Abstract
The transcription factor nuclear factor erythroid-2–related factor 2 (Nrf2) plays a critical role in reducing oxidative stress by promoting the expression of antioxidant genes. Both individuals with diabetes and preclinical diabetes models exhibit evidence of a defect in retinal Nrf2 activation. We recently demonstrated that increased expression of the stress response protein regulated in development and DNA damage 1 (REDD1) is necessary for the development of oxidative stress in the retina of streptozotocin-induced diabetic mice. In the present study, we tested the hypothesis that REDD1 suppresses the retinal antioxidant response to diabetes by repressing Nrf2 function. We found that REDD1 ablation enhances Nrf2 DNA-binding activity in the retina and that the suppressive effect of diabetes on Nrf2 activity is absent in the retina of REDD1-deficient mice compared with WT. In human MIO-M1 Müller cell cultures, REDD1 deletion prevented oxidative stress in response to hyperglycemic conditions, and this protective effect required Nrf2. REDD1 suppressed Nrf2 stability by promoting its proteasomal degradation independently of Nrf2's interaction with Kelch-like ECH-associated protein 1 (Keap1), but REDD1-mediated Nrf2 degradation required glycogen synthase kinase 3 (GSK3) activity and Ser-351/Ser-356 of Nrf2. Diabetes diminished inhibitory phosphorylation of glycogen synthase kinase 3β (GSK3β) at Ser-9 in the retina of WT mice but not in REDD1-deficient mice. Pharmacological inhibition of GSK3 enhanced Nrf2 activity and prevented oxidative stress in the retina of diabetic mice. The findings support a model wherein hyperglycemia-induced REDD1 blunts the Nrf2 antioxidant response to diabetes by activating GSK3, which, in turn, phosphorylates Nrf2 to promote its degradation.
Keywords: diabetes, retina, reactive oxygen species (ROS), oxidative stress, glycogen synthase kinase 3 (GSK-3), nuclear factor 2 (erythroid-derived 2-like factor) (NFE2L2) (Nrf2), post-translational modification (PTM), hyperglycemia, retinopathy
Introduction
Oxidative stress is a major contributing factor to the development and progression of retinal complications caused by diabetes (1, 2). The Diabetes Control and Complications Trial (DCCT) demonstrated that intensive glycemic control is associated with a reduction in both the onset and progression of diabetic retinopathy (DR)2 (3). A unifying mechanism for the pathobiology of diabetic complications links all of the principle pathways responsible for hyperglycemia-induced tissue damage to the accumulation of reactive oxygen species (ROS) (1). Diabetes causes both an increase in ROS production, as well as impairment of the antioxidant defense system (4–6). This imbalance between the production of ROS and the antioxidant defense system results in oxidative stress.
Redox-sensitive activation of the nuclear factor erythroid 2-related factor 2 (Nrf2) antioxidant response is critical in preventing oxidative stress (6, 7). Nrf2 is a basic leucine zipper transcription factor that regulates expression of antioxidant proteins by binding to an antioxidant response element (ARE) in the promoter of target genes (4, 8). There are over 200 members in the Nrf2 gene battery including NAD(P)H quinone oxoreductase 1 (NQO1), heme oxygenase 1 (HO-1), and the GSH cysteine ligase subunits GCLC and GCLM (9). In the absence of oxidative stress, Nrf2 is rapidly ubiquitinated by a protein complex that includes the adaptor protein Kelch-like ECH associated protein 1 (Keap1) and the E3 ubiquitin ligase Cullin 3, targeting the transcription factor for proteasomal degradation (10). Keap1 is redox-sensitive and contains multiple cysteine residues that become oxidized, leading to reduced interaction with Nrf2 (11). When ROS accumulate, Keap1 becomes oxidized and Nrf2 is allowed to translocate into the nucleus to activate ARE-dependent transcription. A number of therapeutics that augment Nrf2 activity by disrupting Keap1-mediated degradation are actively being pursued to combat diseases and complications associated with oxidative stress, including DR (12).
The retinal antioxidant response to diabetes is insufficient to prevent oxidative stress, due at least in part to blunted Nrf2 DNA-binding activity (4, 6, 13). Diabetes enhances oxidative stress in the retina (8, 14), which should promote Nrf2 nuclear translocation. However, Nrf2 nuclear localization is decreased in both the retina of streptozotocin-induced diabetic rats and in retinal endothelial cells cultured under hyperglycemic conditions (4, 6). Curiously, post-mortem retinas from patients with advanced DR exhibit a >5-fold increase in Nrf2 mRNA abundance combined with an ∼1/3 increase in Nrf2 protein content as compared with nondiabetic donor retinas (4). The dramatic difference in up-regulation of Nrf2 mRNA relative to protein suggests a diabetes-induced post-transcriptional defect in Nrf2 regulation. Indeed, Nrf2 binding with Keap1 is paradoxically up-regulated in the retina of diabetic rats (4). Zhong et al. (4) have speculated that this disparity may be due to blunted redox-sensing capacity of Keap1; however, the precise mechanism responsible for blunted Nrf2 DNA-binding in DR remains to be defined.
Since its discovery, the stress response protein regulated in development and DNA damage 1 (REDD1) has been linked to oxidative stress (15–17). REDD1 is transcriptionally up-regulated in response to a variety of adverse physiological events (18, 19), including exposure to hyperglycemic conditions (20). Recently, our laboratory demonstrated that REDD1 plays a necessary role in the development of oxidative stress in the retina of diabetic mice (21). REDD1 acts at least in part by promoting the association of protein phosphatase 2A with Akt, leading to site-specific dephosphorylation of the kinase and altered substrate specificity (22). Retinal Akt kinase activity is attenuated shortly after the onset of streptozotocin-induced diabetes (23). In the retina of diabetic mice, REDD1 expression is increased and is necessary for suppression of retinal Akt kinase activity (24). Importantly, Akt signaling plays an important role in regulating Nrf2-dependent antioxidant function in retinal cells in culture (25). The purpose of this study was to test the hypothesis that REDD1 negatively impacts the retinal antioxidant response to diabetes by suppressing Nrf2 function.
Results
REDD1 deletion increases retinal Nrf2 activity independent of a change in Nrf2 mRNA
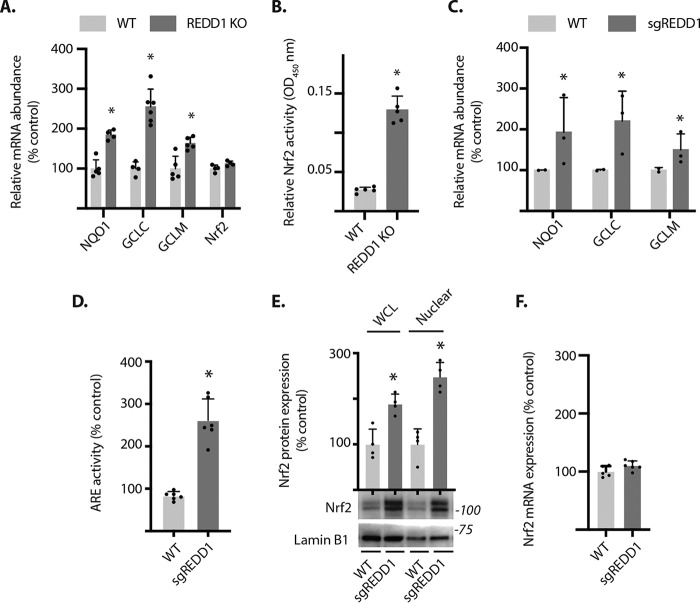
To determine whether REDD1 expression was associated with a change in the Nrf2 antioxidant response, the mRNA abundance of Nrf2-sensitive gene targets was evaluated in the retina of WT and REDD1-deficient mice. The abundances of mRNAs encoding NQO1, GCLC, and GCLM were increased in the retina of mice deficient for REDD1 as compared with WT (Fig. 1A). In contrast, the abundance of the Nrf2 mRNA was not different in the retina of WT and REDD1-deficient mice. Nrf2 activity was quantified in nuclei isolated from whole retina using an ELISA that measures the binding of Nrf2 to an oligonucleotide containing the ARE consensus motif. Nrf2 DNA-binding was dramatically enhanced in the retina of REDD1-deficient mice as compared with WT (Fig. 1B).
Figure 1.
REDD1 deletion enhances Nrf2 activity and protein expression independent of a change in Nrf2 mRNA abundance. A, the abundances of mRNAs encoding NQO1, GCLC, GCLM, and Nrf2 were evaluated in retinal lysates from wildtype (WT) and REDD1 knockout (KO) mice by PCR. B, Nrf2 activity was measured in the nuclear fraction obtained from retinal lysates by DNA-binding ELISA. C-F, REDD1 CRISPR knockout (sgREDD1) was performed in MIO-M1 cell cultures. C, the abundances of mRNAs encoding NQO1, GCLC, and GCLM was evaluated in cell lysates by PCR. D, Nrf2 activity was measured in cells expressing an ARE luciferase reporter. E, Nrf2 and Lamin B1 protein expression were evaluated in whole cell lysates (WCL) or nuclear isolates by Western blotting. Protein molecular mass (in kilodaltons) is indicated at the right of blots. F, Nrf2 mRNA abundance was assessed in cell lysates by PCR. Values are mean ± S.D. *, p < 0.05 versus WT.
REDD1 deletion enhances Nrf2 activity in MIO-M1 cell cultures
To further explore the suppressive effect of REDD1 on Nrf2, we used a previously generated human MIO-M1 retinal Müller cell line wherein CRISPR/Cas9 was employed to knockout REDD1 (26). Retinal Nrf2 expression is especially prominent in Müller glia (27), which play an important role in homeostatic support of the entire retina in part through the synthesis and release of antioxidants like GSH (28–30). Consistent with the findings in retina, the abundance of Nrf2-sensitive mRNAs including GCLC and GCLM was elevated in MIO-M1 cells deficient for REDD1 as compared with WT (Fig. 1C). To evaluate Nrf2 activity, we used an ARE luciferase reporter (25). Nrf2 activity was enhanced in REDD1-deficient cells as compared with WT (Fig. 1D). Nrf2 protein expression was also enhanced in both whole cell lysate and nuclear extracts from REDD1 knockout cells as compared with WT (Fig. 1E). Consistent with the observation in retina, REDD1 deletion did not alter Nrf2 mRNA abundance in MIO-M1 cells (Fig. 1F). These findings support post-transcriptional suppression of Nrf2 by REDD1.
Effect of REDD1 deletion on hyperglycemia-induced ROS is Nrf2-dependent
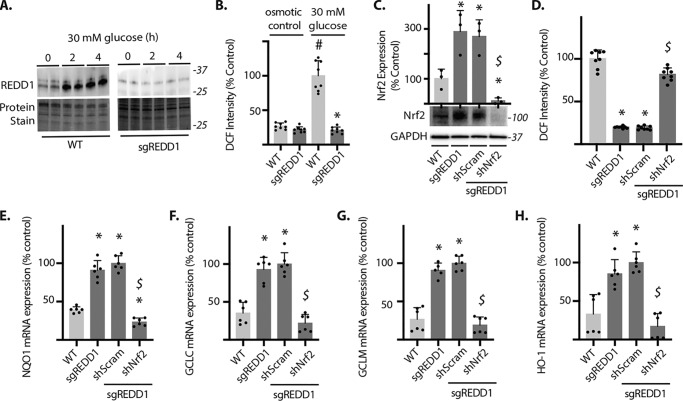
Consistent with our previous report (20), REDD1 expression was enhanced in WT MIO-M1 cells exposed to hyperglycemic conditions (Fig. 2A). We recently demonstrated that REDD1 was necessary for enhanced oxidative stress in retinal R28 cells exposed to hyperglycemic culture conditions (21). To determine whether REDD1 expression had a similar role in MIO-M1 cells, WT and REDD1 knockout MIO-M1 cells were exposed to culture medium containing either 30 mm glucose or 5 mm glucose plus 25 mm mannitol as an osmotic control. Exposure to hyperglycemic conditions enhanced ROS in WT cells as compared with the osmotic control (Fig. 2B). By contrast, REDD1-deficient cells did not exhibit increased ROS levels upon exposure to hyperglycemic conditions. To determine whether the protective effect of REDD1 deletion was mediated by enhanced Nrf2 expression, Nrf2 was knocked down in REDD1-deficient cells (Fig. S1A). Clonal cell lines expressing shRNAs targeting Nrf2 exhibited reduced Nrf2 protein expression (Fig. S1B) and attenuated abundances of Nrf2 target mRNAs (Fig. S1, C–F). In REDD1-deficient MIO-M1 cells exposed to hyperglycemic culture conditions, Nrf2 knockdown reduced Nrf2 protein expression as compared with a scramble shRNA (Fig. 2C). In both the parental REDD1 knockout cell line and REDD1 knockout cells expressing a scramble shRNA, ROS levels were reduced upon exposure to hyperglycemic conditions as compared with WT (Fig. 2D). However, Nrf2 knockdown increased ROS levels in REDD1-deficient cells exposed to hyperglycemic conditions, such that they were no longer different from WT. In contrast to REDD1-deficient cells expressing a scramble shRNA, the abundances of NQO1, GCLC, GCLM, and HO-1 were not enhanced in REDD1-deficient cells expressing a shRNA targeting Nrf2 as compared with WT (Fig. 2, E–H, respectively). To further support that the protective effect of REDD1 knockout on oxidative stress in response to hyperglycemic conditions required enhanced Nrf2 expression, Keap1 was expressed in REDD1-deficient cells. Keap1 reduced Nrf2 protein expression in both WT and REDD1 knockout cells (Fig. S1G). In addition, Keap1 enhanced oxidative stress in REDD1 knockout cells exposed to hyperglycemic culture conditions (Fig. S1H). Together, these findings provide evidence that enhanced Nrf2 activity is necessary for the attenuation of hyperglycemia-induced oxidative stress upon REDD1 deletion.
Figure 2.
Nrf2 is necessary for the attenuation of oxidative stress in REDD1-deficient cells exposed to hyperglycemic conditions. A, WT and REDD1 CRISPR knockout (sgREDD1) MIO-M1 cells were cultured in medium containing 5 mm glucose and exposed to culture medium containing 30 mm glucose. REDD1 protein expression was evaluated in cell lysates by Western blotting. Protein molecular mass (in kilodaltons) is indicated at the right of blots. Protein loading was evaluated by reversible protein stain. B, WT and sgREDD1 cells were exposed to medium containing either 30 mm glucose or 5 mm glucose plus 25 mm mannitol (osmotic control) for 4 h. ROS were visualized with DCFDA and DCF fluorescent intensity was quantified. C-H, Nrf2 was knocked down by stable expression of shRNA (shNrf2). Data in C–H were obtained using sgREDD1 Nrf2 knockdown clone 1 from Fig. S1. Control sgREDD1 cells expressed a scramble shRNA (shScram). All cells in C–H were exposed to medium containing 30 mm glucose for 4 h prior to analysis. C, Western blotting was used to evaluate Nrf2 and GAPDH protein expression in whole cell lysates. D, DCFDA was used to visualize ROS and DCF intensity was quantified. The abundances of mRNAs encoding NQO1, GCLC, GCLM, and HO-1 were evaluated in cell lysates by PCR in E-H, respectively. Values are mean ± S.D. #, p < 0.05 versus osmotic control. *, p < 0.05 versus WT. $, p < 0.05 versus shScram.
REDD1 promotes Nrf2 degradation
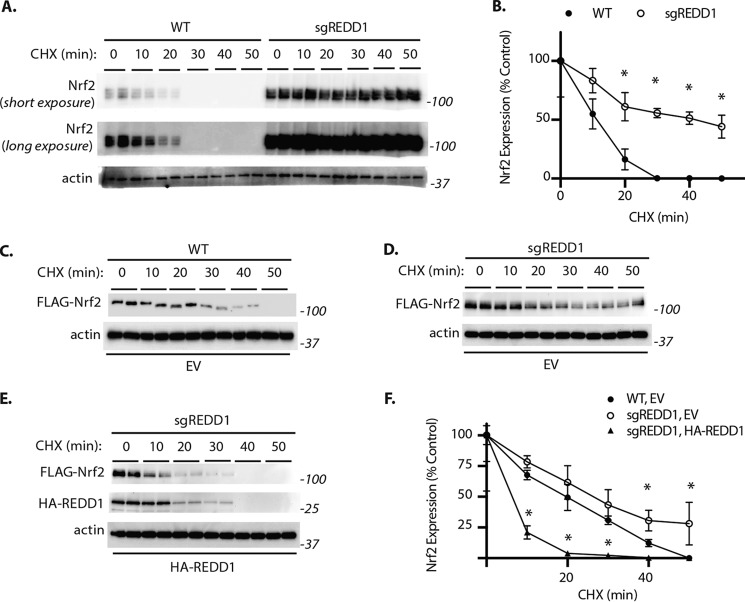
To determine whether REDD1 promoted Nrf2 degradation, a series of cycloheximide chase assays were performed. Upon inhibition of protein synthesis, Nrf2 expression in WT MIO-M1 cells was rapidly diminished (Fig. 3A). As compared with WT cells, Nrf2 expression in sgREDD1 cells was enhanced and upon exposure to cycloheximide the protein was not degraded as quickly (Fig. 3B). Consistent with endogenous Nrf2, the expression of FLAG-tagged Nrf2 was also rapidly diminished in WT cells upon inhibition of protein synthesis, such that it was no longer detected by Western blotting after 50 min (Fig. 3C). In REDD1-deficient cells the rate of FLAG-Nrf2 degradation was reduced as compared with WT, such that it was still detected after 50 min of cycloheximide exposure (Fig. 3D). However, when REDD1 expression was restored in the REDD1 knockout cell line, the rate of Nrf2 degradation was increased as compared with an empty vector control (Fig. 3, E and F). Together these data support that REDD1 promotes Nrf2 destabilization.
Figure 3.
REDD1 suppresses Nrf2 protein stability. Cycloheximide (CHX) was used to inhibit protein synthesis in WT and REDD1 CRISPR knockout (sgREDD1) MIO-M1 cell cultures. A, Nrf2 and actin protein expression was evaluated in cell lysates by Western blotting. B, quantification of Nrf2 expression in A. C–F, FLAG-tagged Nrf2 expression was achieved by transient transfection. FLAG-tagged Nrf2 was co-transfected with either an empty vector (EV, A and B) or a plasmid that facilitates expression of HA-tagged REDD1 (HA-REDD1, C). FLAG-Nrf2, HA-REDD1, and actin protein expression was evaluated in whole cell lysates by Western blotting. D, quantification of FLAG-Nrf2 expression in C–E. Values are mean ± S.D. *, p < 0.05 versus WT.
REDD1 reduces Nrf2 stability independent of Keap1
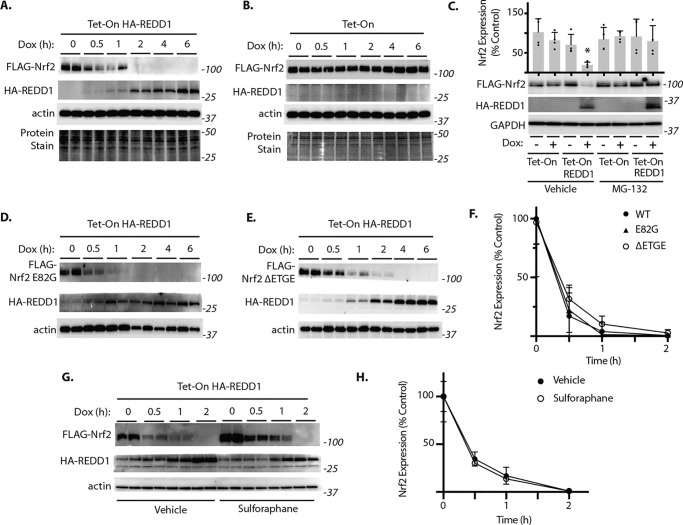
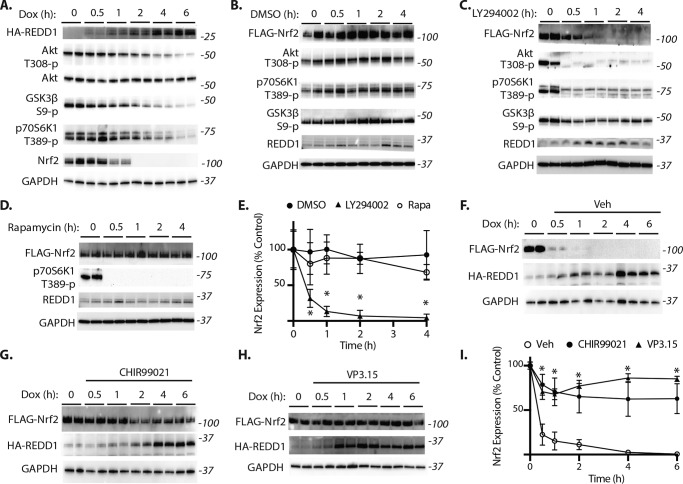
To further explore the impact of REDD1 on Nrf2 stability, FLAG-Nrf2 was expressed in a HEK293 TetON cell line with doxycycline-inducible expression of an HA-tagged REDD1 (22). HA-REDD1 expression in response to doxycycline was inversely associated with Nrf2 protein expression (Fig. 4A). By contrast, doxycycline exposure did not influence Nrf2 expression in a parental HEK293 cell line (Fig. 4B). To determine whether the decrease in Nrf2 was due to proteasome-mediated degradation, REDD1 expression was induced in the presence and absence of the proteasome inhibitor MG-132. Proteasomal inhibition prevented the decrease in Nrf2 expression in response to REDD1 (Fig. 4C). To determine whether proteasomal degradation of Nrf2 in response to REDD1 was mediated by Keap1, we generated mutations within the Neh2 domain of Nrf2 that were previously found to disrupt the high affinity Keap1-binding site and suppress interaction with Nrf2 (32). Remarkably, both the E82G (Fig. 4D) and the ΔETGE (Fig. 4E) variants of Nrf2 failed to resist REDD1-mediated degradation (Fig. 4F). The Nrf2 inducer sulforaphane acts by modifying critical cystine residues of Keap1 to prevent Nrf2 ubiquitination. In cells exposed to sulforaphane, Nrf2 expression was enhanced prior to REDD1 induction; however, sulforaphane was not sufficient to prevent Nrf2 degradation in response to REDD1 (Fig. 4G). Moreover, the rate at which Nrf2 expression was suppressed upon REDD1 induction was similar in the presence and absence of sulforaphane (Fig. 4, G and H). Together, these data support that REDD1 inhibits Nrf2 stability by a mechanism that is independent of Nrf2 interaction with Keap1.
Figure 4.
REDD1 promotes Keap1-independent proteasomal degradation of Nrf2. A–H, FLAG-tagged Nrf2 was expressed in HEK Tet-On HA-REDD1 cells with doxycycline (Dox) inducible expression of HA-REDD1 or a control HEK Tet-On parental cell line (Tet-On). Cells were exposed to Dox as indicated. C, cells were exposed to Dox for 2 h in the presence of a vehicle control or the proteasome inhibitor MG-132. D–F, cells were transfected with plasmids encoding Nrf2 variants deficient for Keap1 binding: E82G or ΔETGE. F, quantification of FLAG-Nrf2 expression in A, D, and E from 0 to 2 h. G, cells were exposed to a vehicle or the Nrf2 inducer sulforaphane for 6 h prior to Dox administration. H, quantification of Nrf2 expression in G. FLAG-Nrf2, HA-REDD1, and actin expression were evaluated by Western blotting. Protein molecular mass (in kilodaltons) is indicated at the right of blots. Protein loading was evaluated by reversible protein stain. Values are mean ± S.D. *, p < 0.05 versus TetOn.
REDD1-induced Nrf2 degradation depends on GSK3 activity
REDD1 acts by promoting dephosphorylation of Akt at Thr-308, leading to decreased phosphorylation of Akt substrates including GSK3 and tuberous sclerosis complex Subunit 2 (TSC2) (22). Akt phosphorylates the N terminus of GSK3 at Ser-9, which reduces the kinase activity of GSK3 by obstructing substrate recognition. Akt also phosphorylates TSC2 and thereby inhibits the tuberous sclerosis complex, to stimulate mTORC1. To evaluate the mechanism responsible for reduced Nrf2 stability in response to REDD1, we explored the impact of these downstream signaling events. Consistent with our previous observation (22), phosphorylation of Akt at Thr-308, GSK3β at Ser-9, and the mTORC1 substrate p70S6K1 at Thr-389 were all suppressed in response to increased REDD1 expression (Fig. 5A). Endogenous Nrf2 expression was correlated with these changes in phosphorylation. Although expression of FLAG-Nrf2 was stable in cells exposed to a vehicle control (Fig. 5, B and E), the PI3K inhibitor LY294002 suppressed Nrf2 expression (Fig. 5, C and E). The decrease in Nrf2 expression in cells exposed to LY294002 was associated with reduced phosphorylation of Akt, p70S6K1, and GSK3β. In contrast to the effect of LY294002, the mTORC1 inhibitor rapamycin was not sufficient to reduce Nrf2 expression (Fig. 5D). This suggests that the suppressive effect of REDD1 on Nrf2 is not mediated by a decline in mTORC1 signaling. To determine whether the impact of REDD1 on Nrf2 expression was mediated by enhanced GSK3 activity, HEK293 TetON cells were exposed to GSK3 inhibitors CHIR99021 or VP3.15 prior to induction of REDD1. GSK3 inhibition suppressed the effect of REDD1 on Nrf2 expression as compared with vehicle control (Fig. 5, F–I). Thus, the suppressive effect of REDD1 on Nrf2 expression required GSK3 activity.
Figure 5.
REDD1-mediated Nrf2 degradation requires GSK3. A, HEK Tet-On HA-REDD1 cells were exposed to Dox as indicated. B–E, FLAG-tagged Nrf2 expression was achieved in HEK293 cell cultures by transient transfection. Cells were exposed to a vehicle (Veh) control (DMSO, B), the PI3K inhibitor LY294002 (C), or the mTORC1 inhibitor rapamycin (D) as indicated. E, quantification of FLAG-Nrf2 expression in B–D. F–I, FLAG-tagged Nrf2 expression was achieved in HEK Tet-On HA-REDD1 cells by transient transfection. Cells were exposed to vehicle (Veh, F) or the GSK3 inhibitors CHIR99021 (G) or VP3.15 (H) prior to Dox administration. I, quantification of FLAG-Nrf2 expression in F–H. Western blotting was used to evaluate expression of HA-REDD1, FLAG-Nrf2, REDD1, Nrf2, Akt, and GAPDH expression, as well as phosphorylation of Akt, GSK3, and p70S6K1. Protein molecular mass (in kilodaltons) is indicated at the right of blots. Values are mean ± S.D. *, p < 0.05 versus DMSO or vehicle.
Neh6 phosphodegron is necessary for the effect of REDD1 on Nrf2 degradation
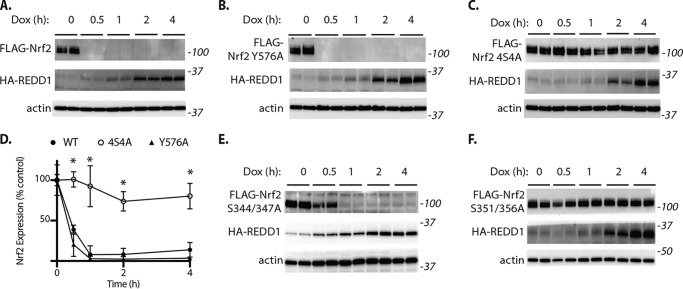
A previous study suggests that GSK3 phosphorylates and activates the tyrosine kinase Fyn, causing it to translocate to the nucleus and phosphorylate Nrf2 at Tyr-576 within the Neh3 domain (33). Fyn-mediated phosphorylation of Nrf2 reportedly stimulates nuclear exclusion of the transcription factor (34). Nrf2 also contains a Keap1-independent degron within the Neh6 domain that is directly phosphorylated by GSK3 leading to enhanced Nrf2 proteasomal degradation (35, 36). To investigate the specific phosphorylation events that mediate the suppressive effect of REDD1 on Nrf2 expression, we generated alanine substitution variants to prevent phosphorylation of Nrf2 within the Neh3 or Neh6 domains. Similar to the effect on WT Nrf2 (Fig. 6A), expression of the Nrf2 Y576A variant was rapidly reduced in response to REDD1 (Fig. 6B). By contrast, four alanine substitutions within the Neh6 phosphodegron prevented REDD1-mediated degradation (Fig. 6, C and D). The Nrf2 S351A/S356A variant was partially resistant to degradation in response to REDD1 (Fig. 6E), whereas disruption of the proline-directed phosphorylation sites at Ser-351 and Ser-356 prevented Nrf2 degradation in response to REDD1 (Fig. 6F). These data support that the suppressive effect of REDD1 on Nrf2 are mediated via the Neh6 domain.
Figure 6.
Nrf2 degradation in response to REDD1 requires the Neh6 phosphodegron. A-F, HEK TetON HA-REDD1 cells were exposed to Dox as indicated. Expression of FLAG-tagged Nrf2 was achieved by transient transfection. Nrf2 alanine substitution variants were generated by site-directed mutagenesis to evaluate the role of Fyn tyrosine phosphorylation (Y576A, B) or the Neh6 phosphodegron (S344A/S347A/S351A/S356A, also known as S4A4; C). D, quantification of FLAG-Nrf2 variant expression in A–C. The impact of REDD1 on the expression of Nrf2 variants containing S344A/S347A or S351A/S356A substitutions was evaluated in E and F, respectively. FLAG-Nrf2, HA-REDD1, and actin expression were evaluated by Western blotting. Protein molecular mass (in kilodaltons) is indicated at the right of blots. Values are mean ± S.D. *, p < 0.05 versus WT.
REDD1-dependent activation of GSK3 is necessary for diabetes-induced oxidative stress and Nrf2 inhibition in mouse retina
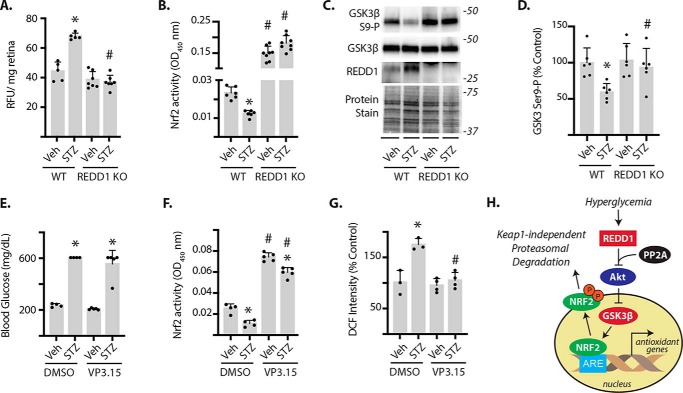
We recently demonstrated that REDD1 deletion was sufficient to prevent diabetes-induced oxidative stress in the retina of mice (21). In that study, ROS were evaluated with 2,7-dichlorofluoroscien (DCF). In support of the previous data, we found that unlike diabetic WT mice, diabetic REDD1-deficient mice did not exhibit an increase in ROS as assessed by dihydroethidium (Fig. 7A). In the retina of streptozotocin (STZ)-induced diabetic mice, Nrf2 activity was attenuated as compared with nondiabetic controls (Fig. 7B). However, in the retina of REDD1-deficient mice, Nrf2 activity was dramatically enhanced as compared with WT and the diabetes-induced suppression that was observed in WT mice was absent (Fig. 7B). GSK3β phosphorylation at Ser-9 was attenuated in the retina of diabetic mice concomitant with increased REDD1 expression (Fig. 7, C and D). Unlike WT mice, REDD1-deficient mice failed to exhibit a diabetes-induced decrease in retinal GSK3β phosphorylation. To determine whether GSK3 inhibition was sufficient to prevent the suppressive effect of diabetes on retinal Nrf2 activity, diabetic mice were treated with VP3.15. Intraperitoneal administration was selected based on the previous demonstration that VP3.15 readily crosses the blood-retina barrier (37). VP3.15 did not alter blood glucose concentrations (Fig. 7E). In contrast to mice receiving a vehicle control, mice receiving VP3.15 exhibited enhanced retinal Nrf2 activity (Fig. 7F). DCF was used to both quantify ROS levels in retinal lysates (Fig. 7G) and visualize ROS in retinal cryosections (Fig. S2). Mice receiving VP3.15 did not exhibit diabetes-induced oxidative stress in retina. Overall, the findings support a model whereby hyperglycemia-induced REDD1 acts to reduce the retinal Nrf2 antioxidant response to diabetes by promoting GSK3 activation (Fig. 7H).
Figure 7.
REDD1-dependent GSK3 activation contributes to diabetes-induced oxidative stress and suppression of Nrf2 activity in the retina. Diabetes was induced in WT and REDD1 knockout (KO) mice by administration of STZ. All analyses were performed 4 weeks after mice were administered STZ or a vehicle control (Veh). A, ROS was quantified in the supernatants from whole retinal lysates using dihydroethidium. B, Nrf2 activity was measured in the nuclear fraction obtained from retinal lysates by DNA-binding ELISA. C, GSK3β phosphorylation, as well as expression of GSK3β and REDD1 in retinal lysates were evaluated by Western blotting. Protein loading was evaluated by reversible protein stain. Protein molecular mass (in kilodaltons) is indicated at the right of blots. D, GSK3β phosphorylation in C was quantified. E–G, mice were treated daily with either VP3.15 or a vehicle control (DMSO) during the 4th week of diabetes. E, blood glucose concentrations were evaluated prior to retinal extraction. F, Nrf2 activity was measured as described in B. G, ROS was quantified in the supernatants from whole retinal lysates using DCF. Values are mean ± S.D. *, p < 0.05 versus Veh. #, p < 0.05 versus WT or DMSO. G, working model for the suppressive effect of REDD1 on Nrf2 stability.
Discussion
Nrf2 is an important factor in regulating the progression of DR as Nrf2-deficient mice exhibit early onset of both vascular and neuronal dysfunction in diabetes (27). The present study investigated regulation of the retinal Nrf2 antioxidant response by REDD1. Retinal REDD1 protein expression is elevated in multiple preclinical models of diabetes (20, 24, 26). Moreover, REDD1 is necessary for the development of oxidative stress in the retina of STZ-diabetic mice (21). Herein, we provided evidence suggesting that REDD1 facilitates the development of oxidative stress in response to diabetes by attenuating the Nrf2 antioxidant response. Nrf2 activity was enhanced in the retina of REDD1-deficient mice relative to WT controls. In the retina of diabetic WT mice, REDD1 protein expression was enhanced concomitant with decreased Nrf2 activity and increased ROS levels. REDD1 ablation not only prevented the suppressive effect of diabetes on retinal Nrf2 activity, but also dramatically enhanced Nrf2 activity relative to nondiabetic controls.
Consistent with the previous report (21), REDD1 ablation was sufficient to prevent enhanced ROS levels in cells exposed to hyperglycemic culture conditions. We found that Nrf2 up-regulation played an important role in the protective effect of REDD1 deletion. Specifically, Nrf2 knockdown enhanced ROS levels in REDD1-deficient cells exposed to hyperglycemic conditions, such that they were similar to those observed in cells expressing REDD1. Importantly, other biologically significant stresses that promote REDD1 expression (e.g. hypoxia, DNA damage, nutrient deprivation) are also likely to negatively impact the Nrf2 antioxidant response. In fact, during preparation of this manuscript, a relationship between REDD1 and Nrf2 was demonstrated in cardiomyocyte cultures (38). In that study, REDD1 knockdown attenuated oxidative stress in cells deprived of oxygen/glucose followed by reperfusion (OGD/R), and the protective effect was blunted by an siRNA targeting Nrf2. OGD/R suppressed expression of the Nrf2 target HO-1, whereas REDD1 knockdown enhanced HO-1 expression in a Nrf2-dependent manner (38). The findings herein extend the previous study by identifying the mechanism responsible for the suppressive effect of REDD1 on Nrf2 activity.
In the absence of oxidative stress, Nrf2 is rapidly targeted for degradation (32). Nrf2's short half-life is attributed to multiple protein complexes that promote proteasomal degradation of the transcription factor, but the most well-studied of these complexes includes the adapter protein Keap1 (10, 32, 39). Redox-sensitive activation of Nrf2 is mediated by the oxidation of Cys-151, Cys-273, and Cys-288 in Keap1, leading to a reduced interaction between Nrf2 and Keap1 (39). The Nrf2 Neh2 domain contains a high affinity Keap1-binding site that is responsible for redox-sensitive Nrf2 turnover (40). Disruption of Neh2 domain in the E82G and ΔETGE Nrf2 variants prevents degradation via Keap1 (41). In the present study, REDD1 promoted proteasome-dependent Nrf2 degradation. However, disruption of the Keap1-binding site, failed to prevent the decrease in Nrf2 expression in response to REDD1. This suggests that REDD1 promotes Nrf2 degradation independent of Keap1.
In addition to the redox-sensitive Neh2 degron, Nrf2 also contains an Neh6 degron that supports Nrf2 turnover even when Keap1 is inactivated (42). GSK3-dependent phosphorylation of Nrf2 within the Neh6 domain leads to nuclear export and increased proteasomal degradation by a protein cluster composed of β-transducin repeat-containing protein (β-TrCP) and the E3 ubiquitin ligase S-phase kinase-associated protein 1-Cullin1–F-box protein (35). The GSK3β isoform contains a nuclear localization signal that allows it to shuttle between the cytosol and nucleus, thus providing access to Nrf2 (43). In addition to the Neh6-phosphodegron, phosphorylation of Nrf2 by the tyrosine kinase Fyn may also contribute to the suppressive effects of GSK3 (33). However, disruption of the Fyn phosphorylation site at Tyr-576 of Nrf2 failed to repress REDD1-mediated degradation. In contrast, disruption of the Neh6 phosphodegron (35) increased Nrf2 stability in response to REDD1.
Structural analysis of the docking interaction between Nrf2 and β-TrCP demonstrates that GSK3 phosphorylation of Ser-344 and Ser-347 is necessary for the turn structure that is recognized by the WD40 domain of β-TrCP (36). Phosphorylation of two adjacent proline-directed sites at Ser-351 and Ser-356 is suspected to prime for phosphorylation at Ser-344 and Ser-347 (36). In a previous study, the Nrf2 S344A/S347A variant failed to co-immunoprecipitate with β-TrCP; however, additional alanine substitutions at S351A/S356A were required to completely prevent GSK3-dependent suppression of ARE activity (36). Consistent with that observation, we found that the Nrf2 S344A/S347A variant was not fully resistant to the effects of REDD1. Rather alanine substitutions at Ser-351 and Ser-356 prevented Nrf2 degradation in response to REDD1. The findings support that the sensitivity of Nrf2 to GSK3 and REDD1 may not be exclusively mediated by β-TrCP binding.
We recently demonstrated that the absence of hyperglycemia-induced oxidative stress in REDD1-deficient cells was prevented by expression of either a dominant-negative Akt variant or a constitutively active GSK3 variant (21). In the present study, REDD1 suppressed phosphorylation of Akt and GSK3 and enhanced proteasomal degradation of Nrf2. The suppressive effect of REDD1 on Nrf2 expression and Akt/GSK3 phosphorylation was recapitulated by PI3K inhibition. Moreover, the GSK3 inhibitors CHIR99021 and VP3.15 prevented the decrease in Nrf2 expression in response to REDD1. CHIR99021 is a highly selective ATP competitive inhibitor of GSK3 (44). VP3.15 is less selective and at higher concentrations also acts to inhibit phosphodiesterase 7 (45). However, it was recently demonstrated that VP3.15 effectively crosses the blood-retinal barrier (37). Thus, VP3.15 was selected for inhibition of GSK3 activity in the retina of STZ-diabetic mice. VP3.15 administration increased Nrf2 activity in the retina of diabetic mice and prevented the diabetes-induced increase in retinal ROS. This supports the potential therapeutic benefit of targeting REDD1-mediated GSK3 inhibition to promote retinal Nrf2 activation in the context of diabetes. Indeed, in the retina of STZ-diabetic rats, GSK3 phosphorylation is attenuated and GSK3 inhibition decreases TUNEL-positive nuclei (23, 46).
Consistent with our previous report (21), REDD1 protein expression was increased in the retina of diabetic mice in association with augmented ROS levels. In contrast to WT mice, REDD1-deficient mice did not exhibit a diabetes-induced increase in retina ROS levels. This demonstrates that REDD1 is necessary for the development of diabetes-induced oxidative stress in the retina. Blunted Nrf2 DNA binding is believed to be at least in part responsible for diabetes-induced oxidative stress, as activity of the transcription factor is reduced in both the retinas of diabetic patients and preclinical models (4, 6, 13). Consistent with the previous observations in STZ-diabetic rats (4, 6, 13), we found that Nrf2 DNA-binding activity was attenuated in the retina of STZ-diabetic mice. In association with the attenuation of Nrf2 activity, the inhibitory phosphorylation of retinal GSK3β at Ser-9 was reduced. In contrast to WT mice, REDD1-deficient mice failed to exhibit diabetes-induced attenuation of GSK3β phosphorylation and Nrf2 activity was dramatically enhanced. Notably, the magnitude of the increase in Nrf2 activity with VP3.15 administration was approximately half of that observed with REDD1 ablation. Although this is likely due to some reduced efficacy of chemical inhibition as compared with genetic knockout, it could indicate that REDD1 is also acting to repress Nrf2 activity through a yet to be identified GSK3-independent mechanism. In this regard, Nrf2 activity is also influenced by variation in the rate of synthesis (47) and other factors are known to impact nuclear localization of the transcription factor (48).
As previously mentioned, Nrf2 binding with Keap1 is paradoxically up-regulated in the retina of diabetic rats (4). It is well-established that diabetes enhances retinal oxidative stress (8, 14), which should lead to Keap1 Cys-oxidation and reduced interaction with Nrf2 (11). The inconsistency may be explained by the impact of REDD1. The activities of GSK3 and Nrf2 are negatively correlated, with GSK3-dependent phosphorylation of Nrf2 promoting nuclear exclusion (49). Indeed, exposure to hyperglycemic conditions is associated with reduced nuclear Nrf2 localization in a variety of cell types (4, 50). Thus, diabetes-induced nuclear exclusion of Nrf2 may be sufficient to increase interaction with Keap1, despite the relatively lower binding affinity under oxidizing conditions.
The present study provides important new evidence for biphasic regulation of the Nrf2 antioxidant response. The Nrf2 Neh6 degron is classically thought of as redox-insensitive (42). However, redox-sensitive REDD1 expression potentially promotes Neh6-mediated degradation. Increasing cellular ROS promotes REDD1 expression, and decreases GSK3β phosphorylation in a manner that is REDD1-dependent (21). Thus, REDD1 likely contributes to a biphasic switch in redox-sensitive regulation of Nrf2. In this new model, an acute rise in oxidative stress would lead to reduced Keap1-binding and increased nuclear translocation of Nrf2; whereas REDD1 would accumulate after prolonged oxidative stress, leading to activation of GSK3β and Nrf2 nuclear export.
The findings here support the conclusion that REDD1 reduces Nrf2 protein stability by promoting Keap1-independent proteasomal degradation of Nrf2. Well-known Nrf2 activators such as sulforaphane, bardoxolone methyl, and trifluoroethyl amide (dh404) have been extensively explored in a number of clinical trials (12). Moreover, dh404 treatment prevents the development of retinal vasculopathy and gliosis in diabetic rats (6). Importantly, all of the present generation of Nrf2 activators target Keap1 to suppress Nrf2-binding or Nrf2-ubiquination. Thus, the dozens of clinical trials that evaluate the benefits of Nrf2 activation, do so by employing Keap1 inhibition. Increasing the pool of Nrf2 by preventing Keap1-mediated degradation is likely beneficial for preventing oxidative stress; however, the data here suggest that this approach may not truly target the diabetes-induced defect in retinal Nrf2 regulation. Indeed, sulforaphane failed to alter the rate of Nrf2 degradation upon REDD1 induction. The data here are consistent with a previous study that attributes a deficit in Nrf2 activity to augmented GSK3-signaling in nonhealing diabetic wounds (51). Thus, new therapeutic approaches targeting REDD1-mediated GSK3 inhibition to promote Nrf2 activation may be beneficial in the context of diabetes.
Experimental procedures
Animals
WT and REDD1 knockout B6;129 mice (52) were administered 50 mg/kg STZ for 5 consecutive days to induce diabetes. Nondiabetic control mice received sodium citrate buffer. Diabetic phenotype was assessed with fasting blood glucose levels >250 mg/dl. C57BL/6N mice were administered STZ as described above in addition to daily intraperitoneal injections of either VP3.15 (10 mg/kg) or vehicle (10% DMSO, 0.9% NaCl) during the 4th week of diabetes. All procedures were approved by the Penn State College of Medicine Institutional Animal Care and Use Committee (IACUC), and were in accordance with the ARVO statement on the ethical use of animals in ophthalmological research.
Cell culture
MIO-M1 human Müller cells were obtained from the UCL Institute of Ophthalmology (London, UK). CRISPR/Cas9 genome editing to generate a stable MIO-M1 cell line deficient in REDD1 (sgREDD1) and development of a HEK293 TetON HA-REDD1 (HEK TetON) advanced stable cell line were previously described (22, 26). All cells were cultured at 37 °C, in 5% CO2 on CellBIND plates (Corning) with Dulbecco's modified Eagle's medium (Invitrogen). MIO-M1 cells were cultured in medium containing 5 mm glucose, 10% heat-inactivated fetal bovine serum, and 1% penicillin/streptomycin. HEK TetON cells were maintained in 25 mm glucose supplemented with Tet System Approved fetal bovine serum (Clontech) and kept under selective pressure using G418 (100 μg/ml) and hygromycin B (200 μg/ml). Where indicated cell culture medium was supplemented with 1 μg/ml of doxycycline (Fisher) to induce HA-REDD1 expression. For studies on the effects of hyperglycemic conditions, cell culture medium contained either 30 mm glucose or 5 mm glucose with 25 mm mannitol as an osmotic control. In specific studies, culture medium was supplemented with 5 μm cycloheximide (Dot Scientific), 20 μm MG-132 (EMD Millipore), 20 μm sulforaphane (Sigma-Aldrich), 50 μm LY294002 (Cell Signaling Technology), 100 nm rapamycin (EMD Millipore), 5 μm CHIR99021 (SelleckChem), or 1 μm VP3.15 (Med Koo Biosciences). Cells were transfected using Lipofectamine 2000 (Life Technologies). Plasmids included pCMV5 vector, pCMV-HA-REDD1 (human), pCMV-FLAG-Nrf2 (human, Addgene number 36971), pRL-CMV (Promega AF025843), and an ARE firefly luciferase reporter (kindly provided by Dr. Jiyang Cai, University of Texas Medical Branch). Nrf2 variants were generated by site-directed mutagenesis using the primers listed in Table S1 and a QuikChange Lightning Kit (Agilent). All plasmids were validated by sequencing analysis.
Nuclear fractionation
Isolation of cell nuclei was performed as previously described (53). Briefly, cells were pelleted, washed with PBS, and re-suspended in ice-cold buffer A (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT) before Dounce homogenization. A fraction of the lysate was combined in an appropriate volume of 5× RIPA buffer and 2× Laemmli buffer to be used as whole cell lysate analysis. Homogenates were centrifuged at 228 × g for 5 min at 4 °C to pellet nuclei. The supernatant was retained as the cytoplasmic fraction. The nuclear pellet was re-suspended in sucrose buffer S1 (0.25 mm sucrose, 10 mm MgCl2) and layered over a cushion of S3 (0.88 mm sucrose, 0.5 mm MgCl2) and centrifuged at 2800 × g for 10 min at 4 °C. The nuclear pellet was suspended in 1× RIPA buffer and combined with 2× Laemmli buffer. Samples were boiled and analyzed by Western blotting.
Short hairpin RNA (shRNA) knockdown of Nrf2
pLKO-shNFE2L2 plasmids for lentiviral shRNA knockdown were obtained from the Penn State College of Medicine shRNA Library Core (Fig. S1A). Lentivirus containing either scramble control shRNA (shScram) (Addgene number 1864) or five separate Nrf2 shRNAs (shNrf2) were prepared via the HEK293FT system and used to infect REDD1-deficient MIO-M1 cells for 48 h. Puromycin (1 μg/ml) was added to the cell culture medium to select for stable clones expressing shRNA. After 2 weeks, the stable cells were verified by Western blotting and quantitative PCR.
ROS detection
Retinas were homogenized in lysis buffer as previously described (24). Lysates were centrifuged at 1500 × g for 3 min and the supernatant was exposed to 10 μm dihydroethidium or 10 μm DCF. Fluorescence was measured using a Spectra Max M5 plate reader (Molecular Devices) (excitation/emission = 480/576 nm). ROS were assessed in cells in culture using a DCFDA Cellular ROS Detection Assay kit (Abcam).
ARE luciferase reporter assay
MIO-M1 WT and REDD1 knockout cells were co-transfected with 500 ng of ARE-Firefly luciferase and 100 ng of pRL-CMV Renilla luciferase. After 24 h, luciferase activity was measured on a FlexStation3 (Molecular Devices) using a Dual-Luciferase Assay Kit (Promega).
Nrf2 DNA-binding ELISA
Nrf2 activity was quantified using a colorimetric Nrf2 DNA-binding ELISA (TransAM Nrf2; Active Motif). Briefly, 10 μg of nuclear protein was isolated from enucleated mouse retinas and incubated for 1 h in the presence of an immobilized oligonucleotide encoding the ARE consensus sequence. Nrf2 binding was quantified using an anti-Nrf2 primary antibody and horseradish peroxidase-conjugated secondary. The resulting signal was measured on a Spectra Max M5 plate reader (Molecular Devices) (Abs/Ref = 450/655 nm).
Western blot analysis
Retinas were flash frozen in liquid nitrogen and homogenized in 250 μl of lysis buffer as previously described (24). Cell lysates and retinal homogenates were combined with Laemmli buffer, boiled, and fractionated using Criterion Precast 4–20% gels (Bio-Rad). Proteins were transferred to a polyvinylidene fluoride membrane, reversibly stained to assess protein loading (Pierce), blocked in 5% milk in TBS Tween 20, and evaluated with the appropriate antibodies (Table S2).
RNA isolation and quantitative PCR
Total RNA was extracted with TRIzol (Invitrogen). RNA (1 μg) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and subjected to quantitative real-time PCR (QuantStudio 12K Flex Real-Time PCR System) using QuantiTect SYBR Green master mix (Qiagen) as previously described (26). Primer sequences are listed in Table S3. Mean cycle threshold (CT) values were determined for control and experimental samples. Changes in mRNA expression were normalized to GAPDH mRNA expression using the 2−ΔΔCT calculations as previously described (31).
Statistical analysis
Data are expressed as mean ± S.D. Data were analyzed overall with either one-way or two-way analysis of variance. Trend test and pairwise comparisons were conducted with the Tukey test for multiple comparisons. Significance was defined as p < 0.05 for all analyses.
Data availability
All data for this publication are included in the manuscript or are available from the corresponding author upon request.
Author contributions
W. P. M., S. S., A. J. B., and M. D. D. conceptualization; W. P. M., S. S., and M. D. D. data curation; W. P. M., S. S., and M. D. D. formal analysis; W. P. M. and M. D. D. funding acquisition; W. P. M., S. S., J. F. G., and A. L. T. investigation; W. P. M. and M. D. D. visualization; W. P. M. and M. D. D. methodology; W. P. M. and M. D. D. writing-original draft; W. P. M., J. F. G., A. L. T., A. J. B., and M. D. D. writing-review and editing; A. L. T. and M. D. D. resources; A. J. B. and M. D. D. supervision; M. D. D. validation.
Supplementary Material
Acknowledgments
We thank Elena Feinstein (Quark Pharmaceuticals) for permission to use the REDD1 knockout mice, Jiyang Cai (University of Oklahoma Health Sciences Center) for generously providing the plasmid encoding the ARE luciferase reporter, and Scot Kimball (Penn State College of Medicine) for critically evaluating the manuscript.
This work was supported by American Diabetes Association Pathway to Stop Diabetes Grant 1-14-INI-04 and National Institutes of Health Grants R01 EY029702 (to M. D. D.) and F31 EY031199 (to W. P. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S3.
- DR
- diabetic retinopathy
- ROS
- reactive oxygen species
- Nrf2
- nuclear factor erythroid 2-related factor 2
- ARE
- antioxidant response element
- shRNA
- short hairpin RNA
- DCF
- 2,7-dichlorofluoroscien
- STZ
- streptozotocin
- HA
- hemagglutinin
- GSK
- glycogen synthase kinase
- REDD1
- regulated in development and DNA damage 1
- PI3K
- phosphatidylinositol 3-kinase
- Dox
- doxycycline
- β-TrCP
- β-transducin repeat-containing protein
- DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HO-1
- heme oxygenase 1
- NQO1
- NAD(P)H quinone oxoreductase 1.
References
- 1. Brownlee M. 2005) The pathobiology of diabetic complications. Diabetes 54, 1615–1625 10.2337/diabetes.54.6.1615 [DOI] [PubMed] [Google Scholar]
- 2. Kowluru R. A., Tang J., and Kern T. S.. 2001) Abnormalities of retinal metabolism in diabetes and experimental galactosemia. Diabetes 50, 1938–1942 10.2337/diabetes.50.8.1938 [DOI] [PubMed] [Google Scholar]
- 3. Diabetes Control and Complications Trial Research Group, Nathan D. M., Genuth S., Lachin J., Cleary P., Crofford O., Davis M., Rand L., and Siebert C.. 1993) The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986 10.1056/NEJM199309303291401 [DOI] [PubMed] [Google Scholar]
- 4. Zhong Q., Mishra M., and Kowluru R. A.. 2013) Transcription factor Nrf2-mediated antioxidant defense system in the development of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 54, 3941–3948 10.1167/iovs.13-11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santos J. M., Mohammad G., Zhong Q., and Kowluru Q. A.. 2011) Diabetic retinopathy, superoxide damage and antioxidants. Curr. Pharm. Biotechnol. 12, 352–361 10.2174/138920111794480507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deliyanti D., Alrashdi S. F., Tan S. M., Meyer C., Ward K. W., de Haan J. B., and Wilkinson-Berka J. L.. 2018) Nrf2 activation is a potential therapeutic approach to attenuate diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 59, 815–825 10.1167/iovs.17-22920 [DOI] [PubMed] [Google Scholar]
- 7. Tonelli C., Chio I. I. C., and Tuveson D. A.. 2087) Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 29, 1727–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ola M. S., and Alhomida A. S.. 2014) Neurodegeneration in diabetic retina and its potential drug targets. Curr. Neuropharmacol. 12, 380–386 10.2174/1570159X12666140619205024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayes J. D., and Dinkova-Kostova A. T.. 2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218 10.1016/j.tibs.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 10. Cullinan S. B., Gordan J. D., Jin J., Harper J. W., and Diehl J. A.. 2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 24, 8477–8486 10.1128/MCB.24.19.8477-8486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., and Talalay P.. 2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 10.1073/pnas.172398899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robledinos-Antón N., Fernández-Ginés R., Manda G., and Cuadrado A.. 2019) Activators and inhibitors of NRF2: a review of their potential for clinical development. Oxid. Med. Cell Longev. 2019, 9372182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mishra M., Zhong Q., and Kowluru R. A.. 2014) Epigenetic modifications of Nrf2-mediated glutamate–cysteine ligase: implications for the development of diabetic retinopathy and the metabolic memory phenomenon associated with its continued progression. Free Radic. Biol. Med. 75, 129–139 10.1016/j.freeradbiomed.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fong D. S., Aiello L. P., Ferris F. L. 3rd, and Klein R.. 2004) Diabetic retinopathy. Diabetes Care. 27, 2540–2553 10.2337/diacare.27.10.2540 [DOI] [PubMed] [Google Scholar]
- 15. Qiao S., Dennis M., Song X., Vadysirisack D. D., Salunke D., Nash Z., Yang Z., Liesa M., Yoshioka J., Matsuzawa S., Shirihai O. S., Lee R. T., Reed J. C., and Ellisen L. W.. 2015) A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat. Commun. 6, 7014 10.1038/ncomms8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horak P., Crawford A. R., Vadysirisack D. D., Nash Z. M., DeYoung M. P., Sgroi D., and Ellisen L. W.. 2010) Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 4675–4680 10.1073/pnas.0907705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellisen L. W., Ramsayer K. D., Johannessen C. M., Yang A., Beppu H., Minda K., Oliner J. D., McKeon F., and Haber D. A.. 2002) REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10, 995–1005 10.1016/S1097-2765(02)00706-2 [DOI] [PubMed] [Google Scholar]
- 18. Whitney M. L., Jefferson L. S., and Kimball S. R.. 2009) ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem. Biophys. Res. Commun. 379, 451–455 10.1016/j.bbrc.2008.12.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan C. Y., and Hagen T.. 2013) mTORC1 dependent regulation of REDD1 protein stability. PLoS ONE 8, e63970–e63970 10.1371/journal.pone.0063970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dennis M. D., Kimball S. R., Fort P. E., and Jefferson L. S.. 2015) Regulated in development and DNA damage 1 is necessary for hyperglycemia-induced vascular endothelial growth factor expression in the retina of diabetic rodents. J. Biol. Chem. 290, 3865–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller W. P., Toro A. L., Barber A. J., and Dennis M. D.. 2019) REDD1 activates a ROS-generating feedback loop in the retina of diabetic mice REDD1 promotes oxidative stress in retina. Invest. Ophthalmol. Vis. Sci. 60, 2369–2379 10.1167/iovs.19-26606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dennis M. D., Coleman C. S., Berg A., Jefferson L. S., and Kimball S. R.. 2014) REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci. Signal. 7, ra68–ra68 10.1126/scisignal.2005103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reiter C. E., Wu X., Sandirasegarane L., Nakamura M., Gilbert K. A., Singh R. S., Fort P. E., Antonetti D. A., and Gardner T. W.. 2006) Diabetes reduces basal retinal insulin receptor signaling. Diabetes 55, 1148–1156 10.2337/diabetes.55.04.06.db05-0744 [DOI] [PubMed] [Google Scholar]
- 24. Miller W. P., Yang C., Mihailescu M. L., Moore J. A., Dai W., Barber A. J., and Dennis M. D.. 2018) Deletion of the Akt/mTORC1 repressor REDD1 prevents visual dysfunction in a rodent model of type 1 Diabetes. Diabetes 67, 110–119 10.2337/db17-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L., Chen Y., Sternberg P., and Cai J.. 2008) Essential roles of the PI3 kinase/Akt pathway in regulating Nrf2-dependent antioxidant functions in the RPE. Invest. Ophthalmol. Vis. Sci. 49, 1671–1678 10.1167/iovs.07-1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai W., Miller W. P., Toro A. L., Black A. J., Dierschke S. K., Feehan R. P., Kimball S. R., and Dennis M. D.. 2018) Deletion of the stress-response protein REDD1 promotes ceramide-induced retinal cell death and JNK activation. FASEB J. 32, 6883–6897 10.1096/fj.201800413RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Z., Wei Y., Gong J., Cho H., Park J. K., Sung E.-R., Huang H., Wu L., Eberhart C., Handa J. T., Du Y., Kern T. S., Thimmulappa R., Barber A. J., Biswal S., and Duh E. J.. 2014) NRF2 plays a protective role in diabetic retinopathy in mice. Diabetologia. 57, 204–213 10.1007/s00125-013-3093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coughlin B. A., Feenstra D. J., and Mohr S.. 2017) Müller cells and diabetic retinopathy. Vision Res. 139, 93–100 10.1016/j.visres.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bringmann A., and Wiedemann P.. 2012) Müller glial cells in retinal disease. Ophthalmologica 227, 1–19 10.1159/000328979 [DOI] [PubMed] [Google Scholar]
- 30. Fletcher E. L., Phipps J. A., and Wilkinson-Berka J. L.. 2005) Dysfunction of retinal neurons and glia during diabetes. Clin. Exp. Optom. 88, 132–145 10.1111/j.1444-0938.2005.tb06686.x [DOI] [PubMed] [Google Scholar]
- 31. Livak K. J., and Schmittgen T. D.. 2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32. Furukawa M., and Xiong Y.. 2005) BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol. 25, 162–171 10.1128/MCB.25.1.162-171.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jain A. K., and Jaiswal A. K.. 2007) GSK-3β acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J. Biol. Chem. 282, 16502–16510 10.1074/jbc.M611336200 [DOI] [PubMed] [Google Scholar]
- 34. Kaspar J. W., and Jaiswal A. K.. 2011) Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression. FASEB J. 25, 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rada P., Rojo A. I., Chowdhry S., McMahon M., Hayes J. D., and Cuadrado A.. 2011) SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 31, 1121–1133 10.1128/MCB.01204-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rada P., Rojo A. I., Evrard-Todeschi N., Innamorato N. G., Cotte A., Jaworski T., Tobón-Velasco J. C., Devijver H., García-Mayoral M. F., Van Leuven F., Hayes J. D., Bertho G., and Cuadrado A.. 2012) Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/β-TrCP Axis. Mol. Cell Biol. 32, 3486–3499 10.1128/MCB.00180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sánchez-Cruz A., Villarejo-Zori B., Marchena M., Zaldivar-Díez J., Palomo V., Gil C., Lizasoain I., de la Villa P., Martínez A., de la Rosa E. J., and Hernández-Sánchez C.. 2018) Modulation of GSK-3 provides cellular and functional neuroprotection in the rd10 mouse model of retinitis pigmentosa. Mol. Neurodegener. 13, 19 10.1186/s13024-018-0251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li P., Lin N., Guo M., Huang H., Yu T., and Zhang L.. 2019) REDD1 knockdown protects H9c2 cells against myocardial ischemia/reperfusion injury through Akt/mTORC1/Nrf2 pathway-ameliorated oxidative stress: an in vitro study. Biochem. Biophys. Res. Commun. 519, 179–185 10.1016/j.bbrc.2019.08.095 [DOI] [PubMed] [Google Scholar]
- 39. He X., and Ma Q.. 2009) NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 76, 1265–1278 10.1124/mol.109.058453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., and Yamamoto M.. 1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 10.1101/gad.13.1.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tong K. I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., and Yamamoto M.. 2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell Biol. 26, 2887–2900 10.1128/MCB.26.8.2887-2900.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McMahon M., Thomas N., Itoh K., Yamamoto M., and Hayes J. D.. 2004) Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279, 31556–31567 10.1074/jbc.M403061200 [DOI] [PubMed] [Google Scholar]
- 43. Shin S. H., Lee E. J., Chun J., Hyun S., Kim Y. I, and Kang S. S.. 2012) The nuclear localization of glycogen synthase kinase 3β is required its putative PY-nuclear localization sequences. Mol. Cells 34, 375–382 10.1007/s10059-012-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ring D. B., Johnson K. W., Henriksen E. J., Nuss J. M., Goff D., Kinnick T. R., Ma S. T., Reeder J. W., Samuels I., Slabiak T., Wagman A. S., Hammond M.-E. W., and Harrison S. D.. 2003) Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52, 588–595 10.2337/diabetes.52.3.588 [DOI] [PubMed] [Google Scholar]
- 45. Medina-Rodríguez E. M., Bribián A., Boyd A., Palomo V., Pastor J., Lagares A., Gil C., Martínez A., Williams A., and de Castro F.. 2017) Promoting in vivo remyelination with small molecules: a neuroreparative pharmacological treatment for multiple sclerosis. Sci. Rep. 7, 43545 10.1038/srep43545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Z., Ma L., Chen X., Li Y., Li S., Zhang J., and Lu L.. 2014) Glycogen synthase kinase-3: a key kinase in retinal neuron apoptosis in early diabetic retinopathy. Chin. Med. J. (Engl.) 127, 3464–3470 [PubMed] [Google Scholar]
- 47. Li W., Thakor N., Xu E. Y., Huang Y., Chen C., Yu R., Holcik M., and Kong A.-N.. 2010) An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 38, 778–788 10.1093/nar/gkp1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Theodore M., Kawai Y., Yang J., Kleshchenko Y., Reddy S. P., Villalta F., and Arinze I. J.. 2008) Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 283, 8984–8994 10.1074/jbc.M709040200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salazar M., Rojo A. I., Velasco D., de Sagarra R. M., and Cuadrado A.. 2006) Glycogen synthase kinase-3β inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 281, 14841–14851 10.1074/jbc.M513737200 [DOI] [PubMed] [Google Scholar]
- 50. Soares M. A., Cohen O. D., Low Y. C., Sartor R. A., Ellison T., Anil U., Anzai L., Chang J. B., Saadeh P. B., Rabbani P. S., and Ceradini D. J.. 2016) Restoration of Nrf2 signaling normalizes the regenerative niche. Diabetes 65, 633–646 10.2337/db15-0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bitar M. S., and Al-Mulla F.. 2011) A defect in Nrf2 signaling constitutes a mechanism for cellular stress hypersensitivity in a genetic rat model of type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 301, E1119–29 10.1152/ajpendo.00047.2011 [DOI] [PubMed] [Google Scholar]
- 52. Brafman A., Mett I., Shafir M., Gottlieb H., Damari G., Gozlan-Kelner S., Vishnevskia-Dai V., Skaliter R., Einat P., Faerman A., Feinstein E., and Shoshani T.. 2004) Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Invest. Ophthalmol. Vis. Sci. 45, 3796–3805 10.1167/iovs.04-0052 [DOI] [PubMed] [Google Scholar]
- 53. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I.. 2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 9, 457–470 10.1074/mcp.M900429-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this publication are included in the manuscript or are available from the corresponding author upon request.