini score (P < .001, r = .430). For clinical characteristics, lncRNA THRIL was positively correlated with diabetes mellitus occurrence (P < .001) and fasting blood glucose (FBG) level (P = .029, r = .147). For inflammation, it was positively associated with CRP (P < .001, r = .374), TNF‐α (P < .001, r = .249), IL‐1β (P = .001, r = .222), IL‐8 (P < .001, r = .254), and IL‐17 (P = .011, r = .172), while negatively correlated with IL‐10 (P < .001, r = −.244). For prognosis, lncRNA THRIL was positively associated with MACE accumulating rate (P = .037) in CHD patients.
Conclusion
Long non‐coding RNA THRIL was increased in CHD patients and well predicted elevated CHD risk. Moreover, it was correlated with enhanced coronary stenosis, systematic inflammation, FBG level, and MACE risk in CHD patients.
Keywords: coronary heart disease, disease risk, inflammation, long non‐coding RNA THRIL, major adverse cardiovascular events
1. INTRODUCTION
Coronary heart disease (CHD) is pathologically characterized by atherosclerosis, which is a chronic pathological process closely related to inflammation.1, 2, 3 Although the management (including pharmacologic therapy such as statins and non‐pharmacologic therapy such as coronary artery bypass grafting) and recognition of risk factors (such as smoking, complication with diabetes mellitus, or hypertension) for CHD have been properly established, CHD is still the leading cause of mortality with 8.9 million cases of death in 2017, and the prognosis is still far from satisfactory.4 Therefore, prediction and prevention of CHD are still of urgent need, and searching for novel biomarkers including long non‐coding RNA (lncRNA) seems a promising solution.5, 6, 7
Long non‐coding RNA tumor necrosis factor (TNF)‐α and heterogenous nuclear ribonucleoprotein L (hnRNPL)‐related immunoregulatory lincRNA (THRIL) are reported to promote inflammation by regulating several pathways (such as TNF‐α and nuclear factor‐kappa B [NF‐κB]).8, 9 Moreover, previous researches revealed that lncRNA THRIL is involved in the hypoxia‐induced injury of cardiomyocytes and participates in the regulation of myocardial infarction, one of the cardiovascular diseases.10, 11 Based on these studies, we hypothesized that lncRNA THRIL might play a critical role in CHD, whereas relevant information is unclear.
In the present study, the predictive value of lncRNA THRIL for CHD risk was evaluated; meanwhile, the correlation of lncRNA THRIL with disease progression and prognosis in CHD patients was investigated.
2. MATERIALS AND METHODS
2.1. Participants
Between July 2015 and June 2017, we consecutively recruited 420 patients who underwent coronary arteriography in our hospital due to unexplained chest pain or suspected symptoms of CHD. The inclusion criteria were as follows: (a) had unexplained chest pain or suspected symptoms of CHD (eg, angina or severe ventricular arrhythmia); (b) age above 18 years; (c) no severe infection, inflammatory, or autoimmune diseases; (d) no history of revascularization (eg, percutaneous coronary intervention [PCI] or coronary artery bypass grafting [CABG]); and (e) without hematologic malignancies or solid tumor. The exclusion criteria were as follows: (a) history of congenital heart disease, cardiomyopathy, or vasospastic angina; (b) had contraindications to coronary arteriography; (c) received immunosuppressive therapy within 3 months; (d) unable to follow up regularly; and (e) pregnant or lactating woman. This study was approved by the Institutional Review Board of our hospital. All participants provided written informed consent before enrollment.
2.2. Date and sample collection
The detailed clinical data including demographic characteristics, CHD risk factors, and biochemical indexes before coronary arteriography were recorded for all participants. The peripheral blood samples of participants were collected before coronary arteriography. After collection, the blood samples were separated at 3000 g for 20 minutes (4℃); then, plasma was isolated and stored at −80℃ until measurement.
2.3. Grouping and Gensini assessment
All participants underwent coronary arteriography, and the coronary angiograms were evaluated according to the AHA/ACC classification,12 and CHD was defined as at least one major epicardial vessel with >50% stenosis. All participants were divided into CHD group (N = 220) and control group (all major epicardial vessels with <50% stenosis, N = 200) according to the evaluation of coronary angiograms. The severity of coronary artery stenosis was assessed by Gensini scoring system, and the Gensini score was including the degree of luminal narrowing score and multiplying factor of principal vascular segment. The luminal narrowing was scored as follows: 1 (25% reduction of lumen diameter), 2 (50% reduction of lumen diameter), 4 (75% reduction of lumen diameter), 8 (90% reduction of lumen diameter), 16 (99% reduction of lumen diameter), and 32 (complete occlusion).13 The multiplying factor of principal vascular segment (such as ×1.0, ×2.5, and so on) was evaluated depending on the functional significance of the area supplied by that segment.13 The total Gensini score was calculated by multiplying the luminal narrowing score and the factor of principal vascular segment.
2.4. LncRNA THRIL detection
For all participants, the level of lncRNA THRIL in plasma was detected by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). RNA extraction was conducted using TRIzol™ Reagent (Thermo). Reverse transcription was performed with a PrimeScript™ RT reagent Kit (Takara) according to the manufacturer's instruction. qPCR was conducted with KOD SYBR® qPCR Mix (Toyobo) according to the manufacturer's guidance. The relative expression of lncRNA THRIL was calculated using the formula, and GAPDH was used as the internal reference. Primers (5′‐3′): LncRNA THRIL forward primer: GAGTGCAGTGGCGTGATCTC, reverse primer: AAAATTAGTCAGGCATGGTGGTG; GAPDH forward primer: GACCACAGTCCATGCCATCAC; reverse primer: ACGCCTGCTTCACCACCTT.
2.5. Inflammatory cytokines measurement
For the CHD patients, the level of inflammatory cytokines (including TNF‐α, interleukin [IL]‐1β, IL‐6, IL‐8, IL‐10, and IL‐17) in plasma was measured using commercial enzyme‐linked immunosorbent assay (ELISA) kits (Thermo) according to manufacturer's instruction.
2.6. Follow‐up
For the CHD patients, regular follow‐up was carried out until 2019/06/30. The median follow‐up duration was 33.0 months, the minimum follow‐up duration was 4.0 months, and the maximum follow‐up duration was 48 months. During the follow‐up, major adverse cardiovascular events (MACE) were recorded, and the MACE was defined as cardiovascular death, myocardial infarction, unplanned coronary revascularization, and hospital admission for cardiovascular cause.14
2.7. Statistical analysis
The normality of continuous variables was checked by Kolmogorov‐Smirnov test, and the variables were displayed as mean ± standard deviation (SD; normally distributed), or the median and interquartile range (IQR), or count and percentage (categorical variables). Comparison was determined by Student's t test, Wilcoxon rank‐sum test, or chi‐square test as appropriate. Correlation was analyzed by Spearman's rank correlation test or Wilcoxon rank‐sum test. The performance of lncRNA THRIL in discriminating different subjects was determined by receiver operating characteristic (ROC) curve. The age‐ and gender‐matched cohort was selected by propensity score matching (PSM). MACE accumulating rate was displayed using Kaplan‐Meier curve, and the difference of MACE accumulating rate was determined by log‐rank test. P value <.05 was considered as statistically significant. Statistical analysis and figure plotting were performed using SPSS 22.0 statistical (SPSS Inc) and GraphPad Prism 7.01 (GraphPad Software Inc).
3. RESULTS
3.1. Study flow
A total of 500 subjects who underwent coronary arteriography were initially screened, and 80 of them were excluded (31 subjects had history of congenital heart disease, cardiomyopathy, or vasospastic angina; 21 subjects had severe infection, inflammatory, or received immunosuppressive therapy within 3 months; 18 subjects refused to provide informed consent; seven subjects were unable to follow up regularly; and three subjects were complicated with hematologic malignancies or solid tumor). Subsequently, 420 subjects were eligible and their plasma samples were collected before they underwent coronary arteriography. After that, 220 subjects were diagnosed as CHD, and the remaining 200 subjects were set as control subjects. LncRNA THRIL in plasma was detected in both CHD patients and control subjects. Besides, in CHD patients, inflammatory cytokines in plasma were measured and MACE was recorded during follow‐up. The detailed study flow was shown in Figure 1.
Figure 1.
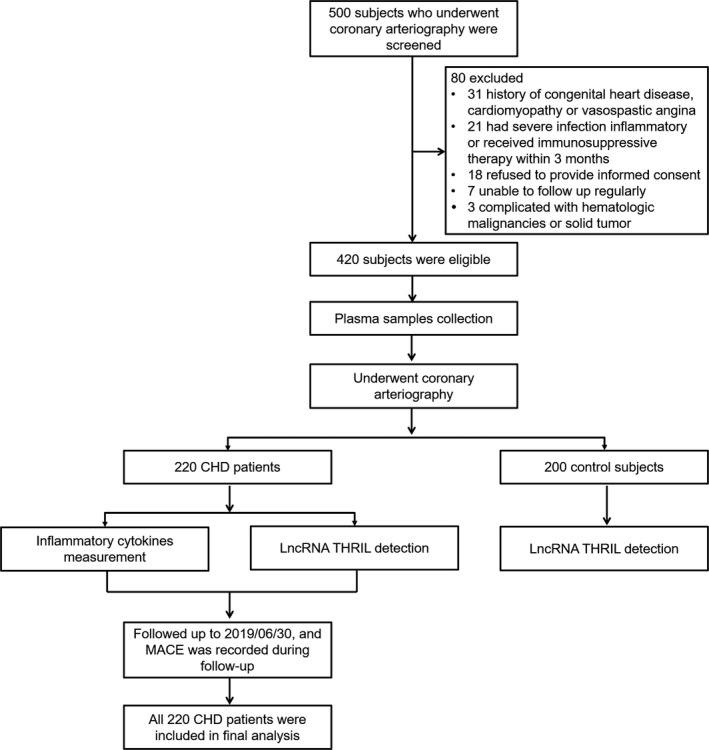
Flow chart. CHD, coronary heart disease; lncRNA, long non‐coding RNA; MACE, major adverse cardiovascular events; THRIL, tumor necrosis factor‐α and heterogenous nuclear ribonucleoprotein L‐related immunoregulatory long intergenic non‐coding RNA
3.2. Clinical characteristics of CHD patients and control subjects
For demographic characteristics, the mean age of CHD patients (62.8 ± 9.4 years) was increased compared with control subjects (59.7 ± 9.0 years; P = .001), while no difference was found in gender(P = .222) or BMI (P = .264) between the two groups. For CHD risk factors, the number of subjects with hypertension in CHD patients (173 [78.6%]) was increased compared to that in control subjects (139 [69.5%]; P = .032), and the number of subjects with diabetes mellitus in CHD patients (53 [24.1%]) was also decreased compared to that in control subjects (29 [14.5%]; P = .013), while no difference was found in smoke (P = .201), family history of CHD (P = .129), hyperlipidemia (P = .151), or hyperuricemia (P = .152) between the two groups. The median Gensini scores of CHD patients (39.5 [19.3‐59.8]) were dramatically increased compared to that of control subjects (1.0 [0.0‐2.0]; P < .001). For biochemical index, FBG (P = .012) and CRP (P < .001) were increased, while HDL‐C (P = .004) was decreased in CHD patients compared with control subjects. However, no difference was observed in TG (P = .111), TC (P = .183), LDL‐C (P = .098), Scr (P = .126), or SUA (P = .145) between the two groups. The detailed clinical characteristics were listed in Table 1.
Table 1.
Clinical characteristics
| Items | CHD patients (N = 220) | Control subjects (N = 200) | P value |
|---|---|---|---|
| Demographic characteristics | |||
| Age (y), mean ± SD | 62.8 ± 9.4 | 59.7 ± 9.0 | .001 |
| Gender, no. (%) | |||
| Female | 37 (16.8) | 43 (21.5) | .222 |
| Male | 183 (83.2) | 157 (78.5) | |
| BMI (kg/m2), mean ± SD | 23.7 ± 2.7 | 23.4 ± 2.9 | .264 |
| CHD risk factors | |||
| Smoke, no. (%) | 96 (43.6) | 75 (37.5) | .201 |
| Family history of CHD, no. (%) | 48 (21.8) | 32 (16.0) | .129 |
| Hypertension, no. (%) | 173 (78.6) | 139 (69.5) | .032 |
| Hyperlipidemia, no. (%) | 110 (50.0) | 86 (43.0) | .151 |
| Hyperuricemia, no. (%) | 84 (38.2) | 63 (31.5) | .152 |
| Diabetes mellitus, no. (%) | 53 (24.1) | 29 (14.5) | .013 |
| Gensini score, median (IQR) | 39.5 (19.3‐59.8) | 1.0 (0.0‐2.0) | <.001 |
| Biochemical indexes | |||
| FBG (mmol/L), mean ± SD | 5.7 ± 1.1 | 5.4 ± 1.1 | .012 |
| TG (mmol/L), mean ± SD | 1.8 ± 0.8 | 1.7 ± 0.8 | .111 |
| TC (mmol/L), mean ± SD | 4.7 ± 1.0 | 4.6 ± 1.0 | .183 |
| LDL‐C (mmol/L), mean ± SD | 3.0 ± 0.7 | 2.9 ± 0.7 | .098 |
| HDL‐C (mmol/L), mean ± SD | 1.1 ± 0.3 | 1.2 ± 0.3 | .004 |
| Scr (μmol/L), mean ± SD | 77.3 ± 15.2 | 75.1 ± 13.9 | .126 |
| SUA (μmol/L), mean ± SD | 346.2 ± 78.1 | 335.3 ± 74.1 | .145 |
| CRP (mg/L), median (IQR) | 16.9 (12.3‐22.6) | 7.3 (3.2‐11.2) | <.001 |
Abbreviations: BMI, body mass index; CHD, coronary heart disease; CRP, C‐reactive protein; FBG, fasting blood glucose; HDL‐C, high‐density lipoprotein cholesterol; IQR, interquartile range; LDL‐C, low‐density lipoprotein cholesterol; Scr, serum creatinine; SD, standard deviation; SUA, serum uric acid; TC, total cholesterol; TG, triglyceride.
3.3. LncRNA THRIL expression and its predictive value for CHD risk
Before adjustment, lncRNA THRIL was increased in CHD patients (median value, 2.156 [1.640‐3.789]) compared with control subjects (median value, 0.969 [0.544‐1.483]; P < .001; Figure 2A); meanwhile, it showed a good predictive value for enhanced CHD risk (AUC: 0.859, 95% CI: 0.824‐0.894; Figure 2B). After adjustment by age/gender, elevated lncRNA THRIL was observed in CHD patients (median value, 2.308 [1.666‐3.846]) compared to control subjects (median value, 0.972 [0.540‐1.489]; P < .001; Figure 2C); also, it presented with a good predictive value for increased CHD risk (AUC: 0.869, 95% CI: 0.834‐0.904; Figure 2D).
Figure 2.
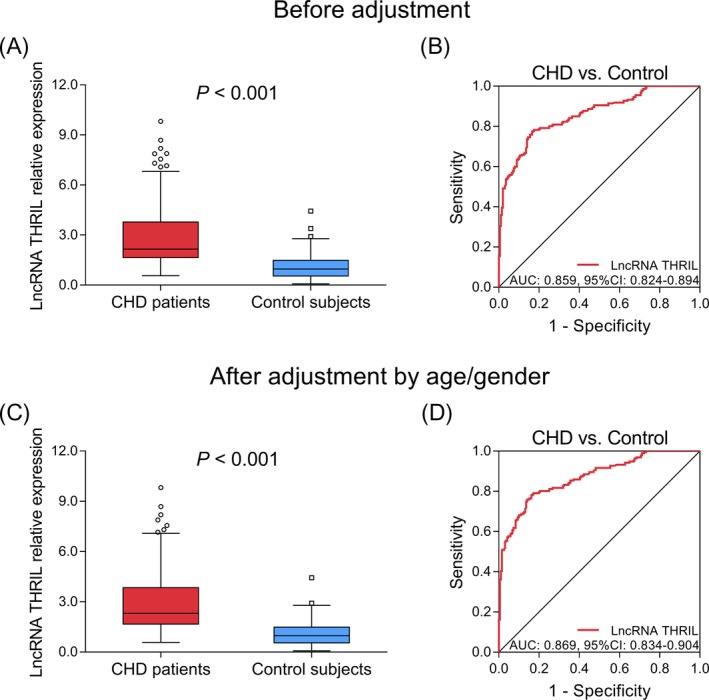
LncRNA THRIL expression and its predictive value for CHD risk before and after adjustment by age/gender. LncRNA THRIL expression (A) and its predictive value for CHD risk (B) before adjustment. LncRNA THRIL expression (C) and its predictive value for CHD risk (D) after adjustment by age/gender. AUC, area under curve; CHD, coronary heart disease; CI, confidence interval; lncRNA, long non‐coding RNA; MACE, major adverse cardiovascular events; THRIL, tumor necrosis factor‐α and heterogenous nuclear ribonucleoprotein L‐related immunoregulatory long intergenic non‐coding RNA
3.4. Correlation of lncRNA THRIL with Gensini score in CHD patients
As shown in Figure 3, lncRNA THRIL was positively correlated with Gensini score in CHD patients (P < .001, r = .430), indicating that CHD patients with increased lncRNA THRIL had enhanced level of coronary artery stenosis.
Figure 3.
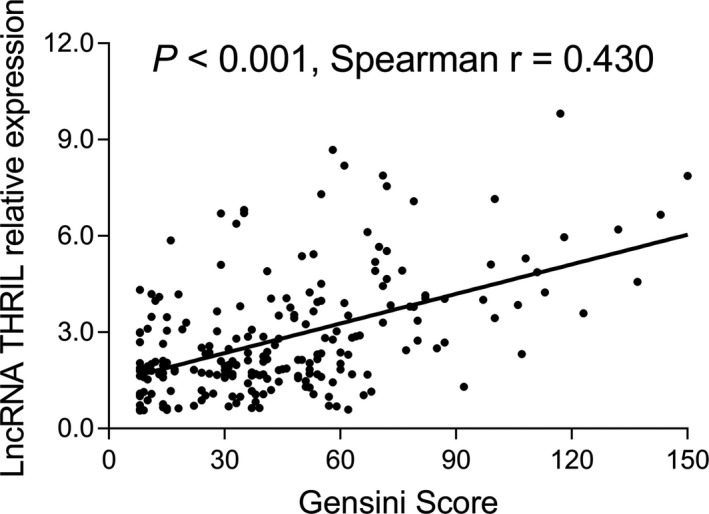
Association between lncRNA THRIL and Gensini score in CHD patients. CHD, coronary heart disease; lncRNA, long non‐coding RNA; THRIL, tumor necrosis factor‐α and heterogenous nuclear ribonucleoprotein L‐related immunoregulatory long intergenic non‐coding RNA
3.5. Correlation of lncRNA THRIL with clinical characteristics in CHD patients
For categorical characteristics, lncRNA THRIL was positively correlated with diabetes mellitus (P < .001), whereas no correlation was found between lncRNA THRIL and gender (P = .662), smoke (P = .681), family history of CHD (P = .225), hypertension (P = .622), hyperlipidemia (P = .241), or hyperuricemia (P = .787) in CHD patients (Table 2). For continuous clinical characteristics, it was positively correlated with FBG (P = .029, r = .147) and CRP (P < .001, r = .374); however, no correlation was observed between lncRNA THRIL and age (P = .557, r = −.040), BMI (P = .821, r = .015), TG (P = .402, r = .057), TC (P = .185, r = .090), LDL‐C (P = .074, r = .121), HDL‐C (P = .455, r = .051), Scr (P = .293, r = −.071), or SUA (P = .348, r = .064) in CHD patients (Table 3).
Table 2.
Correlation of lncRNA THRIL with categorical clinical characteristics in CHD patients
| Items | LncRNA THRIL relative expression | |
|---|---|---|
| Median (IQR) | P value | |
| Gender | ||
| Female | 2.397 (1.744‐3.586) | .662 |
| Male | 2.128 (1.611‐3.816) | |
| Smoke | ||
| No | 2.231 (1.661‐3.902) | .681 |
| Yes | 2.156 (1.607‐3.519) | |
| Family history of CHD | ||
| No | 2.093 (1.607‐3.583) | .225 |
| Yes | 2.361 (1.759‐4.008) | |
| Hypertension | ||
| No | 2.044 (1.469‐3.778) | .622 |
| Yes | 2.250 (1.663‐3.831) | |
| Hyperlipidemia | ||
| No | 2.046 (1.519‐3.608) | .241 |
| Yes | 2.324 (1.664‐3.955) | |
| Hyperuricemia | ||
| No | 2.113 (1.607‐3.797) | .787 |
| Yes | 2.242 (1.691‐3.759) | |
| Diabetes mellitus | ||
| No | 1.861 (1.469‐3.532) | <.001 |
| Yes | 3.033 (2.551‐3.935) | |
Abbreviations: CHD, coronary heart disease; IQR, interquartile range; LncRNA, long non‐coding RNA.
Table 3.
Correlation of lncRNA THRIL with continuous clinical characteristics in CHD patients
| Items | LncRNA THRIL | |
|---|---|---|
| P value | Spearman r | |
| Age | .557 | −.040 |
| BMI | .821 | .015 |
| FBG | .029 | .147 |
| TG | .402 | .057 |
| TC | .185 | .090 |
| LDL‐C | .074 | .121 |
| HDL‐C | .455 | .051 |
| Scr | .293 | −.071 |
| SUA | .348 | .064 |
| CRP | <.001 | .374 |
Abbreviations: BMI, body mass index; CHD, coronary heart disease; CRP, C‐reactive protein; FBG, fasting blood glucose; HDL‐C, high‐density lipoprotein cholesterol; LDL‐C, low‐density lipoprotein cholesterol; lncRNA, long non‐coding RNA; Scr, serum creatinine; SUA, serum uric acid; TC, total cholesterol; TG, triglyceride.
3.6. Correlation of lncRNA THRIL with inflammatory cytokines in CHD patients
Long non‐coding RNA THRIL was positively correlated with pro‐inflammatory cytokines including TNF‐α (P < .001, r = .249), IL‐1β (P = .001, r = .222), IL‐8 (P < .001, r = .254), and IL‐17 (P = .011, r = .172), while negatively correlated with anti‐inflammatory cytokine IL‐10 (P < .001, r = −.244); however, no correlation was found between lncRNA THRIL and IL‐6 (P = .079, r = .119) in CHD patients (Table 4). These data indicated that lncRNA THRIL was associated with increased inflammation in CHD patients.
Table 4.
Correlation of lncRNA THRIL with inflammatory cytokines in CHD patients
| Items | LncRNA THRIL | |
|---|---|---|
| P value | Spearman r | |
| TNF‐α | <.001 | .249 |
| IL‐1β | .001 | .222 |
| IL‐6 | .079 | .119 |
| IL‐8 | <.001 | .254 |
| IL‐10 | <.001 | −.244 |
| IL‐17 | .011 | .172 |
Abbreviations: CHD, coronary heart disease; IL, interleukin; lncRNA, long non‐coding RNA; TNF, tumor necrosis factor.
3.7. Correlation of lncRNA THRIL with MACE risk in CHD patients
According to the median value of lncRNA THRIL in CHD patients, they were further divided into lncRNA THRIL high expression group and lncRNA THRIL low expression group. MACE accumulating rate was increased in lncRNA THRIL high expression group compared with lncRNA THRIL low expression group (P = .037; Figure 4). These data suggested that lncRNA THRIL was correlated with worse prognosis in CHD patients.
Figure 4.

Comparison of MACE accumulating rate between lncRNA THRIL high expression group and lncRNA THRIL low expression group in CHD patients. CHD, coronary heart disease; lncRNA, long non‐coding RNA; MACE, major adverse cardiovascular events; THRIL, tumor necrosis factor‐α and heterogenous nuclear ribonucleoprotein L‐related immunoregulatory long intergenic non‐coding RNA
4. DISCUSSION
Long non‐coding RNA THRIL specifically binds to hnRNPL, and lncRNA THRIL‐hnRNPL complex regulates the transcription of TNF‐α gene by binding to its promoter.9 Several studies reveal that lncRNA THRIL is a vital regulator in inflammation.10, 11, 15, 16 For example, in lipopolysaccharide (LPS)‐induced inflammatory cell model, the overexpression of lncRNA THRIL exacerbates the LPS‐induced inflammatory injury in ATDC5 cells by sponging micro‐RNA (miR)‐125b as well as activating Janus kinase 1 (JAK1)/signal transducer and activator of transcription 3 (STAT3) and NF‐κB pathway.15 Similarly, in an LPS‐induced inflammation HK‐2 cell model, lncRNA THRIL overexpression increases the LPS‐induced apoptosis and pro‐inflammatory cytokines (such as IL‐1β, IL‐6, IL‐8, and TNF‐α) by sponging micro‐RNA (miR)‐34a.16 Moreover, it is reported that lncRNA THRIL regulates cardiomyocytes. In rat cardiomyocytes‐derived H9C2 cell, the overexpression of lncRNA THRIL enhances hypoxia‐induced injury by regulating miR‐99a, while knock‐down of lncRNA THRIL attenuates cell injury, implying that lncRNA THRIL might participate in myocardial infarction.10, 11 Collectively, these studies reveal that lncRNA THRIL not only activates inflammation via several pathways but also regulates cardiomyocytes and take part in the regulation of cardiovascular disease.
The overexpression of lncRNA THRIL in patients with inflammatory‐related diseases is reported by several studies.17, 18, 19, 20 For example, it is reported that lncRNA THRIL is highly expressed in rheumatoid arthritis (RA) patients compared to healthy controls and discriminates RA patients from controls.17 Meanwhile, in multiple sclerosis patients, lncRNA THRIL is overexpressed compared to healthy subjects.18 However, whether lncRNA THRIL would still play a vital role in CHD is unclear. Based on the above‐mentioned information that lncRNA THRIL is a promotor in inflammation and a regulator of cardiomyocytes, we hypothesized that it might be a biomarker for prediction of CHD, as well as for disease progression and prognosis of CHD patients. Therefore, we conducted this study and found that both before and after adjustment by age/gender, lncRNA THRIL was greatly enhanced in CHD patients compared to controls, and it showed good predictive value for increased CHD risk. These data could be explained by that: (a) LncRNA THRIL might regulate several pathways (such as TNF‐α and NF‐κB) to induce inflammation as in systemic lupus erythematosus model and osteoarthritis model, which accelerated the process of atherosclerosis and rose the possibility of CHD incidence21; (b) it is proposed that lncRNA THRIL activated TNF‐α transcription, and TNF‐α is a major cause of local and systemic insulin resistance.1, 9 Thus, it was possible that lncRNA THRIL might up‐regulate TNF‐α to increase the risk of diabetes mellitus, one of the risk factors of CHD, thereby enhanced the incidence of CHD. Therefore, lncRNA THRIL was up‐regulated in CHD patients and well predicted the increased risk of CHD.
To the best of our knowledge, there is no evidence until now regarding the association of lncRNA THRIL with disease severity in cardiovascular disease patients, including CHD patients. In the present study, we discovered that lncRNA THRIL was positively correlated with severity of coronary stenosis, diabetes mellitus occurrence, and FBG level, as well as systematic inflammation in CHD patients. Possible explanations might be that: (a) LncRNA THIRL might sponge miR‐125b,15 which was an inhibitor of atherosclerosis by targeting podocalyxin22; therefore, it was positively associated with severity of coronary stenosis (evaluated by Gensini score) in CHD patients. (b) LncRNA THRIL might increase TNF‐α expression to induce local and systemic insulin resistance and the incidence of diabetes mellitus (mentioned above). Therefore, lncRNA THRIL was positively correlated with diabetes mellitus occurrence and FBG level (one of the most common indicators of diabetes mellitus) in CHD patients. (c) According to previous studies, lncRNA THRIL increases inflammation by activating several pathways including NF‐κB pathway and STAT3 pathway via sponging miR‐34a.15 In this study, LncRNA THRIL might also activate NF‐κB pathway and STAT3 pathway to enhance the systematic inflammation in CHD patients. Therefore, lncRNA THRIL was correlated with increased pro‐inflammatory markers (CRP, TNF‐α, IL‐1β, IL‐8, and IL‐17) but decreased anti‐inflammatory cytokine IL‐10 in CHD patients.
Moreover, we found that patients with lncRNA THRIL high expression had increased MACE accumulating rate in this study, which revealed the potential of lncRNA THRIL as a prognostic factor for MACE risk in CHD patients. These data could be explained by that: (a) LncRNA THRIL was correlated with enhanced severity of coronary stenosis (evaluated by Gensini score) in CHD patients, which elevated the incidence of MACE; (b) LncRNA THRIL was correlated with increased systematic inflammation in CHD patients, which accelerated the progress of atherosclerosis and enhanced MACE incidence; (c) LncRNA THRIL might sponge miR‐125b, which suppressed the development of atherosclerosis,22 thus increased MACE incidence in CHD patients. Therefore, lncRNA THRIL was positively correlated with MACE accumulating rate in CHD patients.
There were several limitations in this study: (a) The follow‐up of this study was relatively short (the maximum follow‐up period was 48 months); therefore, the correlation of lncRNA THRIL with long‐term MACE accumulating rate in CHD patients was unclear, which could be investigated in the future; (b) the underlying mechanisms of lncRNA THRIL in CHD or the process of atherosclerosis were not investigated, and further research could be conducted; (c) in this study, we discovered that lncRNA THRIL was positively correlated with FBG level and diabetes mellitus incidence in CHD patients, indicating that lncRNA THRIL might also be an important biomarker in diabetes mellitus. However, considering that the role of lncRNA THRIL in diabetes was not the main purpose of this study, we did not perform such investigation, which could be conducted in the future; (d) the change of lncRNA THRIL during the disease course and treatment was not investigated in this research, which needed further investigation.
In summary, this study discovered that lncRNA THRIL was up‐regulated in CHD patients and presented with a good predictive value for enhanced CHD risk. Meanwhile, it was correlated with increased artery stenosis, FBG level, inflammation, and MACE risk in CHD patients. Therefore, lncRNA THRIL might be a potential biomarker for the prevention of CHD and prediction of prognosis in CHD patients, which would ameliorate disease management and thereby improving their prognosis.
ACKNOWLEDGMENTS
None.
Qi H, Shen J, Zhou W. Up‐regulation of long non‐coding RNA THRIL in coronary heart disease: Prediction for disease risk, correlation with inflammation, coronary artery stenosis, and major adverse cardiovascular events. J Clin Lab Anal. 2020;34:e23196 10.1002/jcla.23196
Qi and Shen contributed equally to this work.
REFERENCES
- 1. GBD 2017 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789‐1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111(25):3481‐3488. [DOI] [PubMed] [Google Scholar]
- 3. Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685‐1695. [DOI] [PubMed] [Google Scholar]
- 4. GBD 2017 Causes of Death Collaborators . Global, regional, and national age‐sex‐specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cagle SD Jr, Cooperstein N. Coronary artery disease: diagnosis and management. Prim Care. 2018;45(1):45‐61. [DOI] [PubMed] [Google Scholar]
- 6. Malakar AK, Choudhury D, Halder B, et al. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol. 2019;234(10):16812‐16823. [DOI] [PubMed] [Google Scholar]
- 7. Li L, Wang L, Li H, et al. Characterization of LncRNA expression profile and identification of novel LncRNA biomarkers to diagnose coronary artery disease. Atherosclerosis. 2018;275:359‐367. [DOI] [PubMed] [Google Scholar]
- 8. Simion V, Haemmig S, Feinberg MW. LncRNAs in vascular biology and disease. Vascul Pharmacol. 2019;114:145‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111(3):1002‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sheng C, Hu F, Wu L. Geniposide alleviates hypoxia‐induced injury by down‐regulation of lncRNA THRIL in rat cardiomyocytes derived H9c2 cells. Eur J Pharmacol. 2019;854:28‐38. [DOI] [PubMed] [Google Scholar]
- 11. Xia J, Jiang N, Li Y, et al. The long noncoding RNA THRIL knockdown protects hypoxia‐induced injuries of H9C2 cells through regulating miR‐99a. Cardiol J. 2019;26(5):564‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guidelines for Coronary Angiography . A report of the American College of Cardiology/American Heart Association Task Force on Assessment of diagnostic and therapeutic cardiovascular procedures (subcommittee on coronary angiography). J Am Coll Cardiol. 1987;10(4):935‐950. [PubMed] [Google Scholar]
- 13. Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51(3):606. [DOI] [PubMed] [Google Scholar]
- 14. Greenwood JP, Ripley DP, Berry C, et al. Effect of care guided by cardiovascular magnetic resonance, myocardial perfusion scintigraphy, or NICE guidelines on subsequent unnecessary angiography rates: the CE‐MARC 2 randomized clinical trial. JAMA. 2016;316(10):1051‐1060. [DOI] [PubMed] [Google Scholar]
- 15. Liu G, Wang Y, Zhang M, et al. Long non‐coding RNA THRIL promotes LPS‐induced inflammatory injury by down‐regulating microRNA‐125b in ATDC5 cells. Int Immunopharmacol. 2019;66:354‐361. [DOI] [PubMed] [Google Scholar]
- 16. Deng Y, Luan S, Zhang Q, et al. Long noncoding RNA THRIL contributes in lipopolysaccharide‐induced HK‐2 cells injury by sponging miR‐34a. J Cell Biochem. 2019;120(2):1444‐1456. [DOI] [PubMed] [Google Scholar]
- 17. Moharamoghli M, Hassan‐Zadeh V, Dolatshahi E, et al. The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis. Clin Rheumatol. 2019;38(11):3073‐3080. [DOI] [PubMed] [Google Scholar]
- 18. Eftekharian MM, Ghafouri‐Fard S, Soudyab M, et al. Expression analysis of long non‐coding RNAs in the blood of multiple sclerosis patients. J Mol Neurosci. 2017;63(3‐4):333‐341. [DOI] [PubMed] [Google Scholar]
- 19. Safari MR, Komaki A, Arsang‐Jang S, et al. Expression pattern of long non‐coding RNAs in schizophrenic patients. Cell Mol Neurobiol. 2019;39(2):211‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Fu X, Yu B, et al. Long non‐coding RNA THRIL predicts increased acute respiratory distress syndrome risk and positively correlates with disease severity, inflammation, and mortality in sepsis patients. J Clin Lab Anal. 2019;33(6):e22882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geovanini GR, Libby P. Atherosclerosis and inflammation: overview and updates. Clin Sci (Lond). 2018;132(12):1243‐1252. [DOI] [PubMed] [Google Scholar]
- 22. Li X, Yao N, Zhang J, et al. MicroRNA‐125b is involved in atherosclerosis obliterans in vitro by targeting podocalyxin. Mol Med Rep. 2015;12(1):561‐568. [DOI] [PubMed] [Google Scholar]


