Abstract
Background: Prostate-specific membrane antigen (PSMA) has emerged as a promising target for developing radionuclide therapy (RNT) in prostate cancer; however, accumulation of PSMA-RNT in salivary glands can result in irreversible xerostomia. Methods to prevent PSMA-RNT-related xerostomia could be clinically useful; however, little is known about PSMA expression in salivary glands of preclinical animal models. Using [18F]DCFPyL autoradiography/biodistribution, PSMA expression levels were determined in salivary glands of various preclinical monkey and rodent species and compared with humans.
Methods: Binding affinities (Kd) and PSMA levels (Bmax) were determined by in vitro [18F]DCFPyL autoradiography studies. In vivo rodent tissue uptakes (%ID/g) were determined from [18F]DCFPyL biodistributions.
Results: [18F]DCFPyL exhibited low nanomolar Kd for submandibular gland (SMG) PSMA across all the species. PSMA levels in human SMG (Bmax = 60.91 nM) were approximately two-fold lower compared with baboon SMG but were two- to three-fold higher than SMG PSMA levels of cynomolgus and rhesus. Rodents had the lowest SMG PSMA levels, with the mouse being 10-fold higher than the rat. In vivo rodent biodistribution studies confirmed these results.
Conclusions: SMG of monkeys exhibited comparable PSMA expression to human SMG whereas rodents were lower. However, the results suggest that mice are relatively a better small animal preclinical model than rats for PSMA salivary gland studies.
Keywords: autoradiography, PET imaging, PSMA, radionuclide imaging, radionuclide therapy, salivary glands
Introduction
Prostate-specific membrane antigen (PSMA), also known as glutamate carboxypeptidase II or folate hydrolase 1, is encoded by the FOLH1 gene.1 PSMA is overexpressed on the cell membrane of most primary and metastatic prostate cancers (PCa) and in the neovasculature of solid tumors.2,3 PSMA has remarkably high expression levels in PCa cells and low expression in healthy tissues except for kidneys, salivary glands, and lacrimal glands. Hence, PSMA has emerged as a promising target for radionuclide therapy (RNT) for metastatic PCa.4 There is increasing evidence of clinically meaningful responses to PSMA-targeted RNT labeled by using both β′ (177Lutetium; 177Lu) and α (225Actinium; 225Ac) emitting radionuclides.5,6 Several PCa patients, who failed earlier therapies, showed positive responses to 225Ac-labeled PSMA, including radiographic and complete biochemical responses.7,8 Despite this significant progress, PSMA-targeted RNT can cause damage to salivary glands and lacrimal glands, resulting in dry mouth (xerostomia) and dry eyes (xerophthalmia), respectively.9–11 To design effective strategies to protect these tissues from PSMA-targeted RNT, a better understanding of PSMA expression and its regulation in salivary and lacrimal glands is needed.
The salivary complex includes the parotid, submandibular, and sublingual glands, as well as many minor (accessory) salivary glands located throughout the oral cavity and upper airway. The salivary glands are complex organs composed chiefly of two epithelial cell types: acinar and ductal epithelium.12 The PSMA is exclusively expressed on the apical lumen of the acinar epithelium in all types of salivary glands in humans. Although PSMA is well known for its carboxypeptidase and folate hydrolase activities and likely plays a role in secretion, its exact role in salivary gland function remains unknown.13–15 In general, the acinar epithelium is responsible for the production of a plasma-like primary saliva product composed of salivary proteins.16 In humans, the acinar epithelium is particularly sensitive to damage from ionizing radiation. Moreover, acinar cells have poor regenerative capacity and a stem cell niche has not been clearly identified in humans.9,17–19 In patients, PSMA-mediated salivary gland accumulation of RNT agents and the associated ionizing radiation can result in irreversible damage to the acini, leading to reduced secretory function and atrophy of the tissue. The resultant salivary hypofunction from RNT has a profound negative impact on quality of life, including oral function (e.g., difficulty masticating, swallowing, and speaking), impaired sense of taste, tooth decay, and oral infections (e.g., oral candidiasis, periodontal disease).10,11,20–22 Likewise, the permanence of xerostomia caused by PSMA-targeted α-emitting (225Actinium), and β′-emitting radionuclides (177Lutetium) has been shown to be dose dependent.5,10,23,24 This unwanted and often irreversible side-effect has hindered the rapid development of PSMA-targeted alpha RNT. Because of the high clinical importance of PSMA-targeted RNT in treating PCa, there is an urgent need to develop suitable preclinical models to reliably investigate strategies to protect salivary glands from radiation-induced damage.
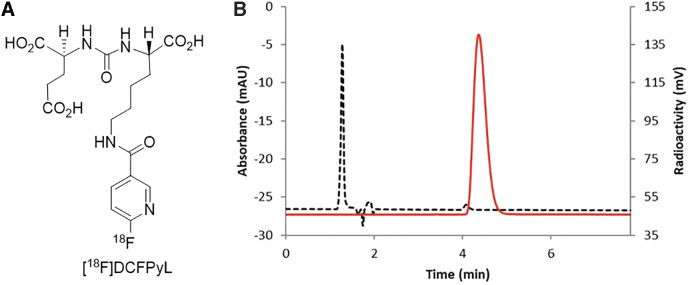
Through proof-of-concept preclinical studies, efficacious clinical studies aimed at preventing uptake of radionuclides may be achievable.25,26 Proper animal models will play a vital role in establishing methods to protect the salivary glands; however, little is known about PSMA expression levels in salivary glands of commonly used laboratory animals. Identification of preclinical animal models with appropriate levels of PSMA will expedite testing of various strategies to protect the salivary glands from PSMA-targeted RNT. Herein, we determined and compared the concentration of PSMA in salivary glands across various species by using 2-(3-{1-carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pentanedioic acid, [18F]DCFPyL27 (Fig. 1A), a synthetic radioligand exhibiting high affinity and specificity for PSMA via autoradiography and rodent biodistribution studies, and correlated these findings with immunofluorescence (IF) confocal microscopy.
FIG. 1.
(A). Structure of [18F]DCFPyL. (B). HPLC analysis of [18F]DcFPyL (HPLC conditions: Agilent Eclipse plus C18 column (4.6 × 150 mm, 3.5 μm), mobile phase: 10% acetonitrile in 0.1 M ammonium formate, with a flow rate of 1.0 mL/min. Solid line: in-line radio detector; dotted line: UV detector at 254 nm.
Materials and Methods
PSMA conservation across common animal models
Protein sequence conservation was compared across human, mouse, rat, cynomolgus, and rhesus species by using a multiple sequence alignment program (Clustal Omega; www.ebi.ac.uk/Tools/msa/clustalo/). Mouse salivary gland RNA-seq data were produced by the Melvin lab at NIDCR28 and are available on the Gene Expression Ontology database under accession GSE96747. Human salivary gland RNA-seq data were acquired from the Human Protein Atlas.29–31
Tissue samples
Fresh healthy salivary glands were collected from two cynomolgus monkeys, two rhesus monkeys, three mice (NCR-nu/nu athymic), and three rats (F344/SAS Fischer); fast frozen; and stored (−80°C) until use. Fresh frozen salivary glands from two humans (Human Cooperative Tissue Network) and two baboons (Bioreclamation IVT) were obtained and stored (−80°C) until use. All human tissues had been examined grossly by a board-certified pathologist (B.M.W.), portioned, either formalin-fixed or embedded in OCT, snap frozen in an isopentane/liquid nitrogen slurry, and stored at −80°C until use. PC3-PSMA tumors (prostate cancer cell line transfected with human PSMA obtained from a mouse xenograft) were fast frozen to be used as a positive control.
Synthesis and radiolabeling of [18F]DCFPyL
[18F]DCFPyL was prepared according to our recently reported literature method with a radiochemical yield of 32%–43% (decay corrected, n > 50) and purity >98% (Fig. 1B) in a 45 min synthesis time.27 The molar activity was 1200–2600 Ci/mmol (44,000–96,200 GBq; end of synthesis).
In vitro autoradiography
Frozen submandibular salivary glands (SMG) of mice, rats, baboons, cynomolgus monkeys, rhesus monkeys, and humans were cryosectioned at 20 μm thickness (Leica CM3050S) and mounted on glass slides followed by air drying. Saturation assays were performed with mounted slides that were pre-incubated in the assay buffer [0.1% BSA, 0.1% aprotinin (100 U/mL), 50 mM Tris (pH 7.5), 1.5 mM EDTA, 150 mM saline (0.9%), 5 mM MgCl2] for 15 min at room temperature, which were incubated (2 h) with increasing concentrations of [18F]DCFPyL (0.3 nM–36 nM) representing total bound activity (Bt). Non-specific binding (Bnsp) was determined by incubating the tissue sections with [18F]DCFPyL (0.3 nM–36 nM) in the presence of the non-radioactive PSMA ligand DCFPyL (1 μM) at the same concentrations. Afterward, incubation slides were washed twice with Tris 50 mM at 4°C for 4 min, dipped once in cold (4°C) distilled water, and allowed to dry. After drying, the slides were exposed to phosphor imaging plates (Fuji BAS-SR2025) for 24 h. After exposure, the plates were scanned by using the Fuji FLA-5100 scanner to produce digital images from which regions of interest [ROIs; phosphostimulated luminescence units per mm2 (PSL/mm2); Image Gauge 4.0)] were drawn to determine specific bound (Bsp; Bt-Bnsp = Bsp). ROIs (PSL/mm2) were converted to molar concentrations by using specific activity of [18F]DCFPyL, as previously described.32 Briefly, selected slides were counted on a gamma counter (PerkinElmer 2480 Wizard3) to determine the radioactive content (counts per minute, CPM) and then exposed to the phosphor imaging plate from which PSL/mm2 units were determined by drawing ROIs around the entire slide. Using the linear relationship of PSL/mm2 to the CPM determined from the entire slide (CPM = slope × PSL/mm2), the ROIs from each tissue section (20-μm thickness) could be converted from PSL/mm2 to CPM/mm2 and used to calculate relative PSMA molar concentrations in the salivary gland sections. Using the molar concentrations (Bsp), the Kd and Bmax were derived for each species. Data were analyzed by using a non-linear regression curve fit, including one-site hyperbola (GraphPad Prism 7).
In vivo biodistribution
Biodistribution studies were performed in rats [F344/SAS Fischer (Strain code: 403, Charles River), female; n = 5] and mice [NCR-nu/nu athymic (Strain code: 553, Charles River), female; n = 5] at 1 h post-injection (intravenous) of [18F]DCFPyL [Rat:3.7 MBq (100 μCi); Mouse:1.85 MBq (50 μCi)]. After 1 h, the animals were euthanized (CO2 asphyxiation); blood/tissues from each animal were excised; the animals were weighed; and radioactive content was determined by using a gamma counter (Counting efficiency 52.4%, 400–1200 Kev). The radioactive content (CPM) of the tissue samples were corrected for background (by counting an empty vial) and all samples were decay corrected to the start time of the sample counting, including the radioactive dose. For each tissue, the percent injected dose per gram (%ID/g) was determined by using the following equations:
-
(1)
%ID/g = {[CPM tissue/CPM total injected dose]/[tissue weight (g)]} × 100
-
(2)
%ID/g (normalized to a 20g mouse) = (%ID/g) × [(body weight of mice)/(20 g)]
-
(3)
%ID/g (normalized to a 125g rat) = (%ID/g) × [(body weight of rat)/(125 g)]
Tissue to blood and tissue to muscle ratios were calculated as Tissue (%ID/g)/Blood (%ID/g) and Tissue (%ID/g)/Muscle (%ID/g), respectively. All animal studies were performed in accordance with IACUC-approved protocols.
Immunofluorescent staining
Sections (5 μm) were cut from PC3-PSMA xenograft, human, baboon, and mouse SMG. Sections were stained with anti-PSMA antibodies (human monoclonal, Novus Biologics) for PC3-PSMA, human, and baboon, and with mouse monoclonal antibody-FITC (LifeSpan Biologicals) for mouse and counterstained with DAPI. Anti-human PSMA monoclonal antibody was detected by using an anti-mouse secondary (Alexa488) at 1:200 concentration. Sections were imaged by using an Olympus Fluoview confocal microscope (15 z-stack, 40 × objective).
Results
PSMA sequence conservation across species
PSMA orthologs from all species demonstrated high sequence identity (81.1%) (Supplementary Fig. S1A and B) compared with human PSMA. Amino acid residues predicted to represent the binding pocket and catalytic domain of the PSMA enzyme exhibited 100% identity, as previously reported for some of the orthologs tested.1 Therefore, a PSMA substrate inhibitor such as DCFPyL would be expected to bind with comparable affinities within the PSMA binding pocket across these species.33
In vitro autoradiography
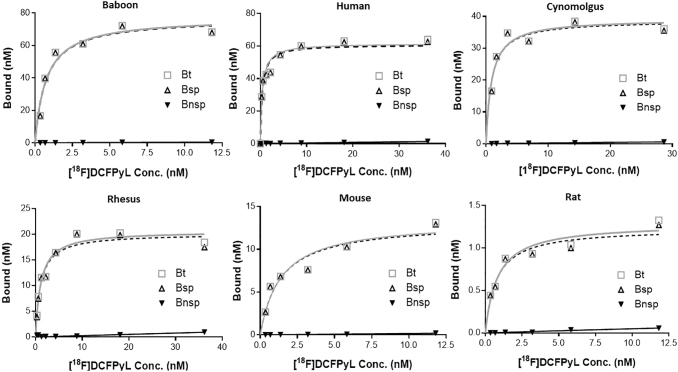
In vitro autoradiography saturation studies (Figs. 2 and 3A) with the SMG gland of mice, rats, baboons, cynomolgus monkeys, rhesus monkeys, and humans were performed to determine: (i) PSMA concentration levels (Bmax) and (ii) binding affinity (Kd) of [18F]DCFPyL to PSMA. SMG sections for all species exhibited high PSMA specific binding (Bsp) ranging from 90% to 95% with high [18F]DCFPyL binding affinities (Kd, 0.42–1.39 nM; Fig. 3B). The highest binding affinity (0.42 nM) was observed for human SMG, with slightly lower affinities observed for cynomolgus monkeys (0.55 nM) and rats [(0.76 nM); Fig. 3B]. Lower binding affinities of two to three-fold compared with human SMG were observed in rhesus monkey (1.08 nM), mouse (1.24 nM), and baboon (1.39 nM). The binding affinity of [18F]DCFPyL for human SMG glands (Kd = 0.42 nM) was comparable to published values for human prostate cancer cells (Kd = 0.49 nM).34 In summary, our studies demonstrated that [18F]DCFPyL exhibits low nanomolar binding affinities for SMG PSMA for all tested species (Fig. 3B).
FIG. 2.
Representative plots of in vitro autoradiography saturation binding studies of human, non-human primates (baboon, cynomolgus, rhesus) and rodent (mouse, rat) submandibular glands; each point (n = 6 points) is derived from ROI analysis. For each plot: Bt = Bound total; Bnsp = Bound non-specific; Bsp = Bound specific (Bt-Bnsp = Bsp). ROI, regions of interest.
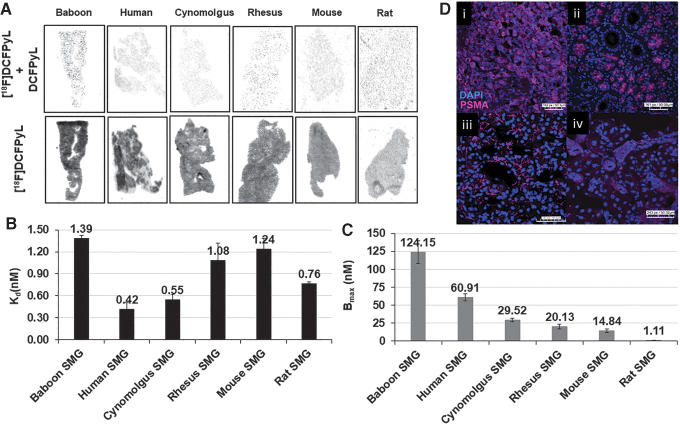
FIG. 3.
(A) Representative autoradiograms showing [18F]DCFPyL binding (Bt:[18F]DCFPyL) and non-specific binding (Bnsp: [18F]DCFPyL+DCFPyL (10−6M)) to PSMA in the SMGs of various species. (B, C) Binding affinity (Kd, nM) of [18F]DCFPyL (B) and PSMA concentrations (C; Bmax, nM) in submandibular glands of various species determined from in vitro autoradiograph saturation plots. Each bar represents mean ± SD, n = 3; (D) Representative immunofluorescence staining of PC3-PSMA tumor (i), human submandibular glands (ii), baboon submandibular glands (iii), and mouse submandibular glands (iv) with PSMA antibody (PSMA expression, pink) and DAPI (nuclear stain, blue). SMG, submandibular gland; PSMA, prostate-specific membrane antigen.
The PSMA concentration (Bmax) in SMG across the species was determined from the saturation assays (Fig. 3C) and ranged from 124.15 nM to 1.11 nM. Baboons expressed the highest level of PSMA concentration with a Bmax value of 124.15 nM and this was ∼2-fold higher than humans (Bmax = 60.91 nM). Cynomolgus and rhesus monkey SMG displayed a Bmax of 29.52 nM and 20.13 nM, ∼2-fold less than that of human SMG. The PSMA level (Bmax) in mice and rat SMG was 14.84 nM and 1.11 nM, respectively. These results indicate that [18F]DCFPyL would be able to detect PSMA in SMG in vivo for all the species tested except for the rat due to low PSMA expression.
Submandibular PSMA expression determined by IF
The protein expression and localization of PSMA in submandibular glands (SMGs) of human, baboon, mouse, and PC3-PSMA tumors were determined by IF (Fig. 3D). PSMA IF demonstrated strong, apical membranous staining exclusively in the acini of SMG in humans and baboons whereas PC3-PSMA tumor cells demonstrated a strong, diffuse cytomembranous pattern of expression. Mouse SMG demonstrated a weak cytomembranous pattern of staining with moderate focal puncta arranged in a perinuclear pattern. These results support the in vitro binding study data demonstrating that humans and baboons exhibit similar protein expression whereas mice have noticeably lower expression. To confirm these results, publicly available RNA sequencing data were queried to determine the human and mouse SMG FOLH1 expression (Supplementary Fig. S2). FOLH1 mRNA is expressed at low levels in the mouse salivary gland, compared with moderate expression of FOLH1 in the human salivary glands. These data agree with in vitro binding data.
In vivo biodistribution in mice and rats
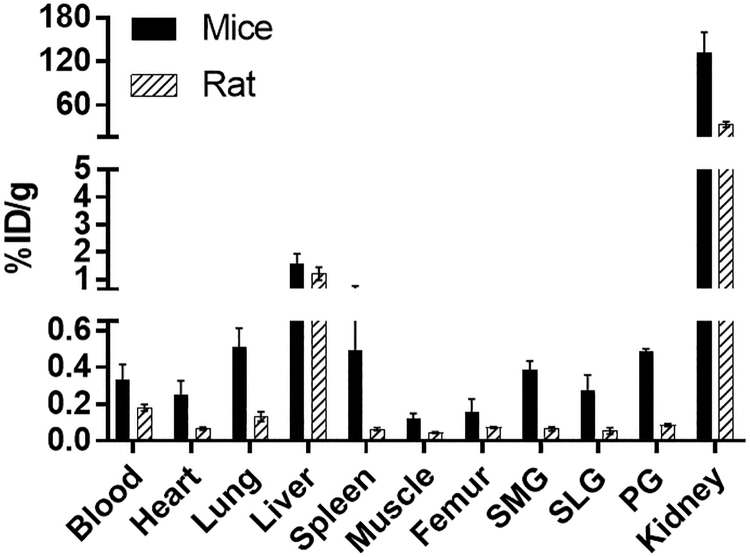
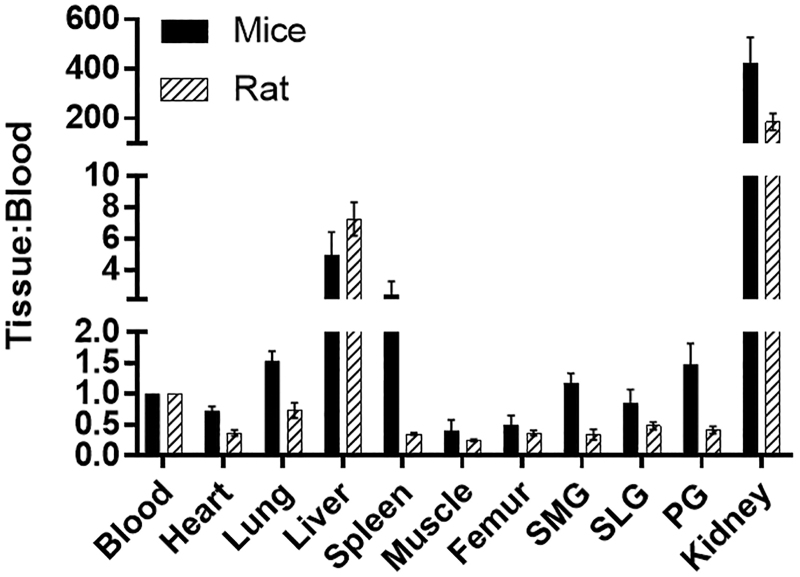
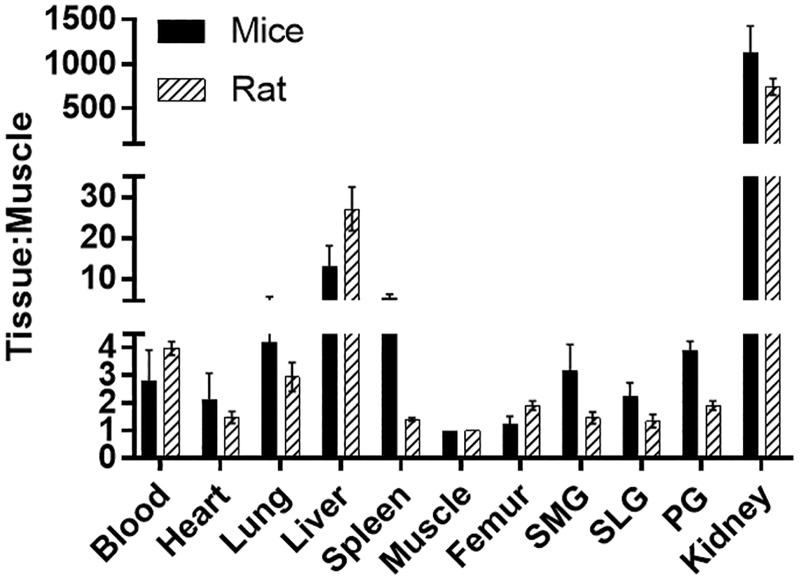
The biodistribution of [18F]DCFPyL was determined in mice and rats at 1 h post-injection (Fig. 4), with the highest uptakes (%ID/g) in the kidney for both mice (132%ID/g) and rats (33%ID/g). The [18F]DCFPyL uptake in mouse submandibular (SMG, 0.37%ID/g), sublingual (SLG, 0.26%ID/g), and parotid (PG, 0.46%ID/g) glands was higher than in rat SMG (0.065%ID/g), SLG(0.054%ID/g), and PG(0.085%ID/g). Further, the uptake in rat salivary glands is similar to the uptake in the muscle (0.04%ID/g), which represents non-target tissue binding. The tissue to blood ratios for all the organs was calculated from the %ID/g of tissue (Fig. 5). The tissue to blood ratio (T:B) of the SMG, SLG, and PG for mice was found to be 1.17, 0.85, and 1.48 whereas the T:B in rat SMG, SLG, and PG was 0.34, 0.48, and 0.41, respectively. All major salivary glands of the mice showed higher T:B than the rats (SMG, 3.4-fold; SLG, 1.7-fold; and PG, 3.6-fold). The rat salivary gland T:Bs were comparable to muscle (0.25), indicating that the uptake observed in salivary glands of rats may not represent PSMA binding but the radioactivity associated with the tissue blood volume. In both mice and rats, the highest uptakes were observed in kidneys with a T:B of 424.5 and 186.4, respectively. Similar to the T:B, the tissue to muscle ratio (T:M) of mouse SMG (3.2), SLG (2.2), and PG (3.9) glands was higher than that of rats (SMG:1.5, SLG:1.3, and PG:1.9; Fig. 6). These higher levels of PSMA observed in mice compared with rats in vivo agree with the in vitro autoradiography results.
FIG. 4.
Comparison of [18F]DCFPyL biodistribution in rats and mice at 1 h after injection of [18F]DCFPyL. Each bar represents %ID/g ± SD (for mice: %ID/g normalized to 20 g mice; for rats: %ID/g normalized to 125 g rat; n = 5 for each group). SMG, SLG, and PG represent submandibular, sublingual, and parotid glands, respectively.
FIG. 5.
Comparison of Tissue:Blood ratios of [18F]DCFPyL in rats and mice at 1 h after injection of [18F]DCFPyL. Each bar represents Tissue:Blood ± SD; n = 5 for each group. SMG, SLG, and PG represent submandibular, sublingual, and parotid glands, respectively.
FIG. 6.
Comparison of Tissue:Muscle ratios of [18F]DCFPyL in rats and mice at 1 h after injection of [18F]DCFPyL. Each bar represents Tissue:Muscle ± SD; n = 5 for each group. SMG, SLG, and PG represent submandibular, sublingual, and parotid glands, respectively.
Discussion
These studies indicate that the binding affinity of [18F]DCFPyL for PSMA was similar across the animal species compared with humans, although the PSMA expression levels varied. Because DCFPyL and other synthetic PSMA ligands have been designed specifically for human PSMA, it is important to first understand the protein similarities across these animal models. Overall, there is high amino acid conservation for PSMA across the tested species, with high similarity between humans and baboons, less so for cynomolgus and rhesus monkeys, and lowest for mice and rats. However, in the predicted binding domain of PSMA, there is 100% identity across the non-human primate and rodent species compared with humans.
Further, not only should the ideal animal salivary gland model have PSMA binding domains that are similar, but also PSMA expression levels in the salivary glands should approximate those found in humans as well. PSMA expression in the salivary glands is highly specific to the apical membrane of acini but overall, protein expression is low when compared with other acini-specific salivary proteins (e.g., AQP5 or NKCC1, www.humanproteinatlas.org). Results obtained from animal models with extremely low levels of target sites may prove to be unreliable due to the presence of high non-specific background radioactivity, as was demonstrated in the rat. In a published study, PSMA levels in pig salivary glands were found to be ∼500-fold lower compared with a PSMA-overexpressing human PCa cell line, thus indicating that the pig would be an unsuitable animal model for PSMA salivary gland studies.35 We have compared the PSMA expression levels among commonly used preclinical animal models and found both a diversity of affinity and expression that could inform as to the most appropriate animal model selection for comparison to humans. Among the mouse, rat, baboon, cynomolgus monkey, rhesus monkey, and human, our results indicated that the SMG of the baboon had the highest expression of PSMA (Bmax = 124.15 nM) whereas that of the rat (Bmax = 1.11 nM) had the lowest. It is important to note that these PSMA levels for the different species may not be reflective of internalized PSMA induced by ligand binding, which has been observed with other small-molecule ligands and antibodies targeting PSMA36–38 Therefore, it might be expected that in vivo salivary gland PSMA expression levels across these species may differ from the in vitro autoradiography results depending on the contribution of the internalized PSMA radioligands to the overall salivary gland uptake. Further, although internalization of the PSMA radioligand may prove to be beneficial for PET imaging applications by lengthening the residence time in the glands of the radioligand, this would not be the case with radiotherapeutic applications in which radiation doses delivered to the salivary glands may be increased.
Despite the potential of internalization to affect PSMA densities, the in vitro autoradiography results compared favorably with the in vivo biodistribution results in which PSMA was found to be greater in mouse salivary glands compared with rats. To determine whether these PSMA concentrations are enough for visualization of the salivary glands in PET imaging studies, it would be important to determine the target to non-target ratios, which can be predicted from the mathematical equation:
where B = Bound concentration, F = Free concentration, Bmax = Maximum receptor concentration, and Kd = Dissociation constant. Using the equation just cited, the target to non-target ratio of 2 to 1 was predicted for mice SMG, indicating that [18F]DCFPyL may be useful to image these glands in vivo. On the other hand, the target to non-target ratio for rat SMG was found to be 0.126, which indicates that in vivo PSMA-specific binding would be indistinguishable from the background. Thus, [18F]DCFPyL may be useful to monitor PSMA-mediated changes in salivary glands in mice while investigating various methods to preserve salivary gland function post–PSMA-RNT. However, because of the decreased density of PSMA in mice salivary glands compared with humans, determining the efficacy of new methods to prevent RNT-mediated toxicity may be limited. It is important to note that blood volumes of the salivary glands and pharmacokinetics of the test agent in humans compared with other species should also be considered when choosing an appropriate animal model to assess PMSMA-RNT-mediated salivary gland toxicity. For instance, the blood volume of the SMGs is greater in mice (110 μL/g of tissue) than rats (81 μL/g), which when applied to this case contributes ∼8% and ∼3%, respectively, to the total radioactivity in the glands.39 Although the blood volume of the SMGs of both the species is different, this does not significantly contribute to the total radioactivity in the glands. Therefore, the majority of the radioactivity in the SMGs represents PSMA-mediated uptake of [18F]DCFPyL and correcting for blood volume would not impact the uptake difference observed in mice and rats. In summary, these results based on PSMA expression levels and affinity suggest that for in vivo studies the mouse would be a better small animal preclinical model than the rat, but non-human primates would provide more reliable and clinically translatable results for investigations related to PSMA-RNT in salivary glands. Alternatively, PSMA-targeted radionuclide probes such as [18F] DCFPyL may prove useful in preliminary in vitro studies to assess responses of salivary glands to PSMA-targeted RNT and methods to prevent xerostomia by using organ-on-a-chip or cadaver tissue from healthy or treated prostate cancer patients40
Conclusion
Our data compare the expression of PSMA in salivary glands of commonly used preclinical animal models. These data should allow researchers to choose the best preclinical animal model for performing salivary gland studies associated with PSMA. Hopefully, this will aid and encourage efforts to develop methods to spare the salivary glands during PSMA-targeted RNT.
Authorship Confirmation Statement
The contributions of each author toward this article are given next.
Jyoti Roy: Study conception and design, acquisition of data, analyzing and interpreting data, drafting of article, and critical revision; Blake M. Warner: Acquisition of data, analyzing and interpreting data, drafting of article, and critical revision; Falguni Basuli: Synthesis of [18F]DCFPyL, critical revision; Xiang Zhang: Synthesis of [18F]DCFPyL, critical revision; Karen Wong: Acquisition of data, critical revision; Thomas Pranzatelli: Acquisition of data, critical revision; Anita T. Ton: Acquisition of data, critical revision; John A. Chiorini: Study conception, critical revision; Peter L. Choyke: Study conception and design, critical revision; Frank I. Lin: Study conception and design, critical revision; Elaine M. Jagoda: Study conception and design, acquisition of data, interpretation of data, and critical revision.
Supplementary Material
Disclosure Statement
There are no potential conflicts of interest to disclose.
Funding Information
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Supplementary Material
References
- 1. Davis MI, Bennett MJ, Thomas LM, et al. Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase. Proc Natl Acad Sci U S A 2005;102:5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sweat SD, Pacelli A, Murphy GP, et al. Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology 1998;52:637. [DOI] [PubMed] [Google Scholar]
- 3. Bostwick DG, Pacelli A, Blute M, et al. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma - A study of 184 cases. Cancer-Am Cancer Soc 1998;82:2256. [DOI] [PubMed] [Google Scholar]
- 4. Rahbar K, Afshar-Oromieh A, Jadvar H, et al. PSMA theranostics: Current status and future directions. Mol Imaging 2018;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakravarty R, Siamof CM, Dash A, et al. Targeted alpha-therapy of prostate cancer using radiolabeled PSMA inhibitors: A game changer in nuclear medicine. Am J Nucl Med Mol Imaging 2018;8:247. [PMC free article] [PubMed] [Google Scholar]
- 6. Kratochwil C, Giesel FL, Stefanova M, et al. PSMA-targeted radionuclide therapy of metastatic castration-resistant prostate cancer with Lu-177-labeled PSMA-617. J Nucl Med 2016;57:1170. [DOI] [PubMed] [Google Scholar]
- 7. Kelly JM, Amor-Coarasa A, Ponnala S, et al. A single dose of (225)Ac-RPS-074 induces a complete tumor response in a LNCaP xenograft model. J Nucl Med 2018;60:649. [DOI] [PubMed] [Google Scholar]
- 8. Sathekge M, Bruchertseifer F, Knoesen O, et al. (225)Ac-PSMA-617 in chemotherapy-naive patients with advanced prostate cancer: A pilot study. Eur J Nucl Med Mol Imaging 2019;46:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langbein T, Chausse G, Baum RP. Salivary gland toxicity of PSMA radioligand therapy: Relevance and preventive strategies. J Nucl Med 2018;59:1172. [DOI] [PubMed] [Google Scholar]
- 10. Taieb D, Foletti JM, Bardies M, et al. PSMA-targeted radionuclide therapy and salivary gland toxicity: Why does it matter? J Nucl Med 2018;59:747. [DOI] [PubMed] [Google Scholar]
- 11. Rathke H, Kratochwil C, Hohenberger R, et al. Initial clinical experience performing sialendoscopy for salivary gland protection in patients undergoing (225)Ac-PSMA-617 RLT. Eur J Nucl Med Mol Imaging 2019;46:139. [DOI] [PubMed] [Google Scholar]
- 12. de Paula F, Teshima THN, Hsieh R, et al. Overview of human salivary glands: Highlights of morphology and developing processes. Anat Rec (Hoboken) 2017;300:1180. [DOI] [PubMed] [Google Scholar]
- 13. Wolf P, Freudenberg N, Buhler P, et al. Three conformational antibodies specific for different PSMA epitopes are promising diagnostic and therapeutic tools for prostate cancer. Prostate 2010;70:562. [DOI] [PubMed] [Google Scholar]
- 14. Klein Nulent TJW, Valstar MH, de Keizer B, et al. Physiologic distribution of PSMA-ligand in salivary glands and seromucous glands of the head and neck on PET/CT. Oral Surg Oral Med Oral Pathol Oral Radiol 2018;125:478. [DOI] [PubMed] [Google Scholar]
- 15. Kondo Y, Nakamoto T, Jaramillo Y, et al. Functional differences in the acinar cells of the murine major salivary glands. J Dent Res 2015;94:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Proctor GB. The physiology of salivary secretion. Periodontol 2000 2016;70:11. [DOI] [PubMed] [Google Scholar]
- 17. Emmerson E, May AJ, Berthoin L, et al. Salivary glands regenerate after radiation injury through SOX2-mediated secretory cell replacement. EMBO Mol Med 2018;10:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ogawa M, Oshima M, Imamura A, et al. Functional salivary gland regeneration by transplantation of a bioengineered organ germ. Nat Commun 2013;4:2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogawa M, Tsuji T. Functional salivary gland regeneration as the next generation of organ replacement regenerative therapy. Odontology 2015;103:248. [DOI] [PubMed] [Google Scholar]
- 20. Jensen SB, Pedersen AM, Vissink A, et al. A systematic review of salivary gland hypofunction and xerostomia induced by cancer therapies: Management strategies and economic impact. Support Care Cancer 2010;18:1061. [DOI] [PubMed] [Google Scholar]
- 21. Jensen SB, Pedersen AML, Vissink A, et al. A systematic review of salivary gland hypofunction and xerostomia induced by cancer therapies: Prevalence, severity and impact on quality of life. Support Care Cancer 2010;18:1039. [DOI] [PubMed] [Google Scholar]
- 22. Villa A, Connell CL, Abati S. Diagnosis and management of xerostomia and hyposalivation. Ther Clin Risk Manag 2015;11:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Begum NJ, Thieme A, Allmann J, et al. The effect of the total tumour volume on the kidneys, salivary glands and tumour BEDs for Lu-177-labelled PSMA ligands. Eur J Nucl Med Mol Imaging 2017;44:S271 [Google Scholar]
- 24. Langbein T, Kulkarni HR, Singh A, et al. Functional imaging of the salivary glands for evaluation of radiation-induced sialadenitis before and after Lu-177 PSMA radioligand therapy. Eur J Nucl Med Mol I 2017;44:S328 [Google Scholar]
- 25. van Kalmthout LWM, Lam MGEH, de Keizer B, et al. Impact of external cooling with icepacks on Ga-68-PSMA uptake in salivary glands. Ejnmmi Res 2018;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baum RP, Langbein T, Singh A, et al. Injection of botulinum toxin for preventing salivary gland toxicity after PSMA radioligand therapy: An empirical proof of a promising concept. Nucl Med Molec Imag 2018;52:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Basuli F, Zhang X, Woodroofe CC, et al. Fast indirect fluorine-18 labeling of protein/peptide using the useful 6-fluoronicotinic acid-2,3,5,6-tetrafluorophenyl prosthetic group: A method comparable to direct fluorination. J Labelled Compd Rad 2017;60:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brooks MJ, Rajasimha HK, Roger JE, et al. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(−/−) retinal transcriptomes. Mol Vis 2011;17:3034. [PMC free article] [PubMed] [Google Scholar]
- 29. Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science 2017;356. [DOI] [PubMed] [Google Scholar]
- 30. Berglund L, Bjorling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 2008;7:2019. [DOI] [PubMed] [Google Scholar]
- 31. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 32. Jagoda EM, Lang LX, Mccullough K, et al. [Br-76]BMK-152, a nonpeptide analogue, with high affinity and low nonspecific binding for the corticotropin-releasing factor type 1 receptor. Synapse 2011;65:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kopka K, Benesova M, Barinka C, et al. Glu-Ureido-based inhibitors of prostate-specific membrane antigen: lessons learned during the development of a novel class of low-molecular-weight theranostic radiotracers. J Nucl Med 2017;58:17S. [DOI] [PubMed] [Google Scholar]
- 34. Rousseau E, Lau J, Kuo HT, et al. Monosodium glutamate reduces (68)Ga-PSMA-11 uptake in salivary glands and kidneys in a preclinical prostate cancer model. J Nucl Med 2018;59:1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tonnesmann R, Meyer PT, Eder M, et al. [(177)Lu]Lu-PSMA-617 salivary gland uptake characterized by quantitative in vitro autoradiography. Pharmaceuticals (Basel) 2019;12:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu T, Wu LY, Kazak M, et al. Cell-Surface labeling and internalization by a fluorescent inhibitor of prostate-specific membrane antigen. Prostate 2008;68:955. [DOI] [PubMed] [Google Scholar]
- 37. Liu H, Rajasekaran AK, Moy P, et al. Constitutive and antibody-induced internalization of prostate-specific membrane antigen. Cancer Res 1998;58:4055. [PubMed] [Google Scholar]
- 38. Anilkumar G, Rajasekaran SA, Wang S, et al. Prostate-specific membrane antigen association with filamin A modulates its internalization and NAALADase activity. Cancer Res 2003;63:2645. [PubMed] [Google Scholar]
- 39. Altman PL, Dittmer DS. Blood and Other Body Fluids: Analysis and Compilation. Washington, DC: Federation of American Societies for Experimental Biology, 1961 [Google Scholar]
- 40. Nikolic M, Sustersic T, Filipovic N. In vitro models and on-chip systems: Biomaterial interaction studies with tissues generated using lung epithelial and liver metabolic cell lines. Front Bioeng Biotechnol 2018;6:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.