Abstract
Heat shock protein 70 (HSP70) chaperones play a central role in protein quality control and are crucial for many cellular processes, including protein folding, degradation, and disaggregation. Human HSP70s compose a family of 13 members that carry out their functions with the aid of even larger families of co-chaperones. A delicate interplay between HSP70s and co-chaperone recruitment is thought to determine substrate fate, yet it has been generally assumed that all Hsp70 paralogs have similar activities and are largely functionally redundant. However, here we found that when expressed in human cells, two highly homologous HSP70s, HSPA1A and HSPA1L, have opposing effects on cellular handling of various substrates. For example, HSPA1A reduced aggregation of the amyotrophic lateral sclerosis–associated protein variant superoxide dismutase 1 (SOD1)–A4V, whereas HSPA1L enhanced its aggregation. Intriguingly, variations in the substrate-binding domain of these HSP70s did not play a role in this difference. Instead, we observed that substrate fate is determined by differential interactions of the HSP70s with co-chaperones. Whereas most co-chaperones bound equally well to these two HSP70s, Hsp70/Hsp90-organizing protein (HOP) preferentially bound to HSPA1L, and the Hsp110 nucleotide-exchange factor HSPH2 preferred HSPA1A. The role of HSPH2 was especially crucial for the HSPA1A-mediated reduction in SOD1-A4V aggregation. These findings reveal a remarkable functional diversity at the level of the cellular HSP70s and indicate that this diversity is defined by their affinities for specific co-chaperones such as HSPH2.
Keywords: proteostasis, amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), 70 kilodalton heat shock protein (Hsp70), heat shock protein (HSP), superoxide dismutase (SOD), protein folding, Hsp110, HSPA4, HSPH2
Introduction
The Hsp70 machinery is a central system of the protein quality control, and it is involved in many different processes including protein folding, degradation, aggregation prevention, and disaggregation (1–3). Hsp70 chaperones are among the most highly conserved proteins in evolution and in humans comprise a family of 13 members (4). They have been reported to interact with a wide range of substrates, both non-native and native, by recognizing exposed hydrophobic motifs found in most proteins (5, 6). Hsp70 proteins consist of an N-terminal nucleotide-binding domain (NBD or N),3 a substrate-binding domain (SBD or S), and a C-terminal domain (CTD or C) that forms a lid stabilizing bound substrates after ATP hydrolysis (1). The Hsp70 activity is based on an ATP-dependent cycle, alternating between the low-substrate-affinity ATP-bound state and the high-substrate-affinity ADP-bound state. However, intrinsic ATPase activity of Hsp70 proteins is too low to function independently; that is why the cycle turnover is aided by the co-chaperones J-domain proteins (JDPs) and nucleotide-exchange factors (NEFs), which stimulate ATP hydrolysis and catalyze ADP/ATP exchange, respectively (7). In addition, the co-chaperones of the large JDP (also referred to as DNAJ) family (53 members in humans) act as recruiters of substrates via interaction with their versatile substrate-binding domains (8). Upon interaction with the Hsp70s, via their conserved J-domain, JDPs together with the substrates stimulate Hsp70–ATPase activity, and substrates are transferred to Hsp70 (9). To promote substrate release, four different types of co-chaperones can stimulate nucleotide exchange in human Hsp70s: BAG-domain proteins (6 members), Hsp110/Grp170 (HSPH, four members), HspBP1/Sil1 (two members), and GrpE (two members) (10). Despite differences in structure and working mechanism, all four types of NEFs interact with the Hsp70-NBD at different but partially overlapping sites. Other co-chaperones of Hsp70 facilitate the handover to other protein quality control systems. These include the Hsc70-interacting protein (HIP, also called ST13) and Hsp70/Hsp90-organizing protein (HOP, also called STIP1) that facilitate the cooperation of the Hsp70 and Hsp90 systems. Finally, co-factors such as the C terminus of Hsp70-interacting protein (CHIP, also called STUB1) are involved in regulating substrate degradation via the ubiquitin proteasome system (11).
Because of their high conservation both in evolution and within the Hsp70 family, (human) Hsp70 proteins are often regarded as largely interchangeable (12–17). However, specificity between Hsp70 machines does exist, and different effects of the various Hsp70s have been reported (6, 8, 18–20). The recognition of substrates by Hsp70 is quite generic, and because there is a lot more variability in JDPs and NEFs, the last two families have been suggested as the ones that confer specificity to the Hsp70 system (8). However, it has not been experimentally explored whether and to what extent (human) Hsp70 are indeed interchangeable. Also, whether different Hsp70s interact with specific co-chaperone partners and, if so, what determines the functional outcome of these different Hsp70 complexes have remained unclear.
Various members of the Hsp70 machinery have been identified as suppressors of protein aggregation (18, 19). From the Hsp70 family, only few members have been tested, mainly HSPA1A (stress-inducible Hsp70) and HSPA8 (constitutive Hsp70, HSC70), but mostly not in a comparative way. In particular, HSPA1A up-regulation has been reported as highly effective in withstanding global protein aggregation induced by unfolding events such as heat shock (21) or aggregation of specific thermosensitive proteins such as luciferase (22). However, neither HSPA1A nor HSPA8 up-regulation is very effective in preventing aggregation of disease-associated amyloidogenic proteins in cells, although results may vary depending on the system or the type of the substrate (18, 19). At least one exception to this is the aggregation of superoxide dismutase 1 (SOD1) mutants that cause amyotrophic lateral sclerosis (ALS) (23). Elevated expression of one Hsp70 (HSPA1A) has been reported to suppress mutant SOD1 aggregation (24).
Here, using two different Hsp70 clients, mutant SOD1 and a folding-impaired, mutant luciferase (25) (R188Q,R261Q referred to as LucDM), we dissect the effects of all cytosolic and nuclear Hsp70 orthologs on protein aggregation in cells. In particular, we found that two highly homologous Hsp70s, HSPA1A and HSPA1L, have opposing effects on the fate of these two clients. Strikingly, this differential activity is explicitly attributed to differences in the NBDs of the two Hsp70s, which subsequently affect their ability to functionally interact with specific co-chaperones. These data suggest another layer of functional diversification within the Hsp70 machines in human cells, which is directed by differential Hsp70–co-chaperone binding.
Results
Diverse effects of various Hsp70s on mutant SOD1 aggregation
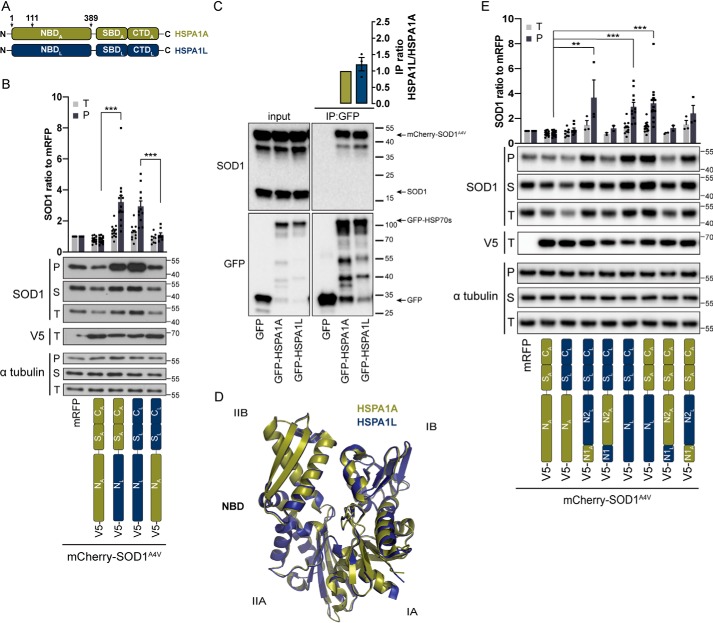
The different cytosolic/nuclear Hsp70 members HSPA1A (stress-inducible HSP70, HSP70–1, HSP72), HSPA1L (HSP70-like), HSPA2 (HSP70–2), HSPA6 (HSP70B), and HSPA8 (constitutive HSP70, HSC70) show high sequence conservation (Fig. S1 and Table S1) and bind similar peptide motifs (5, 6, 26) and hence have often been considered as functionally interchangeable. However, we noticed that the outcome in terms of client handling could differ significantly (27–30). To study this in more detail, we used the well-known aggregating ALS disease–associated SOD1 mutant A4V (SOD1A4V) (23), a reported Hsp70 client (24) as a model substrate. First, we developed a quantifiable fractionation method (Fig. 1A) to monitor aggregation of mCherry-tagged SOD1A4V. mCherry-SOD1A4V formed visible inclusions in cells and was partially detergent-insoluble after fractionation in contrast to mCherry-SOD1WT that showed diffuse expression and remained in the soluble fraction (Fig. 1, B and C, and Fig. S2A). Interestingly, expression of most Hsp70s in HEK293 cells enhanced rather than reduced SOD1A4V aggregation, and only HSPA1A showed a significant aggregation suppressing effect (Fig. 1D and Fig. S2B). Largely similar results were obtained in U2OS cells, with most Hsp70 members having either no effect or enhancing SOD1A4V aggregation and only HSPA1A leading to a significant reduction in SOD1A4V aggregation (Fig. 1E and Fig. S2C). The most striking observation was the consistent opposing effects of two of the closest paralogs HSPA1A and HSPA1L, with the former significantly reducing and the latter greatly enhancing SOD1A4V aggregation in both HEK293 and U2OS cells (Fig. 1, D and E). This opposing effect is not due to a difference in the increased expression because both Hsp70s are similarly (over)expressed (Fig. 1, D–F). SOD1A4V expression did not lead to any growth disadvantages, and neither HSPA1A nor HSPA1L influenced this (Fig. S2D).
Figure 1.
Hsp70 family members show variable effects on SOD1A4V aggregation. A, scheme of Nonidet P-40 fractionation protocol for measuring mutant SOD1 aggregation. Total cell lysates (T) in 1% Nonidet P-40–containing buffer are centrifuged at 20,000 × g and separated into a soluble supernatant (S) and an insoluble pellet (P) fraction. B, Nonidet P-40 fractionations of HEK293 cells expressing mCherry-SOD1WT or mCherry-SOD1A4V for 48 h. Western blots of the P, S, and T fractions using the indicated antibodies are shown. The upper band detected with SOD1 antibody corresponds to full-length mCherry-SOD1, and the lower band indicated with an asterisk is a cleavage product and behaves similarly to the full-length protein. For clarity reasons this band is omitted in the other figures. C, mCherry-SOD1A4V forms aggregates in U2OS cells after 48 h. Fluorescence microscopy images of mCherry-SOD1WT and mCherry-SOD1A4V (left column) and Hoechst (middle column) and the merge (right column, mCherry in red and DNA in blue) are shown. Scale bar, 10 μm. D and E, screen of Hsp70 family members for suppressors of SOD1A4V aggregation in HEK293 (D) and U2OS (E) cells. Western blots of Nonidet P-40 fractionation of cells co-expressing mCherry-SOD1A4V and V5-Hsp70s for 48 h. F, Nonidet P-40 fractions of HEK293 cells expressing GFP-LucWT or GFP-LucDM and the indicated V5-Hsp70s for 24 h. Western blots of the P, S, and T fractions using the indicated antibodies are shown. In D–F, quantification graphs represent total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities normalized to α-tubulin and relative to mRFP control (n = 3–17 for D, n = 3–5 for E, and n = 4 for F). Error bars in D–F indicate S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
Opposing effects of HSPA1A and HSPA1L on mutant luciferase
To explore whether the differential behavior of HSPA1A and HSPA1L is not limited to SOD1A4V aggregation, we investigated the impact of these two Hsp70s on the folding of GFP-tagged double mutant luciferase (GFP-LucDM) in cells (Fig. 1F). This model substrate is partly insoluble (Fig. 1F) and a Hsp70 client (Fig. S2E). When expressed in cells the GFP-LucDM fusion only displays minimal luminescence activity as compared with WT luciferase (Fig. S2F). Co-expression with HSPA1A but not HSPA1L leads to a reduction in the fraction of insoluble luciferase and in parallel to a drop in total protein levels of cellular GFP-LucDM (Fig. 1F) pointing toward a HSPA1A-driven degradation process. Because the total luminescence activity is nearly unaffected by co-expression of either HSPA1A or HSPA1L (Fig. S2F), this implies that the activity per amount of luciferase present in the cell (the “specific activity”) is elevated by the co-expression of HSPA1A (Fig. S2, G and H). Therefore, similar to mutant SOD1, in cells only HSPA1A can assist in the cellular handling of misfolded luciferase.
Differential functioning of HSPA1L and HSPA1A is associated with the nucleotide-binding domain
HSPA1L and HSPA1A are 89% identical in their amino acid sequence, and most differences lie in the substrate-locking C-terminal lid domain (Fig. S1 and Table S1). HSPA1A is one of the most studied human Hsp70s, whereas not much is known about HSPA1L and its cellular functions. In contrast to HSPA1A, HSPA1L lacks a HSF-binding element in its promoter and is indeed less heat stress–inducible (31). HSPA1L is expressed at low levels in most tissues (32). To further investigate why two very similar Hsp70s show such opposing effects on substrate handling, we generated chimeras to identify which part of the protein is responsible for this difference. Exchanging the NBD of HSPA1A with that of HSPA1L (NLSACA) generated a protein with HSPA1L-like activity that enhanced SOD1A4V aggregation (Fig. 2, A and B). Inversely, the chimera with the NBD of HSPA1A and the SBD and CTD of HSPA1L (NASLCL) gained an HSPA1A-like activity in suppressing SOD1A4V aggregation (Fig. 2B). This pointed toward the NBD as being responsible for the opposing effect of HSPA1A and HSPA1L on SOD1A4V aggregation. This was further confirmed because exchanging the individual SBDs or CTDs generated chimeric proteins whose activity fully depended on their NBDs (Fig. S3A). These results indicate that neither the SBD nor the CTD play a role in the differential effect of these two Hsp70s on protein aggregation. Interestingly, the SBD and especially the CTD are the most disparate domains based on the amino acid sequence (Fig. S1). Because the SBD confers substrate binding, this suggests that the difference in substrate fate cannot be attributed to differential SOD1A4V binding. Consistently, mCherry-SOD1A4V co-immunoprecipitated efficiently with both GFP-HSPA1A and GFP-HSPA1L (Fig. 2C). Of note, the GFP-tagged Hsp70s behave similarly to their V5-tagged versions (Fig. S3B). The endogenous WT SOD1 did not co-immunoprecipitate with either Hsp70, despite the similar expression level as mCherry-SOD1A4V (Fig. 2C). This underscores the specificity of both HSPA1A and HSPA1L for the mutant SOD1 protein.
Figure 2.
Nucleotide-binding domains of HSPA1A and HSPA1L are responsible for their opposing activities on mutant SOD1A4V aggregation. A, schematic representation of Hsp70 functional domains. HSPA1A is shown in yellow, and HSPA1L is in blue. Arrows indicate amino acid positions of sub-NBD swaps shown in E. B, effect of NBD chimeras of HSPA1A and HSPA1L on SOD1A4V aggregation. Nonidet P-40 fractionation of HEK293 cells co-expressing mCherry-SOD1A4V and V5-tagged chimeric proteins with swapped NBDs between HSPA1A (yellow) and HSPA1L (blue) for 48 h. Western blots using the indicated antibodies are shown. The quantification graph represents total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities normalized to α-tubulin and relative to mRFP control (n = 8–17). C, interaction of GFP-HSPA1A or GFP-HSPA1L with mCherry-SOD1A4V. GFP Nanotrap was used for native immunoprecipitation (IP) of the GFP-tagged Hsp70s expressed in HEK293 cells for 48 h. Western blots using the indicated antibodies are shown. The quantification graph of binding represents SOD1/GFP signal ratio of IP relative to HSPA1A measurement (n = 3). D, structural alignment between the NBDs of HSPA1A and HSPA1L. Structural superposition of the ribbon models after sequence alignment of HSPA1A-NBD (PDB code 3JXU; yellow) (80) and HSPA1L-NBD (PDB code 3GDQ; blue) (81) with root mean square deviation = 0.438 Å. Subdomains of lobes I and II (IA/B and IIA/B) are marked for orientation. E, effect of N1 (aa 1–111) and N2 (aa 112–389) regions of HSPA1A or N1 (aa 1–113) and N2 (aa 114–391) HSPA1L NBD on SOD1A4V aggregation. Western blots of Nonidet P-40 fractions from HEK293 cells co-expressing for 48 h mCherry-SOD1A4V and V5-tagged chimeric proteins with combinations of N1 and N2 regions of NBDs between HSPA1A (yellow) and HSPA1L (blue). The quantification graph represents total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities normalized to α-tubulin and relative to mRFP control (n = 2–17). In B and E, error bars indicate S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
In agreement with our findings, the importance of the NBD as a driver for functional specificity between Hsp70s has been previously noted for yeast (33) and human Hsp70s (27). The reason for this importance of the NBD is unclear. The NBDs of HSPA1A and HSPA1L share 91% sequence identity (Fig. S1). Structural alignment utilizing previously published data (34) revealed that the NBDs of HSPA1A and HSPA1L are almost identical (Fig. 2D), making such a different impact on a chaperone function really remarkable. Mapping the nonconserved residues between HSPA1A and HSPA1L on HSPA1A-NBD shows that they are spread over the entire NBD structure (Fig. S3C). The ATP/ADP-binding pocket, which resides in the middle of the NBD cleft, is fully conserved between the two Hsp70s (Fig. S3C). Highlighting these nonconserved amino acids pointed out that the accessible surface between the HSPA1A-NBD and HSPA1L-NBD was only slightly different (Fig. S3D). However, there were some subtle differences that could possibly affect the interaction interface with co-chaperones without significantly altering the core structure. Exchanging two subregions, aa 1–111 (N1) and aa 112–389 (N2), of HSPA1A with the homologous regions of HSPA1L (aa 1–113 and aa 114–391 for N1 and N2, respectively) and vice versa revealed that the effects on SOD1A4V aggregation were mainly coupled to the N2 region of the NBD (Fig. 2E).
ATPase cycle and biochemical functionalities of HSPA1A and HSPA1L are indistinguishable
To identify potential intrinsic functional differences in the ATPase cycle and biochemical activity of HSPA1A and HSPA1L, we purified both Hsp70s and NBD swaps. The intrinsic ATPase activity of each Hsp70 alone was low and equal for all four Hsp70 variants tested (Fig. 3A). The addition of increasing amounts of HSPH2 (a canonical NEF) at concentrations used in protein refolding assays accelerated the ATP cycle in a concentration-dependent manner that was similar for all variants as well (Fig. 3A). Consistent with this, the binding affinities of HSPH2 to both HSPA1A and HSPA1L were identical within error (Fig. S4A; HSPA1A, 107 ± 31 nm, and HSPA1L, 85 ± 31 nm). To corroborate this in living cells, we generated glutamate-to-glutamine mutations in the conserved ATP interaction sites Glu175 and Glu177 of HSPA1A and HSPA1L, respectively, that abrogate their nucleotide cycle (35). Expression of either the HSPA1AE175Q or the HSPA1LE177Q mutant in cells dramatically increased SOD1A4V aggregation (Fig. S4B), confirming that a functional Hsp70 ATP hydrolysis step is crucial for SOD1A4V processing by either HSPA1A or HSPA1L.
Figure 3.
HSPA1A and HSPA1L display similar ATPase activity and interact equally well to HSP90. A, nucleotide dissociation rates of recombinant HSPA1A, HSPA1L, and NBD swapped chimeras at 2 μm. The indicated amounts of recombinant HSPH2 were added to measure the effect of a canonical NEF. Error bars indicate S.D. B, interaction of endogenous HIP, HOP, HSP90, or CHIP with GFP-HSPA1A or GFP-HSPA1L in the presence of mCherry-SOD1A4V. GFP Nanotrap was used for native immunoprecipitation (IP) of the GFP-tagged Hsp70s, expressed in HEK293 cells for 48 h. Western blots using the indicated antibodies are shown. C, quantification graphs of co-chaperones binding to HSPA1A or HSPA1L (as in B). Graphs represent ratios of co-immunoprecipitation intensities of HIP (n = 6), HOP (n = 5), HSP90 (n = 7), or CHIP (n = 7). Error bars indicate S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001. D, interaction of GFP-HSPA1L or GFP-HSPA1LΔEEVD with endogenous HOP and DNAJB1 in the presence of mCherry-SOD1A4V. GFP Nanotrap was used for native immunoprecipitation of the GFP-tagged Hsp70s expressed in HEK293 cells for 48 h. Western blots using the indicated antibodies are shown. E, effect of EEVD of HSPA1L on SOD1A4V aggregation. Nonidet P-40 fractionation of HEK293 cells co-expressing mCherry-SOD1A4V and V5-tagged HSPA1L proteins. Western blots using the indicated antibodies are shown.
Moreover, the biochemical activities of HSPA1A and HSPA1L and their NBD swaps were indistinguishable using an in vitro protein refolding assay as readout: both variants lead to similar rates of refolding of heat-denatured luciferase (Fig. S4C). These data confirm that both HSPA1A and HSPA1L are bona fide Hsp70s with similar ATPase and biochemical activities. This implies that the opposing effects on substrate handling observed in cells lies in the cellular context in which these Hsp70s operate.
Preferential binding of HOP to HSPA1L does not affect substrate fate
Handover of certain substrates from the Hsp70 cycle to the Hsp90 system can have dramatic consequences on the fate of substrates (36, 37). Several factors influence this handover of substrates from Hsp70 to Hsp90, the most prominent being the co-chaperone HOP (1). Interestingly, HOP displayed a clear preference for binding to GFP-HSPA1L compared with GFP-HSPA1A (Fig. 3, B and C). However, this higher affinity of HOP to HSPA1L did not result in a detectable change in the interaction with HSP90 for these two Hsp70s in cells (Fig. 3, B and C). The notion that HOP displays preferential binding to HSPA1L is surprising because HOP binds with one of its tetratricopeptide repeat motifs to the extreme C terminus of Hsp70s, the EEVD motif (38, 39). Not only is this region fully conserved between HSPA1A and HSPA1L (Fig. S1), but the entire CTD is interchangeable with respect to the difference in substrate handling of HSPA1A and HSPA1L (Fig. 2). Indeed, deletion of this last part of the CTD (EEVD) of HSPA1L abrogates HOP binding, whereas BAG3 binding is unaffected (Fig. 3D). Moreover, deletion of the EEVD does not influence its effect on SOD1A4V solubility (Fig. 3E). Thus, the increased complex formation between HSPA1L and HOP does not explain the difference in substrate fate and may reflect a more downstream effect.
Inspired by the preferential interaction of HOP to HSPA1L, we wondered whether additional partnerships would display a differential binding to HSPA1A and HSPA1L as well. First, we tested HIP, a co-chaperone that stabilized Hsp70 in the ADP-bound state (40). This co-factor bound to both HSPA1A and HSPA1L in a similar fashion (Fig. 3, B and C). Also, the other negative regulator of the nucleotide cycle CHIP, which has often been suggested to direct Hsp70 clients to proteasomal degradation (41), displayed equal binding to HSPA1A and HSPA1L (Fig. 3, B and C), which is in line with its interaction via the CTD (not NBD) of Hsp70s (42).
Thus, communication with Hsp90 and antagonistic regulators of the Hsp70 cycle does not underlie the differences in substrate handling of HSPA1A and HSPA1L. In cells the N2 region is responsible for the functional differences between HSPA1A and HSPA1L (Fig. 2E). This region contains important interaction sites of JDPs and NEFs (Fig. S1). Therefore, we decided to test whether altered affinity to JDP or NEF in situ was involved in these differential activities.
JDPs deliver mutant SOD1 equally efficient to both HSPA1A and HSPA1L
J-domain proteins interact with Hsp70s through their conserved J-domain and stimulate Hsp70 ATPase activity, a step crucial for substrate transfer to the Hsp70s (8, 43). We first sought to identify which JDPs were involved in SOD1A4V recruitment to the Hsp70s and would therefore be relevant to this activity. Overexpression in HEK293 cells of DNAJA (Fig. 4A) or DNAJB (Fig. 4B) subfamily members had variable effects on SOD1A4V aggregation and revealed DNAJB1, DNAJB2b, and DNAJB7 as the strongest suppressors. Among the JDPs that exhibited a suppressive effect on SOD1A4V, DNAJB1 is one of the best-characterized members for substrate delivery to the Hsp70s and stimulation of their ATPase activity (9, 44). Therefore, we focused on DNAJB1 for the subsequent studies. To first confirm whether DNAJB1 suppresses SOD1A4V aggregation via Hsp70s, we introduced a mutation in the HPD motif of its J-domain (DNAJB1H32Q), which is known to abrogate the ability of the J-domain to stimulate Hsp70s ATPase activity (45, 46). Expressing DNAJB1H32Q together with SOD1A4V led to a massive increase in SOD1A4V aggregation (Fig. 4C), confirming that DNAJB1 requires Hsp70 for this function.
Figure 4.
DNAJB1 acts together with Hsp70s against SOD1A4V aggregation and shows no preference for either HSPA1A or HSPA1L. A and B, screen of DNAJA (A) or DNAJB (B) family members for suppressors of SOD1A4V aggregation; Nonidet P-40 fractionation of HEK293 cells co-expressing mCherry-SOD1A4V and V5-DNAJs for 48 h. Western blots using the indicated antibodies are shown, and the quantification graph represents total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities normalized to α-tubulin and relative to mRFP control (n = 2–3). C, effect of the Hsp70 interaction motif (HPD) mutant of V5-DNAJB1 on SOD1A4V aggregation; Nonidet P-40 fractionation of HEK293 cells co-expressing mCherry-SOD1A4V and V5-DNAJB1 or V5-DNAJB1H32Q for 48 h. Western blots using the indicated antibodies are shown, and the quantification graph represents total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities normalized to α-tubulin and relative to mRFP control (n = 4–6). D, interaction of V5-DNAJB1 with GFP-HSPA1A or GFP-HSPA1L in the presence of SOD1A4V. Transient expression of GFP-HSPA1A or GFP-HSPA1L together with V5-DNAJB1 and mCherry-SOD1A4V in HEK293 cells for 48 h followed by native immunoprecipitation (IP) of Hsp70s using GFP Nanotrap and Western blotting detection. The quantification graph of binding represents V5/GFP signal ratio of IP relative to HSPA1A measurement (n = 3). E, interaction of endogenous DNAJB1 with GFP-HSPA1A or GFP-HSPA1L in the presence of mCherry-SOD1A4V. Native immunoprecipitation after expression for 48 h in HEK293 cells using GFP Nanotrap and Western blotting detection using the indicated antibodies. The quantification graph of binding represents DNAJB1/GFP signal ratio of IP relative to HSPA1A measurement (n = 7). In A–E, error bars indicate S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
Next, we examined whether inadequate delivery via altered DNAJB1-Hsp70 affinities could play a role in the failure of HSPA1L in processing SOD1A4V. Hereto, we immunoprecipitated GFP-HSPA1A or GFP-HSPA1L and examined their co-precipitation with V5-DNAJB1. Both Hsp70s showed a similar ability to interact with V5-DNAJB1 in the presence of SOD1A4V (Fig. 4D). Moreover, binding of endogenous DNAJB1 to GFP-HSPA1A and GFP-HSPA1L in the presence of mutant SOD1A4V was equally similar (Fig. 4E). This argues against the possibility that aggregation-enhancing effects of HSPA1L are a result of inefficient DNAJ interaction. The equal ability of HSPA1A and HSPA1L to interact with SOD1A4V (Fig. 2C) is in line with the similar affinity of these Hsp70s for JDPs (Fig. 4, D and E). The EEVD motif in the CTD of Hsp70s also forms a binding surface for some JDPs including the homolog of DNAJB1, Sis1, in yeast (47). This seems conserved because the EEVD deletion mutant of HSPA1L hardly binds to DNAJB1 (Fig. 3D). The EEVD mutants of both HSPA1L and HSPA1A behave similarly to their respective WT Hsp70s with regard to SOD1A4V processing (Fig. 3E). This implies that the interaction with DNAJB1 is not essential for SOD1A4V processing. Given the vast number of JDPs, substrate delivery to Hsp70 is most likely partly redundant. Thus, together these results point to a step downstream of substrate delivery being fate-determining for SOD1A4V processing by either HSPA1A or HSPA1L.
Differential binding of HSPA1A and HSPA1L to nucleotide-exchange factors
Next, we wondered whether a different substrate binding and release cycle could underlie the difference in substrate handling between HSPA1A and HSPA1L, for example by a reduced substrate dissociation or alternatively by an increased substrate dissociation and a compensatory increased DNAJB1 binding to become retargeted to SOD1A4V in the case of HSPA1L. NEFs play a crucial role in dictating the efficiency of the ATPase cycle of the Hsp70s and the subsequent substrate release (10). Because we found that the intrinsic and stimulated nucleotide cycles of both Hsp70 isoforms were indistinguishable (Fig. 3), we next tested whether the difference in substrate handling by HSPA1A and HSPA1L could be related to differential partnerships with the various NEFs expressed in cells.
To identify potential NEFs relevant to mutant SOD1 processing, we first tested BAG1, BAG3, and BAG4 of the BAG family and HSPH1 (HSP105), HSPH2 (APG2/HSPA4), and HSPH3 (APG1/HSPA4L) of the Hsp110 family for their potential to affect SOD1A4V aggregation. However, upon their sole overexpression, none of the NEFs that we tested inhibited SOD1A4V aggregation; quite to the contrary, some increased SOD1A4V aggregation in both HEK293 or U2OS cells (Fig. S5, A and B). These results are consistent with earlier findings showing that high stoichiometric NEF to Hsp70 ratios have inhibitory effects on Hsp70 cycling and activity (48–52).
Different NEF types have been found to interact with multiple sites in the NBD of Hsp70s (53–56) (Fig. S1). To test whether any of the NEFs has a preferential affinity for HSPA1A or HSPA1L in the presence of SOD1A4V, we performed a series of co-immunoprecipitations. BAG1 and BAG3, used as two representatives of the BAG family, were efficiently co-immunoprecipitated with HSPA1A and HSPA1L (Fig. 5A). Similarly, HSPBP1 exhibited comparable binding to both HSP70s (Fig. 5B). In sharp contrast, all three members of the Hsp110 family of NEFs (HSPH1, HSPH2, and HSPH3) showed a binding preference to HSPA1A over HSPA1L (Fig. 5C). For example, HSPH2 binds four times more to HSPA1A compared with HSPA1L. Co-immunoprecipitation of HSPH2 with the HSPA1A/HSPA1L NBD chimeras confirmed that this differential binding is coupled to the NBDs of the two Hsp70s (Fig. S5C). Human Hsp110s have ∼60% identity (Fig. S6 and Table S2) between them, and Hsp110 contact sites are very broad on Hsp70-NBD interaction surface (Fig. S1). The differential binding of the two Hsp70s to Hsp110 NEFs raised the possibility that functionally different Hsp110–Hsp70 interaction might play a role in the differential ability of HSPA1A and HSPA1L to suppress mutant SOD1 aggregation. In fact, MS analysis of soluble immunoprecipitated mCherry-SOD1A4V revealed endogenous HSPH2 among SOD1A4V interactors (Fig. S5D). Notably, HSPA1A co-expression led to association of all Hsp110 family members with SOD1A4V, something that did not happen upon HSPA1L co-expression, suggesting that HSPA1A attracts all Hsp110 chaperones toward SOD1A4V, and this might be crucial for its aggregation suppressing activity. This preference of Hsp110s over HSPA1A did not seem to be specific for mutant SOD1 as a substrate. Similar results were obtained when we co-immunoprecipitated HSPH2 with HSPA1A or HSPA1L without SOD1A4V co-expression (Fig. S5E), suggesting that this partnership is not limited to SOD1A4V and might be involved in handling other substrates too.
Figure 5.
Hsp110 NEFs show increased interaction with HSPA1A. A–C, interaction of GFP-HSPA1A or GFP-HSPA1L with NEFs of the BAG (A), HSPBP1 (B), or Hsp110 (C) families, in the presence of SOD1A4V. GFP-HSPA1A or GFP-HSPA1L were transiently expressed in HEK293 cells for 48 h with either HA-BAG1 or FLAG-BAG3 (A), untagged-HSPBP1 (B), and V5-tagged HSPH1, HSPH2, or HSPH3 (C). GFP Nanotrap was used for native immunoprecipitation (IP) of the Hsp70s. Western blots using the indicated antibodies are shown. The quantification graph of binding represents ratios of IP intensities of HA/GFP for BAG1 (n = 3) and FLAG/GFP for BAG3 (n = 2) (A), HSPBP1/GFP for HSPBP1 (n = 3) (B), and V5/GFP for HSPH1–3 (n = 3 for each) (C), all relative to HSPA1A measurements. In all graphs, error bars with S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
HSPH2 is necessary for HSPA1A-mediated reduction of mutant SOD1 aggregation
Because the only differences in co-factor interaction detected were with the Hsp110s, we assessed whether any of the Hsp110s is required for the activity of HSPA1A toward mutant SOD1 aggregation. Interestingly, only the loss of HSPH2 in HSPA1A-overexpressing cells strongly diminished the aggregation-suppressing effect of HSPA1A, showing that HSPH2 plays an important role in HSPA1A activity against SOD1A4V aggregation (Fig. 6). Loss of HSPH1 or HSPH3 resulted in a drop in SOD1A4V aggregation irrespective of which of the two Hsp70s was co-expressed. This suggests that these two Hsp110s might compete with HSPH2 for Hsp70 interaction and either are less efficient or lead to a different processing pathway. Importantly, loss of HSPH2 together with HSPA1L overexpression did not result in further increase in SOD1A4V aggregation, suggesting that upon HSPA1L interaction SOD1A4V follows a pathway independent of HSPH2 (Fig. 6). Together, these data suggest that a crucial reason for the functional difference between HSPA1A and HSPA1L in suppressing SOD1A4V aggregation is based on their different functional interaction with HSPH2.
Figure 6.

HSPA1A activity on SOD1A4V aggregation depends on HSPH2 nucleotide-exchange factor. Shown are the effects of Hsp110 knockdown on SOD1A4V together with overexpressing HSPA1A or HSPA1L and Nonidet P-40 fractionation of HEK293 cells co-transfected with mCherry-SOD1A4V, V5-HSPA1A, or V5-HSPA1L (for 48 h) and siRNA for HSPH1–3 or mock (for 72 h). Western blots using the indicated antibodies are shown. The quantification graph represents total (T) or Nonidet P-40–insoluble (P) fraction Western blotting intensities relative to siMock control for each group (n = 6–8). Error bars indicate S.E. *, p = 0.01–0.05; **, p = 0.001–0.01; ***, p < 0.001.
Discussion
Overall, our data show that Hsp70 paralogs, despite being highly conserved, can have different functional outcomes when expressed in cells. This is unrelated to substrate-binding capacity, nucleotide cycling, and biochemical activity. Instead, we find that differential function of Hsp70s in cells depends on the intracellular co-chaperone context and specifically on the differential interaction with NEFs. When expressed in cells, HSPA1A reduces the aggregation of both SOD1A4V and GFP-LucDM, whereas most other Hsp70s enhance the aggregation of these substrates. This is particularly striking because in in vitro assays HSPA1A does not perform better compared with other Hsp70s (Fig. 3 and Refs. 51 and 52). This difference is not only observed with both substrates we tested here but also with the aggregation of ParkinC289G (substitution of Cys289 to Gly, a mutant associated with familial Parkinson's disease) (28) and aggregation and refolding of heat-denatured WT luciferase (27). Although a careful substrate analysis of the different Hsp70s is required, these substrates all form so-called amorphous aggregates. Amorphous aggregates have been associated with disease and general aging (57). For example, one of the substrates tested here, SOD1A4V, is a disease-causing allele, and Hsp70 (i.e. HSPA1A) has been identified as a potential target for disease intervention. Because only HSPA1A reduces mutant SOD1 aggregation, it will be of importance to specifically activate HSPA1A and not any of the other Hsp70s. Because of the high sequence overlap among the Hsp70 paralogs, this might complicate future drug discovery trajectories significantly. Our data suggest that targeting the Hsp110–HSPA1A interaction might provide a potential alternative. Indeed mutant SOD1 has been found to interact with Hsp110 chaperones in an ALS mouse model (58), something we also observed and that seemed to be enhanced by HSPA1A co-expression (Fig. S5D). Moreover, Hsp110s have been reported to extend survival in mutant SOD1 ALS mouse model (59) and rescue transport defects in mutant SOD1-containing squid axoplasm (60).
The drop in aggregation of substrates upon increased expression of HSPA1A and the importance of HSPH2 can be attributed to several activities. For example, Hsp110s together with HSPA1A have been found to be crucial components of a disaggregation machine (51, 61). The differential interaction seen when expressed in living cells might underlie a different disaggregation potential of HSPA1A and HSPA1L. However, disaggregation in mammalian cells strictly depends on DNAJB1 (61). Here we find that the EEVD motif, which is important for the interaction of Hsp70 with DNAJB1, is indispensable for the effect on substrate fate (i.e. the behavior of the EEVD mutants in Fig. 3). This argues against a role of disaggregation in the effects on substrate fate we observe upon expression of HSPA1A and HSPA1L. Moreover, we did not notice any substantial difference with regard to the biochemical properties of the ATPase activities of HSPA1A and HSPA1L. This argues against an intrinsic difference between HSPA1A and HSPA1L with regard to their potential in ATP-dependent (re)folding. Instead prolonged binding of HSPH2 and HSPA1A in cells could reflect more a holdase type of function, perhaps of HSPH2 while in complex with HSPA1A. Both substrates we tested here are intrinsically unstable mutant proteins; therefore an ATP-dependent folding activity might be less effective. Alternatively, we noticed that increasing HSPA1A levels led to a decrease of both insoluble and total levels of SOD1A4V and of GFP-LucDM (Fig. 1), which points toward a degradation pathway for ultimate substrate clearance. The notion that the total amount of active substrate of GFP-LucDM (Fig. S2) is not influenced by either HSPA1A or HSPA1L suggests that it is the nonactive pool of mutant substrate that is specifically degraded. This would speak for a very effective, HSPA1A-specific triage decision of especially mutant proteins. Interestingly, a study in yeast also showed that Hsp110s target Hsp70 substrates for degradation (62), further supporting efficient substrate disposal upon HSPA1A–Hsp110 interaction.
Here we show that in cells, HSPA1A has a higher affinity for Hsp110s as compared with HSPA1L (Fig. 5C). The relevance of this interaction is pointed out by the fact that one of the Hsp110s (i.e. HSPH2) is required for the suppression of substrate aggregation by HSPA1A but not for the enhancing effect on aggregation of HSPA1L (Fig. 6). The difference in affinity to HSPA1A and HSPA1L is specific for Hsp110s because other NEFs, including HSPB1, BAG1, and BAG3, show a similar binding to these Hsp70s (Fig. 5, A and B). Most NEFs bind to the NBD domain in Hsp70. However, co-crystals of the yeast Hsp110 with mammalian Hsp70 revealed that Hsp110s bind not only to the NBD but also to the SBD (53, 54). However, these predicted Hsp110–Hsp70 binding surfaces are mostly conserved between HSPA1A and HSPA1L (Fig. S1) and are not sufficient to explain the differences in HSPA1A and HSPA1L we observe in cells. We also did not observe this preference in vitro using recombinant proteins (Fig. S4). This implies that the differences we observed are most likely induced by the cellular environment. Differences in post-translational modifications (PTMs), competition between (unknown) binding partners, or localization differences are possible underlying mechanisms. Transient modifications via PTMs are well-known to drive and regulate protein interactions. Interestingly, both HSPA1A and HSPA1L have been picked up in numerous PTM screens and are differently modified (information from PhosphoSitePlus (63). Whether such PTMs can influence the Hsp70–Hsp110 interaction is currently unclear.
Although HSPH1, HSPH2, and to a lesser extent HSPH3 show a preference for HSPA1A, SOD1 aggregation suppression is only modulated by HSPH2 interaction. Although HSPH1 and HSPH3 are good binding partners of HSPA1A, at least compared with HSPA1L (Fig. 5C), their partnership with HSPA1A seems not ideal for handling SOD1 aggregation (Fig. 6). This suggests that Hsp110 family members are also functionally diverse. Human Hsp110 has ∼60% sequence identity (Fig. S6 and Table S2), which could allow for such a functional variability. The binding between Hsp70 and Hsp110 is regulated by an acidic intrinsically disordered domain in Hsp110s (64). In vitro this acidic domain stimulates the release of Hsp110s from Hsp70 (i.e. HSPA8) and influences the ATPase cycle of Hsp70 (64). Interestingly, there are differences in the predicted disorder between the acidic loops of Hsp110s. HSPH1 and HSPH2 are predicted to be more disordered than HSPH3 (using prediction software such as IUPred (65)), a pattern that is mirrored in the binding preference to HSPA1A (Fig. 5C). This raises the possibility that in cells HSPA1A influences the intrinsically disordered acidic domain in the SBD of Hsp110s, thereby prolonging its interaction to Hsp110s.
The data presented here reveal that, in living cells, different Hsp70s act with preferred NEFs. This suggests that Hsp70s themselves co-determine substrate fate through a preset selection of co-chaperones and that, at least for certain substrates, binding or delivery to Hsp70 plays a less determining role. Currently it is unclear how such a preset selection in cells is accomplished. A preset coupling of Hsp70 and co-chaperones would imply limitations with regard to flexibility to maximize efficient substrate handling. However, between different cell types or under different conditions (e.g. heat or oxidative stress), the types of substrates or the fates of the substrates can vary, and this might require differently optimized Hsp70–NEF couples. This can be accomplished by changing the relative expression patterns of Hsp70s and NEFs that likely influence co-chaperone context and thus the fate of substrates. There are examples of such a switch. For example, upon various types of stress, the ratio between the NEFs BAG1 and BAG3 is flipped, changing substrate fate from proteasome- to autophagosome-mediated degradation (66). It is well-known that expression of chaperones is mendable, thereby changing the capacity of the system. However, our data explain that relative changes in expression of fixed Hsp70 and NEF pairs not only change the machinery capacity but also change substrate fate. In addition, our data urge for clear specification of Hsp70 member identification in future chaperone studies and careful re-evaluation of possible conflicting existing literature data on Hsp70 functions, especially in cellular or in vivo systems.
Experimental procedures
Chaperone nomenclature
All chaperones mentioned in this study, their commonly mentioned alternative names (gene or protein), and Uniprot accession numbers are listed in Table S3.
Gene cloning, plasmids, and siRNAs
GFP- or V5-HSP70s (HSPAs), V5-JDPs, V5-HSP110 (HSPHs), and HSPBP1 cloned into pcDNA5/FRT/TO (Invitrogen) vector plasmid were previously described (27, 29). HA-BAG1 and FLAG-BAG3 encoding plasmids were previously described (66). GFP-LuciferaseWT and GFP-LuciferaseR188Q, R261Q constructs were previously described (25). pcDNA5-FRT/TO-mCherry-SOD1WT was generated by combining mCherry, which was removed from a pcDNA3.1(+)-mCherry vector (kind gift from Dr. B. Giepmans, University Medical Center, Groningen, The Netherlands), and SOD1WT, which was amplified from pEBB-FLAG-SOD1WT (kind gift from Dr. B. van de Sluis, University Medical Center) previously described (67), into a pcDNA5-FRT/TO backbone plasmid. pcDNA5-FRT/TO-mCherry-SOD1A4V and single or multiple point mutants of FRT/TO-V5-HSPA1A constructs were generated using QuikChange XL site-directed mutagenesis kit (Agilent), according to the manufacturer's instructions. Domain and subdomain swaps between HSPA1A and HSPA1L were constructed by PCR amplification of the domain of interest with flanking restriction sites and re-insertion of the replacing fragment by ligation. All primers used for cloning are listed in Table S4. All generated constructs were verified by sequencing. For gene knockdown, 50 nm of the following siRNAs were used: siGENOME SMARTpool siRNAs (Dharmacon) for HSPH1 (M-004972-00), HSPH2/HSPA4 (M-012636-02), and HSPH3/HSPA4L (M-012636-02); and siGenome nontargeting siRNA (pool 1, D-001206-13, Dharmacon) was used as mock siRNA negative control.
Cell cultures and transfections
HEK293 (human embryonic kidney) stably expressing the tetracycline repressor (Flp-In T-REx HEK293, Invitrogen) and U2OS (human osteosarcoma; a kind gift from Dr. C. Dinant) cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Greiner Bio-One) and penicillin/streptomycin (Gibco). The cells were transiently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Expression of cDNAs cloned in FRT/TO (described above) in Flp-In T-REx HEK293 was induced by 1 μg/ml tetracycline. All cell lines are frequently checked for mycoplasma contamination.
Nonidet P-40 fractionation
48 or 72 h after transfection, the cells were washed with cold (4 °C) PBS and harvested on ice in cold (4 °C) lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 (Igepal-CA 630, Sigma), and complete protease inhibitor mixture (Roche) incubated for 10 min. Cell lysates kept on ice were sonicated at 50% input for 5 s, protein concentrations were measured with DC protein assay (Bio-Rad) and equalized, and a part of each sample was kept as the total (T) fraction representation. The remaining part of the sample was centrifuged at 20,000 × g for 30 min at 4 °C, and supernatant was kept separately as the S fraction. The pellet was washed once with cold (4 °C) lysis buffer; after another centrifugation at 20,000 × g at 4 °C for 30 min, supernatant was discarded, and the pellet was resuspended in one-third of the initial volume lysis buffer with sonication, representing the P fraction. In all three fractions, 4× Laemmli sample buffer (8% SDS, 40% glycerol, 20% 2-mercaptoethanol, 0.001% bromphenol blue) was added, and samples were boiled for 5 min and kept at −20 °C until use.
Immunoprecipitations
For GFP-Trap immunoprecipitation with cross-linking, the cells were washed with cold (4 °C) PBS containing 0.5 mm CaCl2 and 1 mm MgCl2 and harvested in cold (4 °C) PBS, pelleted at 3800 × g at 4 °C for 3 min, and incubated with 1 mm 3,3′-dithiodipropionic acid di(N-hydroxysuccinimide ester) cross-linking reagent (Sigma) in PBS for 30 min on ice. To quench cross-linking, 2 mm cold (4 °C) glycine was added and incubated for 15 min on ice. After centrifugation for 5 min at 3800 × g at 4 °C, the cells were washed once with cold (4 °C) PBS, pelleted again at 3800 × g at 4 °C for 3 min, and snap-frozen in liquid nitrogen. The pellets were resuspended in cold (4 °C) lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 1.5 mm MgCl2, 0.5% Nonidet P-40 (Igepal-CA 630, Sigma), 3% glycerol, 0.9 mm DTT (Sigma), and complete EDTA-free protease inhibitor mixture (Roche), and lysates were homogenized on ice by passing through a 26-gauge needle or by sonication (50% input, 5 s). After spinning twice at 20,000 × g for 10 min at 4 °C to clear lysates from cell debris, a portion of the supernatant was collected for input measurement before adding GFP-Trap® magnetic agarose beads (gtma, Chromotek) to it. Extracts were incubated with beads at 4 °C for 2 h under gentle agitation, followed by one wash with cold (4 °C) lysis buffer without DTT, three or four washes with cold (4 °C) lysis buffer, and one wash with cold (4 °C) lysis buffer containing 300 mm NaCl. Laemmli sample buffer was added to the beads and input, they were boiled for 5 min and kept at −20 °C until use.
Western blotting and antibodies
Equal amounts of proteins were loaded into 10–12% SDS-PAGE gels. Proteins were transferred onto polyvinylidene difluoride membranes and blotted with the primary antibodies: GFP (JL-8, 632381, Clontech); V5 (46-0705, Invitrogen); SOD1 (FL-154, sc-11407, Santa Cruz); FLAG (M2, 035K6196, Sigma); HSPH1/HSP105 (EPR4576, ab109624, Abcam); HSPH2/HSPA4 (EPR14166, ab185962, Abcam); HSPH3/HSPA4L (ab87241, Abcam); α-tubulin (B-5-1-2, T5168, Sigma); β-actin (8H10D10, Cell Signaling); HSPBP1 (1D5, NBP2–01168, Novus Biologicals); Hsp90 (total, 4F3-E8 SMC-149, StressMarq; Hip ([11A6], sc-136175, Santa Cruz); HOP (STI1, D-6; sc-390203, Santa Cruz); CHIP ([C10], sc-133083, Santa Cruz), and HA-peroxidase (12013819001, Roche). The upper band detected with SOD1 antibody corresponds to full-length mCherry-SOD1 (∼45 kDa), and the lower band indicated with an asterisk (∼38 kDa) is a cleavage product of mCherry-SOD1, which is produced after mCherry cleavage under denaturing conditions as described previously (68). After incubation with the appropriate horseradish peroxidase–conjugated secondary antibody (Amersham Biosciences), visualization was performed with enhanced chemiluminescence and Hyperfilm (Amersham Biosciences) or ChemiDoc Imaging System (Bio-Rad). Quantification of Western blots was performed with either ImageJ (National Institutes of Health) or Image Lab (Bio-Rad) software. In all quantifications, each band's intensity was normalized by dividing to the appropriate loading control. For each experiment, each sample value was normalized to a control sample, and ratios were plotted on graphs (control sample ratio = 1). For statistical analysis, one sample t test was performed between the control and each sample for most graphs except graphs on Figs. 2 (B and E) and 4C, for which one-way analysis of variance and Dunnett's multiple comparison test were used together to determine differences between the designated samples. Statistical analysis and graphs were done with GraphPad Prism (GraphPad Software).
Luciferase assay
The cells were transfected with luciferase reporter and control or chaperone constructs. 24 h after transfection, cells from each sample were transferred on ice and lysed with equal volume of BLUC lysis buffer (25 mm Tris/H3PO4, pH 7.8, 10 mm MgCl2, 1% (v/v) Triton X-100, 15% glycerol, and 1 mm EDTA). The samples were then transferred to −80 °C for at least 30 min to complete lysis by freezing and then thawed and kept on ice until measurement. Luciferase activity was measured for 10 s by injecting the substrate (BLUC, 1.25 mm ATP, and 0.087 mg/ml d-luciferin) using a Sirius Luminometer (Berthold Detection Systems). Three measurements from three tubes (technical replicates) were done per condition per sample, and the average was taken as final measurement. For statistical analysis, t tests, one-way analysis of variance and Dunnett's multiple comparison test were used together to determine differences between the designated samples. Statistical analysis and graphs were done with GraphPad Prism (GraphPad Software).
Molecular structure modeling
Sequence alignments were performed with Clustal Omega sequence alignment program (EMBL-EBI). Predicted interaction sites in Figures S1 and S3 were annotated based on previously published data (Ref. 9, 34, 53–56, 70–78). Molecular structure figures were prepared with PyMOL (PyMOL Molecular Graphics System, version 2.0; Schrödinger), and structural alignment between HSPA1A-NBD (PDB code 3JXU) (34, 80) and HSPA1L-NBD (PDB code 3GDQ) (34, 81) was performed using the align command in PyMOL and the script YRB Ref. 79.
Cell viability assay
Cell viability quantification was determined using an MTS colorimetric assay; 0.50 × 104 cells were seeded in 100 μl of Dulbecco's modified Eagle's medium/well in a 96-well plate and grown for 48 h. 20 μl of MTS reagent was added. The samples were incubated at 37 °C for 3 h and measured using a Fluorstar Optima microplate reader with a 485 ± 10 nm laser to determine the absorbance value expressed in optical depth.
Fluorescent microscopy
48 h after transfection, cells grown on coverslips expressing mCherry-SOD1 proteins were fixed with 2% paraformaldehyde in PBS for 15 min, incubated with Hoechst 33342 (Invitrogen) for 5 min to stain nuclei, and mounted on microscopy slides. Microscopy was performed with a TissueFAXS (TissueGnostics) Zeiss AxioObserver Z1-based fluorescence microscope using a Zeiss Plan-Apochromat 63×/1.40 oil objective, and image acquisition was performed with a CMOS-color PL-B623 Pixelink 3.1 Megapixels camera. Brightness/contrast corrections were done with ImageJ (National Institutes of Health).
Mass spectrometry
The cells were washed once with PBS, pelleted at 6000 rpm for 3 min, and snap-frozen in liquid nitrogen. The pellets were resuspended in lysis buffer (50 mm HEPES, pH 7.5, 80 mm KCl, 0.4% Nonidet P-40 (Igepal-CA 630, Sigma), 0.5 mm DTT, 10% glycerol, complete EDTA-free protease inhibitor mixture (Roche)), and lysates were homogenized by passing through a 26-gauge needle. After spinning twice at 20,000 × g for 10 min at 4 °C to clear lysates from cell debris, they were incubated with RFP-Trap® magnetic agarose beads (Chromotek) at 4 °C for 2 h under gentle agitation. The beads were washed four times with lysis buffer before Laemmli sample buffer was added. The samples were boiled for 5 min and were sent to a MS facility for analysis. Mass spectrometry data were analyzed using PEAKS Studio 8.5 (Bioinformatics Solutions Inc.).
Protein purification
HSPH2-FLASH was generated by introduction of a tetracysteine motif (CCPGCC) at the tip of the lid domain (V640) by site-directed mutagenesis and purified as described (69). Purified protein was incubated for 30 min at 22 °C with 10 mm DTT. Reducing agent was removed by desalting column (PD MiniTrapTM G-25, GE Healthcare) and incubated with 3× molar excess of FLASH-EDT2 (Santa Cruz Biotechnology) for 4 h. Excess FLASH was removed by desalting column (PD MiniTrapTM G-25, GE Healthcare) and a labeling efficiency of 95% determined by absorbance at 280 nm (for protein concentration) and 508 nm for label concentration, respectively.
Nucleotide dissociation assay
ADP dissociation rates were determined according to Ref. 70 using the fluorescent analog N8-(4-N′-methylanthraniloylaminobutyl)–8 aminoadenosine 5′-diphosphate. HSPA1A or HSPA1L (2 μm) in complex with 2 μm N8-(4-N′-methylanthraniloylaminobutyl)–8 aminoadenosine 5′-diphosphate were rapidly mixed 1:1 with 0.01–0.3 μm HSPH2 and 2 mm ADP in a stopped flow device (SX-18 M Applied Photophysics, Surrey, UK; λex = 360 nm; cutoff filter, 420 nm). The change in fluorescence was recorded at 30 °C and fitted by a two-phase exponential decay function.
In vitro refolding
Refolding of thermally denatured luciferase was performed as previously described (69). In brief, thermal denaturation was performed by incubating 0.02 μm of native luciferase at 42 °C for 10 min in refolding buffer (40 mm HEPES-KOH, pH 7.5, 50 mm KCl, 5 mm MgCl2, 2 mm DTT, 2 mm ATP, 10 μm BSA) containing the indicated chaperone combinations at the concentrations of 1 μm HSPA1A, 1 μm HSPA1L, 1 μm AL, 1 μm LA, 0.5 μm DNAJB1, and 0.1 μm HSPH2. Luciferase refolding was initiated by adding an ATP-regenerating system (3 mm phosphoenolpyruvate and 20 ng/μl pyruvate kinase) and shifting the reaction temperature to 30 °C. Luciferase reactivation was monitored at the indicated time points with a Lumat LB 9507 luminometer (Berthold Technologies) by transferring 1 μl of sample to 100 μl of assay buffer (25 mm glycylglycine, pH 7.4, 5 mm ATP, pH 7, 100 mm KCl, and 15 mm MgCl2) mixed with 100 μl of 0.25 mm luciferin. For statistical analysis, related-samples Friedman's two-way analysis of variance with post hoc Mann–Whitney test was performed. Statistical analysis and graphs were done with GraphPad Prism (GraphPad Software).
Steady-state fluorescence anisotropy
20 nm HSPH2-FLASH was pre-incubated with indicated concentrations of HSPA1A and HSPA1L for 1 h at 25 °C in 50 mm HEPES-KOH (pH 7.5), 50 mm KCl, 5 mm MgCl2, 2 mm DTT, and 2 mm ATP. Fluorescence intensity was measured using a CLARIOstar plate reader (BMG LABTECH) with excitation at 482 nm and emission recorded at 530 nm. Binding curves were fitted to a one-site binding model using GraphPad Prism (GraphPad Software).
Author contributions
D. S., M. A. W. H. v. W., L. R., A. S. W., N. B. N., M. P. M., and S. B. formal analysis; D. S., M. A. W. H. v. W., L. R., A. S. W., S. L. D., M. J. K., J. M. B., and J. F. B. investigation; D. S., H. H. K., and S. B. writing-original draft; N. B. N., B. B., M. P. M., H. H. K., and S. B. supervision; N. B. N., B. B., and M. P. M. writing-review and editing; B. B., M. P. M., and S. B. resources; D. S., H. H. K., and S. B. conceptualization; H. H. K. and S. B. funding acquisition.
Supplementary Material
Acknowledgments
We thank Drs. Bart van de Sluis (University Medical Center Groningen), Ben Giepmans (University Medical Center Groningen), and Franz-Ulrich Hartl (Max Planck Institute of Biochemistry) for providing reagents. Mass spectrometry was performed at the Interfaculty Mass Spectrometry Center or the University of Groningen and the University Medical Center Groningen. Microscopy was performed at the University Medical Center Groningen Imaging and Microscopy Center.
This work was supported by a grant from the Research School of Behavioral and Cognitive Neurosciences of the University of Groningen (to H. H. K.), Grant ALW 824.15.004 from the open call program of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (to S. B.), a recruitment grant from the Monash University Faculty of Medicine Nursing and Health Sciences (to N. B. N.), and Deutsche Forschungsgemeinschaft Grant MA 1278/4-3 (to M. P. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S6.
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- ALS
- amyotrophic lateral sclerosis
- CTD
- C-terminal domain
- JDP
- J-domain protein
- NEF
- nucleotide-exchange factor
- SOD
- superoxide dismutase
- aa
- amino acid(s)
- PTM
- post-translational modification
- T fraction
- total fraction
- S fraction
- supernatant fraction
- P fraction
- pellet fraction
- HA
- hemagglutinin
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- PDB
- Protein Data Bank.
References
- 1. Mayer M. P., and Bukau B.. 2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U.. 2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 3. Mogk A., Bukau B., and Kampinga H. H.. 2018) Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226 10.1016/j.molcel.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 4. Radons J. 2016) The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones 21, 379–404 10.1007/s12192-016-0676-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rüdiger S., Germeroth L., Schneider-Mergener J., and Bukau B.. 1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 10.1093/emboj/16.7.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M.. 2015) How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayer M. P. 2013) Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 10.1016/j.tibs.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 8. Kampinga H. H., and Craig E. A.. 2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kityk R., Kopp J., and Mayer M. P.. 2018) Molecular mechanism of J-domain–triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4 10.1016/j.molcel.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 10. Bracher A., and Verghese J.. 2015) The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dekker S. L., Kampinga H. H., and Bergink S.. 2015) DNAJs: more than substrate delivery to HSPA. Front. Mol. Biosci. 2, 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warrick J. M., Chan H. Y., Gray-Board G. L., Chai Y., Paulson H. L., and Bonini N. M.. 1999) Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23, 425–428 10.1038/70532 [DOI] [PubMed] [Google Scholar]
- 13. Fernandez-Funez P., Sanchez-Garcia J., de Mena L., Zhang Y., Levites Y., Khare S., Golde T. E., and Rincon-Limas D. E.. 2016) Holdase activity of secreted Hsp70 masks amyloid-β42 neurotoxicity in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, E5212–E5221 10.1073/pnas.1608045113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Auluck P. K., Chan H. Y., Trojanowski J. Q., Lee V. M., and Bonini N. M.. 2002) Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295, 865–868 10.1126/science.1067389 [DOI] [PubMed] [Google Scholar]
- 15. Wong S. L., Chan W. M., and Chan H. Y.. 2008) Sodium dodecyl sulfate-insoluble oligomers are involved in polyglutamine degeneration. FASEB J. 22, 3348–3357 10.1096/fj.07-103887 [DOI] [PubMed] [Google Scholar]
- 16. McLear J. A., Lebrecht D., Messer A., and Wolfgang W. J.. 2008) Combinational approach of intrabody with enhanced Hsp70 expression addresses multiple pathologies in a fly model of Huntington's disease. FASEB J. 22, 2003–2011 10.1096/fj.07-099689 [DOI] [PubMed] [Google Scholar]
- 17. Sojka D. R., Gogler-Pigłowska A., Vydra N., Cortez A. J., Filipczak P. T., Krawczyk Z., and Scieglinska D.. 2019) Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non–small cell lung carcinoma (NSCLC): an implication for NSCLC treatment. Sci. Rep. 10.1038/s41598-019-50840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kakkar V., Meister-Broekema M., Minoia M., Carra S., and Kampinga H. H.. 2014) Barcoding heat shock proteins to human diseases: looking beyond the heat shock response. Dis. Model. Mech. 7, 421–434 10.1242/dmm.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kampinga H. H., and Bergink S.. 2016) Heat shock proteins as potential targets for protective strategies in neurodegeneration. Lancet Neurol. 15, 748–759 10.1016/S1474-4422(16)00099-5 [DOI] [PubMed] [Google Scholar]
- 20. Lotz S. K., Knighton L. E., Nitika Jones G. W., and Truman A. W.. 2019) Not quite the SSAme: unique roles for the yeast cytosolic Hsp70s. Curr. Genet. 65, 1127–1134 10.1007/s00294-019-00978-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stege G. J., Li L., Kampinga H. H., Konings A. W., and Li G. C.. 1994) Importance of the ATP-binding domain and nucleolar localization domain of HSP72 in the protection of nuclear proteins against heat-induced aggregation. Exp. Cell Res. 214, 279–284 10.1006/excr.1994.1259 [DOI] [PubMed] [Google Scholar]
- 22. Nollen E. A., Brunsting J. F., Roelofsen H., Weber L. A., and Kampinga H. H.. 1999) In vivo chaperone activity of heat shock protein 70 and thermotolerance. Mol. Cell Biol. 19, 2069–2079 10.1128/MCB.19.3.2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prudencio M., Hart P. J., Borchelt D. R., and Andersen P. M.. 2009) Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum. Mol. Genet. 18, 3217–3226 10.1093/hmg/ddp260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruening W., Roy J., Giasson B., Figlewicz D. A., Mushynski W. E., and Durham H. D.. 1999) Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J. Neurochem. 72, 693–699 10.1046/j.1471-4159.1999.0720693.x [DOI] [PubMed] [Google Scholar]
- 25. Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F. U., and Raychaudhuri S.. 2011) Firefly luciferase mutants as sensors of proteome stress. Nat. Methods 8, 879–884 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- 26. Fourie A. M., Sambrook J. F., and Gething M. J.. 1994) Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 269, 30470–30478 [PubMed] [Google Scholar]
- 27. Hageman J., van Waarde M. A., Zylicz A., Walerych D., and Kampinga H. H.. 2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 435, 127–142 10.1042/BJ20101247 [DOI] [PubMed] [Google Scholar]
- 28. Kakkar V., Kuiper E. F., Pandey A., Braakman I., and Kampinga H. H.. 2016) Versatile members of the DNAJ family show Hsp70 dependent anti-aggregation activity on RING1 mutant parkin C289G. Sci. Rep. 6, 34830 10.1038/srep34830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hageman J., Rujano M. A., van Waarde M. A., Kakkar V., Dirks R. P., Govorukhina N., Oosterveld-Hut H. M., Lubsen N. H., and Kampinga H. H.. 2010) A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell 37, 355–369 10.1016/j.molcel.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 30. Rujano M. A., Kampinga H. H., and Salomons F. A.. 2007) Modulation of polyglutamine inclusion formation by the Hsp70 chaperone machine. Exp. Cell Res. 313, 3568–3578 10.1016/j.yexcr.2007.07.034 [DOI] [PubMed] [Google Scholar]
- 31. Milner C. M., and Campbell R. D.. 1990) Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics 32, 242–251 [DOI] [PubMed] [Google Scholar]
- 32. Daugaard M., Rohde M., and Jäättelä M.. 2007) The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581, 3702–3710 10.1016/j.febslet.2007.05.039 [DOI] [PubMed] [Google Scholar]
- 33. James P., Pfund C., and Craig E. A.. 1997) Functional specificity among Hsp 70 molecular chaperones. Science 275, 387–389 10.1126/science.275.5298.387 [DOI] [PubMed] [Google Scholar]
- 34. Wisniewska M., Karlberg T., Lehtiö L., Johansson I., Kotenyova T., Moche M., and Schüler H.. 2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70–2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS One 5, e8625 10.1371/journal.pone.0008625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fontaine S. N., Rauch J. N., Nordhues B. A., Assimon V. A., Stothert A. R., Jinwal U. K., Sabbagh J. J., Chang L., Stevens S. M. Jr., Zuiderweg E. R., Gestwicki J. E., and Dickey C. A.. 2015) Isoform-selective genetic inhibition of constitutive cytosolic Hsp70 activity promotes client Tau degradation using an altered co-chaperone complement. J. Biol. Chem. 290, 13115–13127 10.1074/jbc.M115.637595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karras G. I., Yi S., Sahni N., Fischer M., Xie J., Vidal M., D'Andrea A. D., Whitesell L., and Lindquist S.. 2017) HSP90 shapes the consequences of human genetic variation. Cell 168, 856–866.e12 10.1016/j.cell.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morán Luengo T., Kityk R., Mayer M. P., and Rüdiger S. G. D.. 2018) Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 10.1016/j.molcel.2018.03.028 [DOI] [PubMed] [Google Scholar]
- 38. Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A.. 2014) Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 10.1016/j.cell.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alvira S., Cuéllar J., Röhl A., Yamamoto S., Itoh H., Alfonso C., Rivas G., Buchner J., and Valpuesta J. M.. 2014) Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 5, 5484 10.1038/ncomms6484 [DOI] [PubMed] [Google Scholar]
- 40. Li Z., Hartl F. U., and Bracher A.. 2013) Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 20, 929–935 10.1038/nsmb.2608 [DOI] [PubMed] [Google Scholar]
- 41. Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., and Patterson C.. 2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 10.1038/35050618 [DOI] [PubMed] [Google Scholar]
- 42. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., and Patterson C.. 1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 19, 4535–4545 10.1128/MCB.19.6.4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenzweig R., Nillegoda N. B., Mayer M. P., and Bukau B.. 2019) The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680 10.1038/s41580-019-0133-3 [DOI] [PubMed] [Google Scholar]
- 44. Minami Y., Höhfeld J., Ohtsuka K., and Hartl F. U.. 1996) Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J. Biol. Chem. 271, 19617–19624 10.1074/jbc.271.32.19617 [DOI] [PubMed] [Google Scholar]
- 45. Mayer M. P., Laufen T., Paal K., McCarty J. S., and Bukau B.. 1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289, 1131–1144 10.1006/jmbi.1999.2844 [DOI] [PubMed] [Google Scholar]
- 46. Michels A. A., Kanon B., Bensaude O., and Kampinga H. H.. 1999) Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J. Biol. Chem. 274, 36757–36763 10.1074/jbc.274.51.36757 [DOI] [PubMed] [Google Scholar]
- 47. Yu H. Y., Ziegelhoffer T., Osipiuk J., Ciesielski S. J., Baranowski M., Zhou M., Joachimiak A., and Craig E. A.. 2015) Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1. J. Mol. Biol. 427, 1632–1643 10.1016/j.jmb.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rauch J. N., and Gestwicki J. E.. 2014) Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414 10.1074/jbc.M113.521997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nollen E. A., Brunsting J. F., Song J., Kampinga H. H., and Morimoto R. I.. 2000) Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell Biol. 20, 1083–1088 10.1128/MCB.20.3.1083-1088.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., and Hartl F. U.. 2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 10.1038/sj.emboj.7601138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rampelt H., Kirstein-Miles J., Nillegoda N. B., Chi K., Scholz S. R., Morimoto R. I., and Bukau B.. 2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235 10.1038/emboj.2012.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gassler C. S., Wiederkehr T., Brehmer D., Bukau B., and Mayer M. P.. 2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276, 32538–32544 10.1074/jbc.M105328200 [DOI] [PubMed] [Google Scholar]
- 53. Polier S., Dragovic Z., Hartl F. U., and Bracher A.. 2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 10.1016/j.cell.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 54. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., and Sousa R.. 2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 10.1016/j.molcel.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., and Moarefi I.. 2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 10.1126/science.1057268 [DOI] [PubMed] [Google Scholar]
- 56. Shomura Y., Dragovic Z., Chang H.-C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., and Bracher A.. 2005) Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17, 367–379 10.1016/j.molcel.2004.12.023 [DOI] [PubMed] [Google Scholar]
- 57. David D. C., Ollikainen N., Trinidad J. C., Cary M. P., Burlingame A. L., and Kenyon C.. 2010) Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J., Farr G. W., Zeiss C. J., Rodriguez-Gil D. J., Wilson J. H., Furtak K., Rutkowski D. T., Kaufman R. J., Ruse C. I., Yates J. R. 3rd, Perrin S., Feany M. B., and Horwich A. L.. 2009) Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc. Natl. Acad. Sci. U.S.A. 106, 1392–1397 10.1073/pnas.0813045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nagy M., Fenton W. A., Li D., Furtak K., and Horwich A. L.. 2016) Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc. Natl. Acad. Sci. U.S.A. 113, 5424–5428 10.1073/pnas.1604885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song Y., Nagy M., Ni W., Tyagi N. K., Fenton W. A., López-Giráldez F., Overton J. D., Horwich A. L., and Brady S. T.. 2013) Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc. Natl. Acad. Sci. U.S.A. 110, 5428–5433 10.1073/pnas.1303279110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shorter J. 2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6, e26319 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kandasamy G., and Andréasson C.. 2018) Hsp70–Hsp110 chaperones deliver ubiquitin dependent and independent substrates to the 26S proteasome for proteolysis. J. Cell Sci. 131, jcs210948 10.1242/jcs.210948 [DOI] [PubMed] [Google Scholar]
- 63. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E.. 2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cabrera Y., Dublang L., Fernández-Higuero J. A., Albesa-Jové D., Lucas M., Viguera A. R., Guerin M. E., Vilar J. M. G., Muga A., and Moro F.. 2019) Regulation of human Hsc70 ATPase and chaperone activities by Apg2: role of the acidic subdomain. J. Mol. Biol. 431, 444–461 10.1016/j.jmb.2018.11.026 [DOI] [PubMed] [Google Scholar]
- 65. Mészáros B., Erdos G., and Dosztányi Z.. 2018) IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 10.1093/nar/gky384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Minoia M., Boncoraglio A., Vinet J., Morelli F. F., Brunsting J. F., Poletti A., Krom S., Reits E., Kampinga H. H., and Carra S.. 2014) BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta. Autophagy 10, 1603–1621 10.4161/auto.29409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vonk W. I., Wijmenga C., Berger R., van de Sluis B., and Klomp L. W.. 2010) Cu,Zn superoxide dismutase maturation and activity are regulated by COMMD1. J. Biol. Chem. 285, 28991–29000 10.1074/jbc.M110.101477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gross L. A., Baird G. S., Hoffman R. C., Baldridge K. K., and Tsien R. Y.. 2000) The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U.S.A. 97, 11990–11995 10.1073/pnas.97.22.11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nillegoda N. B., Stank A., Malinverni D., Alberts N., Szlachcic A., Barducci A., De Los Rios P., Wade R. C., and Bukau B.. 2017) Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife 6, e24560 10.7554/eLife.24560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Theyssen H., Schuster H. P., Packschies L., Bukau B., and Reinstein J.. 1996) The second step of ATP binding to DnaK induces peptide release. J. Mol. Biol. 263, 657–670 10.1006/jmbi.1996.0606 [DOI] [PubMed] [Google Scholar]
- 71. Kityk R., Kopp J., Sinning I., and Mayer M. P.. 2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 72. Flaherty K. M., DeLuca-Flaherty C., and McKay D. B.. 1990) Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 10.1038/346623a0 [DOI] [PubMed] [Google Scholar]
- 73. Flaherty K. M., Wilbanks S. M., DeLuca-Flaherty C., and McKay D. B.. 1994) Structural basis of the 70-kilodalton heat shock cognate protein. J. Biol. Chem. 269, 12899–12907 [PubMed] [Google Scholar]
- 74. Sriram M., Osipiuk J., Freeman B., Morimoto R., and Joachimiak A.. 1997) Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5, 403–414 10.1016/S0969-2126(97)00197-4 [DOI] [PubMed] [Google Scholar]
- 75. Arakawa A., Handa N., Shirouzu M., and Yokoyama S.. 2011) Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 20, 1367–1379 10.1002/pro.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Qi R., Sarbeng E. B., Liu Q., Le K. Q., Xu X., Xu H., Yang J., Wong J. L., Vorvis C., Hendrickson W. A., Zhou L., and Liu Q.. 2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 10.1038/nsmb.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Arakawa A., Handa N., Ohsawa N., Shida M., Kigawa T., Hayashi F., Shirouzu M., and Yokoyama S.. 2010) The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure 18, 309–319 10.1016/j.str.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 78. Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., and Misra S.. 2008) Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317 10.1038/nsmb.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hagemans D., van Belzen I. A., Morán Luengo T., and Rüdiger S. G.. 2015) A script to highlight hydrophobicity and charge on protein surfaces. Front. Mol. Biosci. 2, 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wisniewska M. M., Karlberg T., Arrowsmith C. H., Berglund H., Bountra C., Collins R., Edwards A. M., Flodin S., Flores A., Graslund S., Hammarstrom M., Johansson A., Johansson I., Kallas A., Kraulis P., et al. (2009) Crystal structure of the human 70 kDa heat shock protein 1A (Hsp70-1) ATPase domain in complex with ADP and inorganic phosphate. Protein Data Bank, 10.2210/pdb3JXU/pdb. [DOI]
- 81. Wisniewska M., Moche M., Arrowsmith C. H., Berglund H., Bountra C., Collins R., Dahlgren L. G., Edwards A. M., Flodin S., Flores A., Graslund S., Hammarstrom M., Johansson A., Johansson I., Karlberg, et al. Crystal structure of the human 70 kDa heat shock protein 1-like ATPase domain in complex with ADP and inorganic phosphate. Protein Data Bank, 10.2210/pdb3GDQ/pdb. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.