Abstract
Background
Renal tumors are highly heterogeneous, and identification of tumor heterogeneity is an urgent clinical need for effective treatment. Mass cytometry (MC) can be used to perform high‐dimensional single‐cell proteomics analysis of heterogeneous samples via cytometry by time‐of‐flight (CyTOF), in order to achieve more accurate observation and classification of phenotypes within a cell population. This study aimed to develop a high‐dimensional MC method for the detection and analysis of heterogeneity in renal tumors.
Materials and Methods
We collected tissue samples from 8 patients with different types of renal tumors. Single‐cell suspensions were prepared and stained using a panel of 28 immune cell‐centric antibodies and a panel of 21 stem‐like cell‐centric antibodies. The stained cells were detected using CyTOF.
Result
Renal tumors were divided into 25 immune cell subsets (4 CD4+ T cells, 7 CD8+ T cells, 1 B cells, 8 macrophages, 1 dendritic cells, 2 natural killer (NK) cells, 1 granulocyte, and 1 other subset) and 7 stem‐like cells subsets (based on positivity of vimentin, CD326, CD34, CD90, CD13, CD44, and CD47). Different types of renal tumors have different cell subsets with significantly different characteristics.
Conclusion
High‐dimensional single‐cell proteomics analysis using MC aids in the discovery and analysis of renal tumors heterogeneity. Additionally, it can be used to accurately classify the immune cell population and analyze the expression of stem cell‐related markers in renal tumors. Our findings provide a valuable resource for deciphering tumor heterogeneity and might improve the clinical management of patients with renal tumors.
Keywords: cancer stem cells, mass cytometry, renal tumors, tumor heterogeneity, tumor microenvironment
1. BACKGROUND
Renal cell carcinoma (RCC) is the most common type of renal tumors, and it is derived from the epithelium of the renal tubules.1 Several subtypes of RCC have been defined. Clear cell RCC (ccRCC) is the most common subtype,2 which accounts for approximately 70% of all RCC cases and is associated with poor prognosis due to its high potential for metastasis and recurrence.3 Papillary RCC (pRCC), the second most common subtype, comprises of 15%‐20% of RCC and is associated with high 5‐year survival rate (80%‐90%). Hence, the prognosis of pRCC is better than that of ccRCC.4 Chromophobe RCC (chRCC) accounts for 6%‐11% of all RCC cases and has a good prognosis and low metastasis rate.5 The frequency of occurrence of other rare types of RCC is less than 1%.6 Metanephric adenoma (MA) is an uncommon benign type of renal tumors, and it is derived from the residual renal organization during embryonic development.7 In addition, although uncommon, urothelial carcinoma (UC) of the renal pelvis is classified as renal tumors and is characterized by high malignancy and poor prognosis.8 The differences between these histological subtypes of renal tumors are important as they emphasize that renal tumors should not be treated as a single disease and in a uniform manner. In addition, renal tumors are highly heterogeneous. The heterogeneity of tumors introduces significant challenges in prediction of therapeutic effect as well as for classifying patients that might benefit from specific therapies.9 Hence, the study of renal tumor heterogeneity is an urgent clinical need for effective treatment.
Tumor microenvironment is one of the main causes of renal tumor heterogeneity. The tumor microenvironment exerts selective pressure in distinct regions of the tumor, generating intra‐tumor heterogeneity,10 which is the key to the treatment and prognosis of tumors. Tumor‐infiltrating immune cells are important cellular components of tumor microenvironment.11 It has been linked to prognosis and response to immunotherapy. For instance, tumor‐associated macrophages are significant for promoting or blocking tumor progression.12 In pRCC, M1 macrophages were associated with a favorable outcome, while M2 macrophages indicated a worse outcome.13 In addition, CD8+ T cells have been associated with improved clinical outcomes and response to immunotherapy. However, due to the limitations of traditional research methods, the phenotypes of many tumor‐infiltrating immune subpopulations are not well described. Therefore, we need a suitable approach to achieve more accurate observation and classification of phenotypes within a cell population, which is of great significance for revealing the heterogeneity.
Cancer stem cells (CSCs) are another important cause of renal tumor heterogeneity. Cancer stem cells are a small population of neoplastic cells within a tumor which sustains tumor growth through self‐renewal and differentiation.1 In the CSCs model, a stem‐like cells population contributes to metastasis (tumorigenicity), treatment resistance, and recurrence.14 Therefore, CSCs are the most optimal target populations of therapy and essential for clinical targeting.15 For a long time, many researchers have been committed to look for specific surface markers on tumor stem cells. So far, different approaches have been developed in order to isolate the CSCs.16, 17 Consequently, specific markers such as CD105, ALDH1, CD44, CD133, and CXCR4 have been found in RCC‐derived cancer stem‐like cells.16, 18, 19, 20 However, a single marker cannot be used for identifying all the CSCs,21 and therefore, we need to find an appropriate method to discover novel biomarkers and reveal the heterogeneity of CSCs. This can help in clarifying the role of CSCs in the occurrence, development, recurrence, metastasis, and multidrug resistance of renal tumors and can enable more personalized treatment strategies to establish novel therapeutic targets.
Currently, single‐cell profiling is an important means to elucidate tumor heterogeneity, and22, 23 the main methods involved in this are single‐cell sequencing and cytology. These methods allow analysis of multiple markers in a single tumor cell that have been isolated from fresh or fixed primary and metastatic tumors.24 Single‐cell RNA sequencing provides high‐dimensional, single‐cell data.25 However, high cost and instability of the RNA samples make the technique unsuitable for analyzing a large number of samples. Although flow cytometry is the most commonly used single‐cell technique, the overlap of fluorescence emission spectra causes mutual interference between the channels and limits the increase in the number of channels.26 Mass cytometry (MC) is a single‐cell detection technology that uses non‐radioactive heavy metal isotopes for antibody labeling rather than fluorophores, which allows the simultaneous detection of a large number of parameters with negligible overlap at the single‐cell level.27 The emergence of MC or cytometry by time‐of‐flight (CyTOF) has revolutionized single‐cell proteomics, enabling a comprehensive understanding of cell phenotypes, tumor heterogeneity, signaling pathways, and function.28 Using single‐cell MC, Wagner et al described the single‐cell atlas of breast cancer cells and immune cells.29 Through in‐depth immune analysis of 73 cases of ccRCC by MC, Chevrier et al identified 17 tumor‐related macrophage phenotypes and 22 T cell phenotypes, demonstrating the detailed human atlas of the immune cells in the tumor microenvironment in this disease.30 In this study, we present an MC‐based atlas of immune and stem cells in tumor samples from 2 patients with chRCC, 3 patients with ccRCC, 1 patient each with pRCC, MA, and UC. This unprecedented MC data can provide valuable information for the study of tumor heterogeneity in different types of renal tumors.
2. MATERIALS AND METHODS
2.1. Sample processing and storage
A total of 8 renal tumors patients (2 chRCC, 3 ccRCC, 1 pRCC, 1 MA, and 1 UC) were harvested from patients in the First Affiliated Hospital of Guangxi Medical University between Jan 2018 and July 2018. These patients voluntarily undergone curative surgery and were pathologically diagnosed with renal tumors. Patients undergoing chemo‐ or radiotherapy before resection were excluded. All samples were obtained with informed consent from patients in accordance with the study protocols approved by the review board of the First Affiliated Hospital of Guangxi Medical University.
Tumor tissues were obtained and disposed as described.31 After surgical resection, tissue samples were collected in pre‐cooling transfer buffer with DMEM, 2% human serum, 1% penicillin‐streptomycin, and 0.2% fluconazole and were shipped at ice box. Firstly, tumor tissue was cut into small pieces using surgical scalpels. Then, tumor pieces were incubated for 30 minutes at 37°C in HBSS dissociation cocktail containing 20 µg/mL DNAse I (Sigma) and 2 mg/mL Collagenase Type II (Gibco), after which the tumor was dispersed to single cells using the GentleMACS dissociator (Miltenyi) with a company installed program (h_tumor_01). Attach C tube upside down onto the sleeve of the GentleMACS dissociator, run the GentleMACS program (h_tumor_02) again, and incubated for another 30 minutes at 37°C. Suck out 80% of the cell suspension for later use and run the program (m_imptumor_01) with a separator for the remaining cells. Next, put on a 70 µm cell strainer (Falcon) to obtain a single‐cell suspension following centrifugation, erythrocytes were removed with red blood cell lysis buffer (Solarbio) on ice for ten minutes. Cells were resuspended in PBS after centrifuged, counted using Trypan blue exclusion (Solarbio), and cryopreserved at approximately 3 million cells/vial. Finally, 9:1 as cryopreserved with FBS and DMSO solution to preserve the cell suspension in liquid nitrogen for subsequent testing. All tumor samples were dissociated as above.
2.2. Antibody conjugation
Purified antibodies were purchased from the companies listed in Table 1 and Table 2. These antibodies be used for MC were conjugated to isotopic tags using a MaxPar X8 Antibody Labeling Kit according to the manufacturer's instruction. The polymer was mounted on the preset lanthanide metal of choice by co‐incubation at 37°C for 30 minutes. Separately, antibodies were performed buffer exchange in R‐buffer with a 50 kDa filter and partially reduced in 4 mmol/L TCEP‐R‐Buffer bond breaker solution at 37°C for 30 minutes. After purification of the polymer and antibodies, the recovery antibody conjugated with metal‐loaded polymers that concentrated in the 50 kDa filter and incubated at 37°C for 90 minutes. After antibody binding, the unbound polymer and metal were eluted with buffer W, quantified by measuring absorbance at 280 nm on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific), resuspended at a concentration of 0.5 mg/mL in antibody stabilization buffer (Candor Bioscience GmbH), and stored long term at 4°C. All antibodies were titrated before use.
Table 1.
Purified antibodies about the immune cell‐centric panel
| Immune cell‐centric panel | ||||
|---|---|---|---|---|
| Antibodies | Metal | Clone | Source | Identifier |
| CD19 | 142Nd | HIB19 | Fluidigm | 3142001B |
| CD163 | 145Nd | GHI/61 | Fluidigm | 3145010B |
| CD14 | 148Nd | RMO52 | Fluidigm | 3148010B |
| CD11c | 146Nd | 3.9 | Fluidigm | 3146014B |
| CD196/CCR6 | 176Yb | G034E3 | Fluidigm | 3176022A |
| CD161 | 164Dy | HP‐3G10 | Fluidigm | 3164009B |
| CD27 | 150Nd | LG.3A10 | Fluidigm | 3150017B |
| CD206/MMR | 168Er | 43511 | Fluidigm | 3168008B |
| CD25/IL‐2R | 149Sm | 2A3 | Fluidigm | 3149010B |
| CD3e | 154Sm | UCHT1 | Fluidigm | 3154003B |
| CD326/EpCAM | 141Pr | 9C4 | Fluidigm | 3141006B |
| CD4 | 174Yb | SK3 | Fluidigm | 3174004B |
| CD45 | 89Y | HI30 | Fluidigm | 3089003B |
| CD45RA | 170Er | HI100 | Fluidigm | 3170010B |
| CD66b | 162Dy | 80H3 | Fluidigm | 3162023B |
| HLA‐DR | 173Yb | L243 | Fluidigm | 3173005B |
| CD86 | 156Gd | IT2.2 | Fluidigm | 3156008B |
| Foxp3 | 159Tb | 259D/C7 | Fluidigm | 3159028A |
| CD197/CCR7 | 167Er | G043H7 | Fluidigm | 3167009A |
| Granzyme B | 171Yb | GB11 | Fluidigm | 3171002B |
| CD279/PD‐1 | 155Gd | EH12.2H7 | Fluidigm | 3155009B |
| Ki‐67 | 172Yb | B56 | Fluidigm | 3172024B |
| TGF‐β | 163Dy | TW4‐6H10 | Fluidigm | 3163010B |
| TNF‐α | 152Sm | Mab11 | Fluidigm | 3152002B |
| CD20 | 161Dy | 2H7 | Biolegend | 302302 |
| CD38 | 143Nd | HIT2 | Biolegend | 303502 |
| CD45RO | 151Eu | UCHL1 | Biolegend | 304202 |
| CD8a | 144Nd | RPA‐T8 | Biolegend | 301 002 |
Table 2.
Purified antibodies about the stem‐like cell‐centric panel
| Stem‐like cell‐centric panel | ||||
|---|---|---|---|---|
| Antibodies | Metal | Clone | Source | Identifier |
| CD47 | 209Bi | CC2C6 | Fluidigm | 3209004B |
| c‐Myc | 176Yb | 9E10 | Fluidigm | 3176012B |
| CD274 | 175Lu | 29E.2A3 | Fluidigm | 3175017B |
| CD44 | 171Yb | IM7 | Fluidigm | 3171003B |
| CD54 | 170Er | HA58 | Fluidigm | 3170014B |
| Met | 167Er | D1C2 | Fluidigm | 3167017A |
| CD24 | 166Er | ML5 | Fluidigm | 3166007B |
| Notch2 | 165Ho | MHN225 | Fluidigm | 3165026B |
| DNMT3B | 164Dy | 832121 | Fluidigm | 3164021B |
| CD13 | 160Gd | WM15 | Fluidigm | 3160014B |
| p21 | 159Tb | 12D1 | Fluidigm | 3159026A |
| Vimentin | 156Gd | RV202 | Fluidigm | 3156023A |
| p53 | 143Nd | 7F5 | Fluidigm | 3143018A |
| CD326 | 141Pr | 9C4 | Fluidigm | 3141006B |
| CD45 | 89Y | HI30 | Fluidigm | 3089003B |
| CD90 | 158Gd | 5E10 | biolegend | 328102 |
| CK19 | 162Dy | A53‐B/A2 | biolegend | 628502 |
| MUC1 | 168Er | SM3 | abcam | ab22711 |
| OV6 | 152Sm | OV‐6 | R&D | MAB2020 |
| CD325 | 148Nd | 8C11 | biolegend | 350802 |
| LGR5 | 155Gd | SA222C5 | biolegend | 373802 |
2.3. Living cell barcoding and antibody labeling
The cells were removed from liquid nitrogen and rapidly dissolved. Taking 4 samples for barcoding as an example, 1.5 million cells were taken from each sample and washed twice with 2 mL PBS. For viability staining, cells were stained with cisplatin (Fluidigm) to a final concentration of 5 mmol/L. To minimize inter‐sample staining variation, Samples were barcoded by adding three unique metal isotopes after fixation. A 20‐well barcoding group was composed of unique combinations of six barcoding metals (102 Pd, 104 Pd, 105 Pd, 106 Pd, 108 Pd, and 110 Pd. Fluidigm) were used for this study. Cells were washed once with 1× Barcode Perm Buffer (Fluidigm) before incubated in 200 ml barcoding cocktail for 30 minutes at room temperature. Then, Cells were washed four times with Cell Staining Buffer and combined all barcoded samples into one tube. Then, cell suspensions (50 mL) were incubated with 50 μL of human Fc receptor blocking solution (Biolegend) for 10 minutes and incubated surface antibody cocktail for 1 hour. The cells were then washed and incubated with 1 mL Nuclear Antigen Staining Buffer working solution for 30 minutes at room temperature. The intracellular antibody cocktail was added into cell suspension for 1 hour. After washing, we incubated samples with a fresh solution of 1.6% paraformaldehyde at room temperature for 10 minutes and stained with DNA intercalator (Fluidigm) overnight at 4°C. Before acquired cells on a CyTOF2 instrument (Fluidigm), cells were prepared with subsequent washes in Cell Staining Buffer and deionized water to remove buffer salts. Finally, cells resuspended with 10% EQ™ Four Element Calibration Beads.
2.4. CyTOF debarcode and analysis
Samples were analyzed on a CyTOF2 (Fluidigm) equipped with a pneumatic introduction system at an event rate less than 500 events/second. After samples data acquisition, FCS files were normalized and concatenated using CyTOF software v6.7. After files standardization, cells were manually debarocoded. Briefly, bar‐code file was modified unique sample names for debarcode. Using the normalized bar‐coded FCS files, debug BcS values to the point where the vertical red line is before the sharp decline. Each individual file contained the corresponding debarcoded cell events. Prior data analysis, we manually gated single cells, living cells for each file using online software Cytobank (https://www.cytobank.org/).
For SPADE analysis, we used 25 target numbers of nodes and 100% downsampled events target. The clustering channels were selected based on whether they were lineage markers and which cell population to be clustered.
A t‐SNE map was generated by the t‐distribution stochastic neighbor embedding (t‐SNE) analysis that makes a pairwise comparison of cellular phenotypes to optimally plot similar cells close to each other and reduces multiple parameters into two dimensions (tSNE1 and tSNE2). For most analysis, we selected equal events for each sample. Channel (markers) selection was variable depending on cell populations to be clustered. We chose the default parameters settings (perplexity, 30; iterations, 1000; theta, 0.5). Different cell populations were visualized and quantified.
For heat map, transformed ratio of medians intensity corresponds to a logical data scale. The colors in the heat map represent the measured means intensity value of a given marker in a given cluster. Four‐color scale was used with blue‐white indicating low expression values, white‐yellow indicating medium intensity expressed markers, and red representing highly expressed markers.
3. RESULTS
3.1. Preprocessing of tumor samples and data acquisition
We recruited 2 patients with chRCC, 3 patients with ccRCC, 1 patient with pRCC, 1 patient with MA, and 1 patient with UC in the First Affiliated Hospital of Guangxi Medical University and obtained tumor tissues by surgery. A standard operating procedure was used to generate single‐cell suspension from all tissue samples. These samples were stained with 49 antibodies including a panel of 28 immune cell‐centric antibodies and a panel of 21 stem‐like cell‐centric antibodies. The stained cells were detected using CyTOF. Application of our workflow yielded 1.5 million single‐cell profiles from 8 patients and measured in 49 protein epitope dimensions (Figure 1).
Figure 1.
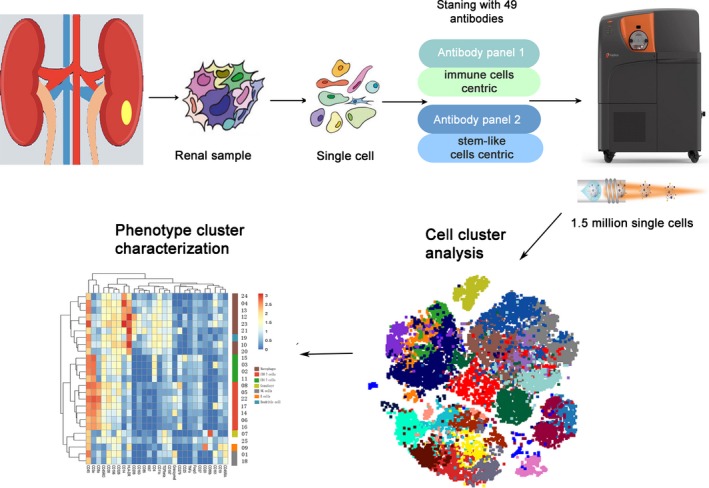
Workflow processing of samples of renal tumors and analytical methods for mass cytometry
3.2. Analysis of immuno‐phenotypes of renal tumors samples by MC
In the tumor microenvironment, tumor and immune cells interact and strengthen each other.29 The nature and degree of the tumor‐infiltrating immune cell greatly influences the treatment and prognosis of renal tumors.32 Therefore, understanding the composition and difference of immune cell subsets of different types of renal tumors provides necessary information for improving the level of immunotherapy and predicting prognosis. To visualize the phenotypic diversity of immune cells in different types of renal tumors, we use viSNE analysis to identify and characterize immune cell subsets. We identified 25 distinct cell subsets in CD45+ cells from renal tumors based on phenotypic similarity. Meanwhile, the 25 cell subsets were further divided into CD4+ T cell subsets, CD8+ T cell subsets, B cell subsets, macrophage subsets, dendritic cell subsets, NK cell subsets, and granulocyte subsets according to 13 clustering markers (CD3e, CD4, CD8a, CD20, CD25, FOXF3, CD38, CD161, CD14, HLA‐DR, CD206, CD19, and CD11c) (Figure 2A,B). Compared with the panorama, the distribution of 25 subsets of CD45+ cells in chRCC, pRCC, MA, and UC were obviously uneven and dominated by only some subsets, while the distribution of 25 subsets of CD45+ cells in ccRCC was relatively rich and uniform compared with other types of renal tumors. The composition of immune cells in the tumor microenvironment is obviously different in different types of renal tumors, which reflects the complexity of the tumor microenvironment.
Figure 2.
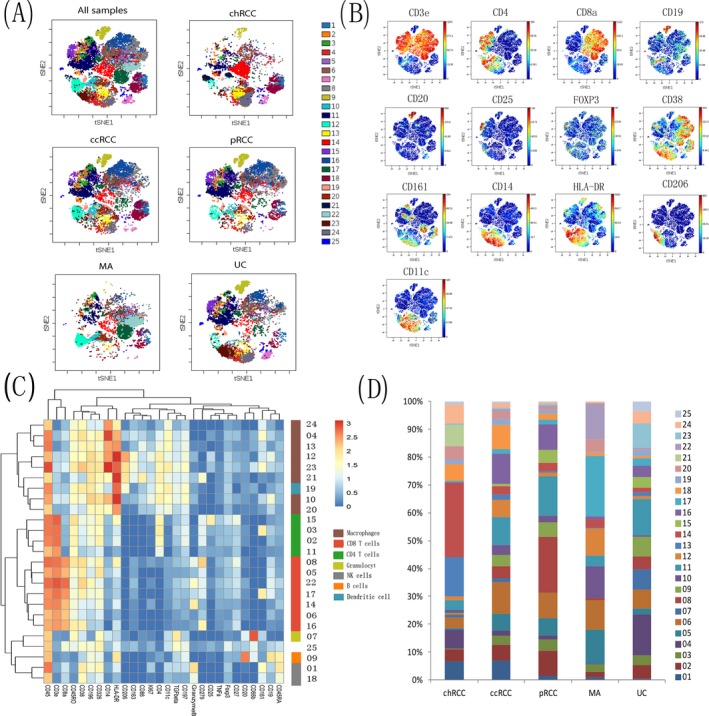
Immune landscape of various types of renal tumors. A, The figure shows the t‐SNE descending dimension map of the samples of various types of renal tumors. About 12 000 CD45+ immune cells were grouped into 25 clusters, the samples were classified and analysed, and colors represent different clusters of immune cells. B, The figure shows that the expression level and distribution of main cluster markers on the t‐SNE dimensionality reduction map. Bar on the right represents the median expression intensity of each marker. C, The heat map shows the differential expression of immune markers in the 25 subsets. Certain clusters were identified as known cell types according to typically expressed markers. Cluster ids and relative intensity were shown as bars on the right. D, The stacked graph represents the relative content of each cluster in different renal tumors, and the Y‐axis is the percentage of each cluster in CD45+ cells
To further explore the differences in the immune microenvironment and its mechanism in different types of renal tumors, we identified four CD4+ T cell subsets (02, 03, 11, and 15); seven CD8+ T cell subsets (05, 06, 08, 14, 16, 17, and 22); one B cell subset (09); eight macrophage subsets (04, 10, 12, 13, 20, 21, 23, and 24); one dendritic cell subset (19); two NK cell subsets (01 and 18); one granulocyte subset (07); and one other type of cell subset by observing the expression of markers according to the heat map (Figure 2C). Next, we explored the relative content of each immune cell subset in the tumor microenvironment. Interestingly, some tumor immune cell types had a significant numerical advantage in specific renal tumors subtypes (Figure 2D). Clusters 13 (14.05%) and 14 (26.53%) mainly appeared in chRCC, while it was few or virtually absent in other renal tumor subtypes. Similarly, cluster 06 (11.32%), cluster 11 (10.10%), and cluster 16 (10.86%) were found predominantly in ccRCC. Cluster 08 (19.84%) was mainly distributed in pRCC. Cluster 10 (11.28%), cluster 17 (21.6%), and cluster 22 (12.81%) were higher in MA while they were less numerous in others subtypes. Cluster 04 (14.79%) was mainly distributed in UC. In general, the immune landscape of renal tumors was significantly differenct, especially with regards to the subsets of CD8+ T cells, CD4+ T cells, and macrophages.
Further analysis of some clusters shows that PD‐1+ (CD279) cells were observed in both the CD8+ and CD4+ subsets. The clusters 08 and 16, which were characterized by the highest level of PD‐1 (CD279) expression among all the CD8+ cells, were also positive for the activation marker CD38. CD38 was found to be a potential exhaustion marker of T cells in ccRCC.33 Overlapping expressions of CD38 and PD‐1 (CD279) is a predictor of exhaustion of T cells in ccRCC,30 which suggest that some subsets may play an important role in renal tumor immune response. Taken together, this data reflected that there is significant heterogeneity in the immune microenvironment in different renal tumors. Hence, in this study, we performed MC to identify the differences in the immune microenvironment among different renal tumors by elucidating the immune cell landscape. This may be helpful in grouping renal tumors patients for implementing precision therapy.
3.3. Characterization of cancer stem cell‐related markers in various types of renal tumors
Increasing evidence suggests that a subset of cancer cells with stem cell‐like properties, known as CSCs, are capable of self‐renewal and differentiation.34 Although CSCs have been determined using a variety of surface markers, their isolation from different types of tumors might require different markers.35, 36 Many studies have attempted to establish a unique individual biomarker to identify the CSCs populations in renal tumors.16, 17, 37 However, due to the heterogeneity of stem cells, there is no universal marker that can identify CSCs in various types of renal tumors. In order to investigate the heterogeneity of CSCs in renal tumors and the co‐expression of CSCs related markers in different types of renal tumors, the expression profiles of these markers were expressed as heat map and t‐SNE (Figure 3A,B). According to the heat map, we found that 7 stem‐like cell‐centric markers (vimentin, CD13, CD90, CD34, CD326, CD44, and CD47) were expressed in all types of renal tumors. In addition, the expression level of the same marker was different among different renal tumors (Figure 3A). The expression of vimentin in UC was significantly stronger than that of other types of renal tumors. CD326 had the highest expression in chRCC, while the pRCC samples had the lowest. The expression of CD44 was strongest in pRCC and UC while CD47 was highly expressed in all types of renal tumors. In fact, these markers have been reported in literatures as putative stem cell markers for renal tumors or other types of cancer. For instance, CD44, CD90, and CD47 are recognized as UC stem cell markers.38 Meanwhile, in gastric cancer, CD44 and CD90 are considered to be specific biomarkers that can be used to identify and isolate metastatic and tumorigenic CSCs.39 This suggests that the same stem cell marker may be expressed in different types of cancer although their expression levels may be different. The expression of these stem cell markers in renal tumors cells can be used as specific markers to screen the stem cells in renal tumors. In order to further study the co‐expression of the tumor stem cell markers in different types of renal tumors using t‐SNE analysis, pairwise comparison of cell phenotypes was conducted to optimize the drawing of similar cells close to each other, and multiple parameters were reduced to a two‐dimensional display. Renal tumors were divided into 7 stem‐like cells subsets (based on positivity of vimentin, CD326, CD34, CD90, CD13, CD44, and CD47), and some stem‐like cells subsets have the characteristics of co‐expression (Figure 3B). We found two co‐expression cell populations that were characterized by CD326+ CD47+ and CD326+ CD44+ in chRCC. The difference was that MA only has CD326+ CD44+ co‐expression cells population. Similarly, two co‐expression cells population characterized by CD326+ CD44+ and vimentin+ CD13+ CD44+ were found in ccRCC. The co‐expression cells in pRCC and UC were CD13+ CD44+ CD47+ and vimentin+ CD13+ CD44+ cells, respectively. This suggests that different stem cell populations may exist in the same as well as different forms of cancers.40 Thus, this detailed data might provide further clues to explore stem cell markers in renal tumors.
Figure 3.
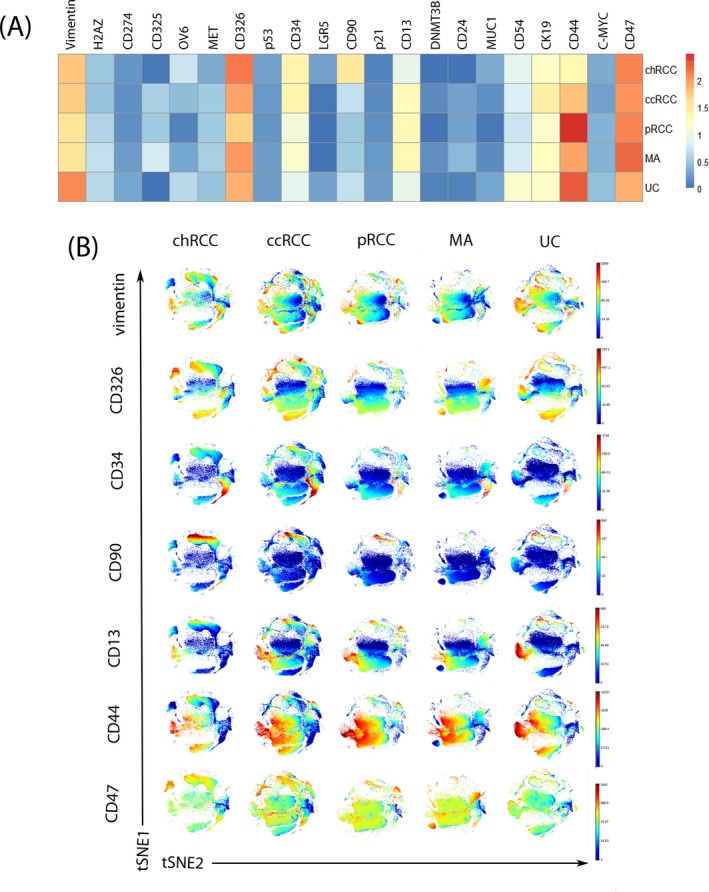
Expression level and co‐localization of stem cell markers in renal tumors samples. A, Heat map depicting differential expression for stem cell markers in renal tumors. B, t‐SNE plots of marker expression of 70 000 living cells from each tumor
4. DISCUSSION
Renal tumors have high intratumoral and intertumoral heterogeneity.14 The main reason for cancer treatment failure is that treatment regimens are not designed to address tumor heterogeneity. As a result, renal tumors tend to have a worse response to standard treatment regimens and therefore worse prognoses than other types of cancer. The heterogeneity of cancer cells presents significant challenges to the use of molecular prognostic markers and the identification of patients who may benefit from targeted treatments. Therefore, the study of heterogeneous characteristics will help to improve the clinical management of patients with renal tumors.
Single‐cell analysis is one of the methods to effectively solve the problem of characterizing tumor heterogeneity.22, 23 Mass cytometry simultaneously allows detection and quantification of dozens of markers in a single cell, and hence, it is uniquely suited for multi‐parametric analyses of heterogeneous samples and to understand the detailed phenotype of cell populations. Mass cytometry has been successfully applied in a number of areas, such as mapping the phenotypic heterogeneity in leukemia41 and assessing the effects of drugs on immune cells,42 revealing the composition of many cancer immune microenvironments, which enables the determination of the best method to enhance the anti‐tumor immune response.43, 44 Zhang et al studied the heterogeneity of the immune microenvironment in hepatocellular carcinoma (HCC) using MC and identified three different HCC subtypes with immunocompetent, immunodeficient, and immunosuppressed characteristics, respectively, by clustering immune cells in the HCC microenvironment. This classification can be used to evaluate the prognosis of patients with HCC and to provide theoretical support for the choice of treatment.24 Currently, there is little knowledge about the role of heterogeneity in the development of renal tumors due to the limitations in the research methods. This study aimed to establish a high‐dimensional MC‐based method to detect and analyze the heterogeneity of renal tumors of microenvironment and stem cells.
In this study, we present an MC‐based atlas of the immune landscape. Based on clustering of data generated using a panel of antibodies, we identified 25 major immune cell phenotypes including four CD4+ T cell subsets (02, 03, 11, and 15); seven CD8+ T cell subsets (05, 06, 08, 14, 16, 17, and 22); one B cell subset (09); eight macrophage subsets ( 04, 10, 12, 13, 20, 21, 23, and 24); one dendritic cell subset (19); two NK cell subsets (01 and 18); one granulocyte subset (07); and one other type of cell subset. It is well known that CD4+ helper T cells and cytotoxic CD8+ T cells inhibit tumor growth by targeting antigenic tumor cells.30 However, the prognostic significance of T cells in tumors is controversial as the T cell subsets may be positively or negatively correlated with the patient's prognosis.45 In addition, due to the limitations of traditional research methods, the phenotype of T cells in many tumors has not been well described. The influence of malignant cells on the functional characteristics of known T cell subsets, such as depleted T cells and T helper cells, has not been widely studied. In our study, we performed accurate immune typing of tumor cell populations and determined whether they consisted of typical or novel cell subsets, which can potentially provide useful information for the study of immune cell subsets and prediction of tumors prognosis.
To further analyze the phenotypic and functional associations between immune cell subpopulations, we also used a two‐dimensional graph embedded with a dimension reducing t‐distributed random adjacency to characterize how these immune cell clusters are related.41 We found that the degree of infiltration of immune cells in the tumor microenvironment were significantly different in different tumor tissues, especially with regards to subsets of CD8+ T cells, CD4+ T cells, and macrophages. In T cells, we focused on the differences in the expression of T cell depletion induced factors such as PD‐1 and CD38 in different renal tumor tissues. Overlapping expressions of CD38 and PD‐1 (CD279) are a predictor of exhaustion of T cells in ccRCC,30 It indicates that T cell subpopulations with unique functional characteristics exhibit unique phenotypic characteristics. We also found that the relative content of T cells, with overlapping expressions of CD38 and PD‐1 (CD279) in ccRCC and chRCC, was higher than that in other types of renal tumors. Taken together, this data reflected that there is significant heterogeneity in the immune microenvironment in different renal tumors. Our results showed the unique cytological characteristics and revealed the differences in the immune cell populations between different renal tumors, which enable the potential development of individualized immunotherapy.
Overcoming the heterogeneity of CSCs is also the key to treat renal tumors, and these cells can be identified through the expression of cell surface markers or functional markers. Several of these types of markers are derived from the markers that have been established in studies with normal hematopoietic stem cells or embryonic stem cells, such as CD133 and CD44 molecules.46 We studied the expression of 21 stem‐like cell‐centric markers using antibodies in renal tumors using MC. We found 7 stem‐like cells subsets in the renal tumors (based on the expression of vimentin, CD326, CD34, CD90, CD13, CD44, and CD47). Our data suggested that the expression of these stem cell markers in renal tumors could be used as specific markers for screening renal CSCs. Although the distribution of these tumor stem‐like cells subsets in different types of renal tumors was roughly the same, their expression levels were different. To further investigate the co‐expression of the different stem cell markers in renal tumors, we used the t‐SNE method for dimensional reduction analysis. It is noteworthy that we also found some co‐expression tumor stem‐like cells subsets, such as two co‐expression cell populations that were characterized by CD326+ CD47+ and CD326+ CD44+ in chRCC. The difference was that MA only has CD326+ CD44+ co‐expression cells population. Similarly, two co‐expression cells population characterized by CD326+ CD44+ and vimentin+ CD13+ CD44+ were found in ccRCC. The co‐expression cells in pRCC and UC were CD13+ CD44+ CD47+ and vimentin+ CD13+ CD44+ cells, respectively. This suggest that different stem cell populations may exist in the same as well as different forms of cancers40 owing to the fact that the self‐renewal and differentiation of CSCs.47 Our analysis showed that renal tumors have different stem‐like cells subsets, which means that more precise and specific stem‐like cells subsets can be distinguished by identifying the co‐expression of tumor stem cell‐like subsets. The discovery of these stem‐like cells subsets provides the basis for screening drugs for targeted therapy of renal tumors stem cells, thus greatly improving RCC diagnosis and treatment.
Having said that, our study does have certain limitations. Firstly, MC relies on the usage of high‐quality antibodies, and the expression of some markers might not be detected efficiently as the current options for commercially available high‐quality tumor antibodies are limited. Secondly, the presented immune cell phenotypes are based on single‐cell measurements coupled with computational analysis. Although our approach recapitulated the known immune landscape, follow‐up studies are required to define the roles of the phenotypes identified here.30 Thirdly, whether these stem‐like cells subsets have the characteristics of tumor stem cells as well as their clinical significance still needs to be experimentally verified and analyzed in combination with the clinical data. Finally, due to the limited sample size, much of the MC data were descriptive, and the tendency for the increased expression of some markers or subsets requires further validation.48 These limitations will be addressed in our future work, which will aim at increasing the sample size and further screening for high‐quality antibodies in order to verify the experimental results.
5. CONCLUSION
High‐dimensional single‐cell proteomics analysis using MC aids in the discovery and analysis of renal tumors heterogeneity. Additionally, it can be used to accurately classify the immune cell population and analyze the expression of stem‐like cells in renal tumors, thus, aiding in the discovery of novel disease‐related subsets. Our findings provide a valuable resource for deciphering tumor heterogeneity and might improve the clinical management of patients with renal tumors.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Li Z, Hu J, Qin Z, et al. High‐dimensional single‐cell proteomics analysis reveals the landscape of immune cells and stem‐like cells in renal tumors. J Clin Lab Anal. 2020;34:e23155 10.1002/jcla.23155
Li, Hu and Qin Contributed equally.
Funding information
National Natural Science Foundation of China (81760454, 31471271, 31560311). Guangxi Natural Science Foundation (2017JJA140260z, 2016JJB140183, AD17195090, 2016GXNSFGA38006). Promotion of Basic Ability of Young and Middle‐aged Teachers in Universities in Guangxi (KY2016LX055). Talents Highland of Emergency and Medical Rescue of Guangxi Province in China (GXJZ201611).
Contributor Information
Qiuyan Wang, Email: qiuyanwang510@yahoo.com.
Tianyu Li, Email: 547370852@qq.com, Email: qiuyanwang510@yahoo.com.
REFERENCES
- 1. Yuan ZX, Mo J, Zhao G, Shu G, Fu HL, Zhao W. Targeting strategies for renal cell carcinoma: from renal cancer cells to renal cancer stem cells. Front Pharmacol. 2016;7:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin ZZ, Ming DS, Chen YB, et al. KMT5A promotes metastasis of clear cell renal cell carcinoma through reducing cadherin‐1 expression. Oncol Lett. 2019;17(6):4907‐4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mydlo JH, Bard RH. Analysis of papillary renal adenocarcinoma. Urology. 1987;30(6):529‐534. [DOI] [PubMed] [Google Scholar]
- 5. Volpe A, Novara G, Antonelli A, et al. Chromophobe renal cell carcinoma (RCC): oncological outcomes and prognostic factors in a large multicentre series. BJU Int. 2012;110(1):76‐83. [DOI] [PubMed] [Google Scholar]
- 6. Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67(1):85‐97. [DOI] [PubMed] [Google Scholar]
- 7. Sun Z, Kan S, Zhang L, et al. Immunohistochemical phenotype and molecular pathological characteristics of metanephric adenoma. Int J Clin Exp Pathol. 2015;8(6):6031‐6036. [PMC free article] [PubMed] [Google Scholar]
- 8. Osmanov YI, Gaibov ZA, Kogan EA, Radenska‐Lopovok SG, Tursunov KZ. Comparative morphological characteristics and immunophenotype of urothelial carcinomas of the renal pelvis and bladder. Arkh Patol. 2018;80(5):23‐32. [DOI] [PubMed] [Google Scholar]
- 9. Tellez‐Gabriel M, Ory B, Lamoureux F, Heymann MF, Heymann D. Tumour heterogeneity: the key advantages of single‐cell analysis. Int J Mol Sci. 2016;17(12):E2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McAllister SS, Weinberg RA. Tumor‐host interactions: a far‐reaching relationship. J Clin Oncol. 2010;28(26):4022‐4028. [DOI] [PubMed] [Google Scholar]
- 12. Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15(2):73‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S, Zhang E, Long J, et al. Immune infiltration in renal cell carcinoma. Cancer Sci. 2019;110(5):1564‐1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275‐291. [DOI] [PubMed] [Google Scholar]
- 15. Moch H. An overview of renal cell cancer: pathology and genetics. Semin Cancer Biol. 2013;23(1):3‐9. [DOI] [PubMed] [Google Scholar]
- 16. Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G. Identification of a tumor‐initiating stem cell population in human renal carcinomas. FASEB J. 2008;22(10):3696‐3705. [DOI] [PubMed] [Google Scholar]
- 17. Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlow B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS ONE. 2014;9(4):e94621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Florek M, Haase M, Marzesco AM, et al. Prominin‐1/CD133, a neural and hematopoietic stem cell marker, is expressed in adult human differentiated cells and certain types of kidney cancer. Cell Tissue Res. 2005;319(1):15‐26. [DOI] [PubMed] [Google Scholar]
- 19. Gassenmaier M, Chen D, Buchner A, et al. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma‐initiating cells and predicts metastasis. Stem Cells. 2013;31(8):1467‐1476. [DOI] [PubMed] [Google Scholar]
- 20. Ueda K, Ogasawara S, Akiba J, et al. Aldehyde dehydrogenase 1 identifies cells with cancer stem cell‐like properties in a human renal cell carcinoma cell line. PLoS ONE. 2013;8(10):e75463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu J, Guan W, Liu P, et al. Endoglin is essential for the maintenance of self‐renewal and chemoresistance in renal cancer stem cells. Stem Cell Reports. 2017;9(2):464‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pestrin M, Salvianti F, Galardi F, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9(4):749‐757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single‐cell sequencing. Nature. 2011;472(7341):90‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Q, Lou Y, Yang J, et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. 2019;68(11):2019‐2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single‐cell RNA‐seq. Science. 2016;352(6282):189‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tung JW, Heydari K, Tirouvanziam R, et al. Modern flow cytometry: a practical approach. Clin Lab Med. 2007;27(3):453‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang ZZ, Kim HJ, Villasboas JC, et al. Cytometry analysis reveals that specific intratumoral CD4(+) T cell subsets correlate with patient survival in follicular lymphoma. Cell Rep. 2019;26(8):2178.e3‐2193.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blair TA, Michelson AD, Frelinger AL 3rd. Mass cytometry reveals distinct platelet subtypes in healthy subjects and novel alterations in surface glycoproteins in Glanzmann Thrombasthenia. Sci Rep. 2018;8(1):10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner J, Rapsomaniki MA, Chevrier S, et al. A single‐cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177(5):1330.e18‐1345.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chevrier S, Levine JH, Zanotelli VRT, et al. Immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736.e18‐749.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santegoets SJ, van Ham VJ, Ehsan I, et al. The anatomical location shapes the immune infiltrate in tumors of same etiology and affects survival. Clin Cancer Res. 2019;25(1):240‐252. [DOI] [PubMed] [Google Scholar]
- 32. Wang T, Fu X, Jin T, et al. Aspirin targets P4HA2 through inhibiting NF‐kappaB and LMCD1‐AS1/let‐7g to inhibit tumour growth and collagen deposition in hepatocellular carcinoma. EBioMedicine. 2019;45:168‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mayo L, Jacob‐Hirsch J, Amariglio N, et al. Dual role of CD38 in microglial activation and activation‐induced cell death. J Immunol. 2008;181(1):92‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313‐319. [DOI] [PubMed] [Google Scholar]
- 35. Teng YD, Wang L, Kabatas S, Ulrich H, Zafonte RD. Cancer stem cells or tumor survival cells? Stem Cells Dev. 2018;27(21):1466‐1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khan MI, Czarnecka AM, Helbrecht I, Bartnik E, Lian F, Szczylik C. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Res Ther. 2015;6:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grosse‐Gehling P, Fargeas CA, Dittfeld C, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol. 2013;229(3):355‐378. [DOI] [PubMed] [Google Scholar]
- 38. Hatina J, Parmar HS, Kripnerova M, Hepburn A, Heer R. Urothelial carcinoma stem cells: current concepts, controversies, and methods. Methods Mol Biol. 2018;1655:121‐136. [DOI] [PubMed] [Google Scholar]
- 39. Shu X, Liu H, Pan Y, et al. Distinct biological characterization of the CD44 and CD90 phenotypes of cancer stem cells in gastric cancer cell lines. Mol Cell Biochem. 2019;459(1‐2):35‐47. [DOI] [PubMed] [Google Scholar]
- 40. Vermeulen L, Todaro M, de Sousa MF, et al. Single‐cell cloning of colon cancer stem cells reveals a multi‐lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105(36):13427‐13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amir el‐AD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single‐cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Atkuri KR, Stevens JC, Neubert H. Mass cytometry: a highly multiplexed single‐cell technology for advancing drug development. Drug Metab Dispos. 2015;43(2):227‐233. [DOI] [PubMed] [Google Scholar]
- 43. Subrahmanyam PB, Dong Z, Gusenleitner D, et al. Distinct predictive biomarker candidates for response to anti‐CTLA‐4 and anti‐PD‐1 immunotherapy in melanoma patients. J Immunother Cancer. 2018;6(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clancy T, Hovig E. Profiling networks of distinct immune‐cells in tumors. BMC Bioinformatics. 2016;17(1):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laurent C, Muller S, Do C, et al. Distribution, function, and prognostic value of cytotoxic T lymphocytes in follicular lymphoma: a 3‐D tissue‐imaging study. Blood. 2011;118(20):5371‐5379. [DOI] [PubMed] [Google Scholar]
- 46. Fang D, Kitamura H. Cancer stem cells and epithelial‐mesenchymal transition in urothelial carcinoma: Possible pathways and potential therapeutic approaches. Int J Urol. 2018;25(1):7‐17. [DOI] [PubMed] [Google Scholar]
- 47. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755‐768. [DOI] [PubMed] [Google Scholar]
- 48. Guo R, Zhang T, Meng X, et al. Lymphocyte mass cytometry identifies a CD3‐CD4+ cell subset with a potential role in psoriasis. JCI Insight. 2019;4(6):125306. [DOI] [PMC free article] [PubMed] [Google Scholar]


