Abstract
Vascular calcification (VC) is characterized by calcium deposition inside arteries and is closely associated with the morbidity and mortality of atherosclerosis, chronic kidney disease, diabetes, and other cardiovascular diseases (CVDs). VC is now widely known to be an active process occurring in vascular smooth muscle cells (VSMCs) involving multiple mechanisms and factors. These mechanisms share features with the process of bone formation, since the phenotype switching from the contractile to the osteochondrogenic phenotype also occurs in VSMCs during VC. In addition, VC can be regulated by epigenetic factors, including DNA methylation, histone modification, and noncoding RNAs. Although VC is commonly observed in patients with chronic kidney disease and CVD, specific drugs for VC have not been developed. Thus, discovering novel therapeutic targets may be necessary. In this review, we summarize the current experimental evidence regarding the role of epigenetic regulators including histone deacetylases and propose the therapeutic implication of these regulators in the treatment of VC.
Keywords: histone deacetylase, histone modifiers, epigenetic regulator, vascular calcification, vascular smooth muscle cells, post-translational modification
1. Introduction
Vascular calcification (VC) is common in patients with chronic kidney disease, atherosclerosis, and diabetes. It is characterized by the accumulation of calcium phosphate products inside the vascular walls [1]. VC is an independent risk factor for cardiovascular diseases (CVDs) [2] and is highly associated with CVD mortality. VC can be categorized into two types: intimal calcification and medial calcification. Intimal calcification develops in the intimal layer of the vascular walls in patients with atherosclerosis, whereas medial calcification occurs in the medial layer of the vascular walls in patients with chronic kidney disease, diabetes, hypertension, and osteoporosis [3].
Previously, VC, an elevation of calcium phosphate crystals in the vascular walls, was considered to be the passive result of VSMC death. However, it is now known to be an active process involving various regulators of bone formation [4]. During the progression of VC, the expression of contractile markers such as smooth muscle 22 alpha (SM22α) and alpha smooth muscle actin (α-actin) are downregulated, whereas the expression of bone-related factors such as runt-related transcription factor 2 (RUNX2), msh homeobox (MSX), bone morphogenetic proteins (BMPs), and osteocalcin are upregulated [1]. Thus, smooth muscle cells (SMCs) undergo phenotype switching from the contractile phenotype to the osteochodrogenic phenotype [5]. Although VC could be treated with conventional therapeutics used for chronic kidney disease, CVD, and osteoporosis [3], it is still common in patients with chronic kidney disease and atherosclerosis. Thus, it is critical to find novel key regulators in the pathogenesis of VC and new therapeutic targets.
Previously, epigenetic regulators were reported to be involved in the progression of diverse diseases such as cancer, autoimmune diseases, and neurological and psychological diseases [6] and showed potential as therapeutic targets [7]. Epigenetic refers to a heritable phenotype resulting from an alteration in gene expression without a change in DNA sequence [8]. Epigenetic regulations such as DNA methylation, histone modification, and noncoding RNAs (ncRNAs) [9] can be involved in the development of VC. In this review, we will focus on these three regulators with an emphasis on the role of histone modification.
2. Mechanisms of Vascular Calcification
VC is a complex and interactive process involving various calcification-related factors, apoptosis, mitochondrial dysfunction, and senescence [4,5]. Procalcifying factors such as BMP2 and RUNX2 can promote VC, whereas anticalcifying factors such as OPG and OPN may inhibit VC. Moreover, various signaling pathways such as BMP signaling and the Wnt/β-catenin pathway are involved in the development of VC [10]. Additionally, age-related factors including cell death and mitochondrial metabolism may affect VC.
2.1. Procalcifying Factors
The BMPs are members of the transforming growth factor-β (TGF-β) family that are reported to be involved in embryogenesis, organogenesis, and osteoblast differentiation [11]. Although there are more than 30 different types of BMPs [12], we will focus on BMP-2, which is well-known for its procalcifying properties. BMP-2 may promote VC by activating muscle segment homeobox2 (MSX2) and inhibiting matrix Gla protein (MGP). It may also promote apoptosis of vascular smooth muscle cells (VSMCs) [13]. Derwall et al. found that suppressing BMP-2 inhibited the formation of atheromas and VC in low-density lipoprotein receptor-deficient (LDLR−/−) mice [14]. On the other hand, VC was promoted in BMP-2 transgenic mice [15]. BMP signaling is activated when BMP-2 binds to type I and II BMP-2 receptor and phosphorylates SMAD (small mothers against decapentaplegic) 1/5/8. Phosphorylated SMAD 1/5/8 can enter the nucleus and further activate downstream calcification genes, such as RUNX2 and MSX2 [12,13]. Additionally, it was revealed that BMP-2 and MSX2 can activate the Wnt/β-catenin pathway and induce VSMC calcification [16,17]. The Wnt/β-catenin pathway is one of the major osteoinductive signaling pathways in VC [10]. WNT, a ligand protein, binds to the cell membrane receptors of the lipoprotein receptor-related protein 5/6 and Frizzled family and activates β-catenin. β-catenin translocates to the nucleus and activates downstream target genes, including calcification genes [18].
RUNX2 is a transcription factor involved in osteoblast differentiation and bone formation [19]. Although the expression of RUNX2 is low in normal vessels, it is highly expressed in calcified vessels, indicating that RUNX2 plays an important role in VC [20]. RUNX2 has been shown to induce calcification in VSMCs in vitro and was found to be critical in VSMC calcification induced by oxidative stress [21].
MSX2 is an essential transcription factor for bone formation and organogenesis [22,23]. MSX2 is upregulated in calcified arteries of diabetic mice, patients with diabetes, dyslipidemia, and vascular disease. On the other hand, downregulating MSX2 and MSX1 inhibits VC in diabetic LDLR mice [24].
Alkaline phosphatase (ALP) is a metalloenzyme and another key player of osteogenesis. It is widely expressed in various tissue, but is highly expressed in liver, kidney, and bone. Tissue-nonspecific ALP is activated by BMP2 and vitamin D agents, and this activation of ALP results in osteogenic transdifferentiation of VSMCs. ALP can act as a pyrophosphatase or can catalyze the hydrolysis of phosphomonoesters releasing inorganic phosphate (Pi). Elevated ALP levels alter osteoblasts and cause bone disease [25].
2.2. Anticalcifying Factors
Osteoprotegerin (OPG), which is a receptor for tumor necrosis factor, is reported to be involved in bone resorption [26]. OPG is expressed in diverse tissues such as heart, lung, and kidney and is involved in regulating the immune system [27]. OPG knockout (KO) mice exhibit severe osteoporosis and medial calcification [28], whereas transgenic mice overexpressing OPG show reduced osteoclast differentiation and enhanced bone mass [26]. The serum OPG level positively correlates with the severity of VC [29]. OPG acts as a decoy and interrupts the binding of receptor activator of NF-κB ligand (RANKL) to RANK, which ultimately inhibits osteoclast differentiation [27].
Osteopontin (OPN) is a phosphoprotein that is generally found in bone and teeth [30,31]. Although OPN is not commonly present in normal blood vessels, it is abundant in calcified atherosclerotic plaques and aortic valves [32,33,34]. Speer et al. revealed that MGP and OPN KO mice (MGP−/−, OPN−/−) develop more severe VC than MGP KO mice (MGP−/−, OPN+/+), which suggests that OPN has an inhibitory role in VC [35].
MGP, a vitamin K-dependent protein, is expressed in endothelial cells and VSMCs of normal blood vessels. Expression of MGP was reported to be downregulated in calcified vessels [36]. The severity of coronary artery calcification is negatively correlated with MGP levels in patients with suspected coronary artery diseases [37]. In addition, MGP levels are reduced in an in vitro VSMC calcification model [38], and MGP KO mice exhibit severe calcification [39]. These findings suggest that MGP acts as an inhibitor of VC. Moreover, MGP can interrupt BMP2 signaling by binding directly to BMP2, which can prevent BMP2 from binding to BMP receptors [39].
Fetuin-A, a glycoprotein expressed in adipose tissue and liver, is another key regulator in bone formation and resorption [40]. It inhibits VC by binding to calcium phosphate in the blood [41]. Fetuin-A KO mice demonstrate severe calcification in heart, kidney, skin, and tongue [42]. Coronary artery calcification scores are directly associated with fetuin-A levels [43,44]. In addition, administration of fetuin-A reduces calcification in vitro [45].
2.3. Other Factors Affecting VC
Apoptosis is another factor that directly influences VC. After cell death, the cell releases cellular DNA that precipitates calcium phosphate products and results in VC. Apoptosis occurs early in VSMC before VSMC calcification. Apoptotic bodies produced after cell death are high in calcium and are ultimately included in the formation of calcium phosphate crystals [46]. Suppression of apoptosis through caspase inhibitors reduces calcification and vesicles including calcium [47].
The phenotype of VSMCs can be influenced by changes in mitochondrial metabolism such as increased proliferation in pulmonary artery SMCs [48]. Mitochondrial dysfunction can contribute to the development of atherosclerosis [49]. Mitochondrial damage results in reduced adenosine triphosphate production and impaired respiratory chain function, which ultimately causes cellular malfunction and initiate atherosclerosis, a progressive disease that may lead to VC. Mitochondrial dysfunction also induces VC through the apoptotic pathway by promoting casapase-9, which releases cytochrome c [50].
VC was reported to be associated with aging and VSMC senescence [51]. Senescent cells release cytokines, proteases, and growth factors and stop mitosis. Osteogenic markers such as RUNX2, collagen 1, and ALP are increased in senescent VSMC cells in vitro, and medial calcification and upregulated RUNX2 levels are observed in the in vivo aging mouse model [52]. It is well known that senescence-related VC also increases microvesicles, a subset of extracellular vesicles. Microvesicles carry calcium, calcification nucleation-related proteins, and calcium-binding annexins and promote osteoblast transformation of VSMCs to induce VC [53]
Endothelial cell can transform into mesenchymal stem cells, multipotent cells, by undergoing endothelial-to-mesenchymal transition (EndoMT). When mesenchymal stem cells are exposed to stimuli, they can differentiate into fibroblast, adipocyte, osteoblast, and chondrocyte [54]. Recent studies reported that TGF-β or BMP2-stimulated valvular endothelial cells undergo EndoMT process. These TGF-β or BMP2-stimulated valvular endothelial cells transform into osteoblast-like cells by increasing ALP expression during EndoMT, which eventually result in VC [55]. In contrast, suppression of EndoMT in valvular endothelial cells can inhibit VC [56].
3. Epigenetic Regulation in Vascular Smooth Muscle Cells
Chromatin is composed mostly of DNA and protein and is regulated by epigenetic mechanisms [57]. Epigenetic regulation can alter gene expression during transcription, post-transcription, translation, and post-translation. Thus, DNA, DNA-binding proteins, and histones can be modified by epigenetic mechanisms [6]. In the sections below, we discuss three major epigenetic regulations: DNA methylation, histone modification, and ncRNAs.
3.1. DNA Methylation
DNA methylation is processed by a group of DNA methyltransferase (DNMTs), such as DNMT1, DNMT3a, and DNMT3b. DNA methylation occurs when a methyl group is added to cytosine in cytosine phosphate guanine (CpG) islands. Transcriptional repressors bind with the methylated CpGs and hinder gene transcription. Several reports have demonstrated that DNA methylation plays a critical role in various diseases including atherosclerosis [58,59], vascular remodeling [60], and cardiovascular diseases [61]. For example, Dunn et al. showed that inhibiting DNA methylation could attenuate atherosclerosis by ameliorating endothelial dysfunction and restoring the mechanosensitive endothelial gene expression induced by oscillatory shear stress [62]. VSMCs can undergo phenotype switching from the contractile phenotype to the synthetic phenotype by environmental causes such as stress. Such phenotype switching is manifest as increased matrix stiffness and inflammation. Recently, the effects of matrix stiffness in VSMC phenotype and function were reported. Xie et al. reported that DNMT1 is reduced in arterial stiffness models such as acute aortic injury, the chronic kidney failure model, and calcified atherosclerotic lesions in human carotid arteries [63]. Inhibition of DNMT1 facilitates arterial stiffening in vivo and promotes osteogenic transdifferentiation and cellular stiffening of VSMCs in vitro. Mechanistically, DNMT1 elicits effects on contractility by regulating the promoter activities of smooth muscle α-actin and SM22α. Zhang et al. reported that DNMT3 is a direct target of miR-143, and DNMT3a regulates the hypermethylation of miR-143 promoter in homocysteine-induced VSMC proliferation [64]. These results show that DNA methylation plays important roles in vascular smooth muscle remodeling.
The role of DNA methylation in VC has been extensively investigated. Montes de Oca et al. found that DNMT activity and methylation of the promoter region of the SMC-specific protein SM22α are increased in high-phosphate-induced calcified VSMCs [65]. Azechi et al. [66] and Xie et al. [63] also demonstrated that downregulating DNMT1 expression facilitates Pi-induced arterial calcification by upregulating ALP expression and downregulating the DNA methylation level of the ALP promoter region. Lin et al. showed that miR-204 is decreased in human aortic VSMCs with high Pi treatment and that it acts as a negative regulator during the process of osteoblastic differentiation [67]. Additionally, the methylation level of miR-204 is regulated by DNMT3a. DNMT3a was revealed as a direct target of miR-204 and to be involved in the process of human aortic SMCs differentiation. These results suggest that arterial calcification is regulated by the miR-204/DNMT3a regulatory circuit. Another study reported that downregulation of miR-34b in calcified VSMCs and arteries is associated with hypermethylation of CpG sites in the upstream region of miR-34b DNA regulated by DNMT3a. Lin et al. showed that miR-34b targets Notch1 in the regulation of VC [68]. Recently, da Silva et al. found that treatment of human aortic SMCs with calcium and phosphate increases DNMT3b, resulting in hypermethylation of osterix and bone sialoprotein (BSP), which ultimately leads to VC [69] (Table 1).
Table 1.
Role of DNA methyltransferases (DMNTs) in vascular calcification.
3.2. Histone Post-Translational Modification
The post-translational modifications (PTMs) of histones are important epigenetic regulatory mechanisms because the gene expression patterns can be inherited. PTMs can occur in histone proteins, which are located inside the nucleosome of chromatin. The N-terminal tails of histones are unstructured and can be modified [70]. The types of modifications that can occur in histone tails include acetylation, ADP-ribosylation, deimination, isomerization, methylation, phosphorylation, ubiquitination, and sumoylation [71]. Histone modifications have vital roles in transcriptional regulation, such as DNA replication, alternative splicing [72], DNA repair [73], and chromosome condensation [67]. Furthermore, not only histones but also other multiple proteins can undergo PTMs, resulting in changes in biological and pathological functions. In particular, the lysine residues of proteins undergo various PTMs such as methylation, ubiquitination, sumoylation, and acetylation. Lysine acetylation is a reversible PTM that can be conserved among different species. Lysine acetylation alters the charge on lysine residues and modifies the protein structure, thereby influencing enzyme activity, DNA-binding affinity, and protein stability [74,75].
3.3. Noncoding RNA
NcRNAs are functional RNAs that generally do not translate protein, and they are variable in length and structure. In addition to microRNAs (miRNAs), one of the extensively investigated ncRNAs, long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) have recently emerged as epigenetic regulators [76,77]. MiRNAs are single-stranded, endogenous, small noncoding RNAs of 16–25 nucleotides in length. They bind to the 3′-untranslated region of target mRNAs and mediate the translational repression or degradation of target transcripts. They play key roles in cellular processes such as differentiation, proliferation, migration, and apoptosis [78]. LncRNAs are longer than 200 nucleotides which are mostly transcribed by RNA polymerase II. They are spliced, capped at the 5′ end, and polyadenylated at the 3′ end [79]. Function of lncRNAs can vary depending on their localization. Nuclear lncRNAs can regulate gene transcription, whereas cytoplasmic lncRNAs may act as miRNA sponges [80]. CircRNAs are mainly produced by back-splicing reaction and are variable in length. CircRNAs are usually localized in cytoplasm and work as miRNA sponges [81]. To date, many studies have reported that ncRNAs act as both positive and negative regulators of the progression of VC.
Several reports demonstrated that ncRNAs positively regulate VC. For example, Gui et al. reported that miR-135a*, miR-712*, miR-714, and miR-762 promote matrix mineralization in Ca/Pi-stimulated VSMCs by inhibiting calcium efflux proteins, NCX1, PMCA1, and NCKX4 [82]. In addition, Xia et al. revealed that miR-2861 and miR-3960 increase matrix mineralization in β-glycerophosphate-stimulated VSMCs by suppressing HDAC5 and Hoxa2 [83]. Moreover, Panizo et al. found that miR-29b enhances VC and downregulates the osteoblast differentiation inhibitors ACVR2A, CTNNBIP1, and HDAC4 in Ca/Pi-stimulated VSMCs [84]. Liu et al. revealed that miR-32 inhibits PTEN through the activation of PI3K signaling, thereby increasing RUNX2 in β-glycerophosphate-stimulated VSMCs [85]. LncRNA-ES3 was reported to increase expression of calcification-related genes and BMF by acting as a miR-34c-5p sponge in high-glucose-stimulated human aorta VSMCs [86].
On the other hand, ncRNAs can also act as negative regulators of VC. miR-125b was revealed to downregulate osterix in osteogenic medium-stimulated HCASMCs [87]. Chao et al. showed that serum miR-125b can be used to predict the severity of VC [88]. miR-204 was reported to inhibit RUNX2 in β-glycerophosphate-stimulated VSMCs [89]. Yanagawa et al. reported that miR-141 inhibits BMP-2 in TGF-β-induced valvular interstitial cells (VICs) [90]. Additionally, miR-30b and miR-30c were revealed to inhibit RUNX2 in BMP-2 or β-glycerophosphate-stimulated VSMCs [91]. Jeong et al. reported that lncRNA Lrrc75a-as1 inhibits osteoblast-related genes in Pi-stimulated RVSMCs [92]. Circ-Samd4a was revealed to reduce VC by acting as miRNA sponges and regulating Camsap2 and Flna in Pi-stimulated RVSMCs [93]. These studies imply that discovering novel ncRNAs that control VC is important and that modulating these ncRNAs can be considered in the prevention and treatment of VC.
4. Role of Histone Modification in Vascular Calcification
Lysine acetylation of proteins is catalyzed by lysine acetyltransferases (KATs), which transfer the acetyl group of acetyl-coA to the ε-amino group of a lysine residue [94,95]. The reverse reaction is catalyzed by lysine deacetylases (KDACs), which comprise histone deacetylases (HDACs) and sirtuins (class III HDACs). Typically, acetylation of lysine in the tails of histones in the nucleosome weakens the interaction of these histones in the DNA by neutralization and reduction of the positive charge of the lysine residue, which then induces the activation of gene transcription [96]. Conversely, histone deacetylation alters the electrostatic properties of chromatin in a manner that favors the repression of gene transcription [97].
4.1. Histone Acetyltransferase in Vascular Calcification
Mammalian KATs can be categorized into two types, type A and type B, depending on their cellular localization. Type A KATs are nuclear KATs, whereas type B are cytoplasmic KATs [98]. Type A KATs mainly regulate the transcription of genes and are divided into five subgroups: MYST, GANT, p300/CBP, nuclear receptor coactivator family, and basal transcription factors [99,100]. Compared with type A KATs, only a few type B KATs have been reported, including KAT1 and KAT4 [101]. Type B KATs acetylate free histones and cytoplasmic substrates. The acetylation of lysines modifies the function of the protein by altering their structure or affinity to other binding partners. Thus, the acetylation of lysines regulates a variety of diseases, including neurological disorders [102], cancer [103], and CVDs [104]. Of the KATs, p300 has been extensively studied. p300 is a transcriptional coactivator that regulates gene expression by scaffolding, bridging, or activating intrinsic KAT [105].
Sierra et al. reported that p300 binds to RUNX2, resulting in an increase in osteocalcin promoter activity to induce osteogenesis in osteoblasts [106]. In aortic valvular calcification models, activation of p300 increases the acetylation of histones (H3 and H4) or RUNX2, thereby upregulating osteoblast-related genes such as osteocalcin and ALP. By contrast, C646, the most potent inhibitor of p300, reduces aortic valve calcification by suppressing the acetylation of H3 and H4. C646 is a promising anticancer agent owing to its ability to promote terminal differentiation and to induce growth arrest in cancer [107,108]. This suggests that C646 can also be used for the treatment of VC.
4.2. Histone Deacetylase in Vascular Calcification
Histone deacetylation is mediated by a family of HDACs that consists of 18 different HDAC molecules. Depending on their structural similarities and substrates, those 18 HDACs are divided into four different classes. HDAC1, 2, 3, and 8 belong to class I, whereas HDAC4, 5, 6, 7, 9, and 10 are class II. SIRT 1-7 (also known as HDAC12-18), class III HDACs, differ from the other HDACs in that their activities are dependent on NAD+. Lastly, there is only one HDAC in class IV, which is HDAC11 [109]. The deacetylase domain is conserved in these HDACs [110]. It is noteworthy that the HDACs themselves can undergo diverse PTMs including phosphorylation, ubiquitination, and even acetylation [109]. HDACs have been revealed to regulate multiple pathological processes, including cancer [111,112,113], apoptosis [114], inflammation [115], VC [116], and CVD processes [117]. Our group also reported the role of HDACs in the development of cardiac hypertrophy and heart failure [118,119,120,121,122,123]. Although the roles of HDACs in VC have not been extensively investigated, it is likely that HDACs work as transcriptional repressors of their targets and that the overall effects of those HDACs may depend on the properties of their target genes.
4.2.1. Class I HDACs
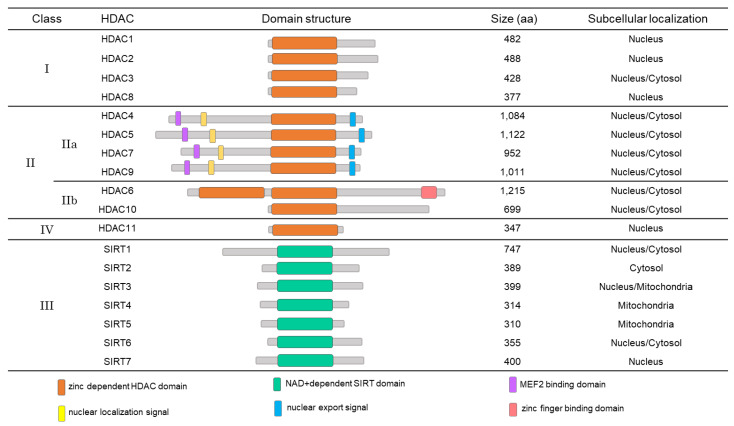
HDAC1, 2, and 8 are mainly localized in the nucleus. On the other hand, HDAC3 is sometimes detected in the cytoplasm (Figure 1). Although class I HDACs have short regulatory domains compared with those of the class IIa HDACs, class I HDACs are more actively modified than are the other classes of HDACs. Among the class I HDACs, HDAC1 and HDAC2 have similar sequences and structures [109]. Many studies have revealed that HDACs are correlated with the development of CVD or CVD risk factors, including hypertension, cardiomyopathy [118,122,124], myocardial infarction [125], and heart failure [126,127]. Among the class I HDACs, we discovered that HDAC1 is involved in the development of VC in both high-phosphate-treated VSMCs and vitamin D3-injected mice [116]. In diverse VC models, the degradation of HDAC1 protein is increased and protein amount is significantly reduced. HDAC1 K74 is a ubiquitination target residue, and polyubiquitination of K74 mediates the VC-induced protein degradation. We also found that MDM2 E3 ligase is involved in HDAC1 polyubiquitination of VC; overexpression of MDM2 promotes VC, whereas knockdown of MDM2 inhibits VC because degradation of HDAC1 is suppressed. In addition, we observed that MDM2 is upregulated in the intimal and medial layers of calcified human coronary arteries. It is noteworthy that RG 7112, an MDM2 inhibitor, completely inhibits VC. These results suggested that the MDM2/HDAC1 axis is a novel signaling pathway that regulates VC in VSMCs, and that targeting this axis can be considered to prevent VC [116] (Table 2). Like C646, considering that RG 7112 is under extensive investigation as an anticancer drug [128,129], the extension of its use to VC would be of great interest.
Figure 1.
Classification and domain structure of the histone deacetylases (HDACs). The colored boxes on the domain structure are indicated as follows: orange, zinc-dependent HDAC domain; mint, NAD+-dependent SIRT domain; purple, MEF2 binding domain; yellow, nuclear localization signal; blue, nuclear export signal; and peach, zinc finger binding domain. HDAC, histone deacetylase, SIRT, sirtuin; and MEF2, myocyte enhancer factor 2.
Table 2.
Role of histone deacetylases (HDACs) in vascular calcification.
| Type of Class | HDACs | Expression | PTMs | Regulator | Function | Reference |
|---|---|---|---|---|---|---|
| Class I | HDAC1 | Downregulation | Ubiquitination | MDM2 | Decrease Runx2 | [116] |
| HDAC2 | N.C. | |||||
| HDAC3 | N.C. | |||||
| HDAC8 | N.C. | |||||
| Class IIa | HDAC4 | Upregulation | Phosphorylation | SIK and Pdlim7 | Increase Runx2 and OPN | [130] |
| HDAC5 | Downregulation | miR-2861 | Decrease Runx2 | [83] | ||
| HDAC7 | N.C. | |||||
| HDAC9 | Upregulation | Increase Runx2 | [131] | |||
| Class IIb | HDAC6 | Downregulation | ATF4 | Decrease Runx2 | [132] | |
| HDAC10 | N.D. | |||||
| Class III | SIRT1 | Downregulation | p21 | Decrease Runx2 | [133] | |
| Class IV | HDAC11 | N.D. |
Abbreviation: N.C., not changed; N.D., not detectable; MDM2, Mouse double minute 2 homolog; PTM, post-translational modification; SIK, salt-inducible kinase; and Pdlim7, protein ENIGMA.
4.2.2. Class II HDACs
The class II HDACs can be divided into two subclasses: class IIa (HDAC4, 5, 7, and 9) and class IIb (HDAC6 and HDAC10). The class IIa HDACs are characterized by a long N-terminal domain extension in addition to their catalytic domain, whereas the class IIb HDACs such as HDAC6 and HDAC10 contain two catalytic domains [134]. The class IIa HDACs differ from the other HDACs because they contain binding sites for myocyte enhancer factor 2 (MEF2), a nuclear localization signal in the N-terminal domain and a nuclear export signal in the C-terminal domain (Figure 1). The expression of the class II HDACs is more tissue-specific compared with that of the other classes of HDACs. Like the class I HDACs, the functions of the class II HDACs are also regulated by PTMs, including phosphorylation, acetylation, ubiquitination, and sumoylation. Class II HDACs have crucial roles in the development of the heart, vasculature, muscle, bone, brain, and immune system [135].
HDAC4 is a key regulator of cell growth [136], differentiation [137], proliferation [138], and migration [139] in various cell types and participates in physiologic and pathologic processes such as proliferation and migration during neointimal hyperplasia [138]. Recently, Zheng et al. reported that suppression of HDAC4 inhibits the proliferation of VSMCs induced by platelet-derived growth factor-BB [140]. They demonstrated that HDAC4 plays a key role in cellular processes. Ren et al. found that HDAC4 is required for the proliferation and migration of VSMCs to regulate neointimal hyperplasia. In addition, HDAC4 was shown to have important functions in bone formation and cartilage development. Whole-body deletion of HDAC4 by conventional knocking out in mice results in premature ossification as a result of ectopic hypertrophy of chondrocytes [141]. Moreover, double knockout of HDAC4 and RUNX2 causes low bone mass. Thus, HDAC4 is critical in bone formation. Since VC and bone formation share similar features, one can easily assume that HDAC4 may regulate VC as well. Recently, Abend et al. reported that HDAC4 is upregulated early in VC, which significantly increases the expression of RUNX2 and OPN, the markers of VC [130]. Knocking-down of HDAC4 in VSMCs reduces VC. Class IIa HDAC4 can be exported from the nucleus in response to activation of stimulus-dependent kinase. Likewise, the salt-inducible kinase (SIK)-mediated phosphorylation of HDAC4 keeps HDAC4 in the cytoplasm. In the cytoplasm, HDAC4 binds with adaptor protein ENIGMA (Pdlim7) to promote VC. Conversely, pan-SIK inhibitors translocate HDAC4 from the cytoplasm to the nucleus, which results in the inhibition of VC (Table 2).
HDAC5 undergoes nuclear-cytoplasmic shuttling and acts as an important regulator in cellular and epigenetic processes that underlie the progression of human disease, including cardiac diseases [142,143,144,145], tumorigenesis [146], and VC [83]. Xia et al. reported that HDAC5, a direct target of miR-2861, inhibits RUNX2 expression in β-glycerophosphate-induced osteogenic transdifferentiation of VSMCs [83]. Several studies revealed that HDAC4 or HDAC5, class II HDACs, promote Smad ubiquitin regulatory factor 1-mediated degradation of RUNX2, which induces BMP-2-mediated osteoblast differentiation and bone formation [147,148]. We also identified that miR-mediated inhibition of HDAC5 can induce VC by decreasing the inhibition of RUNX2 in high-phosphate-stimulated VSMCs (unpublished data). These studies show that the pathogenesis of VC is similar to the process of bone formation.
HDAC9, as a member of the HDAC IIa family, regulates diverse diseases including neurodegenerative diseases [149,150], cancer [151,152], and cardiovascular diseases [153,154]. Recently, Malhotra et al. revealed that HDAC9 is associated with abdominal aortic calcification using genome-wide association meta-analysis [131]. HDAC9 was increased in osteogenic medium-induced human aortic SMCs, which increased RUNX2 expression and reduced contractility. Using siRNA, knocking-down of HDAC9 significantly reduced VC by inhibiting RUNX2. Furthermore, HDAC9 and MGP KO mice (HDAC9−/− and MGP−/−) show reduced VC compared with MGP KO mice (MGP−/−). These results demonstrate that HDAC9 may function as an activator of atherosclerosis and VC. However, further studies need to be considered to elucidate the precise mechanism of HDAC9 in the regulation of VC.
Several reports revealed that HDAC6, a member of the HDAC IIb subfamily, regulates the progression of diverse diseases including cancer [155], bone remodeling [156], and vascular remodeling [157]. Fu et al. showed that HDAC6 was significantly reduced in human calcified aortic valves, and knocking-down of HDAC6 increased RUNX2 expression and induced aortic calcification in VICs [132]. Rao and colleagues found that HDAC6, the regulator of endoplasmic reticulum stress in breast cancer cells, inhibits endoplasmic reticulum stress through deacetylation of glucose-related protein 78 (GRP 78) and suppression of activating transcription factor 4 (ATF4)-mediated osteogenesis [158]. Previous studies reported that endoplasmic reticulum stress is related to aortic valve calcification through ATF signaling [159,160]. ATF4 is a sensor of endoplasmic reticulum stress and also works as a transcription factor that involves the osteogenic pathway [161]. According to previous reports, it is likely that pulmonary arterial hypertension, an elevation of pulmonary arterial pressure due to heart, lung, or systemic disorders [162], may be related to VC [163,164]. Ruffenach et al. claimed that VC may be associated with pulmonary artery stiffness in patients with pulmonary arterial hypertension [163]. Boucherat et al. recently reported that expression of HDAC6 protein is increased in response to upregulated HSP90 in pulmonary artery SMCs isolated from patients with pulmonary arterial hypertension [165]. Hsp90 is a central coordinator involved in many diverse cellular pathways upon stress [166]. Weisell et al. reported that Hsp90 is downregulated in patients with calcified aortic valve diseases [167]. Taken together, these results suggest that HDAC6 may regulate VC in aortic valve disease and pulmonary arterial hypertension.
4.2.3. Class III HDACs
Compared with class I and II HDACs, sirtuins (SIRTs), class III HDACs, are nicotinamide adenine dinucleotide (NAD+)-dependent deacetylating enzymes that were first identified in yeast. Sirtuins have highly conserved NAD+-binding domains with an ~250 amino acid core and variable amino- and carboxy-terminal extensions. Mammals possess seven sirtuins with different properties of subcellular localization, enzymatic activity, and binding targets. SIRT1 and SIRT6 are mainly localized in the nucleus, but they can translocate into the cytoplasm under specific conditions [168]. SIRT2 resides mostly in the cytoplasm, whereas SIRT3, SIRT4, and SIRT5 are localized in mitochondria. In contrast, SIRT7 is localized in the nucleus (Figure 1). SIRTs regulate essential molecular pathways in eubacteria, archaea, and eukaryotes and are involved in stress resistance, apoptosis, aging, senescence, and inflammation. SIRT1 and SIRT6 are characterized for their protective roles against inflammation, vascular aging, heart disease, and atherosclerotic plaque development [169]. SIRT1 antagonizes p53-induced cellular senescence in mouse embryo fibroblast (MEF) and suppresses oxidative stress-mediated premature senescence in vascular endothelial cells [170]. Senescent cells are also detected in Pi-induced VSMCs and in the calcified areas of an adenine-induced chronic renal failure rat model [133,171]. Therefore, it seems that cellular senescence is positively correlated with VC. Also, Takemura et al. reported that high-phosphate-induced senescence is related to downregulation of SIRT1 expression, leading to p21 activation. Further evidence showed that knocking-down of SIRT1 accelerates the high-phosphate-induced cellular senescence and VC in SMCs [133,171]. These results suggest that SIRT1 induces arterial calcification by inhibiting osteoblastic transdifferentiation such as RUNX2 expression in conditions such as aging and diabetes (Table 2).
5. Crosstalk between Osteogenic Transcription Factors and HDACs in Noncalcified Smooth Muscle Cells
Since the mechanism of VC is similar to the process of osteoblast differentiation, for a better understanding of the roles of HDACs in VC, in this section, we review studies of HDACs in ossification other than in SMCs. Osteogenic transcription factors such as RUNX2, MSX2, OPG, and OPN are expressed in multiple tissues and regulate the progression of diverse diseases such as cancer [172,173], fibrosis [174,175], and bone diseases [176]. Moreover, HDACs act as important epigenetic regulators in various diseases and play a critical role in cellular processes [109]. To date, many studies have reported that there is crosstalk between HDACs and other osteogenic transcription factors in various cells. Manzotti et al. revealed that HDAC1 and HDAC6 activate RUNX2 transcription in cancer cells, such as thyroid TPC1 cells and MDA-MB231 cells by binding to the RUNX2 P1 promoter. In particular, HDAC6 interacts with RUNX2 to regulate downstream targets such as TERF1 and platelet-derived growth factor β in thyroid TPC1 cells [155]. In addition, Huang et al. reported that both HDAC1 and HDAC2 are required for RUNX2 to increase TGF-β-induced OPN expression, which promotes epithelial-to-mesenchymal transition-mediated invasion in nonsmall cell lung cancer cells [177]. Among the class I HDACs, HDAC1 and HDAC3 interact with RUNX2, respectively, during osteoblast differentiation and inhibit RUNX2-mediated osteogenesis, including osteocalcin in osteogenic cells like MC3T3-E1, ROS12/2.8, and primary bone marrow cells [178,179]. In addition, Yoshizawa et al. reported that MSX2 interacts with RUNX2/Osf2 and prevents ossification by recruiting HDAC1 in mesenchymal progenitor C3H10T1/2 fibroblastic cells [180]. In studies of class II HDACs, miR-29b-mediated suppression of HDAC4 was shown to promote osteoblast differentiation by activating RUNX2 in MC3T3-E1 cells [181]. Another study conducted by Westendorf et al. revealed that the binding between HDAC6 and RUNX2 in the nucleus induces osteoblast differentiation [156]. On the other hand, the interaction between HDAC7 and RUNX2 suppresses osteoblast maturation by inhibiting RUNX2 activity in osteoblast lineage cells such as multipotent progenitor C2C12 cells [182].
6. HDAC Inhibitor/Activator and Therapeutic Application in Vascular Calcification
Since diverse diseases are regulated by HDACs, HDAC inhibitors may be potential therapeutic agents. Various HDAC inhibitors have been evaluated as therapeutic agents for myelodysplastic syndromes, advanced leukemia, neurologic diseases, immune disorders, and CVDs [98,183]. To date, six HDAC inhibitors such as vorinostat, belinostat, romidepsin, sodium butyrate, valproic acid (VPA), and panobinostat are approved by the Food and Drug Administration as drugs for diverse diseases. However, studies of HDAC inhibitors in VC and CVDs are still in preclinical phases.
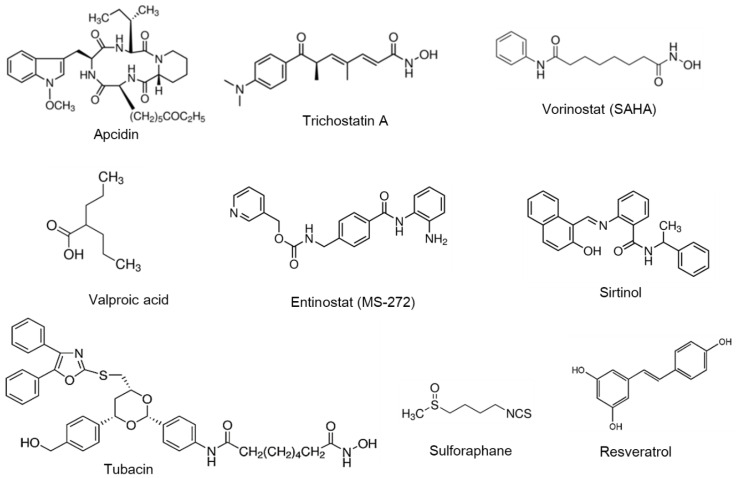
Most HDAC inhibitors act as pan-HDAC inhibitors that block more than two HDACs. HDAC inhibitors can be classified into four different classes: cyclic peptide (e.g., apicidin), hydroxamic acid (e.g., vorinostat and trichostatin A), short-chain fatty acid (e.g., VPA), and benzamide (e.g., entinostat and sirtinol). Other novel classified chemical classes are as follows: isothiocyanate (e.g., sulforaphane) and stilbenoid (e.g., resveratrol) (Figure 2). In general, it is widely accepted that HDAC inhibitors may be used in the treatment of epilepsy, angiogenesis, cardiac hypertrophy, heart failure, fibrosis, and myocardial infarction [98]. Some HDAC inhibitors including apicidin, trichostatin A, vorinostat, tubacin, and sirtinol are reported to aggravate VC [107,116,132,171,184], whereas other inhibitors such as entinostat, VPA, and sulforaphane have shown potential as therapeutic agents in VC [185,186,187]. Since studies of these HDAC inhibitors in VC are at the preclinical level (Table 3), further studies are necessary to determine their use as therapeutic agents.
Figure 2.
Structure of HDAC inhibitors and activators. The chemical structures of the HDAC inhibitors and activators discussed in this review are depicted.
Table 3.
Characteristics of HDAC inhibitors and activators in vascular calcification.
| HDAC Modulator | Chemical Classification | HDAC Specificity | Effect on VC | Study Model | Cell Type | Mechanism | References |
|---|---|---|---|---|---|---|---|
| HDAC inhibitor | |||||||
| Apicidin | Cyclic peptide | Class I | Promote | 2 mM Pi | RVSMCs | Reduces HDAC1 and increases Runx2 | [116] |
| Trichostatin A | Hydroxamic acid | Class I, II, IV | Promote | 3 mM Pi | HASMCs | Reduces HDAC1 and increases Runx2 | [184] |
| Vorinostat | Hydroxamic acid | Class I, II, IV | Promote | 1.5 mM Ca2+ and 2 mM Pi | pVICs | Enhances H4 acetylation, Runx2, and OPN | [107] |
| Tubacin | Hydroxamic acid | HDAC6 | Promote | 10 mM β-GP, 10 nM DM, 4 μg/mL vitamin D3, and 8 mM CaCl2 | hVICs | Activates ER stress and increases Runx2 | [132] |
| Sirtinol | Benzamide | SIRT1, SIRT2 | Promote | 5 mM β-GP and 2.6 mM CaCl2 | HCASMCs | Increase senescence and promotes Runx2 and osteocalcin | [171] |
| Entinostat | Benzamide | Class I, IV | Inhibit | 10 nM DM, 10 mM 10 mM β-GP, and 50 mg/mL AA | pVICs | Reduce Wnt signaling, Runx2, and ALP | [185] |
| Valproic acid | Short-chain fatty acids | Class I, IIa | Inhibit | 3mM Pi | BASMCs | Enhances autophagy and reduces Runx expression | [186] |
| Sulforaphane | Isothiocyanate | Class III | Inhibit | 770 mg β-GP, 11 mg L-AA, and 10 nM DM | RVSMCs | Reduces oxidative stress and Runx2 | [187] |
| HDAC activator | |||||||
| Resveratrol | Stilbenoid | SIRT1, SIRT2 | Inhibit | 3.2mM Pi or 770 mg β-GP, 11 mg L-AA, and 10 nM DM | HASMCs or RVSMCs | Reduces senescence and Runx2 and increases Nrf2 | [133,188] |
Pi, inorganic phosphate; β-GP, β-glycerophosphate; DM, dexamethasone; AA, ascorbic acid; RVSMCs, rat vascular smooth muscle cells; HASMCs, human aortic smooth muscle cells; pVICs, porcine aortic valvular interstitial cells; hVICs, human aortic valvular interstitial cells; HCASMCs, human coronary artery smooth muscle cells; BASMCs, bovine aortic smooth muscle cells; and Nrf2, nuclear factor-erythroid related factor 2.
6.1. HDAC Regulators that May Exaggerate VC
Trichostatin A, which is a pan-HDAC inhibitor, and apicidin, which is a class I HDAC inhibitor, increase VC by activating RUNX2 in Pi-induced VSMCs [116,184]. Additionally, TSA enhances VC in a vitamin D3-induced in vivo VC model. As mentioned above, the E3 ligase MDM2 is dramatically increased after the induction of VC, which enhances polyubiquitination and results in the subsequent degradation of HDAC1. The loss of HDAC1 activity plays a crucial role in the progression of VC. Hence, HDAC inhibitors may accelerate VC in vivo [116].
Vorinostat (also known as SAHA) is as a pan-HDAC inhibitor that is widely used as a treatment for epilepsy, cancer, and other diseases [189,190]. Vorinostat is reported to increase osteogenic differentiation through H4 acetylation and modulation of insulin/Akt/FoxO1 signaling in osteoblast cells [191]. Additionally, vorinostat increases acetylation of H3 and H4, which increases RUNX2 and ALP in high-calcium/phosphate-induced porcine aortic VICs (pVICs) [107].
Tubacin is an HDAC6 inhibitor that was reported to promote aortic valvular calcification by activating GRP78 acetylation in the endoplasmic reticulum. GRP78 acetylation induces osteogenic differentiation signaling, including ATF4 and RUNX2 expression in osteogenic medium-induced human aortic VICs (hVICs) [132]. This study demonstrated that tubacin has a procalcification role and suppresses HDAC6.
Sirtinol, an inhibitor of Sirt1 and Sirt2, reduces DNA replication and transcription in the hepatitis B virus. Recently, Takemura et al. reported the effect of sirtinol, which promotes high-glucose-induced VC by activating the expression of RUNX2 and OCN [133]. Moreover, sirtinol also increases mineralization as shown in alizarin red S staining of osteogenic-medium-induced SMCs [171].
6.2. HDAC Modulators that Can Block VC
Entinostat (also known as MS275) is a potent class I and IV HDAC inhibitor. It is currently in clinical trials for the treatment of cancers and has therapeutic effects on bone defects [192] and cardiovascular diseases including hypertension [193,194], cardiac hypertrophy [195], and heart failure [196]. In the osteogenic-medium-induced calcification models, Li et al. reported that entinostat reduces RUNX2 expression and ALP activity by downregulating Wnt signaling, which interacts with GSK-3β, β-catenin, and p-SMAD1/5/8 in calcified VICs [185].
VPA, a class I HDAC inhibitor, is used for the treatment of epilepsy and is in clinical trials for cancer [197]. Dai et al. showed that calcification stress-induced reactive oxygen species products potentiate autophagy. Interestingly, VPA inhibits calcium deposition by upregulating autophagy in phosphate-induced VSMCs and in the aortic walls of adenine-induced chronic kidney disease rats [186]. This result was the opposite of the previously reported phenomenon that inhibition of autophagy reduces VC. The authors explained that this finding may be caused by an increase in release of matrix vesicles. Thus, this study demonstrated that VPA acts as an inducer of autophagy and counteracts calcification by inhibiting matrix vesicle release [186].
Sulforaphane is an isothiocyanate derived from broccoli [198] that acts as an inhibitor of class I HDACs and HDAC6 in diverse cell types such as HCT116 [199,200], prostate cell lines [201], and human peripheral blood mononuclear cells [202]. Recently, Zhang et al. reported that sulforaphane decreased reactive oxygen species production by increasing nuclear factor-erythroid2-related factor-2 (NRF-2) expression and subsequently inhibiting RUNX2 expression in β-glycerophosphate-induced rat VSMCs [187]. These results demonstrated that sulforaphane may have an anticalcification role in oxidative stress-induced VC.
Resveratrol [203] is a polyphenol phytoalexin that is present in plants and that works as a SIRT1 (also known as HDAC 12) activator. It may prevent the progression of diverse diseases including neurodegenerative diseases, cancer, and cardiovascular diseases [204,205]. Takemura et al. reported that resveratrol also has protective effects on calcification by decreasing osteogenesis and has antioxidative effects by increasing NRF2 mRNA levels in a hyperphosphatemia-induced calcification model [133]. NRF2 has a protective role on calcification by counteraction of inflammation through the inhibition of oxidative stress and pro-inflammatory cytokines [206]. Many researchers have reported that oxidative stress can activate osteogenic signals such as RUNX2 in VSMCs [21]. Zhang et al. found that resveratrol effectively preserves oxidative injury by inhibiting VC through the regulation of SIRT-1 and NRF2 in rat aortic VSMCs [188]. However, further studies may be necessary to understand the regulatory mechanism of SIRT1 and NRF2 in VC.
7. Conclusions
VC is an interactive and complex process involving multiple factors such as pro- and anti-calcifying factors, apoptosis, mitochondrial dysfunction, cellular senescence, and endoMT. Since VC is an important risk factor for CVD, investigation of novel regulators may be needed to resolve VC. In the current review, we summarized and discussed the roles of epigenetic regulators including DNA methylation, HDACs, and ncRNAs and proposed therapeutic applications of the HDACs in the treatment of VC. DNA methylation and ncRNAs including miRNAs, lncRNAs, and circRNAs can act as positive or negative regulator of VC and may serve as novel therapeutic targets. Furthermore, these epigenetic regulators may crosstalk with each other to regulate VC. In addition, HDACs have several different classes and many subtypes, and their action may vary in the development of VC. Although some HDACs play pro-calcification roles, others demonstrate anti-calcification effects. Since the mechanism of action of HDAC inhibitors and activators may differ depending on the type of HDACs, further studies are required to understand the precise role of individual HDACs and to validate the effects of HDAC inhibitors or activators and to determine the optimal route of medication administration. Here, we summarized the roles of many epigenetic regulators in the development of VC. Although multiple complex mechanisms await further extensive investigations, this review will add new insights for the development of new therapeutics against VC.
Abbreviations
| ALP | alkaline phosphatase |
| ATF4 | activating transcription factor 4 |
| BMP | bone morphogenetic protein |
| BSP | bone sialoprotein |
| circRNA | circular RNA |
| CpG | cytopsin phosphate guanine |
| CVD | cardiovascular disease |
| DNMT | DNA methyltransferase |
| EndoMT | endothelial-to-mesenchymal transition |
| GRP 78 | glucose-related protein 78 |
| HDAC | histone deacetylase |
| KAT | lysine acetyl-transferase |
| KDAC | lysine deacetylase |
| KO | knockout |
| lncRNA | long noncoding RNA |
| MGP | matrix Gla protein |
| miRNA | microRNAs |
| MSX | msh homeobox |
| ncRNA | noncoding RNA |
| NRF | nuclear factor-erythroid2-related factor |
| OPG | Osteoprotegerin |
| OPN | Osteopontin |
| Pdlim7 | protein ENIGMA |
| Pi | inorganic phosphate |
| PTM | post-translational modification |
| RUNX2 | runt-related transcription factor 2 |
| SIK | salt-inducible kinase |
| SIRT | Sirtuin |
| SM22a | smooth muscle 22 alpha |
| SMC | smooth muscle cell |
| VC | vascular calcification |
| VIC | valvular interstitial cell |
| VPA | valproic acid |
| VSMC | vascular smooth muscle cells |
Author Contributions
Conceptualization, Y.-K.K. and H.K.; investigation, D.-H.K. and J.R.; writing—original draft preparation, D.-H.K. and J.R.; writing—review and editing, Y.-K.K. and H.K.; visualization, D.-H.K. and J.R.; supervision and project administration, Y.-K.K. and H.K.; and funding acquisition, D.-H.K. and H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) from the Ministry of Science, ICT and Future Planning (NRF- 2018R1A2B3001503, NRF-2019R1A4A1028534 and NRF-2020R1C1C1006507). The funder has no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bardeesi A.S.A., Gao J., Zhang K., Yu S., Wei M., Liu P., Huang H. A novel role of cellular interactions in vascular calcification. J. Transl. Med. 2017;15:95. doi: 10.1186/s12967-017-1190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sage A.P., Tintut Y., Demer L.L. Regulatory mechanisms in vascular calcification. Nat. Rev. Cardiol. 2010;7:528–536. doi: 10.1038/nrcardio.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M., Rementer C., Giachelli C.M. Vascular calcification: An update on mechanisms and challenges in treatment. Calcif. Tissue Int. 2013;93:365–373. doi: 10.1007/s00223-013-9712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizobuchi M., Towler D., Slatopolsky E. Vascular calcification: The killer of patients with chronic kidney disease. J. Am. Soc. Nephrol. Jasn. 2009;20:1453–1464. doi: 10.1681/ASN.2008070692. [DOI] [PubMed] [Google Scholar]
- 5.Durham A.L., Speer M.Y., Scatena M., Giachelli C.M., Shanahan C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018;114:590–600. doi: 10.1093/cvr/cvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moosavi A., Motevalizadeh Ardekani A. Role of Epigenetics in Biology and Human Diseases. Iran. Biomed. J. 2016;20:246–258. doi: 10.22045/ibj.2016.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heerboth S., Lapinska K., Snyder N., Leary M., Rollinson S., Sarkar S. Use of epigenetic drugs in disease: An overview. Genet. Epigenet. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg A.D., Allis C.D., Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Cancer Epigenetics for the 21st Century: What’s Next? Genes Cancer. 2011;2:604–606. doi: 10.1177/1947601911423096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voelkl J., Lang F., Eckardt K.U., Amann K., Kuro O.M., Pasch A., Pieske B., Alesutan I. Signaling pathways involved in vascular smooth muscle cell calcification during hyperphosphatemia. Cell Mol. Life Sci. 2019;76:2077–2091. doi: 10.1007/s00018-019-03054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D., Zhao M., Mundy G.R. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 12.Cai J., Pardali E., Sanchez-Duffhues G., ten Dijke P. BMP signaling in vascular diseases. Febs. Lett. 2012;586:1993–2002. doi: 10.1016/j.febslet.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 13.Hruska K.A., Mathew S., Saab G. Bone morphogenetic proteins in vascular calcification. Circ. Res. 2005;97:105–114. doi: 10.1161/01.RES.00000175571.53833.6c. [DOI] [PubMed] [Google Scholar]
- 14.Derwall M., Malhotra R., Lai C.S., Beppu Y., Aikawa E., Seehra J.S., Zapol W.M., Bloch K.D., Yu P.B. Inhibition of bone morphogenetic protein signaling reduces vascular calcification and atherosclerosis. Arter. Thromb. Vasc. Biol. 2012;32:613–622. doi: 10.1161/ATVBAHA.111.242594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa Y., Ikeda K., Akakabe Y., Koide M., Uraoka M., Yutaka K.T., Kurimoto-Nakano R., Takahashi T., Matoba S., Yamada H., et al. Paracrine osteogenic signals via bone morphogenetic protein-2 accelerate the atherosclerotic intimal calcification in vivo. Arter. Thromb. Vasc. Biol. 2010;30:1908–1915. doi: 10.1161/ATVBAHA.110.206185. [DOI] [PubMed] [Google Scholar]
- 16.Rong S., Zhao X., Jin X., Zhang Z., Chen L., Zhu Y., Yuan W. Vascular calcification in chronic kidney disease is induced by bone morphogenetic protein-2 via a mechanism involving the Wnt/beta-catenin pathway. Cell Physiol. Biochem. 2014;34:2049–2060. doi: 10.1159/000366400. [DOI] [PubMed] [Google Scholar]
- 17.Shao J.S., Cheng S.L., Pingsterhaus J.M., Charlton-Kachigian N., Loewy A.P., Towler D.A. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. J. Clin. Investig. 2005;115:1210–1220. doi: 10.1172/JCI24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson M.L., Rajamannan N. Diseases of Wnt signaling. Rev. Endocr. Metab. Disord. 2006;7:41–49. doi: 10.1007/s11154-006-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R., et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 20.Engelse M.A., Neele J.M., Bronckers A.L., Pannekoek H., de Vries C.J. Vascular calcification: Expression patterns of the osteoblast-specific gene core binding factor alpha-1 and the protective factor matrix gla protein in human atherogenesis. Cardiovasc. Res. 2001;52:281–289. doi: 10.1016/S0008-6363(01)00375-3. [DOI] [PubMed] [Google Scholar]
- 21.Byon C.H., Javed A., Dai Q., Kappes J.C., Clemens T.L., Darley-Usmar V.M., McDonald J.M., Chen Y. Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J. Biol. Chem. 2008;283:15319–15327. doi: 10.1074/jbc.M800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satokata I., Ma L., Ohshima H., Bei M., Woo I., Nishizawa K., Maeda T., Takano Y., Uchiyama M., Heaney S., et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 23.Wilkie A.O., Tang Z., Elanko N., Walsh S., Twigg S.R., Hurst J.A., Wall S.A., Chrzanowska K.H., Maxson R.E., Jr. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat. Genet. 2000;24:387–390. doi: 10.1038/74224. [DOI] [PubMed] [Google Scholar]
- 24.Cheng S.L., Behrmann A., Shao J.S., Ramachandran B., Krchma K., Bello Arredondo Y., Kovacs A., Mead M., Maxson R., Towler D.A. Targeted reduction of vascular Msx1 and Msx2 mitigates arteriosclerotic calcification and aortic stiffness in LDLR-deficient mice fed diabetogenic diets. Diabetes. 2014;63:4326–4337. doi: 10.2337/db14-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoppet M., Shanahan C.M. Role for alkaline phosphatase as an inducer of vascular calcification in renal failure? Kidney Int. 2008;73:989–991. doi: 10.1038/ki.2008.104. [DOI] [PubMed] [Google Scholar]
- 26.Simonet W.S., Lacey D.L., Dunstan C.R., Kelley M., Chang M.S., Luthy R., Nguyen H.Q., Wooden S., Bennett L., Boone T., et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/S0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 27.Collin-Osdoby P. Regulation of vascular calcification by osteoclast regulatory factors RANKL and osteoprotegerin. Circ. Res. 2004;95:1046–1057. doi: 10.1161/01.RES.0000149165.99974.12. [DOI] [PubMed] [Google Scholar]
- 28.Bucay N., Sarosi I., Dunstan C.R., Morony S., Tarpley J., Capparelli C., Scully S., Tan H.L., Xu W., Lacey D.L., et al. Osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12:1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jono S., Ikari Y., Shioi A., Mori K., Miki T., Hara K., Nishizawa Y. Serum osteoprotegerin levels are associated with the presence and severity of coronary artery disease. Circulation. 2002;106:1192–1194. doi: 10.1161/01.CIR.0000031524.49139.29. [DOI] [PubMed] [Google Scholar]
- 30.Giachelli C.M., Steitz S. Osteopontin: A versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19:615–622. doi: 10.1016/S0945-053X(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 31.Valdivielso J.M. Vascular calcification: Types and mechanisms. Nefrologia. 2011;31:142–147. doi: 10.3265/Nefrologia.pre2010.Nov.10754. [DOI] [PubMed] [Google Scholar]
- 32.Fitzpatrick L.A., Severson A., Edwards W.D., Ingram R.T. Diffuse calcification in human coronary arteries. Association of osteopontin with atherosclerosis. J. Clin. Investig. 1994;94:1597–1604. doi: 10.1172/JCI117501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirota S., Imakita M., Kohri K., Ito A., Morii E., Adachi S., Kim H.M., Kitamura Y., Yutani C., Nomura S. Expression of osteopontin messenger RNA by macrophages in atherosclerotic plaques. A possible association with calcification. Am. J. Pathol. 1993;143:1003–1008. [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda T., Shirasawa T., Esaki Y., Yoshiki S., Hirokawa K. Osteopontin mRNA is expressed by smooth muscle-derived foam cells in human atherosclerotic lesions of the aorta. J. Clin. Investig. 1993;92:2814–2820. doi: 10.1172/JCI116901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speer M.Y., McKee M.D., Guldberg R.E., Liaw L., Yang H.Y., Tung E., Karsenty G., Giachelli C.M. Inactivation of the osteopontin gene enhances vascular calcification of matrix Gla protein-deficient mice: Evidence for osteopontin as an inducible inhibitor of vascular calcification in vivo. J. Exp. Med. 2002;196:1047–1055. doi: 10.1084/jem.20020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyson K.L., Reynolds J.L., McNair R., Zhang Q., Weissberg P.L., Shanahan C.M. Osteo/chondrocytic transcription factors and their target genes exhibit distinct patterns of expression in human arterial calcification. Arter. Thromb. Vasc. Biol. 2003;23:489–494. doi: 10.1161/01.ATV.0000059406.92165.31. [DOI] [PubMed] [Google Scholar]
- 37.Jono S., Ikari Y., Vermeer C., Dissel P., Hasegawa K., Shioi A., Taniwaki H., Kizu A., Nishizawa Y., Saito S. Matrix Gla protein is associated with coronary artery calcification as assessed by electron-beam computed tomography. Thromb. Haemost. 2004;91:790–794. doi: 10.1160/TH03-08-0572. [DOI] [PubMed] [Google Scholar]
- 38.Mori K., Shioi A., Jono S., Nishizawa Y., Morii H. Expression of matrix Gla protein (MGP) in an in vitro model of vascular calcification. Febs. Lett. 1998;433:19–22. doi: 10.1016/S0014-5793(98)00870-9. [DOI] [PubMed] [Google Scholar]
- 39.Jono S., Nishizawa Y., Shioi A., Morii H. 1,25-Dihydroxyvitamin D3 increases in vitro vascular calcification by modulating secretion of endogenous parathyroid hormone-related peptide. Circulation. 1998;98:1302–1306. doi: 10.1161/01.CIR.98.13.1302. [DOI] [PubMed] [Google Scholar]
- 40.Schinke T., Amendt C., Trindl A., Poschke O., Muller-Esterl W., Jahnen-Dechent W. The serum protein alpha2-HS glycoprotein/fetuin inhibits apatite formation in vitro and in mineralizing calvaria cells. A possible role in mineralization and calcium homeostasis. J. Biol. Chem. 1996;271:20789–20796. doi: 10.1074/jbc.271.34.20789. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds J.L., Skepper J.N., McNair R., Kasama T., Gupta K., Weissberg P.L., Jahnen-Dechent W., Shanahan C.M. Multifunctional roles for serum protein fetuin-a in inhibition of human vascular smooth muscle cell calcification. J. Am. Soc. Nephrol. Jasn. 2005;16:2920–2930. doi: 10.1681/ASN.2004100895. [DOI] [PubMed] [Google Scholar]
- 42.Schafer C., Heiss A., Schwarz A., Westenfeld R., Ketteler M., Floege J., Muller-Esterl W., Schinke T., Jahnen-Dechent W. The serum protein alpha 2-Heremans-Schmid glycoprotein/fetuin-A is a systemically acting inhibitor of ectopic calcification. J. Clin. Investig. 2003;112:357–366. doi: 10.1172/JCI17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odamaki M., Shibata T., Takita T., Kumagai H. Serum fetuin-A and aortic calcification in hemodialysis patients. Kidney Int. 2005;68:2915. doi: 10.1111/j.1523-1755.2005.00583_13.x. [DOI] [PubMed] [Google Scholar]
- 44.Mehrotra R., Westenfeld R., Christenson P., Budoff M., Ipp E., Takasu J., Gupta A., Norris K., Ketteler M., Adler S. Serum fetuin-A in nondialyzed patients with diabetic nephropathy: Relationship with coronary artery calcification. Kidney Int. 2005;67:1070–1077. doi: 10.1111/j.1523-1755.2005.00172.x. [DOI] [PubMed] [Google Scholar]
- 45.El-Abbadi M., Giachelli C.M. Mechanisms of vascular calcification. Adv. Chronic Kidney Dis. 2007;14:54–66. doi: 10.1053/j.ackd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Coscas R., Bensussan M., Jacob M.P., Louedec L., Massy Z., Sadoine J., Daudon M., Chaussain C., Bazin D., Michel J.B. Free DNA precipitates calcium phosphate apatite crystals in the arterial wall in vivo. Atherosclerosis. 2017;259:60–67. doi: 10.1016/j.atherosclerosis.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds J.L., Joannides A.J., Skepper J.N., McNair R., Schurgers L.J., Proudfoot D., Jahnen-Dechent W., Weissberg P.L., Shanahan C.M. Human vascular smooth muscle cells undergo vesicle-mediated calcification in response to changes in extracellular calcium and phosphate concentrations: A potential mechanism for accelerated vascular calcification in ESRD. J. Am. Soc. Nephrol. Jasn. 2004;15:2857–2867. doi: 10.1097/01.ASN.0000141960.01035.28. [DOI] [PubMed] [Google Scholar]
- 48.Chiong M., Cartes-Saavedra B., Norambuena-Soto I., Mondaca-Ruff D., Morales P.E., Garcia-Miguel M., Mellado R. Mitochondrial metabolism and the control of vascular smooth muscle cell proliferation. Front. Cell Dev. Biol. 2014;2:72. doi: 10.3389/fcell.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu E., Calvert P.A., Mercer J.R., Harrison J., Baker L., Figg N.L., Kumar S., Wang J.C., Hurst L.A., Obaid D.R., et al. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation. 2013;128:702–712. doi: 10.1161/CIRCULATIONAHA.113.002271. [DOI] [PubMed] [Google Scholar]
- 50.Kim H., Kim H.J., Lee K., Kim J.M., Kim H.S., Kim J.R., Ha C.M., Choi Y.K., Lee S.J., Kim J.Y., et al. Alpha-Lipoic acid attenuates vascular calcification via reversal of mitochondrial function and restoration of Gas6/Axl/Akt survival pathway. J. Cell. Mol. Med. 2012;16:273–286. doi: 10.1111/j.1582-4934.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton D.G., Matsubara H., Ikeda K. Pathophysiology of vascular calcification: Pivotal role of cellular senescence in vascular smooth muscle cells. Exp. Gerontol. 2010;45:819–824. doi: 10.1016/j.exger.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Nakano-Kurimoto R., Ikeda K., Uraoka M., Nakagawa Y., Yutaka K., Koide M., Takahashi T., Matoba S., Yamada H., Okigaki M., et al. Replicative senescence of vascular smooth muscle cells enhances the calcification through initiating the osteoblastic transition. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1673–H1684. doi: 10.1152/ajpheart.00455.2009. [DOI] [PubMed] [Google Scholar]
- 53.Alique M., Ruiz-Torres M.P., Bodega G., Noci M.V., Troyano N., Bohorquez L., Luna C., Luque R., Carmona A., Carracedo J., et al. Microvesicles from the plasma of elderly subjects and from senescent endothelial cells promote vascular calcification. Aging. 2017;9:778–789. doi: 10.18632/aging.101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medici D., Kalluri R. Endothelial-mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin. Cancer Biol. 2012;22:379–384. doi: 10.1016/j.semcancer.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osman L., Yacoub M.H., Latif N., Amrani M., Chester A.H. Role of human valve interstitial cells in valve calcification and their response to atorvastatin. Circulation. 2006;114:I547–I552. doi: 10.1161/CIRCULATIONAHA.105.001115. [DOI] [PubMed] [Google Scholar]
- 56.Hjortnaes J., Shapero K., Goettsch C., Hutcheson J.D., Keegan J., Kluin J., Mayer J.E., Bischoff J., Aikawa E. Valvular interstitial cells suppress calcification of valvular endothelial cells. Atherosclerosis. 2015;242:251–260. doi: 10.1016/j.atherosclerosis.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibney E.R., Nolan C.M. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 58.Hiltunen M.O., Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arter. Thromb. Vasc. Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 59.Tabaei S., Tabaee S.S. DNA methylation abnormalities in atherosclerosis. Artif. Cells Nanomed. Biotechnol. 2019;47:2031–2041. doi: 10.1080/21691401.2019.1617724. [DOI] [PubMed] [Google Scholar]
- 60.Gluckman P.D., Hanson M.A., Buklijas T., Low F.M., Beedle A.S. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat. Rev. Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y., Zeng C. Role of DNA methylation in cardiovascular diseases. Clin. Exp. Hypertens. 2016;38:261–267. doi: 10.3109/10641963.2015.1107087. [DOI] [PubMed] [Google Scholar]
- 62.Dunn J., Qiu H., Kim S., Jjingo D., Hoffman R., Kim C.W., Jang I., Son D.J., Kim D., Pan C., et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Investig. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie S.A., Zhang T., Wang J., Zhao F., Zhang Y.P., Yao W.J., Hur S.S., Yeh Y.T., Pang W., Zheng L.S., et al. Matrix stiffness determines the phenotype of vascular smooth muscle cell in vitro and in vivo: Role of DNA methyltransferase 1. Biomaterials. 2018;155:203–216. doi: 10.1016/j.biomaterials.2017.11.033. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H.P., Wang Y.H., Cao C.J., Yang X.M., Ma S.C., Han X.B., Yang X.L., Yang A.N., Tian J., Xu H., et al. A regulatory circuit involving miR-143 and DNMT3a mediates vascular smooth muscle cell proliferation induced by homocysteine. Mol. Med. Rep. 2016;13:483–490. doi: 10.3892/mmr.2015.4558. [DOI] [PubMed] [Google Scholar]
- 65.Montes de Oca A., Madueno J.A., Martinez-Moreno J.M., Guerrero F., Munoz-Castaneda J., Rodriguez-Ortiz M.E., Mendoza F.J., Almaden Y., Lopez I., Rodriguez M., et al. High-phosphate-induced calcification is related to SM22alpha promoter methylation in vascular smooth muscle cells. J. Bone Min. Res. 2010;25:1996–2005. doi: 10.1002/jbmr.93. [DOI] [PubMed] [Google Scholar]
- 66.Azechi T., Sato F., Sudo R., Wachi H. 5-aza-2’-Deoxycytidine, a DNA methyltransferase inhibitor, facilitates the inorganic phosphorus-induced mineralization of vascular smooth muscle cells. J. Atheroscler Thromb. 2014;21:463–476. doi: 10.5551/jat.20818. [DOI] [PubMed] [Google Scholar]
- 67.Lin X., Xu F., Cui R.R., Xiong D., Zhong J.Y., Zhu T., Li F., Wu F., Xie X.B., Mao M.Z., et al. Arterial Calcification Is Regulated Via an miR-204/DNMT3a Regulatory Circuit Both In Vitro and in Female Mice. Endocrinology. 2018;159:2905–2916. doi: 10.1210/en.2018-00320. [DOI] [PubMed] [Google Scholar]
- 68.Lin X., Li F., Xu F., Cui R.R., Xiong D., Zhong J.Y., Zhu T., Shan S.K., Wu F., Xie X.B., et al. Aberration methylation of miR-34b was involved in regulating vascular calcification by targeting Notch1. Aging. 2019;11:3182–3197. doi: 10.18632/aging.101973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Da Silva R.A., da S., Feltrana G., da C. Fernandes C.J., Zambuzzi W.F. Osteogenic gene markers are epigenetically reprogrammed during contractile-to-calcifying vascular smooth muscle cell phenotype transition. Cell. Signal. 2020;66:109458. doi: 10.1016/j.cellsig.2019.109458. [DOI] [PubMed] [Google Scholar]
- 70.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 71.Wu S.S., Lin X., Yuan L.Q., Liao E.Y. The Role of Epigenetics in Arterial Calcification. Biomed. Res. Int. 2015;2015:320849. doi: 10.1155/2015/320849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O.M., Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huertas D., Sendra R., Munoz P. Chromatin dynamics coupled to DNA repair. Epigenetics. 2009;4:31–42. doi: 10.4161/epi.4.1.7733. [DOI] [PubMed] [Google Scholar]
- 74.Glozak M.A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 75.Verdin E., Ott M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 76.Wei J.W., Huang K., Yang C., Kang C.S. Non-coding RNAs as regulators in epigenetics (Review) Oncol. Rep. 2017;37:3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- 77.Abbaszadeh-Goudarzi K., Radbakhsh S., Pourhanifeh M.H., Khanbabaei H., Davoodvandi A., Fathizadeh H., Sahebkar A., Shahrzad M.K., Mirzaei H. Circular RNA and Diabetes: Epigenetic Regulator with Diagnostic role. Curr. Mol. Med. 2020 doi: 10.2174/1566524020666200129142106. [DOI] [PubMed] [Google Scholar]
- 78.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 79.Li C.H., Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int. J. Biochem. Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 80.Fatica A., Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 81.Lasda E., Parker R. Circular RNAs: Diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gui T., Zhou G., Sun Y., Shimokado A., Itoh S., Oikawa K., Muragaki Y. MicroRNAs that target Ca(2+) transporters are involved in vascular smooth muscle cell calcification. Lab. Investig. 2012;92:1250–1259. doi: 10.1038/labinvest.2012.85. [DOI] [PubMed] [Google Scholar]
- 83.Xia Z.Y., Hu Y., Xie P.L., Tang S.Y., Luo X.H., Liao E.Y., Chen F., Xie H. Runx2/miR-3960/miR-2861 Positive Feedback Loop Is Responsible for Osteogenic Transdifferentiation of Vascular Smooth Muscle Cells. Biomed. Res. Int. 2015;2015:624037. doi: 10.1155/2015/624037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Panizo S., Naves-Diaz M., Carrillo-Lopez N., Martinez-Arias L., Fernandez-Martin J.L., Ruiz-Torres M.P., Cannata-Andia J.B., Rodriguez I. MicroRNAs 29b, 133b, and 211 Regulate Vascular Smooth Muscle Calcification Mediated by High Phosphorus. J. Am. Soc. Nephrol. Jasn. 2016;27:824–834. doi: 10.1681/ASN.2014050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu J., Xiao X., Shen Y., Chen L., Xu C., Zhao H., Wu Y., Zhang Q., Zhong J., Tang Z., et al. MicroRNA-32 promotes calcification in vascular smooth muscle cells: Implications as a novel marker for coronary artery calcification. PLoS ONE. 2017;12:e0174138. doi: 10.1371/journal.pone.0174138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin X., Zhan J.K., Zhong J.Y., Wang Y.J., Wang Y., Li S., He J.Y., Tan P., Chen Y.Y., Liu X.B., et al. lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating high-glucose-induced calcification/senescence of VSMCs. Aging. 2019;11:523–535. doi: 10.18632/aging.101758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goettsch C., Rauner M., Pacyna N., Hempel U., Bornstein S.R., Hofbauer L.C. miR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 2011;179:1594–1600. doi: 10.1016/j.ajpath.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chao C.T., Liu Y.P., Su S.F., Yeh H.Y., Chen H.Y., Lee P.J., Chen W.J., Lee Y.M., Huang J.W., Chiang C.K., et al. Circulating MicroRNA-125b Predicts the Presence and Progression of Uremic Vascular Calcification. Arter. Thromb. Vasc. Biol. 2017;37:1402–1414. doi: 10.1161/ATVBAHA.117.309566. [DOI] [PubMed] [Google Scholar]
- 89.Cui R.R., Li S.J., Liu L.J., Yi L., Liang Q.H., Zhu X., Liu G.Y., Liu Y., Wu S.S., Liao X.B., et al. MicroRNA-204 regulates vascular smooth muscle cell calcification in vitro and in vivo. Cardiovasc. Res. 2012;96:320–329. doi: 10.1093/cvr/cvs258. [DOI] [PubMed] [Google Scholar]
- 90.Yanagawa B., Lovren F., Pan Y., Garg V., Quan A., Tang G., Singh K.K., Shukla P.C., Kalra N.P., Peterson M.D., et al. miRNA-141 is a novel regulator of BMP-2-mediated calcification in aortic stenosis. J. Thorac. Cardiovasc. Surg. 2012;144:256–262. doi: 10.1016/j.jtcvs.2011.10.097. [DOI] [PubMed] [Google Scholar]
- 91.Balderman J.A., Lee H.Y., Mahoney C.E., Handy D.E., White K., Annis S., Lebeche D., Hajjar R.J., Loscalzo J., Leopold J.A. Bone morphogenetic protein-2 decreases microRNA-30b and microRNA-30c to promote vascular smooth muscle cell calcification. J. Am. Heart Assoc. 2012;1:e003905. doi: 10.1161/JAHA.112.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeong G., Kwon D.H., Shin S., Choe N., Ryu J., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcification. Sci. Rep. 2019;9:5848. doi: 10.1038/s41598-019-42283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ryu J., Kwon D.H., Choe N., Shin S., Jeong G., Lim Y.H., Kim J., Park W.J., Kook H., Kim Y.K. Characterization of Circular RNAs in Vascular Smooth Muscle Cells with Vascular Calcification. Mol. Nucleic Acids. 2020;19:31–41. doi: 10.1016/j.omtn.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weinert B.T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S.A., Magnes C., Zechner R., Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol. Syst. Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 96.Koprinarova M., Schnekenburger M., Diederich M. Role of Histone Acetylation in Cell Cycle Regulation. Curr. Top. Med. Chem. 2016;16:732–744. doi: 10.2174/1568026615666150825140822. [DOI] [PubMed] [Google Scholar]
- 97.Bush E.W., McKinsey T.A. Protein acetylation in the cardiorenal axis: The promise of histone deacetylase inhibitors. Circ. Res. 2010;106:272–284. doi: 10.1161/CIRCRESAHA.109.209338. [DOI] [PubMed] [Google Scholar]
- 98.Li P., Ge J., Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2020;17:96–115. doi: 10.1038/s41569-019-0235-9. [DOI] [PubMed] [Google Scholar]
- 99.Menzies K.J., Zhang H., Katsyuba E., Auwerx J. Protein acetylation in metabolism—Metabolites and cofactors. Nat. Rev. Endocrinol. 2016;12:43–60. doi: 10.1038/nrendo.2015.181. [DOI] [PubMed] [Google Scholar]
- 100.Trisciuoglio D., Di Martile M., Del Bufalo D. Emerging Role of Histone Acetyltransferase in Stem Cells and Cancer. Stem Cells Int. 2018;2018:8908751. doi: 10.1155/2018/8908751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tafrova J.I., Tafrov S.T. Human histone acetyltransferase 1 (Hat1) acetylates lysine 5 of histone H2A in vivo. Mol. Cell. Biochem. 2014;392:259–272. doi: 10.1007/s11010-014-2036-0. [DOI] [PubMed] [Google Scholar]
- 102.Dompierre J.P., Godin J.D., Charrin B.C., Cordelieres F.P., King S.J., Humbert S., Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J. Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saba N.F., Magliocca K.R., Kim S., Muller S., Chen Z., Owonikoko T.K., Sarlis N.J., Eggers C., Phelan V., Grist W.J., et al. Acetylated tubulin (AT) as a prognostic marker in squamous cell carcinoma of the head and neck. Head Neck Pathol. 2014;8:66–72. doi: 10.1007/s12105-013-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McLendon P.M., Ferguson B.S., Osinska H., Bhuiyan M.S., James J., McKinsey T.A., Robbins J. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc. Natl. Acad. Sci. USA. 2014;111:E5178–E5186. doi: 10.1073/pnas.1415589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chan H.M., La Thangue N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 106.Sierra J., Villagra A., Paredes R., Cruzat F., Gutierrez S., Javed A., Arriagada G., Olate J., Imschenetzky M., Van Wijnen A.J., et al. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 2003;23:3339–3351. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gu J., Lu Y., Deng M., Qiu M., Tian Y., Ji Y., Zong P., Shao Y., Zheng R., Zhou B., et al. Inhibition of acetylation of histones 3 and 4 attenuates aortic valve calcification. Exp. Mol. Med. 2019;51:79. doi: 10.1038/s12276-019-0272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li S.J., Kao Y.H., Chung C.C., Chen W.Y., Cheng W.L., Chen Y.J. Activated p300 acetyltransferase activity modulates aortic valvular calcification with osteogenic transdifferentiation and downregulation of Klotho. Int. J. Cardiol. 2017;232:271–279. doi: 10.1016/j.ijcard.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 109.Eom G.H., Kook H. Posttranslational modifications of histone deacetylases: Implications for cardiovascular diseases. Pharmacol. Ther. 2014;143:168–180. doi: 10.1016/j.pharmthera.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 110.Kao H.Y., Downes M., Ordentlich P., Evans R.M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 111.Wilson A.J., Byun D.S., Popova N., Murray L.B., L’Italien K., Sowa Y., Arango D., Velcich A., Augenlicht L.H., Mariadason J.M. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 112.Huang B.H., Laban M., Leung C.H., Lee L., Lee C.K., Salto-Tellez M., Raju G.C., Hooi S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 113.Song J., Noh J.H., Lee J.H., Eun J.W., Ahn Y.M., Kim S.Y., Lee S.H., Park W.S., Yoo N.J., Lee J.Y., et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 114.Peart M.J., Tainton K.M., Ruefli A.A., Dear A.E., Sedelies K.A., O’Reilly L.A., Waterhouse N.J., Trapani J.A., Johnstone R.W. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2003;63:4460–4471. [PubMed] [Google Scholar]
- 115.Kee H.J., Kwon J.S., Shin S., Ahn Y., Jeong M.H., Kook H. Trichostatin A prevents neointimal hyperplasia via activation of Kruppel like factor 4. Vasc. Pharm. 2011;55:127–134. doi: 10.1016/j.vph.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 116.Kwon D.H., Eom G.H., Ko J.H., Shin S., Joung H., Choe N., Nam Y.S., Min H.K., Kook T., Yoon S., et al. MDM2 E3 ligase-mediated ubiquitination and degradation of HDAC1 in vascular calcification. Nat. Commun. 2016;7:10492. doi: 10.1038/ncomms10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kook H., Lepore J.J., Gitler A.D., Lu M.M., Wing-Man Yung W., Mackay J., Zhou R., Ferrari V., Gruber P., Epstein J.A. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Investig. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kee H.J., Eom G.H., Joung H., Shin S., Kim J.R., Cho Y.K., Choe N., Sim B.W., Jo D., Jeong M.H., et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ. Res. 2008;103:1259–1269. doi: 10.1161/01.RES.0000338570.27156.84. [DOI] [PubMed] [Google Scholar]
- 119.Kee H.J., Kook H. Kruppel-like factor 4 mediates histone deacetylase inhibitor-induced prevention of cardiac hypertrophy. J. Mol. Cell. Cardiol. 2009;47:770–780. doi: 10.1016/j.yjmcc.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 120.Kee H.J., Sohn I.S., Nam K.I., Park J.E., Qian Y.R., Yin Z., Ahn Y., Jeong M.H., Bang Y.J., Kim N., et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 121.Cho Y.K., Eom G.H., Kee H.J., Kim H.S., Choi W.Y., Nam K.I., Ma J.S., Kook H. Sodium valproate, a histone deacetylase inhibitor, but not captopril, prevents right ventricular hypertrophy in rats. Circ. J. 2010;74:760–770. doi: 10.1253/circj.CJ-09-0580. [DOI] [PubMed] [Google Scholar]
- 122.Eom G.H., Cho Y.K., Ko J.H., Shin S., Choe N., Kim Y., Joung H., Kim H.S., Nam K.I., Kee H.J., et al. Casein kinase-2alpha1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- 123.Yoon S., Kook T., Min H.K., Kwon D.H., Cho Y.K., Kim M., Shin S., Joung H., Jeong S.H., Lee S., et al. PP2A negatively regulates the hypertrophic response by dephosphorylating HDAC2 S394 in the heart. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoon S., Kim M., Min H.K., Lee Y.U., Kwon D.H., Lee M., Lee S., Kook T., Joung H., Nam K.I., et al. Inhibition of heat shock protein 70 blocks the development of cardiac hypertrophy by modulating the phosphorylation of histone deacetylase 2. Cardiovasc. Res. 2019;115:1850–1860. doi: 10.1093/cvr/cvy317. [DOI] [PubMed] [Google Scholar]
- 125.Zhao T.C., Cheng G., Zhang L.X., Tseng Y.T., Padbury J.F. Inhibition of histone deacetylases triggers pharmacologic preconditioning effects against myocardial ischemic injury. Cardiovasc. Res. 2007;76:473–481. doi: 10.1016/j.cardiores.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 126.Nural-Guvener H.F., Zakharova L., Nimlos J., Popovic S., Mastroeni D., Gaballa M.A. HDAC class I inhibitor, Mocetinostat, reverses cardiac fibrosis in heart failure and diminishes CD90+ cardiac myofibroblast activation. Fibrogenesis Tissue Repair. 2014;7:10. doi: 10.1186/1755-1536-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma J., Luo T., Zeng Z., Fu H., Asano Y., Liao Y., Minamino T., Kitakaze M. Histone Deacetylase Inhibitor Phenylbutyrate Exaggerates Heart Failure in Pressure Overloaded Mice independently of HDAC inhibition. Sci. Rep. 2016;6:34036. doi: 10.1038/srep34036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tovar C., Graves B., Packman K., Filipovic Z., Higgins B., Xia M., Tardell C., Garrido R., Lee E., Kolinsky K., et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 129.Andreeff M., Kelly K.R., Yee K., Assouline S., Strair R., Popplewell L., Bowen D., Martinelli G., Drummond M.W., Vyas P., et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 2016;22:868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abend A., Shkedi O., Fertouk M., Caspi L.H., Kehat I. Salt-inducible kinase induces cytoplasmic histone deacetylase 4 to promote vascular calcification. EMBO Rep. 2017;18:1166–1185. doi: 10.15252/embr.201643686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Malhotra R., Mauer A.C., Lino Cardenas C.L., Guo X., Yao J., Zhang X., Wunderer F., Smith A.V., Wong Q., Pechlivanis S., et al. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat. Genet. 2019;51:1580–1587. doi: 10.1038/s41588-019-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fu Z., Li F., Jia L., Su S., Wang Y., Cai Z., Xiang M. Histone deacetylase 6 reduction promotes aortic valve calcification via an endoplasmic reticulum stress-mediated osteogenic pathway. J. Thorac. Cardiovasc. Surg. 2019;158:408–417. doi: 10.1016/j.jtcvs.2018.10.136. [DOI] [PubMed] [Google Scholar]
- 133.Takemura A., Iijima K., Ota H., Son B.K., Ito Y., Ogawa S., Eto M., Akishita M., Ouchi Y. Sirtuin 1 retards hyperphosphatemia-induced calcification of vascular smooth muscle cells. Arter. Thromb. Vasc. Biol. 2011;31:2054–2062. doi: 10.1161/ATVBAHA.110.216739. [DOI] [PubMed] [Google Scholar]
- 134.Kwon D.H., Kim Y.K., Kook H. New Aspects of Vascular Calcification: Histone Deacetylases and Beyond. J. Korean Med. Sci. 2017;32:1738–1748. doi: 10.3346/jkms.2017.32.11.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ren G., Zhang G., Dong Z., Liu Z., Li L., Feng Y., Su D., Zhang Y., Huang B., Lu J. Recruitment of HDAC4 by transcription factor YY1 represses HOXB13 to affect cell growth in AR-negative prostate cancers. Int. J. Biochem. Cell Biol. 2009;41:1094–1101. doi: 10.1016/j.biocel.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 137.Huang Q.K., Qiao H.Y., Fu M.H., Li G., Li W.B., Chen Z., Wei J., Liang B.S. MiR-206 Attenuates Denervation-Induced Skeletal Muscle Atrophy in Rats Through Regulation of Satellite Cell Differentiation via TGF-beta1, Smad3, and HDAC4 Signaling. Med. Sci. Monit. 2016;22:1161–1170. doi: 10.12659/MSM.897909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Usui T., Morita T., Okada M., Yamawaki H. Histone deacetylase 4 controls neointimal hyperplasia via stimulating proliferation and migration of vascular smooth muscle cells. Hypertension. 2014;63:397–403. doi: 10.1161/HYPERTENSIONAHA.113.01843. [DOI] [PubMed] [Google Scholar]
- 139.Liu J., Zhou X., Li Q., Zhou S.M., Hu B., Hu G.W., Niu X., Guo S.C., Wang Y., Deng Z.F. Role of Phosphorylated HDAC4 in Stroke-Induced Angiogenesis. Biomed. Res. Int. 2017;2017:2957538. doi: 10.1155/2017/2957538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zheng X., Wu Z., Xu K., Qiu Y., Su X., Zhang Z., Zhou M. Interfering histone deacetylase 4 inhibits the proliferation of vascular smooth muscle cells via regulating MEG3/miR-125a-5p/IRF1. Cell Adh. Migr. 2019;13:41–49. doi: 10.1080/19336918.2018.1506653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J.M., Richardson J.A., et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 142.Hu T., Schreiter F.C., Bagchi R.A., Tatman P.D., Hannink M., McKinsey T.A. HDAC5 catalytic activity suppresses cardiomyocyte oxidative stress and NRF2 target gene expression. J. Biol. Chem. 2019;294:8640–8652. doi: 10.1074/jbc.RA118.007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eom G.H., Nam Y.S., Oh J.G., Choe N., Min H.K., Yoo E.K., Kang G., Nguyen V.H., Min J.J., Kim J.K., et al. Regulation of acetylation of histone deacetylase 2 by p300/CBP-associated factor/histone deacetylase 5 in the development of cardiac hypertrophy. Circ. Res. 2014;114:1133–1143. doi: 10.1161/CIRCRESAHA.114.303429. [DOI] [PubMed] [Google Scholar]
- 144.Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stathopoulou K., Cuello F., Candasamy A.J., Kemp E.M., Ehler E., Haworth R.S., Avkiran M. Four-and-a-half LIM domains proteins are novel regulators of the protein kinase D pathway in cardiac myocytes. Biochem. J. 2014;457:451–461. doi: 10.1042/BJ20131026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hsieh T.H., Hsu C.Y., Tsai C.F., Long C.Y., Wu C.H., Wu D.C., Lee J.N., Chang W.C., Tsai E.M. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol. Ther. 2015;23:656–666. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li H., Xie H., Liu W., Hu R., Huang B., Tan Y.F., Xu K., Sheng Z.F., Zhou H.D., Wu X.P., et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Investig. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jeon E.J., Lee K.Y., Choi N.S., Lee M.H., Kim H.N., Jin Y.H., Ryoo H.M., Choi J.Y., Yoshida M., Nishino N., et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 149.Lu S., Li H., Li K., Fan X.D. HDAC9 promotes brain ischemic injury by provoking IkappaBalpha/NF-kappaB and MAPKs signaling pathways. Biochem. Biophys. Res. Commun. 2018;503:1322–1329. doi: 10.1016/j.bbrc.2018.07.043. [DOI] [PubMed] [Google Scholar]
- 150.Shroff N., Ander B.P., Zhan X., Stamova B., Liu D., Hull H., Hamade F.R., Dykstra-Aiello C., Ng K., Sharp F.R., et al. HDAC9 Polymorphism Alters Blood Gene Expression in Patients with Large Vessel Atherosclerotic Stroke. Transl. Stroke Res. 2019;10:19–25. doi: 10.1007/s12975-018-0619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Salgado E., Bian X., Feng A., Shim H., Liang Z. HDAC9 overexpression confers invasive and angiogenic potential to triple negative breast cancer cells via modulating microRNA-206. Biochem. Biophys. Res. Commun. 2018;503:1087–1091. doi: 10.1016/j.bbrc.2018.06.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ma Z., Liu D., Di S., Zhang Z., Li W., Zhang J., Xu L., Guo K., Zhu Y., Li X., et al. Histone deacetylase 9 downregulation decreases tumor growth and promotes apoptosis in non-small cell lung cancer after melatonin treatment. J. Pineal Res. 2019;67:e12587. doi: 10.1111/jpi.12587. [DOI] [PubMed] [Google Scholar]
- 153.Lino Cardenas C.L., Kessinger C.W., Cheng Y., MacDonald C., MacGillivray T., Ghoshhajra B., Huleihel L., Nuri S., Yeri A.S., Jaffer F.A., et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun. 2018;9:1009. doi: 10.1038/s41467-018-03394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liao P.P., Liu L.H., Wang B., Fang X., Zhou S.Q., Li W., Zhang Y.Q., Guan S.M. Correlation between Histone Deacetylase 9 and Regulatory T Cell in Patients with Chronic Heart Failure. Curr. Med. Sci. 2018;38:199–203. doi: 10.1007/s11596-018-1866-5. [DOI] [PubMed] [Google Scholar]
- 155.Manzotti G., Torricelli F., Donati B., Sancisi V., Gugnoni M., Ciarrocchi A. HDACs control RUNX2 expression in cancer cells through redundant and cell context-dependent mechanisms. J. Exp. Clin. Cancer Res. 2019;38:346. doi: 10.1186/s13046-019-1350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Westendorf J.J., Zaidi S.K., Cascino J.E., Kahler R., van Wijnen A.J., Lian J.B., Yoshida M., Stein G.S., Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu H., Cheng X.W., Hu L., Takeshita K., Hu C., Du Q., Li X., Zhu E., Huang Z., Yisireyili M., et al. Cathepsin S Activity Controls Injury-Related Vascular Repair in Mice via the TLR2-Mediated p38MAPK and PI3K-Akt/p-HDAC6 Signaling Pathway. Arter. Thromb. Vasc. Biol. 2016;36:1549–1557. doi: 10.1161/ATVBAHA.115.307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rao R., Nalluri S., Kolhe R., Yang Y., Fiskus W., Chen J., Ha K., Buckley K.M., Balusu R., Coothankandaswamy V., et al. Treatment with panobinostat induces glucose-regulated protein 78 acetylation and endoplasmic reticulum stress in breast cancer cells. Mol. Cancer. 2010;9:942–952. doi: 10.1158/1535-7163.MCT-09-0988. [DOI] [PubMed] [Google Scholar]
- 159.Cai Z., Li F., Gong W., Liu W., Duan Q., Chen C., Ni L., Xia Y., Cianflone K., Dong N., et al. Endoplasmic reticulum stress participates in aortic valve calcification in hypercholesterolemic animals. Arter. Thromb. Vasc. Biol. 2013;33:2345–2354. doi: 10.1161/ATVBAHA.112.300226. [DOI] [PubMed] [Google Scholar]
- 160.Furmanik M., Shanahan C.M. ER stress regulates alkaline phosphatase gene expression in vascular smooth muscle cells via an ATF4-dependent mechanism. BMC Res. Notes. 2018;11:483. doi: 10.1186/s13104-018-3582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Duan X.H., Chang J.R., Zhang J., Zhang B.H., Li Y.L., Teng X., Zhu Y., Du J., Tang C.S., Qi Y.F. Activating transcription factor 4 is involved in endoplasmic reticulum stress-mediated apoptosis contributing to vascular calcification. Apoptosis. 2013;18:1132–1144. doi: 10.1007/s10495-013-0861-3. [DOI] [PubMed] [Google Scholar]
- 162.Montani D., Gunther S., Dorfmuller P., Perros F., Girerd B., Garcia G., Jais X., Savale L., Artaud-Macari E., Price L.C., et al. Pulmonary arterial hypertension. Orphanet J. Rare Dis. 2013;8:97. doi: 10.1186/1750-1172-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ruffenach G., Chabot S., Tanguay V.F., Courboulin A., Boucherat O., Potus F., Meloche J., Pflieger A., Breuils-Bonnet S., Nadeau V., et al. Role for Runt-related Transcription Factor 2 in Proliferative and Calcified Vascular Lesions in Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2016;194:1273–1285. doi: 10.1164/rccm.201512-2380OC. [DOI] [PubMed] [Google Scholar]
- 164.Mao M., Zhang M., Ge A., Ge X., Gu R., Zhang C., Fu Y., Gao J., Wang X., Liu Y., et al. Granzyme B deficiency promotes osteoblastic differentiation and calcification of vascular smooth muscle cells in hypoxic pulmonary hypertension. Cell Death Dis. 2018;9:221. doi: 10.1038/s41419-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Boucherat O., Chabot S., Paulin R., Trinh I., Bourgeois A., Potus F., Lampron M.C., Lambert C., Breuils-Bonnet S., Nadeau V., et al. HDAC6: A Novel Histone Deacetylase Implicated in Pulmonary Arterial Hypertension. Sci. Rep. 2017;7:4546. doi: 10.1038/s41598-017-04874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brown M.A., Zhu L., Schmidt C., Tucker P.W. Hsp90--from signal transduction to cell transformation. Biochem. Biophys. Res. Commun. 2007;363:241–246. doi: 10.1016/j.bbrc.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weisell J., Ohukainen P., Napankangas J., Ohlmeier S., Bergmann U., Peltonen T., Taskinen P., Ruskoaho H., Rysa J. Heat shock protein 90 is downregulated in calcific aortic valve disease. BMC Cardiovasc. Disord. 2019;19:306. doi: 10.1186/s12872-019-01294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yanagisawa S., Baker J.R., Vuppusetty C., Koga T., Colley T., Fenwick P., Donnelly L.E., Barnes P.J., Ito K. The dynamic shuttling of SIRT1 between cytoplasm and nuclei in bronchial epithelial cells by single and repeated cigarette smoke exposure. PLoS ONE. 2018;13:e0193921. doi: 10.1371/journal.pone.0193921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.D’Onofrio N., Servillo L., Balestrieri M.L. SIRT1 and SIRT6 Signaling Pathways in Cardiovascular Disease Protection. Antioxid. Redox Signal. 2018;28:711–732. doi: 10.1089/ars.2017.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ota H., Akishita M., Eto M., Iijima K., Kaneki M., Ouchi Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. J. Mol. Cell. Cardiol. 2007;43:571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 171.Bartoli-Leonard F., Wilkinson F.L., Schiro A., Inglott F.S., Alexander M.Y., Weston R. Suppression of SIRT1 in Diabetic Conditions Induces Osteogenic Differentiation of Human Vascular Smooth Muscle Cells via RUNX2 Signalling. Sci. Rep. 2019;9:878. doi: 10.1038/s41598-018-37027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Barnes G.L., Javed A., Waller S.M., Kamal M.H., Hebert K.E., Hassan M.Q., Bellahcene A., Van Wijnen A.J., Young M.F., Lian J.B., et al. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003;63:2631–2637. [PubMed] [Google Scholar]
- 173.Treskova I., Topolcan O., Windrichova J., Simanek V., Slouka D., Treska V., Kucera R. OPG, OPN, EGF and VEGF Levels at Individual Breslow Score Stages in Malignant Melanoma. Anticancer Res. 2018;38:4907–4911. doi: 10.21873/anticanres.12806. [DOI] [PubMed] [Google Scholar]
- 174.Chen J., Lin Y., Sun Z. Deficiency in the anti-aging gene Klotho promotes aortic valve fibrosis through AMPKalpha-mediated activation of RUNX2. Aging Cell. 2016;15:853–860. doi: 10.1111/acel.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang Y., Michiels T., Setroikromo R., van Merkerk R., Cool R.H., Quax W.J. Creation of RANKL mutants with low affinity for decoy receptor OPG and their potential anti-fibrosis activity. Febs. J. 2019;286:3582–3593. doi: 10.1111/febs.14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Matsubara T., Kida K., Yamaguchi A., Hata K., Ichida F., Meguro H., Aburatani H., Nishimura R., Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J. Biol. Chem. 2008;283:29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang J., Chang S., Lu Y., Wang J., Si Y., Zhang L., Cheng S., Jiang W.G. Enhanced osteopontin splicing regulated by RUNX2 is HDAC-dependent and induces invasive phenotypes in NSCLC cells. Cancer Cell Int. 2019;19:306. doi: 10.1186/s12935-019-1033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee H.W., Suh J.H., Kim A.Y., Lee Y.S., Park S.Y., Kim J.B. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 179.Schroeder T.M., Kahler R.A., Li X., Westendorf J.J. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 180.Yoshizawa T., Takizawa F., Iizawa F., Ishibashi O., Kawashima H., Matsuda A., Endo N., Kawashima H. Homeobox protein MSX2 acts as a molecular defense mechanism for preventing ossification in ligament fibroblasts. Mol. Cell. Biol. 2004;24:3460–3472. doi: 10.1128/MCB.24.8.3460-3472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jensen E.D., Schroeder T.M., Bailey J., Gopalakrishnan R., Westendorf J.J. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J. Bone Min. Res. 2008;23:361–372. doi: 10.1359/jbmr.071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Falkenberg K.J., Johnstone R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 184.Azechi T., Kanehira D., Kobayashi T., Sudo R., Nishimura A., Sato F., Wachi H., Trichostatin A. An HDAC class I/II inhibitor, promotes Pi-induced vascular calcification via up-regulation of the expression of alkaline phosphatase. J. Atheroscler Thromb. 2013;20:538–547. doi: 10.5551/jat.15826. [DOI] [PubMed] [Google Scholar]
- 185.Li S.J., Kao Y.H., Chung C.C., Cheng W.L., Chen Y.J. HDAC I inhibitor regulates RUNX2 transactivation through canonical and non-canonical Wnt signaling in aortic valvular interstitial cells. Am. J. Transl. Res. 2019;11:744–754. [PMC free article] [PubMed] [Google Scholar]
- 186.Dai X.Y., Zhao M.M., Cai Y., Guan Q.C., Zhao Y., Guan Y., Kong W., Zhu W.G., Xu M.J., Wang X. Phosphate-induced autophagy counteracts vascular calcification by reducing matrix vesicle release. Kidney Int. 2013;83:1042–1051. doi: 10.1038/ki.2012.482. [DOI] [PubMed] [Google Scholar]
- 187.Zhang W., Li Y., Ding H., Du Y., Wang L. Hydrogen peroxide prevents vascular calcification induced ROS production by regulating Nrf-2 pathway. Ren. Fail. 2016;38:1099–1106. doi: 10.1080/0886022X.2016.1194143. [DOI] [PubMed] [Google Scholar]
- 188.Zhang P., Li Y., Du Y., Li G., Wang L., Zhou F. Resveratrol Ameliorated Vascular Calcification by Regulating Sirt-1 and Nrf2. Transpl. Proc. 2016;48:3378–3386. doi: 10.1016/j.transproceed.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 189.Gryder B.E., Sodji Q.H., Oyelere A.K. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 2012;4:505–524. doi: 10.4155/fmc.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim H.J., Bae S.C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011;3:166–179. [PMC free article] [PubMed] [Google Scholar]
- 191.Dudakovic A., Evans J.M., Li Y., Middha S., McGee-Lawrence M.E., van Wijnen A.J., Westendorf J.J. Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J. Biol. Chem. 2013;288:28783–28791. doi: 10.1074/jbc.M113.489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bae H.S., Yoon W.J., Cho Y.D., Islam R., Shin H.R., Kim B.S., Lim J.M., Seo M.S., Cho S.A., Choi K.Y., et al. An HDAC Inhibitor, Entinostat/MS-275, Partially Prevents Delayed Cranial Suture Closure in Heterozygous Runx2 Null Mice. J. Bone Min. Res. 2017;32:951–961. doi: 10.1002/jbmr.3076. [DOI] [PubMed] [Google Scholar]
- 193.Lee H.A., Kang S.H., Kim M., Lee E., Cho H.M., Moon E.K., Kim I. Histone deacetylase inhibition ameliorates hypertension and hyperglycemia in a model of Cushing’s syndrome. Am. J. Physiol. Endocrinol. Metab. 2018;314:E39–E52. doi: 10.1152/ajpendo.00267.2017. [DOI] [PubMed] [Google Scholar]
- 194.Ryu Y., Kee H.J., Sun S., Seok Y.M., Choi S.Y., Kim G.R., Kee S.J., Pflieger M., Kurz T., Kim H.S., et al. Class I histone deacetylase inhibitor MS-275 attenuates vasoconstriction and inflammation in angiotensin II-induced hypertension. PLoS ONE. 2019;14:e0213186. doi: 10.1371/journal.pone.0213186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cavasin M.A., Demos-Davies K., Horn T.R., Walker L.A., Lemon D.D., Birdsey N., Weiser-Evans M.C., Harral J., Irwin D.C., Anwar A., et al. Selective class I histone deacetylase inhibition suppresses hypoxia-induced cardiopulmonary remodeling through an antiproliferative mechanism. Circ. Res. 2012;110:739–748. doi: 10.1161/CIRCRESAHA.111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Freundt J.K., Frommeyer G., Spieker T., Wotzel F., Grotthoff J.S., Stypmann J., Hempel G., Schafers M., Jacobs A.H., Eckardt L., et al. Histone deacetylase inhibition by Entinostat for the prevention of electrical and structural remodeling in heart failure. BMC Pharm. Toxicol. 2019;20:16. doi: 10.1186/s40360-019-0294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kramer O.H., Zhu P., Ostendorff H.P., Golebiewski M., Tiefenbach J., Peters M.A., Brill B., Groner B., Bach I., Heinzel T., et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fahey J.W., Zhang Y., Talalay P. Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Myzak M.C., Karplus P.A., Chung F.L., Dashwood R.H. A novel mechanism of chemoprotection by sulforaphane: Inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 200.Dickinson S.E., Rusche J.J., Bec S.L., Horn D.J., Janda J., Rim S.H., Smith C.L., Bowden G.T. The effect of sulforaphane on histone deacetylase activity in keratinocytes: Differences between in vitro and in vivo analyses. Mol. Carcinog. 2015;54:1513–1520. doi: 10.1002/mc.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Myzak M.C., Hardin K., Wang R., Dashwood R.H., Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27:811–819. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Myzak M.C., Tong P., Dashwood W.M., Dashwood R.H., Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp. Biol. Med. 2007;232:227–234. [PMC free article] [PubMed] [Google Scholar]
- 203.Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G., Zipkin R.E., Chung P., Kisielewski A., Zhang L.L., et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 204.Baur J.A., Sinclair D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 205.Juhasz B., Varga B., Gesztelyi R., Kemeny-Beke A., Zsuga J., Tosaki A. Resveratrol: A multifunctional cytoprotective molecule. Curr. Pharm. Biotechnol. 2010;11:810–818. doi: 10.2174/138920110793262079. [DOI] [PubMed] [Google Scholar]
- 206.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]