Abstract
N-Acyl-phosphatidylethanolamine phospholipase D (NAPE-PLD) (EC 3.1.4.4) catalyzes the final step in the biosynthesis of N-acyl-ethanolamides. Reduced NAPE-PLD expression and activity may contribute to obesity and inflammation, but a lack of effective NAPE-PLD inhibitors has been a major obstacle to elucidating the role of NAPE-PLD and N-acyl-ethanolamide biosynthesis in these processes. The endogenous bile acid lithocholic acid (LCA) inhibits NAPE-PLD activity (with an IC50 of 68 μm), but LCA is also a highly potent ligand for TGR5 (EC50 0.52 μm). Recently, the first selective small-molecule inhibitor of NAPE-PLD, ARN19874, has been reported (having an IC50 of 34 μm). To identify more potent inhibitors of NAPE-PLD, here we used a quenched fluorescent NAPE analog, PED-A1, as a substrate for recombinant mouse Nape-pld to screen a panel of bile acids and a library of experimental compounds (the Spectrum Collection). Muricholic acids and several other bile acids inhibited Nape-pld with potency similar to that of LCA. We identified 14 potent Nape-pld inhibitors in the Spectrum Collection, with the two most potent (IC50 = ∼2 μm) being symmetrically substituted dichlorophenes, i.e. hexachlorophene and bithionol. Structure–activity relationship assays using additional substituted dichlorophenes identified key moieties needed for Nape-pld inhibition. Both hexachlorophene and bithionol exhibited significant selectivity for Nape-pld compared with nontarget lipase activities such as Streptomyces chromofuscus PLD or serum lipase. Both also effectively inhibited NAPE-PLD activity in cultured HEK293 cells. We conclude that symmetrically substituted dichlorophenes potently inhibit NAPE-PLD in cultured cells and have significant selectivity for NAPE-PLD versus other tissue-associated lipases.
Keywords: phospholipase D, endocannabinoid, high-throughput screening (HTS), lipid signaling, protein–lipid interaction, bile acid, metabolic disease, dichlorophene, N-acyl-ethanolamide, N-acyl-phosphatidylethanolamine, NAPE-PLD, TGR5
Introduction
N-Acyl-ethanolamide (NAE)2 biosynthesis and signaling appear to play important roles in regulating energy balance, inflammation, stress responses, and addiction (1–6). Saturated and monounsaturated NAEs like N-palmitoyl-ethanolamide (C16:0NAE) and N-oleoyl-ethanolamide (C18:1NAE) act on GPR55 and GPR119, respectively, as well as on peroxisome proliferator-activated receptor α (7–13). Pharmacological administration of these compounds enhances the resolution of inflammation, induces satiety, and protects against the development of obesity on high-fat diet (14–17). In contrast, N-arachidonoyl-ethanolamide (C20:4NAE, anandamide) is a polyunsaturated NAE that acts at endocannabinoid receptors (CB1 and CB2) to exert pleotropic effects on food intake, anxiety, nociception, inflammation, locomotion, and memory (7, 18).
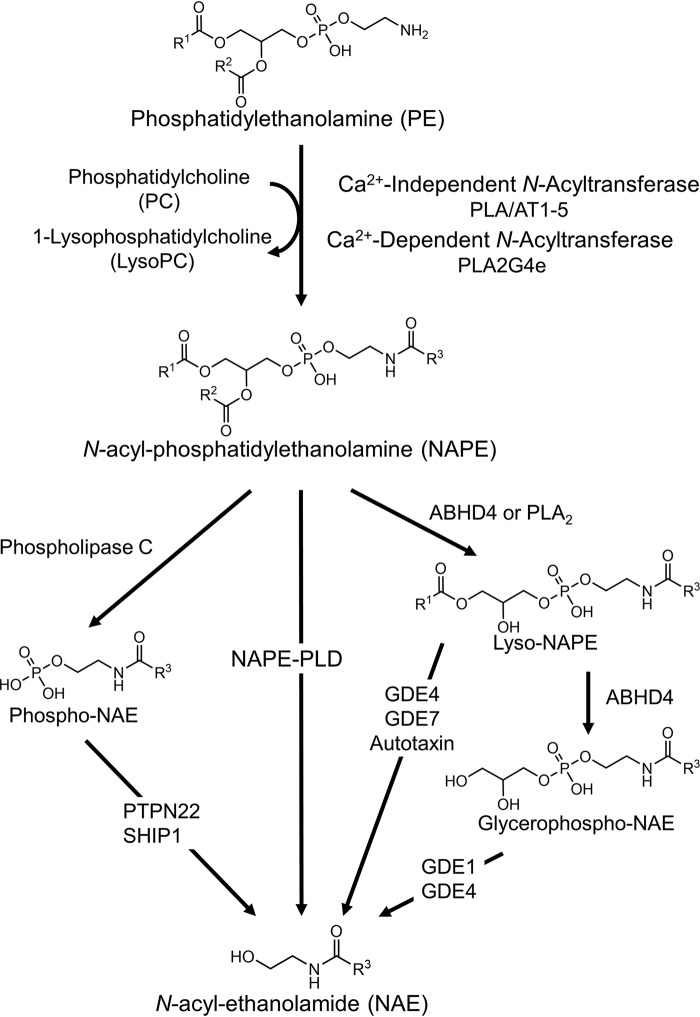
Even with recent advances, much about the regulation of NAE levels and their contributions to biological processes remains poorly understood. The first step of NAE biosynthesis requires the transfer of the appropriate O-acyl chain from phosphatidylcholine to the ethanolamine headgroup of a phosphatidylethanolamine (PE) to form N-acyl-phosphatidylethanolamines (NAPEs) (19). Five calcium-independent PE N-acyltransferases (PLAAT1–5) and one calcium-dependent PE N-acyltransferase (PLA2G4e) that catalyze this transfer have been identified in humans (19–20). The second step of NAE biosynthesis, conversion of NAPE to NAEs, can be directly catalyzed by NAPE phospholipase D, a member of the zinc metallo-β-lactamase family (Fig. 1) (21–23). NAPE-PLD homologs are found in mammals, reptiles, worms, and yeast, suggesting that it has highly conserved functions in normal physiology (12, 21, 24, 25). Surprisingly, global deletion of the Nape-pld in mice from birth causes marked changes in multiple lipid pathways yet only partially reduces NAE levels in most tissues (26–29). Several alternative pathways for the conversion of NAPE to NAE have now been identified (Fig. 1) (24, 30, 31). Despite the identification of these enzymes, how biosynthesis of individual NAEs is controlled remains unclear. For instance, refeeding of rodents leads to an increase in jejunal C18:1NAE levels but also leads to a decrease in jejunal C20:4NAE levels (32), despite the notion that both NAEs theoretically share the same biosynthetic machinery. Modulating the NAE-biosynthetic enzymes with appropriate tool compounds should help elucidate the contribution of individual enzymes to the biosynthesis of individual NAEs under various physiological and pathophysiological conditions and could lead to the development of effective therapies for a range of clinical conditions including obesity, inflammation, chronic pain, and addiction (33, 34).
Figure 1.
Pathways of NAE biosynthesis from NAPE. ABH4, α/β-hydrolase 4; GDE, glycerophosphodiester phosphodiesterase; PLA/AT, phospholipase A/acyltransferase; PLA2, phospholipase A2; PLA2G4e, PLA2 group 1Ve; PTPN22, protein-tyrosine phosphatase nonreceptor type 22; R1–3, acyl chain substituents; SHIP-1, SH-2 containing inositol 5′ polyphosphatase 1.
There is currently a lack of specific inhibitors or activators to directly assess the contribution of NAPE-PLD to NAE biosynthesis under various conditions. One endogenous bile acid, lithocholic acid (LCA), has been reported to inhibit NAPE-PLD (IC50 = 68 μm) (35). Although its bioavailability and relatively low toxicity allow LCA to be administered in vivo, LCA is also a potent ligand for the bile acid receptor Tgr5, with an EC50 of 0.53 μm (36), so it cannot be used as a selective inhibitor of NAPE-PLD. More recently, ARN19874 was reported to be a selective NAPE-PLD inhibitor, with an IC50 of 34 μm (37). The absorbance, distribution, metabolism, and excretion properties of this compound and whether it has efficacy in vivo have not been reported. We therefore performed high-throughput screening (HTS) for modulators of recombinant mouse Nape-pld using a small chemical library consisting of a collection of American and European drugs with known absorbance, distribution, metabolism, and excretion characteristics (the Spectrum Collection) and identified 14 Nape-pld inhibitors with IC50 <20 μm. The two most potent compounds were symmetrically substituted dichlorophenes that showed at least 75-fold specificity toward Nape-pld over other lipases tested and effectively inhibited NAPE-PLD in HEK293 cells.
Results and discussion
An optimal HTS assay for changes in NAPE-PLD activity requires high reproducibility for replicate samples and sufficient dynamic range to reliably detect activators and inhibitors. Fluorogenic lipid substrates have been previously used successfully in HTS assays for modulators of lipase activity (38, 39). PED-A1 is a quenched fluorogenic NAPE analog that has previously been used to measure phospholipase A1 activity in vitro and in tissue samples (40). To test whether PED-A1 could be used as a substrate for NAPE-PLD, we incubated PED-A1 with hexahistidine-tagged recombinant mouse Nape-pld and measured the development of fluorescence. Incubation of PED-A1 with active Nape-pld resulted in a rapid rise in fluorescence, whereas incubation with Nape-pld that had been inactivated by boiling did not (Fig. S1A). The Km of recombinant Nape-pld for PED-A1 (3.0 ± 0.5 μm) was determined by varying the PED-A1 concentration incubated with 0.1 μm Nape-pld (Fig. S1B). To ensure that the assay would tolerate small variations in buffer composition, we determined the concentration range of N-octyl-β-d-glucoside (NOG, used to stabilize Nape-pld) and DMSO (used as vehicle for PED-A1 and for screening compounds) in which Nape-pld activity did not differ by more than 10% from maximal activity. For NOG, this was 0.2–0.6% (w/v), and for DMSO, this was 1.0–3.7% (v/v) (data not shown). Based on these results, a final concentration of 0.4% NOG (w/v) and 1.6% DMSO (v/v) was used in all subsequent assays. To assess the assay's ability to reproducibly distinguish between normal and inhibited signals across replicated wells in 384-well plate, 100 μm LCA or vehicle was added in checkerboard fashion to a 384-well plate, and initial rate of fluorescence change (t = 30–100 s) and steady-state fluorescence (t = 419–420 s) was determined (Fig. S2). The Z′ factor calculated for LCA (0.45–0.47) demonstrated that the signal windows were sufficiently separated to observe hits in the screen, regardless of plate position.
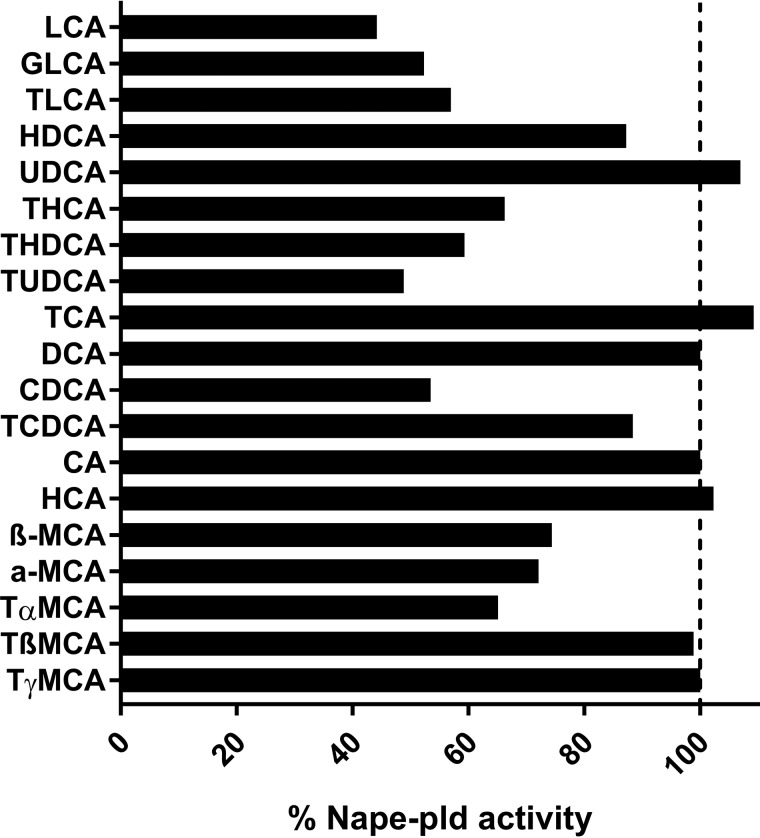
LCA is currently the only known endogenous inhibitor of NAPE-PLD (35). There are a variety of primary and secondary bile acids besides LCA, and these acids differ from each other in respect to the number, position, and stereochemistry of their hydroxyl groups, as well as their conjugated moieties. To determine whether other bile acids serve as endogenous inhibitors of Nape-pld, we screened a panel of 19 primary or secondary bile acids (50 μm each) (Fig. 2). The LCA conjugates glycolithocholic and taurolithocholic acid were slightly less potent Nape-pld inhibitors than LCA, which suggests that there is some flexibility in the length of the negatively charged moiety needed for inhibitory interactions with Nape-pld. Taurine conjugates of deoxycholic acid (tauroursodeoxycholic acid and taurohyodeoxycholic acid) were nearly as potent as LCA, whereas their nonconjugated forms (deoxycholic acid and ursocholic acid) had no inhibitory effects. Finally, several muricholic acids (α-, β-, and tauro-α-muricholic acids) were slightly less potent than LCA. These results identify a number of new endogenous inhibitors of Nape-pld that may be relevant to Nape-pld activity in the intestinal tract where bile acid concentrations are high.
Figure 2.
Effect of individual bile acid species (50 μm) on recombinant mouse Nape-pld activity measured by hydrolysis of PED-A1. Each bar represents the average of two replicates normalized to the activity of the DMSO negative control. GLCA, glycolithocholic acid; TLCA, taurolithocholic acid; HDCA, hyodeoxycholic acid; UDCA, ursodeoxycholic acid; THCA, taurohyocholic acid; THDCA, taurohyodeoxycholic acid; TUDCA, tauroursodeoxycholic acid; TCA, taurocholic acid; DCA, deoxycholic acid; CDCA, chenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; CA, cholic acid; HCA, hyocholic acid; MCA, muricholic acid; TαMCA, tauro-α-muricholic acid.
We next screened 2,388 biologically active and structurally diverse compounds (the Spectrum Collection compound library) at 10 μm final concentration of each compound. For all compounds, a B score (41) for both initial rate of fluorescence change and steady-state fluorescence value was calculated. Compounds with a B score 3 standard deviations above (activators) or below (inhibitors) the mean (or 3 median absolute deviations from the median) were classified as potential hits after confirmation by manual inspection (Fig. S3). Structures known to potentially interfere with the assay because of fluorescence or quenching were eliminated from further study. Hits were then reassayed to confirm the original findings. Although all 14 of the potential inhibitors demonstrated reproducible inhibition in the confirmation assay, none of the 12 potential activator compounds demonstrated reproducible activation.
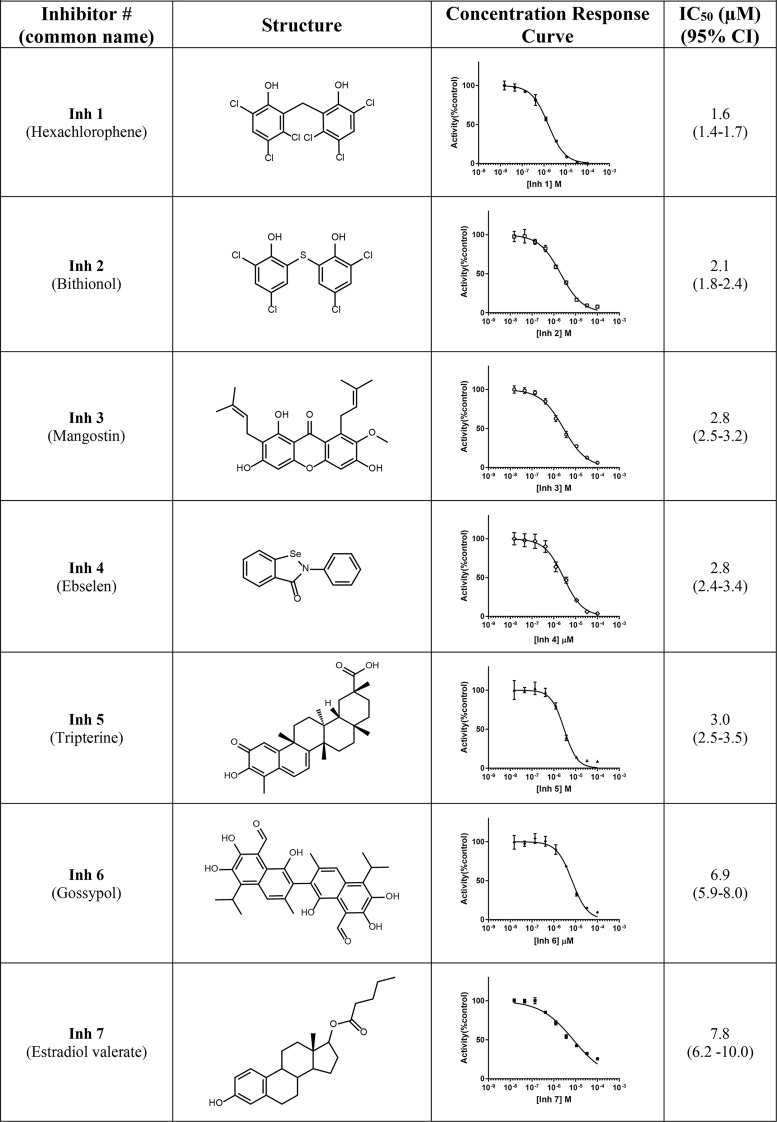
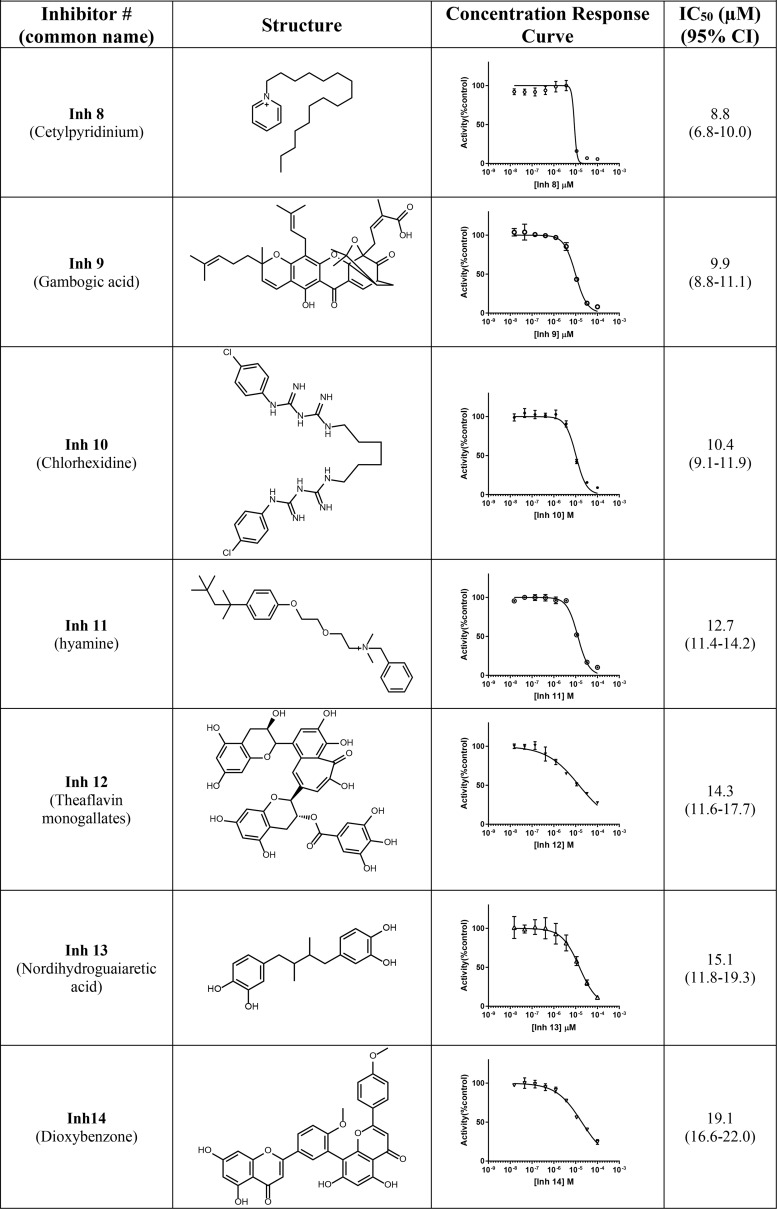
To determine whether the reduction in fluorescence by any of the 14 potential inhibitor compounds was due to direct quenching of BODIPY fluorescence rather than inhibition of Nape-pld, these compounds were incubated with BODIPY FL C5, the phospholipase A1 cleavage product of PED-A1. For each hit compound, the fluorescence of BODIPY FL C5 after treatment with 10 μm compound for 30 min was compared with vehicle treatment. None of the 14 hit compounds altered fluorescence by more than 10% (Fig. S4), indicating they were bona fide inhibitors of Nape-pld. All 14 inhibitor compounds (Inh 1–14) were then purchased from commercial sources to validate the putative library compound and analyzed by MS to confirm identity and purity, and concentration-response curve studies were performed to establish potency. The IC50 values of these compounds ranged from 1.6 to 19.1 μm, so that each was potentially more potent in vitro than ARN19874 (Table 1) (37).
Table 1.
Compounds identified from HTS screen of spectrum collection
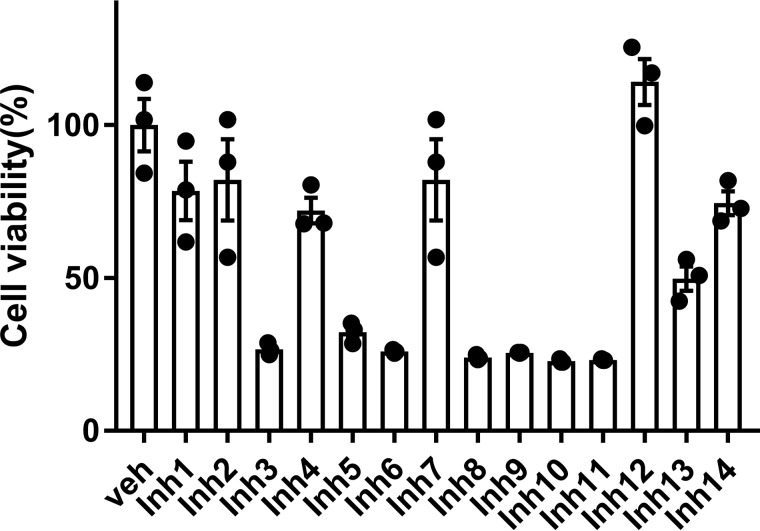
The effects of Inh 1 to 14 on cell viability were tested in HEK293 cells. The HEK293 cell line was chosen because it has been extensively used as a model for transfection and inhibition studies. Cytotoxicity of each compound was tested at a concentration five times that of their calculated IC50 in Table 1 (5× IC50). At these concentrations, six of the fourteen compounds gave cell viability >70%: Inh 1 (hexachlorophene), Inh 2 (bithionol), Inh 4 (ebselen), Inh 7 (estradiol valerate), Inh 12 (theaflavin mongallates), and Inh 14 (dioxybenzone) (Fig. 3).
Figure 3.
Effect of inhibitors on cell viability. HEK293 cells were incubated for 24 h with a concentration of inhibitor representing five times the IC50 value for recombinant Nape-pld for that compound. The resulting cell viability was measured by MTT assay and normalized to that of vehicle-treated cells (Veh). Three replicate wells of 24-well plate were used for each compound. The means ± S.E. are shown for each compound.
Our finding that estradiol valerate (Inh 7, IC50 = 7.8 μm) inhibited Nape-pld activity is of interest both because it shares significant structural homology to bile acids such as LCA and because estradiol valerate is a widely used birth control agent because of its potent estrogen receptor agonist activity. Although this finding suggests the possibility that medicinal chemistry could be used to modify estradiol valerate to improve potency and reduce estrogen receptor activity, it is unclear what further modifications of this compound could be undertaken to this end. Several closely related estradiol analogs, including estradiol, estradiol diacetate, estradiol cypionate, and estratriol, were tested in the original screen of 2,388 compounds but did not show significant inhibitory activity at 10 μm. Therefore, development of estradiol valerate was not pursued further.
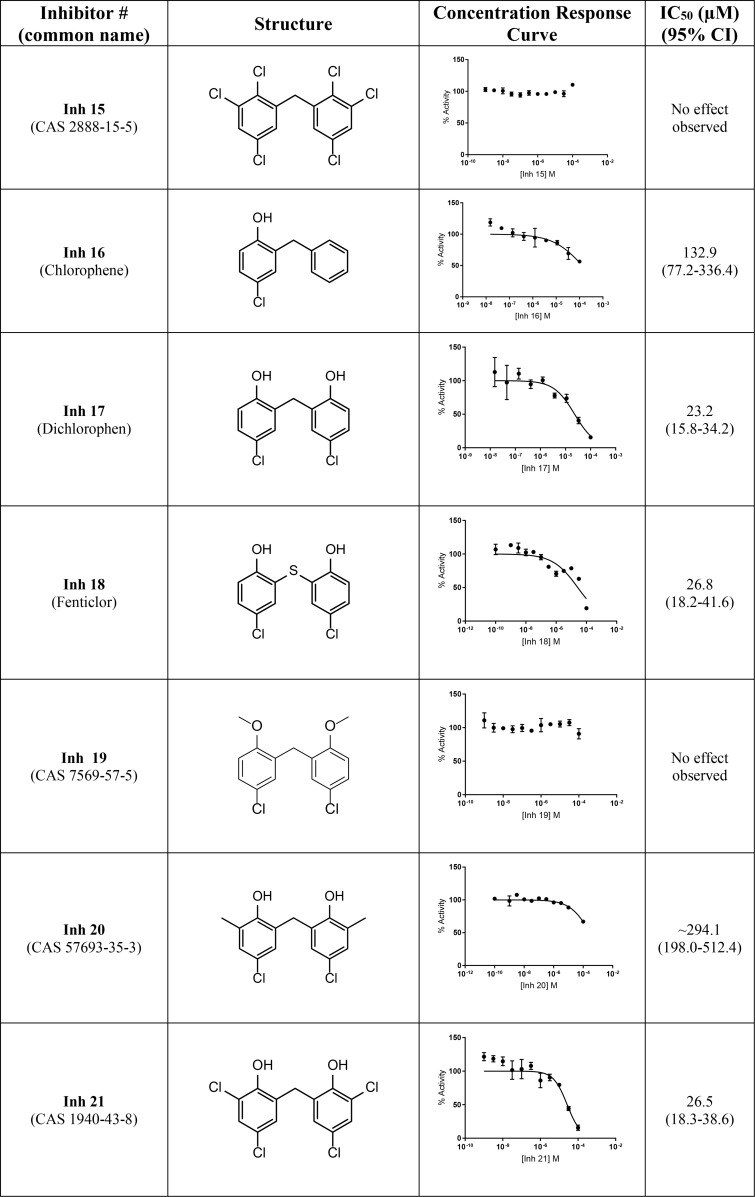
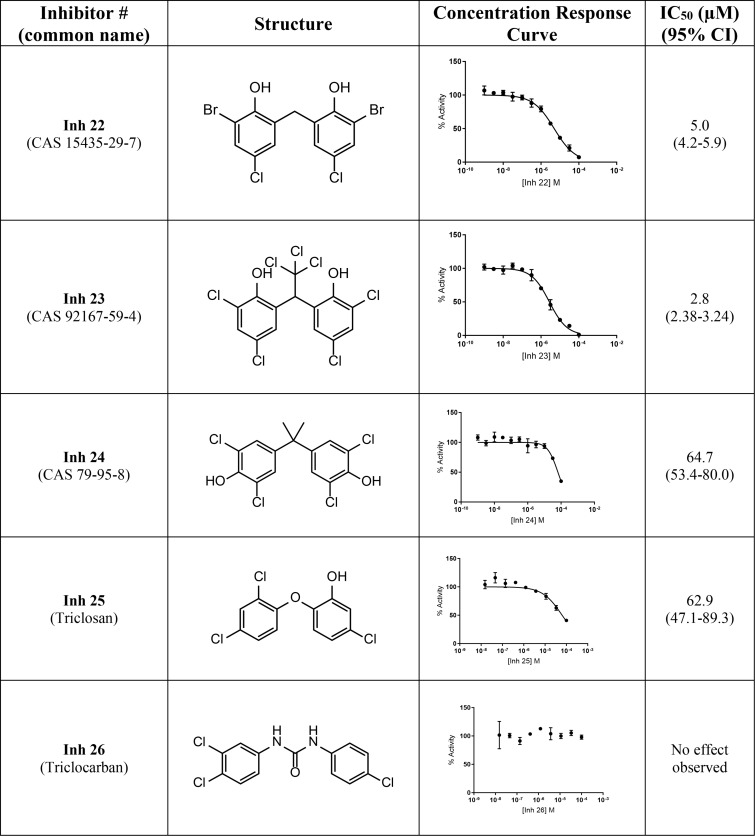
Our two most potent inhibitors, Inh 1 (hexachlorophene) and Inh 2 (bithionol), share a symmetrically substituted dichlorophene structure, with Inh 2 having a thioether linker rather than an alkane linker, as well as lacking the 5/5′-chloro groups of Inh 1. Because Nape-pld acts as a homodimer, the potency of these symmetrical compounds may arise from acting at the dimer interface (e.g. by binding similar residues on both Nape-pld monomers). We used commercially available dichlorophene analogs to carry out limited structure–activity relationship studies. We first tested whether the two hydroxyl groups contributed to inhibition. Inh 15, in which chloro groups replace the hydroxyl groups, had no inhibitory activity (Table 2). Inh 16 (chlorophene), with a hydroxyl and a chloro group on only one of the two aromatic rings had only very weak inhibitory activity. Inh 17 (dichlorophen) and Inh 18 (fenticlor) have a hydroxyl and chloro group on both aromatic rings, and both these compounds were ∼10-fold less potent than Inh 1 and Inh 2. Inh 19, in which both hydroxyl groups of Inh 17 are replaced with methoxy groups, gave no measurable inhibitory activity, supporting the need for symmetrical hydroxyl groups. Symmetrical addition of methyl (Inh 20), chloro (Inh 21), or bromo groups (Inh 22) at the 2 and 2′ positions to Inh 17 resulted in less, equal, or more potent inhibition of Nape-pld, respectively, than for Inh 17, suggesting that addition of strong electronegative groups at this position enhances inhibitory activity. The addition of a trichloromethane group to the carbon bridging the two aromatic rings of Inh 17 (Inh 23) increased potency ∼10-fold. Inh 24, in which the 1/1′ hydroxy groups of Inh 17 are at the 3/3′ positions instead and there is dimethyl substitution of the carbon bridging the two aromatic rings, is ∼3-fold less potent than Inh 17. Inh 25 (triclosan) also showed ∼3-fold less potency than Inh 17, whereas Inh 26 (triclocarban) showed no inhibitory activity. Together, these results suggest that symmetrical halide substitution of the two phenolic groups is critical for potent inhibition. Although Inh 22 and Inh 23 showed similar potency of inhibition of recombinant Nape-pld as Inh 1 and Inh 2, both of them showed significantly greater cytotoxicity in HEK293 cells (data not shown), so we pursued further mechanistic testing only with Inh 1 and 2.
Table 2.
Structure–activity relationships for substituted chlorophene analogs
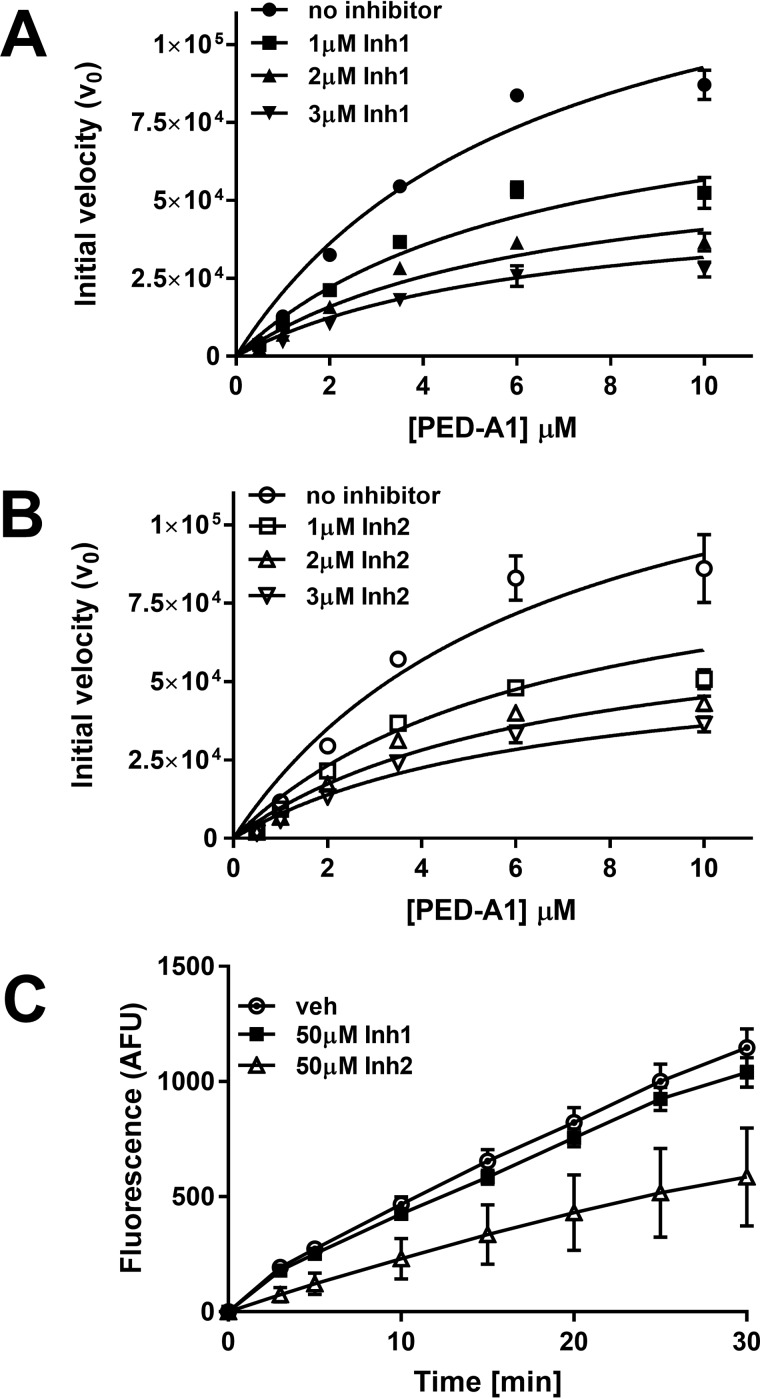
Inhibition of recombinant Nape-pld with Inh 1 and Inh 2 altered Vmax but not Km, consistent with a noncompetitive mechanism of inhibition. Apparent KI for both Inh 1 and Inh 2 was ∼2 μm (Fig. 4, A and B). Inhibition by Inh 1 was completely lost after 100-fold dilution of the enzyme-inhibitor complex, indicating a noncovalent, highly reversible interaction (Fig. 4C). In contrast, inhibition by Inh 2 persisted even after 100-fold dilution of the complex, indicating tight binding or potentially a covalent interaction. Thus, despite their chemical similarities, these two inhibitors appear to interact with Nape-pld by somewhat different mechanisms.
Figure 4.
Mechanism of inhibition by Inh 1 and Inh 2. A, recombinant mouse Nape-pld was incubated with 0–3 μm Inh1, then 0–10 μm PED-A1 was added, and the initial velocity of PED-A1 hydrolysis was determined for each (means ± S.E., n = 3). B, recombinant Nape-pld was incubated with 0–3 μm Inh 2, then 0–10 μm PED-A1 was added, and the initial velocity of PED-A1 hydrolysis was determined for each (means ± S.E., n = 3). C, a rapid dilution assay for Inh 1 and Inh 2. Recombinant Nape-pld was incubated with DMSO (veh) or 50 μm Inh 1 or Inh 2 for 1 h, then samples were diluted 100-fold with buffer immediately prior to addition of 4 μm PED-A1, and the resulting fluorescence was measured as arbitrary fluorescence units (AFU) (means ± S.E., n = 9).
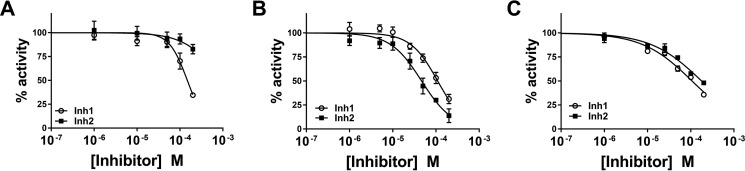
Selectivity of the two inhibitors for Nape-pld versus other lipases was first examined using Streptomyces chromofuscus phospholipase D (ScPld), which is a broad-spectrum phospholipase D that catalyzes the hydrolysis of the headgroup of a variety of lipid substrates including phosphatidylcholine, PE, NAPE, and other N-modified phospholipids (21, 22, 42). As with recombinant Nape-pld, incubation of ScPld with PED-A1 resulted in a rapid increase in fluorescence intensity. Both Inh 1 and Inh 2 inhibited ScPld activity, but only at concentrations ∼100-fold higher (IC50 values of 149 and >200 μm, respectively) than required to inhibit Nape-pld (Fig. 5A). Potential inhibition of serum lipases was examined using 1-arachidonyl-thiolglycerol (1-AT) as a general lipase substrate, with released thiolglycerol quantified by fluorescence resulting from reaction with 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin. The IC50 values for serum lipase activity were 110 and 46 μm for Inh 1 and Inh 2, respectively (Fig. 5B). Because Nape-pld is a member of the zinc-dependent metalloenzyme family, we determined whether Inh 1 and Inh 2 also inhibited other zinc metalloenzymes using carbonic anhydrase as a model enzyme. p-Nitrophenylacetate was used as substrate to measure carbonic anhydrase activity. The IC50 values for carbonic anhydrase activity were 107 and 178 μm for Inh 1 and Inh 2, respectively (Fig. 5C). Thus, both Inh 1 and Inh 2 showed relatively selective inhibition of Nape-pld when compared with their effects on other lipases or zinc-dependent metalloenzymes.
Figure 5.
Selectivity of Inh 1 and Inh 2. A, effect on S. chromofuscus phospholipase D activity (means ± S.E., n = 6). B, effect on serum lipase activity (means ± S.E., n = 9). C, effect on serum carbonic anhydrase activity (means ± S.E., n = 6).
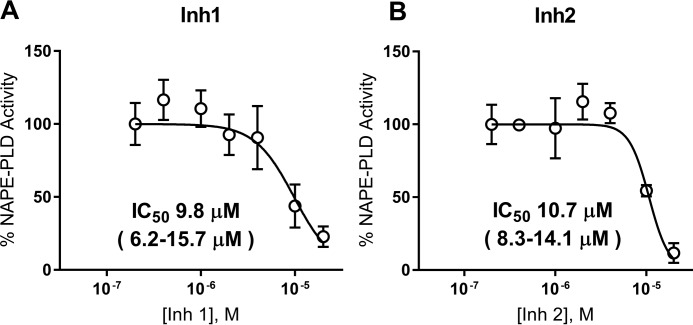
HEK293 endogenously express NAPE-PLD (37, 43) and were therefore used to assess the ability of these inhibitors to inhibit the NAPE-PLD activity of intact cells. To determine the maximum concentration of Inh 1 and Inh 2 that could be used for cellular inhibition studies without cytotoxicity, HEK293 were treated with 0–100 μm inhibitor, and cytotoxicity was measured as before. Both inhibitors showed cytotoxicity at 100 μm (Fig. S5), so 20 μm was chosen as the maximum concentration for inhibition studies. HEK293 cells were treated with 0–20 μm of each inhibitor for 30 min, and then NAPE-PLD activity was measured by adding PED-A1. Inh 1 and Inh 2 inhibited NAPE-PLD activity with IC50 values of 9.8 and 10.7 μm, respectively (Fig. 6, A and B).
Figure 6.
Inh 1 and Inh 2 inhibit the NAPE-PLD activity of HEK293 cells. 0–20 μm of Inh1 (A) or Inh 2 (B) were added to confluent wells of HEK293 cells plated in 96-well plates and incubated for 30 min. PED-A1 was then added, and NAPE-PLD activity was determined by change in fluorescence (excitation/emission, 488/530 nm) over 25 min and then normalized to control. The points represent means ± S.E. (n = 6, three wells on two replicate days). IC50 and 95% confidence interval calculated using log(inhibitor) versus normalized response–variable slope analysis.
From these studies, we conclude that symmetrically substituted dichlorophene are potent inhibitors of NAPE-PLD in cultured cells and show significant selectivity for NAPE-PLD versus other tissue lipases. Both Inh 1 and Inh 2 can therefore be used for cultured cell experiments to test the effects of NAPE-PLD inhibition. Although both inhibitors are relatively selective for NAPE-PLD compared with other lipases, both are known to have other off-target effects not related to lipases, so care must be taken to ensure that nontoxic concentrations are used because both inhibitors are known anti-helminth, antibacterial, and antifungal agents.
Experimental procedures
Reagents
Recombinant hexahistidine-tagged mouse Nape-pld was expressed and purified as described previously (43). N-((6-(2,4-DNP)amino)hexanoyl)-1-(BODIPY™ FL C5)-2-hexyl-sn-glycero-3-phosphoethanolamine (PED-A1) and BODIPYTM FL C5 were purchased from Thermo Fisher Scientific. Rabbit anti-NAPE-PLD primary antibody was obtained from Abcam. Human embryonic kidney 293 (HEK293) cells were purchased from American Type Culture Collection (Manassas, VA). The 14 hit inhibitor compounds used for confirmation were purchased from MicroSource Discovery Systems, Inc. Dulbecco's modified Eagle's medium (DMEM), optiMEM (1×) reduced serum medium, and heat-inactivated fetal bovine serum (HI-FBS) were from Gibco Chemicals. S. chromofuscus PLD (ScPld), p-nitrophenylacetate, lithocholic acid, 2-acetazolamide, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Millipore–Sigma. 1-AT, NOG, and 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin were obtained from Cayman Chemicals (Ann Arbor, MI). Standard protein molecular weight marker was purchased from Invitrogen. Nickel nitrilotriacetic acid–agarose was from GE Healthcare. Deidentified, surplus human serum was provided by the Vanderbilt Clinical Pharmacology Phlebotomy Core. The Spectrum Collection compound library was provided by the Vanderbilt University High-Throughput Screening Core facility and was originally obtained from MicroSource (Gaylordville, CT). The collection includes a wide range of biologically active and structurally diverse compounds consisting of active drugs (50%), natural products (30%), and other bioactive components (20%).
Initial Nape-pld activity assay
Recombinant mouse Nape-pld or boiled Nape-pld was diluted in assay buffer containing 50 mm Tris-HCl, pH 8.0, and 1% NOG to a final concentration of 1.0 μm and incubated with 10 μm PED-A1. Fluorescence (excitation/emission, 488/530 nm; fixed bandwidth, 16 nm) was measured at 1-min time points for 2 h in 96-well plate reader (Synergy H1 hybrid multimode reader, BioTeK).
NAPE-PLD activity assay optimization
Km for PED-A1 hydrolysis by recombinant mouse Nape-pld was determined by enzyme kinetics. Briefly, Nape-pld was diluted in assay buffer to a final concentration of 4.5 μg ml−1, and enzyme kinetics was performed in a 384-well plate using 0–20 μm PED-A1. Fluorescence (excitation, 482 ± 18 nm; emission, 536 ± 20 nm) was measured for 15 min using a Wavefront Panoptic kinetic imaging plate reader. The effects of varying concentrations of NOG (0–1%) were measured using final concentration of 4 μm PED-A1 and 4.5 μg ml−1 Nape-pld. For the DMSO tolerance test, DMSO (35–900 nl) was transferred onto an empty 384-well plate using an Echo liquid handler. Then 30 μl of 4.5 μg ml−11 enzyme diluted in assay buffer containing 0.4% NOG (w/v) was dispensed into the DMSO-containing plate using a Bravo liquid handler robot, followed by 5 μl of PED-A1 (final concentration, 4 μm), and fluorescence kinetics (excitation/emission, 482/536 nm) were measured for 15 min. The consistency of replicates across the 384-plate was determined using the final optimized concentrations of DMSO (1.6%, v/v), NOG (0.4%, w/v), and 4.5 μg ml−1 Nape-pld, with replicates of the NAPE-PLD inhibitor, LCA, and dispersed in checkerboard pattern across the plate (“Checkerboard Assay”), and then 5 μl of PED-A1 (final concentration, 4 μm) was added. The Z′ factor for LCA inhibition across the plate was calculated as Z = 1 − [3(X + X′)/(Y − Y′)], where Y and Y′ are the mean values of PED-A1 produced fluorescence in the absence and presence, respectively, of LCA as a control inhibitor at initial rate of reaction. X and X′ are standard deviations of PED-A1–produced fluorescence.
High-throughput screen
Screening was performed on 2,388 compounds. 35 nl of 10 mm test compounds and 140 nl of 100% DMSO were combined in black-walled, clear-bottomed 384-well plates using an Echo Acoustic liquid handler, resulting in a final compound concentration of 10 μm for the primary screen. 175 nl of 20 mm LCA or 175 nl of DMSO were added to control wells as inhibited and uninhibited signal controls, respectively. 30 μl of 4.5 μg ml−1 Nape-pld enzyme in buffer solution with 0.4% NOG (w/v) was dispensed in each well using an Agilent Bravo robotic liquid handler. The plates were incubated at 37 °C for 1 h. The assay was initiated by the Panoptic instrument's internal Bravo robotic liquid handler when it dispensed 5 μl of PED-A1 (final concentration, 4 μm) in buffer solution (1.6% DMSO final, v/v) to each well. Changes in fluorescence (excitation/emission, 482/536 nm) were measured for 15 min. Initial slope measurements used slope from t = 30–100 s.
Concentration-response curve
The IC50 for each hit compound was determined using similar conditions as for screening except that the concentration of inhibitor was varied from 0 to 200 μm. The averaged initial slope measurements for NAPE-PLD activity in the absence of inhibitor were set as 100% activity, and the normalized percentage of activity was calculated by dividing the initial slope observed with each concentration of inhibitor by this value. Fitting of the response curve and calculation of the IC50 and 95% confidence interval were performed using log(inhibitor) versus normalized response–variable slope analysis in GraphPad Prism version 7.04.
Rapid dilution assay
This assay was performed using the methods of Castellani et al. (37). Briefly, 81 μg ml−1 recombinant Nape-pld was preincubated with Inh 1 or Inh 2 at 50 μm for 1 h, and then the samples were diluted 100-fold with 50 mm Tris-HCl (pH 8.0) immediately prior to addition of 4 μm PED-A1. Fluorescence was measured at 1-min intervals for 30 min.
Cytotoxicity
Effect of compounds on viability of HEK293 cells were determined using MTT assay to measure changes in cellular redox state. Briefly, 10 × 104 HEK293 cells/well were seeded in 24-well plates for 24 h. The cells were treated with a concentration of each compound that represented 500% of the IC50 concentration determined in the concentration-response curve assay (5× IC50) prepared in opti-MEM reduced serum medium containing 1% HI-FBS for 24 h. After treatment, the medium was removed, and 300 μl of 0.5 mg ml−1 MTT solution was added. After 3 h, the medium was removed, the purple crystals were dissolved in 100 μl of DMSO, 80 μl of dissolved crystal solution was transferred to 96-well plate, and absorbance was measured at 590 nm. The percentage of viability was normalized to cells treated with vehicle (DMSO) only.
Cellular NAPE-PLD activity
HEK293 cells in DMEM buffer containing 4.5 g liter−1 glucose, 4 mm l-glutamine, 1 mm sodium pyruvate, and phenol red and supplemented with 10% HI-FBS were seeded at ∼20,000 cells/well in tissue culture-treated, clear-bottomed, black-walled, 96-well plates. After reaching ∼95% confluency (48 h), the medium was removed and replaced with similar DMEM buffer (70 μl/well) except without phenol red or HI-FBS. 5 μl of inhibitor or vehicle solutions were then added to each well to generate final concentrations of inhibitors of 0.2–20 μm. 30 min after addition of inhibitors, 5 μl of 56 μm PED-A1 was added to each well (final concentration, 3.5 μm), and fluorescence signal (excitation/emission, 488/530 nm) was recorded every minute for 25 min in Synergy H1 plate reader. NAPE-PLD activity was measured as the increase in fluorescence from time = 1–25 min and normalized to average activity found in absence of inhibitor. For each concentration of inhibitor, three replicate wells on two separate days were measured (n = 6 wells total), and the means ± S.E. were determined.
Counter-screens and specificity screens
All assays were carried out in 96-well plates. To assess effect of compounds on BODIPY fluorescence, 4 μm of BODIPY-FL C5 was incubated with or without 10 μm of each compound in total 80 μl of 50 mm Tris-HCl (pH 8.0), and fluorescence (excitation/emission, 482/536 nm) was measured for 30 min. To assess the effects of compound on S. chromofuscus phospholipase D (ScPld) activity, 0.1 μm ScPLD was incubated with 0–200 μm of Inh 1 or Inh 2 in 70 μl of 50 mm Tris-HCl (pH 8.0) for 1 h, PED-A1 was added to 4 μm, and fluorescence (excitation/emission, 488/530 nm) was measured by Synergy H1 plate reader for 15 min. For assays assessing the effect of compounds on serum enzymes, deidentified surplus human serum was obtained from the Vanderbilt Clinical Pharmacology Phlebotomy Core and diluted 1:9 (i.e. 10-fold dilution factor) using 50 mm Tris-HCl (pH 8.0) to generate a diluted serum stock solution. Pan-lipase activity was measured using a lipase assay kit (Cayman Chemical, 700640), which uses 1-AT as substrate (final concentration, 23 μm), and the released thioglycerol was detected using thiol detection agent (7-dethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin; final concentration, 50 μm). Recombinant mouse Nape-pld (4.5 mg liter−1) did not hydrolyze 1-AT to any appreciable extent, whereas the bovine milk lipoprotein lipase supplied in the assay kit (diluted according to manufacturer's recommendation of 10 μl of supplied stock diluted with 140 μl of assay buffer) gave robust 1-AT hydrolysis (Fig. S6A). To determine the Km for 1-AT hydrolysis activity in diluted serum, 1-AT concentration was varied from 0 to 90 μm. This gave an estimated Km of 21.5 ± 3.4 μm (Fig. S6B). To measure effect of inhibitors on serum lipase activity, 5 μl of Inh 1, Inh 2, or vehicle (DMSO) (final inhibitor concentration, 0–200 μm) was added to each well containing 60 μl of 50 mm Tris-HCl (pH 8.0), and 10 μl of diluted serum stock solution and was incubated together for 30 min. 5 μl of a solution containing 1-AT and thiol detection agent (7-dethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin; final concentrations, 23 and 50 μm, respectively) was then added, and the change in fluorescence (excitation/emission, 380/515 nm) was monitored using Synergy H1 plate reader for 30 min. 100% activity was set as the fluorescence detected for vehicle only. Serum carbonic anhydrase activity was measured using a carbonic anhydrase assay kit (Biovision Inc., K472), which uses p-nitrophenylacetate (1 mm) as substrate, and the released p-nitrophenol was measured by absorbance at 405 nm. The positive control carbonic anhydrase provided in the assay kit robustly hydrolyzed p-nitrophenylacetate, whereas recombinant Nape-pld (4.5 mg liter−1) did not to any appreciable extent (Fig. S7A). Varying p-nitrophenylacetate concentration from 0 to 5 mm gave an estimated Km of 2 mm using diluted serum (Fig. S7B). A known carbonic anhydrase inhibitor, acetazolamide (1 mm), robustly inhibited p-nitrophenylacetate hydrolysis activity of diluted serum (Fig. S7C). To assess the effect of inhibitors on serum carbonic anhydrase activity, 5 μl of Inh 1, Inh 2, or vehicle (DMSO) (final inhibitor concentration, 0–200 μm) was added to 60 μl of 50 mm Tris-HCl (pH 8.0) and 10 μl of diluted serum stock solution per well in 96-well plate and incubated together for 30 min. 5 μl of p-nitrophenylacetate (final concentration, 1 mm) was then added, and hydrolysis by carbonic anhydrase was measured as the change in absorbance at 405 nm in Synergy H1 plate reader.
Statistical analysis
In all reactions, the IC50 was calculated using log(inhibitor) versus normalized response–variable slope analysis in GraphPad Prism version 7.04.
Author contributions
G. A., C. R. F., P. V., C. D. W., and S. S. D. conceptualization; G. A., P. V., and S. S. D. data curation; G. A., J. E. Z., P. V., and S. S. D. formal analysis; G. A., J. E. Z., and Z. M. investigation; G. A., P. V., C. D. W., and S. S. D. methodology; G. A., J. E. Z., and S. S. D. writing-original draft; J. E. Z., C. R. F., P. V., C. D. W., and S. S. D. writing-review and editing; C. R. F. and C. D. W. resources; P. V. and S. S. D. supervision; P. V. and S. S. D. project administration; C. D. W. software; S. S. D. funding acquisition.
Supplementary Material
Acknowledgments
The WaveFront Biosciences Panoptic kinetic imaging plate reader is housed and managed within the Vanderbilt High-Throughput Screening Core Facility, an institutionally supported core, and was funded by National Institutes of Health Shared Instrumentation Grant 1S10OD021734. The Microsource Spectrum Collection was distributed by the Vanderbilt High-Throughput Screening Core Facility with assistance from Corbin Whitwell. The High-Throughput Screening Core receives support from the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center, which in turn receives support through National Institutes of Health Grant P30 CA68485.
This work was supported by a Discovery Grant from the Vanderbilt Diabetes Center (to S. S. D.) and by National Institutes of Health Grants DK105847 (to C. R. F.) and T32GM065086 (J. E. Z.). C. D. W. is an owner of WaveFront Biosciences, the manufacturer of the Panoptic kinetic imaging plate reader used in these studies. P. V. receives compensation from the sales of the Panoptic through WaveFront Biosciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7.
- NAE
- N-acyl-ethanolamide
- NAPE
- N-acyl-phosphatidylethanolamine
- PLD
- phospholipase D
- LCA
- lithocholic acid
- PE
- phosphatidylethanolamine
- HTS
- high-throughput screening
- NOG
- N-octyl-β-d-glucoside
- Inh
- inhibitor compound
- 1-AT
- 1-arachidonyl-thiolglycerol
- DMEM
- Dulbecco's modified Eagle's medium
- HI-FBS
- heat-inactivated fetal bovine serum
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Chanda D., Neumann D., and Glatz J. F. C.. 2019) The endocannabinoid system: overview of an emerging multi-faceted therapeutic target. Prostaglandins Leukot. Essent. Fatty Acids 140, 51–56 10.1016/j.plefa.2018.11.016 [DOI] [PubMed] [Google Scholar]
- 2. Brown J. D., Karimian Azari E., and Ayala J. E.. 2017) Oleoylethanolamide: a fat ally in the fight against obesity. Physiol. Behav. 176, 50–58 10.1016/j.physbeh.2017.02.034 [DOI] [PubMed] [Google Scholar]
- 3. Tsuboi K., Uyama T., Okamoto Y., and Ueda N.. 2018) Endocannabinoids and related N-acylethanolamines: biological activities and metabolism. Inflamm. Regen. 38, 28 10.1186/s41232-018-0086-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hansen H. S. 2014) Role of anorectic N-acylethanolamines in intestinal physiology and satiety control with respect to dietary fat. Pharmacol. Res. 86, 18–25 10.1016/j.phrs.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 5. Wei D., Allsop S., Tye K., and Piomelli D.. 2017) Endocannabinoid signaling in the control of social behavior. Trends Neurosci. 40, 385–396 10.1016/j.tins.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DiPatrizio N. V., and Piomelli D.. 2015) Intestinal lipid-derived signals that sense dietary fat. J. Clin. Invest. 125, 891–898 10.1172/JCI76302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Marzo V. 2008) Targeting the endocannabinoid system: to enhance or reduce? Nat. Rev. Drug. Discov. 7, 438–455 10.1038/nrd2553 [DOI] [PubMed] [Google Scholar]
- 8. Godlewski G., Offertáler L., Wagner J. A., and Kunos G.. 2009) Receptors for acylethanolamides-GPR55 and GPR119. Prostaglandins Other Lipid Mediat. 89, 105–111 10.1016/j.prostaglandins.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodríguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., and Piomelli D.. 2003) Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-α. Nature 425, 90–93 10.1038/nature01921 [DOI] [PubMed] [Google Scholar]
- 10. Lo Verme J., Gaetani S., Fu J., Oveisi F., Burton K., and Piomelli D.. 2005) Regulation of food intake by oleoylethanolamide. Cell Mol. Life Sci. 62, 708–716 10.1007/s00018-004-4494-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thabuis C., Tissot-Favre D., Bezelgues J. B., Martin J. C., Cruz-Hernandez C., Dionisi F., and Destaillats F.. 2008) Biological functions and metabolism of oleoylethanolamide. Lipids 43, 887–894 10.1007/s11745-008-3217-y [DOI] [PubMed] [Google Scholar]
- 12. Piomelli D. 2013) A fatty gut feeling. Trends Endocrinol. Metab. 24, 332–441 10.1016/j.tem.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romano A., Tempesta B., Provensi G., Passani M. B., and Gaetani S.. 2015) Central mechanisms mediating the hypophagic effects of oleoylethanolamide and N-acylphosphatidylethanolamines: different lipid signals? Front. Pharmacol. 6, 137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodríguez de Fonseca F., Navarro M., Gómez R., Escuredo L., Nava F., Fu J., Murillo-Rodríguez E., Giuffrida A., LoVerme J., Gaetani S., Kathuria S., Gall C., and Piomelli D.. 2001) An anorexic lipid mediator regulated by feeding. Nature 414, 209–212 10.1038/35102582 [DOI] [PubMed] [Google Scholar]
- 15. Nielsen M. J., Petersen G., Astrup A., and Hansen H. S.. 2004) Food intake is inhibited by oral oleoylethanolamide. J. Lipid Res. 45, 1027–1029 10.1194/jlr.C300008-JLR200 [DOI] [PubMed] [Google Scholar]
- 16. Terrazzino S., Berto F., DalleCarbonare M., Fabris M., Guiotto A., Bernardini D., and Leon A.. 2004) Stearoylethanolamide exerts anorexic effects in mice via down-regulation of liver stearoyl-coenzyme A desaturase-1 mRNA expression. FASEB J. 18, 1580–1582 10.1096/fj.03-1080fje [DOI] [PubMed] [Google Scholar]
- 17. Diep T. A., Madsen A. N., Holst B., Kristiansen M. M., Wellner N., Hansen S. H., and Hansen H. S.. 2011) Dietary fat decreases intestinal levels of the anorectic lipids through a fat sensor. FASEB J. 25, 765–774 10.1096/fj.10-166595 [DOI] [PubMed] [Google Scholar]
- 18. Ueda N., Okamoto Y., and Tsuboi K.. 2005) Endocannabinoid-related enzymes as drug targets with special reference to N-acylphosphatidylethanolamine-hydrolyzing phospholipase D. Curr. Med. Chem. 12, 1413–1422 10.2174/0929867054020918 [DOI] [PubMed] [Google Scholar]
- 19. Uyama T., Ikematsu N., Inoue M., Shinohara N., Jin X. H., Tsuboi K., Tonai T., Tokumura A., and Ueda N.. 2012) Generation of N-acylphosphatidylethanolamine by members of the phospholipase A/acyltransferase (PLA/AT) family. J. Biol. Chem. 287, 31905–31919 10.1074/jbc.M112.368712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogura Y., Parsons W. H., Kamat S. S., and Cravatt B. F.. 2016) A calcium-dependent acyltransferase that produces N-acyl phosphatidylethanolamines. Nat. Chem. Biol. 12, 669–671 10.1038/nchembio.2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okamoto Y., Morishita J., Tsuboi K., Tonai T., and Ueda N.. 2004) Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem. 279, 5298–5305 10.1074/jbc.M306642200 [DOI] [PubMed] [Google Scholar]
- 22. Pettinati I., Brem J., Lee S. Y., McHugh P. J., and Schofield C. J.. 2016) The chemical biology of human metallo-β-lactamase fold proteins. Trends Biochem. Sci. 41, 338–355 10.1016/j.tibs.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Okamoto Y., Morishita J., Tsuboi K., Miyatake A., and Ueda N.. 2006) Functional analysis of the purified anandamide-generating phospholipase D as a member of the metallo-β-lactamase family. J. Biol. Chem. 281, 12325–12335 10.1074/jbc.M512359200 [DOI] [PubMed] [Google Scholar]
- 24. Hussain Z., Uyama T., Tsuboi K., and Ueda N.. 2017) Mammalian enzymes responsible for the biosynthesis of N-acylethanolamines. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1546–1561 10.1016/j.bbalip.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 25. Elphick M. R., and Egertová M.. 2005) The phylogenetic distribution and evolutionary origins of endocannabinoid signalling. In Cannabinoids (Pertwee R. G., ed) pp. 283–297, Springer, Berlin: [DOI] [PubMed] [Google Scholar]
- 26. Leishman E., Mackie K., Luquet S., and Bradshaw H. B.. 2016) Lipidomics profile of a NAPE-PLD KO mouse provides evidence of a broader role of this enzyme in lipid metabolism in the brain. Biochim. Biophys. Acta 1861, 491–500 10.1016/j.bbalip.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leung D., Saghatelian A., Simon G. M., and Cravatt B. F.. 2006) Inactivation of N-acyl phosphatidylethanolamine phospholipase D reveals multiple mechanisms for the biosynthesis of endocannabinoids. Biochemistry 45, 4720–4726 10.1021/bi060163l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inoue M., Tsuboi K., Okamoto Y., Hidaka M., Uyama T., Tsutsumi T., Tanaka T., Ueda N., and Tokumura A.. 2017) Peripheral tissue levels and molecular species compositions of N-acyl-phosphatidylethanolamine and its metabolites in mice lacking N-acyl-phosphatidylethanolamine-specific phospholipase D. J. Biochem. 162, 449–458 10.1093/jb/mvx054 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z., Zhang Y., Guo L., Dosoky N., de Ferra L., Peters S., Niswender K. D., and Davies S. S.. 2017) Leptogenic effects of NAPE require activity of NAPE-hydrolyzing phospholipase D. J. Lipid Res. 58, 1624–1635 10.1194/jlr.M076513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okamoto Y., Wang J., Morishita J., and Ueda N.. 2007) Biosynthetic pathways of the endocannabinoid anandamide. Chem. Biodivers. 4, 1842–1857 10.1002/cbdv.200790155 [DOI] [PubMed] [Google Scholar]
- 31. Simon G. M., and Cravatt B. F.. 2010) Characterization of mice lacking candidate N-acyl ethanolamine biosynthetic enzymes provides evidence for multiple pathways that contribute to endocannabinoid production in vivo. Mol. Biosyst. 6, 1411–1418 10.1039/c000237b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petersen G., Sørensen C., Schmid P. C., Artmann A., Tang-Christensen M., Hansen S. H., Larsen P. J., Schmid H. H., and Hansen H. S.. 2006) Intestinal levels of anandamide and oleoylethanolamide in food-deprived rats are regulated through their precursors. Biochim. Biophys. Acta 1761, 143–150, discussion 141–142 10.1016/j.bbalip.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 33. Di Marzo V. 2018) New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 17, 623–639 10.1038/nrd.2018.115 [DOI] [PubMed] [Google Scholar]
- 34. Orio L., Alen F., Pavón F. J., Serrano A., and García-Bueno B.. 2018) Oleoylethanolamide, neuroinflammation, and alcohol abuse. Front. Mol. Neurosci. 11, 490 10.3389/fnmol.2018.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Margheritis E., Castellani B., Magotti P., Peruzzi S., Romeo E., Natali F., Mostarda S., Gioiello A., Piomelli D., and Garau G.. 2016) Bile acid recognition by NAPE-PLD. ACS Chem. Biol. 11, 2908–2914 10.1021/acschembio.6b00624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., and Tanaka K.. 2002) Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298, 714–719 10.1016/S0006-291X(02)02550-0 [DOI] [PubMed] [Google Scholar]
- 37. Castellani B., Diamanti E., Pizzirani D., Tardia P., Maccesi M., Realini N., Magotti P., Garau G., Bakkum T., Rivara S., Mor M., and Piomelli D.. 2017) Synthesis and characterization of the first inhibitor of N-acylphosphatidylethanolamine phospholipase D (NAPE-PLD). Chem. Commun. (Camb.) 53, 12814–12817 10.1039/c7cc07582k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitnaul L. J., Tian J., Burton C., Lam M.-H., Zhu Y., Olson S. H., Schneeweis J. E., Zuck P., Pandit S., Anderson M., Maletic M. M., Waddell S. T., Wright S. D., Sparrow C. P., and Lund E. G.. 2007) Fluorogenic substrates for high-throughput measurements of endothelial lipase activity. J. Lipid Res. 48, 472–482 10.1194/jlr.D600041-JLR200 [DOI] [PubMed] [Google Scholar]
- 39. Miceli M., Casati S., Ottria R., Di Leo S., Eberini I., Palazzolo L., Parravicini C., and Ciuffreda P.. 2019) Set-up and validation of a high throughput screening method for human monoacylglycerol lipase (MAGL) based on a new red fluorescent probe. Molecules 24, E2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Darrow A. L., Olson M. W., Xin H., Burke S. L., Smith C., Schalk-Hihi C., Williams R., Bayoumy S. S., Deckman I. C., Todd M. J., Damiano B. P., and Connelly M. A.. 2011) A novel fluorogenic substrate for the measurement of endothelial lipase activity. J. Lipid Res. 52, 374–382 10.1194/jlr.D007971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brideau C., Gunter B., Pikounis B., and Liaw A.. 2003) Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 8, 634–647 10.1177/1087057103258285 [DOI] [PubMed] [Google Scholar]
- 42. Brown H. A., Thomas P. G., and Lindsley C. W.. 2017) Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat. Rev. Drug Discov. 16, 351–367 10.1038/nrd.2016.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo L., Gragg S. D., Chen Z., Zhang Y., Amarnath V., and Davies S. S.. 2013) Isolevuglandin-modified phosphatidylethanolamine is metabolized by NAPE-hydrolyzing phospholipase D. J. Lipid Res. 54, 3151–3157 10.1194/jlr.M042556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.