Abstract
What began with a sign of pneumonia-related respiratory disorders in China has now become a pandemic named by WHO as Covid-19 known to be caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). The SARS-CoV-2 are newly emerged β coronaviruses belonging to the Coronaviridae family. SARS-CoV-2 has a positive viral RNA genome expressing open reading frames that code for structural and non-structural proteins. The structural proteins include spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins. The S1 subunit of S protein facilitates ACE2 mediated virus attachment while S2 subunit promotes membrane fusion. The presence of glutamine, asparagine, leucine, phenylalanine and serine amino acids in SARS-CoV-2 enhances ACE2 binding. The N protein is composed of a serine-rich linker region sandwiched between N Terminal Domain (NTD) and C Terminal Domain (CTD). These terminals play a role in viral entry and its processing post entry. The NTD forms orthorhombic crystals and binds to the viral genome. The linker region contains phosphorylation sites that regulate its functioning. The CTD promotes nucleocapsid formation. The E protein contains a NTD, hydrophobic domain and CTD which form viroporins needed for viral assembly. The M protein possesses hydrophilic C terminal and amphipathic N terminal. Its long-form promotes spike incorporations and the interaction with E facilitates virion production. As each protein is essential in viral functioning, this review describes the insights of SARS-CoV-2 structural proteins that would help in developing therapeutic strategies by targeting each protein to curb the rapidly growing pandemic.
Key Words: SARS-CoV-2, COVID-19, Pandemic, Structural protein, Nucleocapsid
Graphical abstract
Introduction
Just before the dawn of 2020 was making its way to the earth, China witnessed a flare of pneumonia-like conditions in Wuhan, which now has turned to be a global pandemic. During its early outbreak, it was termed as the Wuhan virus going by the location of its origin but eventually the World Health Organization (WHO) officially declared it as COVID-19. Under the guidelines framed by the WHO, the World Organization for Animal Health (OIE) and the United Nations Food and Agricultural Organization (FAO), the coronavirus study group (CSG) of the International Committee on Taxonomy of Viruses declared the name of the virus as SARS-CoV-2. Therefore SARS-CoV-2 is said to cause COVID-19 (1). The conditions caused by the coronavirus have been named by WHO as COVID-19 expanding as CO- Corona, VI-virus, D -Disease, and “19” as the year of emergence (2). The CSG has proposed to create better identification of patients by coding the cases as, SARS-CoV-2 followed by isolate, host, date and location of patients (1). The cause of the viral outbreak is not yet identified but is suspected to be originated from the horseshoe bats (3). It showed to be closely related to the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) sequence obtained from bat strain RaTG13 (4). During the initial outbreak, China was seen to be profoundly affected. However, in the current scenario to date the regions like the United States of America, Spain, Italy, Germany, France and Iran have been tremendously affected.
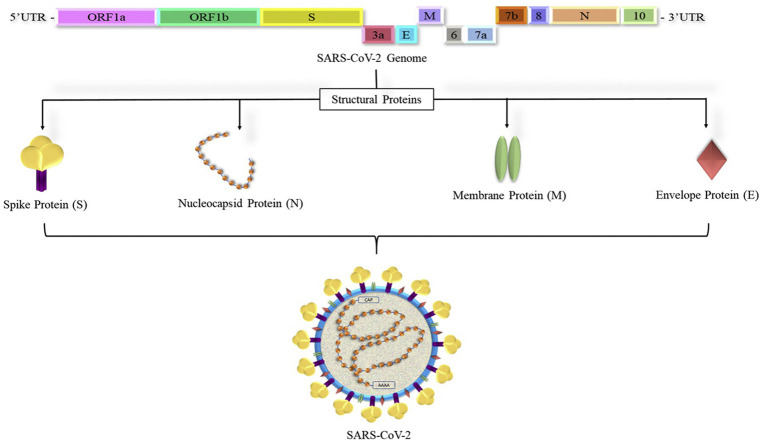
Microscopic imaging has shown that SARS-CoV-2 has crown-like surface projections that indicates it belongs to a family of coronaviruses (5). As SARS-CoV-2 belongs to a group of a previously known family of coronaviruses, it retains the important structural proteins namely Spike Proteins, Nucleocapsid Proteins, Envelope Proteins and Membrane Proteins. These proteins form an essential part in the viral genome production, replication, virion-receptor attachment, virion and viroporin formation that ultimately promote its entry into a host organism and proliferate, thus spreading infection. This review attempts to describe in detail the organization of structure and functional characteristics of the structural proteins of SARS-CoV-2.
Coronaviruses
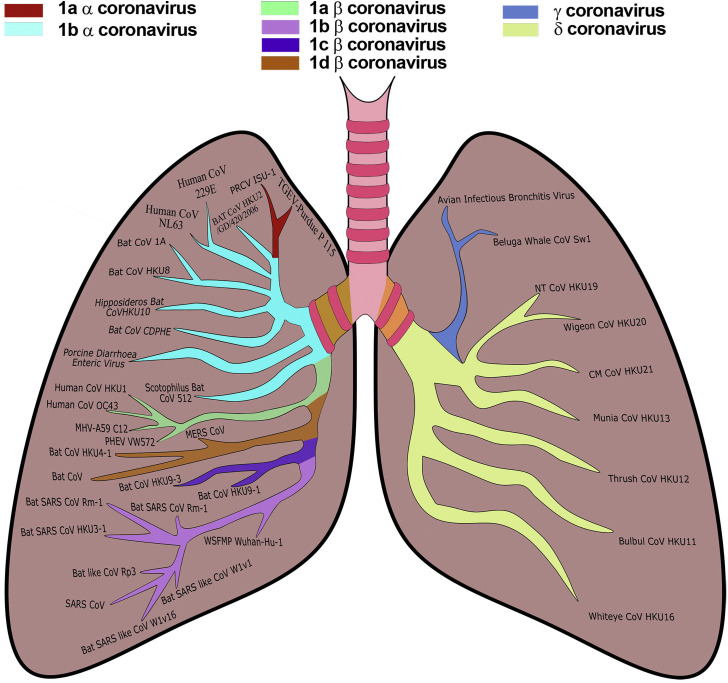
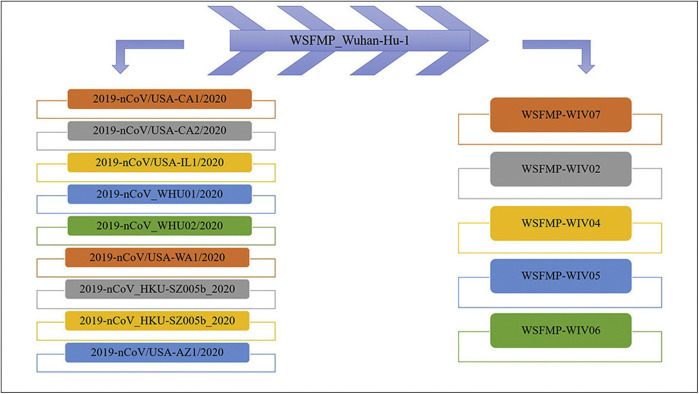
Coronaviruses are the members of the Coronaviridae family and order Nidovirales. They are categorized into four genera, namely α coronavirus, β coronavirus, γ coronavirus, and lastly δ coronavirus (6) as shown in Figure 1 . Their hosts mainly porcine, bovines, humans, avians, etc. suffer from infections like pneumonia, diarrhea, enteric indications, and kidney failure. The α coronavirus and β coronavirus affect mammalians while γ coronavirus, and δ coronavirus affect birds (8). Previously six coronaviruses were identified as the ones to infect humans, namely Human Coronavirus-229E and Human Coronavirus-NL63 belonging to α coronaviruses and Human Coronavirus-HKU1, Human Coronavirus-OC43, Severe Acute Respiratory Syndrome (SARS-CoV) and Middle East Respiratory Syndrome (MERS-CoV) belonging to β coronaviruses (9). Recently new strains of SARS-CoV-2 have been identified as shown in Figure 2 .
Figure 1.
Family of Coronaviruses. Coronaviruses are divided into four genera namely α coronavirus, β coronavirus, γ coronavirus, and δ coronavirus. The α coronavirus is further divided into sub groups a and b, while the β coronavirus is divided into a, b, c, and d (7). TGEV-Transmittable Gastroenteritis Virus TGEV, PRCV-Porcine Respiratory Coronavirus, CDPHE-Colorado Department of Public Health and Environment, MHV-Murine Hepatitis Virus, PHEV-Porcine Hemagglutinating Encephalomyelitis Virus, MERS-CoV-Middle East Respiratory Syndrome Coronavirus, SARS-CoV-Severe Acute Respiratory Syndrome Coronavirus, WSFMP-Wuhan Seafood Market Pneumonia.
Figure 2.
Newly Identified Family of SARS-CoV-2 (WSFMP_Wuhan-Hu-1 is used as a reference). WSFMP- Wuhan Seafood Market Pneumonia, 2019-nCoV- 2019-novel Coronavirus.
Structure of SARS-CoV-2
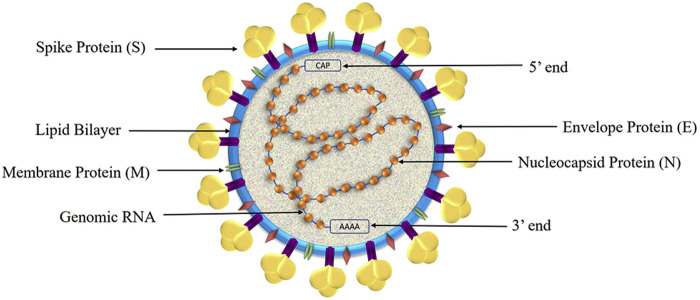
The nucleic acid sequence of the SARS-CoV-2 is identified as that of β- coronavirus (10). The SARS-CoV-2 is composed of a large positive-stranded RNA genome of 29891 nucleotides with 9860 amino acids (11). This genome is present inside circular nucleocapsid proteins and further encapsulated by an envelope (6). SARS-CoV-2 genome consists of 10 Open Reading Frames (ORF) (12). In the first ORF (ORF1a/b) about two-thirds of viral RNA is present that encodes for polyprotein1a and polyprotein 1b and 1–16 non-structural protein (13). Remaining ORFs encodes for structural proteins like S, M, E, and N and accessory proteins (14). The genomic sequence of SARS-CoV-2 in shown in Figure 3 and its structure is shown in Figure 4 . Some corona virions additionally contain hemagglutinin-esterase (HE) proteins but in SARS-CoV-2 it is seen to be lost may be in efforts to recent adaptations (9).
Figure 3.
Genomic sequence of SARS-CoV-2. ORF–Open Reading Frame, UTR-Untranslated region, S-Spike protein, M-Membrane protein, E-Envelope protein, N-Nucleocapsid protein.
Figure 4.
Schematic representation of SARS-CoV-2 structure.
The genomic RNA expresses the gene in a characteristic sequence in the form of 5′-replicase gene – S–E-M–N -3′ flanked by untranslated regions on both the ends. The rep gene codes for the non-structural proteins, membrane and envelope proteins that are essential in viral assembly and nucleocapsid protein important for RNA synthesis (15). The spike protein is responsible for the attachment of the virus to the host cell and its subsequent entry into it (6,15,16).
Recently, mutations in the ORF1, ORF8, N region of SARS-CoV-2 were observed (17). The mutations in non-structural proteins (nsp) 2 and nsp 3 could be the reasons for its unique mechanism of action as compared to SARS. The presence of glutamine, serine, and proline at various positions in the sequence of SARS-CoV-2 is believed to affect its properties as shown in Table 1, Table 2, Table 3 (18).
Table 1.
Comparison of amino acids at position 501 in the different strains of coronaviruses
| Coronavirus strain | Amino acid at position 501 | Predicted significance of the amino acid substitutions |
|---|---|---|
| SARS-CoV-2 | Glutamine | Due to the presence of a longer side chain, higher polarity, and stronger ability to form H bonds, glutamine could possibly provide increased stability to protein (18). |
| Bat like SARS-CoV | Threonine | |
| SARS-CoV | Alanine |
SARS-CoV-2, Severe Acute Respiratory Coronavirus-2; SARS-CoV, Severe Acute Respiratory Coronavirus.
Table 2.
Comparison of amino acids at position 723 in the different strains of coronaviruses
| Coronavirus strain | Amino acid at position 723 | Predicted significance of the amino acid substitutions |
|---|---|---|
| SARS-CoV-2 | Serine | Serine is believed to promote rigorousness to the polypeptide chain due to the steric effect and its capability to form H bonds. At the active site of enzymes, it may also act as a nucleophile (18). |
| Bat like SARS-CoV | Glycine | |
| SARS-CoV | Glycine |
SARS-CoV-2, Severe Acute Respiratory Coronavirus-2; SARS-CoV, Severe Acute Respiratory Coronavirus.
Table 3.
Comparison of amino acids at position 501 in the different strains of coronaviruses
| Coronavirus strain | Amino acid at position 1010 | Predicted significance of the amino acid substitutions |
|---|---|---|
| SARS-CoV-2 | Proline | Proline is expected to create a steric bulk and rigorousness due to which the structure of SARS-CoV-2 may undergo a change in its conformation (18). |
| Bat like SARS-CoV | Histidine | |
| SARS-CoV | Isoleucine |
SARS-CoV-2, Severe Acute Respiratory Coronavirus-2; SARS-CoV, Severe Acute Respiratory Coronavirus.
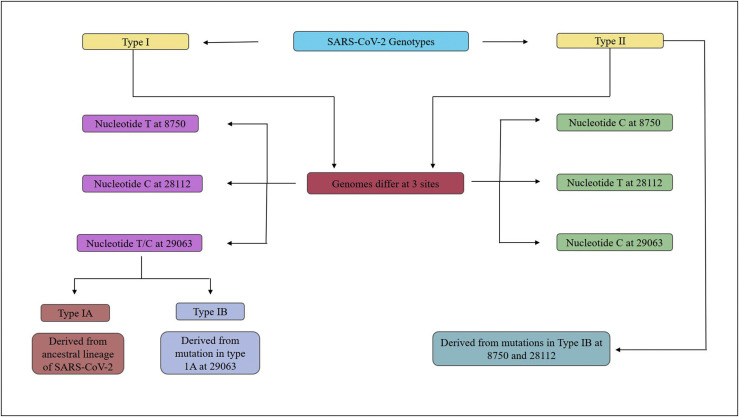
Only 3.8 % variability was seen in the genome sequence of SARS-CoV-2 and the strain of coronavirus obtained from bats i.e. RaTG13, thereby indicating 96.2 % similarity between the two. These genomes of SARS-CoV-2 were seen to be expressed in two types, namely L and S type. Recently genomes of 103 patients infected with SARS-CoV-2 were analysed, where-in 101 of them showed a linkage in polymorphism for single nucleotide. Among these 72 of them expressed L type, which is named due to involvement of Leucine codon and 29 strains showed S type named due to involvement of Serine codon. This change was observed at the site of 8,728 and 28,144 of the sequence (19).Similar studies of SARS-CoV-2 genome analysis showed Type I and Type II SARS-CoV-2 genotypes that differed at sites 8750, 29063 and 28112 (20) as shown in Figure 5 and SARS-CoV-2 genome types A (ancestral genome), B (obtained by mutations at C28144T and T8782C in type A) and C (obtained by mutation at G26144T in type B) (21). Full genome sequencing of various SARS-CoV-2 isolates have shown multiple variations in different ORFs corresponding to the sequence of SARS-CoV-2 obtained from bat RaTG13 which shows that as the virus having spread globally, has evolved with numerous mutations (22) as shown in Supplementary Table 1 that could place the medical fraternity at an even more alarming situation.
Figure 5.
Types of SARS-CoV-2 genotypes.
Structural Proteins of SARS-CoV-2
Spike Protein
The envelope of corona-virion contains protruding projections from its surface called the large surface glycoproteins or spike proteins responsible for recognizing the host's receptor followed by its binding to it and fusing with its membrane (23). Due to the crown-shaped appearance of these projections it has been named coronavirus {corona-a crown (Latin)} (24).
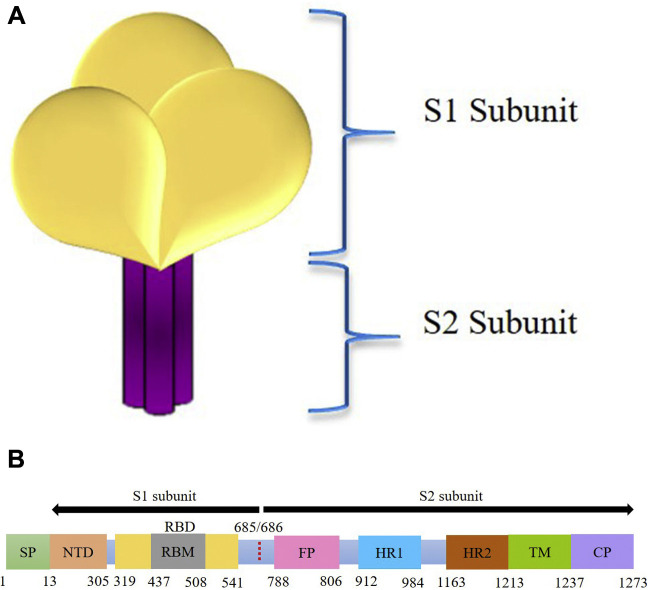
The amino acid sequence of the SARS-CoV-2 S protein has ∼75 % homology with SARS-CoV spike protein (25). Other findings have reported a 70% similarity of S1 subunit and 99% similarity of S2 subunit of SARS-CoV-2 with SARS-CoV (11). Its molecular weight is about 141178 kDa and contains 1273 amino acids (26). Generally, a coronavirus particle may contain about 50–100 timers of spikes (27). The spike protein consists of an ectodomain element, transmembrane moiety and a short intracellular C fragment. The viral ectodomain inherits two subunits, namely S1 that facilitates receptor binding and S2 that facilitates membrane fusion. It is structured like a clove built from three S1 subunits and S2 stem formed of a trimer (10) as shown in Figure 6 A. The genomic sequence of S proteins with its amino acids is shown in Figure 6B. The functional components of spike protein were mapped according to the amino acid positions as shown in Table 4 .
Figure 6.
A. Schematic Illustration of the Structure of Spike Protein. B. Genomic Sequence of Spike Protein (28). SP-Signal Peptide, NTD-N terminal Domain, RBM-Receptor Binding Motif, RBD-Receptor Binding Domain, FP-Fusion Peptide, HR1-Heptat Repeat 1, HR2-Heptad Repeat 2, TM-Transmembrane Domain, CP-Cytoplasm Domain.
Table 4.
| Sr. No. | Functional components of spike protein | Amino acid position |
|---|---|---|
| 1 | N terminal domain | 14–305 |
| 2 | Receptor Binding Motif | 437–508 |
| 3 | Receptor Binding Domain | 319–541 |
| 4 | S1 Subunit | 14–685 |
| 5 | S2 Subunit | 686–1273 |
| 6 | Fusion Peptide | 788–806 |
| 7 | Heptad Repeat 1 | 912–984 |
| 8 | Heptad Repeat 2 | 1163–1213 |
| 9 | Transmembrane Domain | 1214–1237 |
| 10 | Cytoplasmic Domain | 1238–1273 |
S proteins also help in promoting adhesion of infected cells with adjacent non-infected cells that enhance the spreading of the virus (29). The additional 12 nucleotides towards the arginine cleaving site correlate to a cleavage site similar to furin and this site is known to be cleaved during virus activation (24).
The Angiotensin-Converting Enzyme 2 (ACE2) and Type II transmembrane Serine Protease (TMPRSS2) are co-expressed in type II pneumocytes. TMPRSS2 initiates cleavage followed by activation of spike protein that promotes membrane fusion and viral entry into the cells and also the transmission of infection to neighbouring cells, specifically in the conditions of low pH (30). The S protein of SARS-CoV-2 undergoes a structural transformation for the binding of the viral membrane to the host membrane to occur. A Receptor Binding Domain (RBD) is present in the S1 subunit that recognizes and binds to the human ACE2 with 10–20 times more affinity than the SARS-CoV domain (31) possibly due to polymorphism at 501T thus enhancing the receptor-spike interactions and promoting infectivity (32). A unique feature is seen in the S1 and S2 sites of SARS-CoV-2 where both these sites form a prolonged loop which lies towards the outer end of the trimer thus rendering SARS-CoV-2 to more proteolytic interaction by the proteases of host cells (4).
The S1 subunit occurs in the prefusion trimer phase. However, upon binding to the host cell receptor, it destabilizes and sheds itself and causes the S2 subunit to transform into a stable postfusion state. This spike protein can display itself in an upward phase (available for a receptor) or downward phase (unavailable for a receptor).In the downward phase of S protein, the RBD lies near the trimer's central pocket (31).
The S2 subunit comprises a single fusion peptide (FP), an additional proteolytic site with an internal fusion peptide, and just prior to the transmembrane domain, a two heptad-repeat (24). Recent analysis has suggested four novel inserts in the spike protein that are unique to SARS-CoV2. In the S1 subunit, the insert I hints about the N terminal domain while the C terminal domain has been implicated by the insert II and insert III. At the junction of the subdomain 1 and subdomain 2 of the S1 subunit lies the insert IV. Interestingly the inserts I, II, and III have shown remarkable homology with the HIV-1 gp120 while insert IV to Gag proteins (33).
Another study revealed the characteristic features of certain amino acids in S protein that has rendered SARS-CoV-2 with high affinity to ACE2 receptor. The presence of glutamine 493, asparagine 501, leucine 455, phenylalanine 486 and serine 494 amino acids have shown to provide a boost in ACE2 binding (32).
Therefore, the S protein plays a crucial role in viral entry into the host cells and the structural abilities in this newly discovered SARS-CoV-2 boost its intended actions. The fact that these protruding spikes are the first point of contact with host receptors, therapeutic strategies can be applied to prevent its binding to target receptors and prevent viral entry into host cells.
Nucleocapsid Protein
The N protein is the most abundant viral protein and is expressed in host samples during the early stages of infection. It is known to bind to viral RNA to form a core of a ribonucleoprotein which helps in its host cell entry and interaction with cellular processes following the fusion of virus (34). The sequence of SARS-CoV-2 N protein shows ∼90 % similarity with the N protein of SARS-CoV (25). The RTC i.e. the Replication Transcription Complexes formed by the nsp play an essential part in viral genome synthesis (35). The N protein genome consists of a serine (SR) rich linker region sandwiched between an N terminal domain (NTD) and a C terminal domain (CTD) as shown in Figure 7 .
Figure 7.
Genomic sequence of SARS-CoV-2 N protein (36).
The N protein in SARS-CoV promotes the activation of Cyclooxygenase-2 (COX-2) leading to inflammation in the lungs (36). It is also involved in the inhibition of phosphorylation of B23 phosphoprotein that is essential in the progression of the cell cycle during the duplication of centrosome (37). The N protein interacts with the p42 proteasome subunit, which is known to degrade the viral proteins (38). It also inhibits type I Interferon (IFN) causing restrictions in immune responses generated by the body due to viral infections (39). Cell line studies showed that due to the inhibitory action of N protein on the cyclin-cyclin-dependent kinase complex (C-CDK) the progression of S-Phase was reduced (40) Another cell line study showed reduced proliferation of cells due to inhibition of cytokinesis and protein translation due to N protein-induced aggregation of a translation factor named as Human elongation factor 1 α (HEF1 α) (41). The viral RNA synthesis is known to increase when N protein is seen to interact with Heterogeneous nuclear ribonucleoprotein (hnRNPA1) (42).
The structural elucidation of SARS-CoV-2 N protein NTD showed that a single asymmetric confirmation of four N protein NTD depicted a structural alignment of an orthorhombic crystal. Basically, it looks like a wrist made up of acidic moieties with a palm of basic components and the core of β sheet extending like fingers (43). The NTD of N protein is known to bind to the RNA genome where the sequencing of different ordered and disordered regions of N protein showed that RNA binding occurs at the N45-181 region which exists as a monomer (44). It was also reported that the presence of amino acids Arginine at position 94 and Tyrosine at 122 might be essential for the binding of SARS-CoV RNA (45). But further studies proved that the combination of the linker region and the NTD and even the CTD of N protein was essential in enhanced binding capacity to the viral RNA where the latter showed about 6–8 times more binding performance than the earlier, as CTD is dimeric in nature with 2 disordered regions around it as compared to the single disordered region around NTD (46).
The central linker region is rich in serine and arginine residues (SR region) possessing essential phosphorylation sites that may regulate the N protein functioning (47). This site forms the major phosphorylation site that enhances the interactions in proteins, localization of linker proteins within the cell, and mediates the desired activity. Higher alanine substitutions reduces the amount of N protein phosphorylation (48). In the linker region, the interactions of SR moieties with the central region lead to the formation of dimers of N proteins essential for activity (49).
The CTD is hydrophobic in nature, rich in helix, and is also known as the domain where dimerization occurs. The reason for this is that it contains residues that self-associate to form homodimers (45). Structurally every single asymmetric moiety of CTD consists of four individual homodimers joined to form an octamer. The arrangement of these moieties appears in the shape of the letter “X” that forms symmetric fold structures, perpendicular to the midpoint of the structure. Due to the positive charges in this region, the N terminus appears to be basic that could hint it to be a site for the binding of nucleic acid (50).
The domain components possess large forces of repulsion that prevents interactions within the domains. This provides an electrostatically larger binding surface and prevents oligomer and nucleocapsid formation. Therefore when nucleic acid binds to it, it causes neutralization of the charges provoking accumulation of protein molecules to come closer and oligomerize thus forming nucleocapsids (46).
The N protein showcases various activities essential in the functioning and proliferation of the virus and thus is another essential component after spike proteins. Along with S protein , the N protein shows a promising area in the field of developing effective therapeutics to prevent the proliferation of viral progenies.
Envelope Protein
The E protein is a tiny integral membrane protein composed of an NTD, hydrophobic domain, and a chain at C terminal (51) having 76–109 amino acids (52) and weighing 8–12 kDa in size (53).
The N terminal stretches from 1st–9th amino acids, the hydrophobic region ranges from 10th–37th position and C terminal from 38th–76th position in the structural sequence (54). The amino acids in the 1st –11th position are present in virion while the hydrophobic tail is towards the cytoplasm (55). The hydrophobic region oligomerises to form an ionic pore across the membranes. Structural elucidation of this protein shows its pentameric form containing 35 α-helical regions and 40 looped regions. Both these structures showed randomized movements that modulated the normal activity of the ion channels, thus enhancing the viral pathogenicity (56). In contrast, interactions within the C terminal domain may affect this pentameric conformation (57).
A novel feature is seen in the 69th position of the E protein sequence of SARS-CoV-2 where Arginine has been replaced by alanine, glutamine and aspartate compared in other similar coronaviruses. Also, at positions 55–56 threonine and valine have been identified (58). After translation E can undergo palmitoylation at cysteine residues or N mediated glycosylation at aspartate amino acid but may not be necessary for virus like particles (VLP) formation (59).
This protein form viroporins that are proteins of hydrophobic nature and small in architecture. These viroporins are essential for viral assembly, along with its release. They also mediate pathogenic processes and induce cytotoxicity (60). The interactions of nsp2 and nsp3 heterotypically, are essential in inducing the desired curvature in the Endoplasmic Reticulum (ER) membrane and M protein and E protein co-expression facilitates the production of spherical virulent particles (29).
The tail of SARS-CoV E protein that lies in cytoplasm, targets cis-Golgi complex region with the help of proline residues incorporated in it. The N terminal of E protein also contains additional Golgi complex associating elements due to which mutations in the tail region do not affect the Golgi complex targeting process (61). The ionic gradient may be dissipated in the Endoplasmic Reticulum Golgi Intermediate Compartment (ERGIC) and Golgi compartment by the E protein that may lead to the exit of virion (52). The final four amino acids present in the C terminal of E protein hosts a motif named as postsynaptic density protein/Disc Large/Zonula occludans-1 (PDZ) binding motif (PDM). Therefore, E protein may facilitate disruption of the epithelium of the lungs due to binding of Protein Associated with Caenorhabditis elegans Lin-7 protein 1 (PALS1) to the PDM (62). Thus,targetting of E protein could aslo lead to improved therapeutics against SARS-CoV-2.
Membrane Protein
The M protein is present in high amounts out of all proteins in coronaviruses (63). Its length spans to about 220–260 amino acids with a short length N terminal domain, attached to triple transmembrane domains that are further connected to a carboxyl-terminal domain and belong to the N-linked glycosylated proteins with a conserved domain of 12 amino acids (64).
Structural analysis of this protein shows that it exists in two forms, namely long and compact form. These two forms are originally homodimers of N terminal ectodomain and C terminal endodomain that are different in conformation. The endodomain may undergo elongation or compression due to which it is named as a long and compact form. Spike protein can be seen over these both forms but predominantly at the long-form that may suggest this form promotes spike installation. The tyrosine residues at 211 may be essential in the stability of the long form of M. This form bends the membrane that creates a spherical structure encircling the ribonucleoprotein. (65).
The M protein is organized in a 2D lattice and provides a scaffold in viral assembly (27). They undergo translation on the polysomes bound to the membranes, fused in the ER, and are carried towards the Golgi complex where they interact with E proteins to generate virions. Out of the three TDM, the first one is capable enough to encourage self-association of M proteins, improved membrane affinity, and retention in Golgi (66).
NFκB (Nuclear Factor Kappa B) activation is necessary to generate immune responses against the pathogens. The M protein is known to inhibit NFκB through interactions with IKKβ (I Kappa B Kinase) and reduces levels of COX-2, thus enhancing the proliferation of the viral pathogen (67). The 3-phosphoinositide-dependent protein kinase 1 (PDK1) is a critical kinase of Protein Kinase B (PKB) that is known to slow down the process of apoptosis. The C terminal of M hinders the interaction of PDK1 and PKB and leads to the release of caspases 8 and 9, ultimately causing cell death or apoptosis (68). The SARS CoV M also leads to the activation of β interferons (IFN-β) in cell lines (69).
With such indications of M protein in viral life cycle, it can be looked upon as a therapeutic option to inhibit the virion formation and to prevent inflammatory reactions in host cells.
Conclusion
The SARS-CoV-2 is a β coronavirus belonging to the Coronaviridae family known to cause Covid-19. It consists of ORFs that code for structural, non-structural, and accessory proteins. The S, N, M, E form the structural proteins that play a vital role in the life cycle of the viral particles. The S protein is shaped like a clove with two subunits S1 and S2 which promotes receptor binding and membrane fusion respectively. The N protein consists of an NTD, serine-rich linker and CTD. It enhances viral entry and performs post-fusion cellular processes necessary for viral survival in the host. The E protein promotes virion formation and viral pathogenicity while M protein forms ribonucleoproteins and mediates inflammatory responses in hosts.
The elucidation of SARS-CoV-2 structural proteins in our review will help in exploring these proteins and gaining new insights on them. Since it is a rapidly evolving pandemic, these insights would be a helping aid to the upcoming research on structural proteins of SARS-CoV-2. This would lead to better understanding of each protein ultimately helping in making effective therapeutics to curb COVID-19.
(ARCMED_2020_649)
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.arcmed.2020.05.012.
Funding
This review did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declarations of Interest
None.
Supplementary Data
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Novel coronavirus (2019-nCoV) Situation Report-22. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200211-sitrep-22-ncov.pdf?sfvrsn=fb6d49b1_2 pp. 1–7.
- 3.MacKenzie J.S., Smith D.W. COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiol Aust. 2020;41:45–50. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaimes J.A., André N.M., Chappie J.S. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically-Sensitive Activation Loop. J Mol Biol. 2020:1–17. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasad S., Potdar V., Cherian S. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020;151:241–243. doi: 10.4103/ijmr.IJMR_577_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shereen M.A., Khan S., Kazmi A. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Q., Song Y., Shi M. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci Rep. 2015;5:1–8. doi: 10.1038/srep17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakkers M.J.G., Lang Y., Feitsma L.J. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell Host Microbe. 2017;21:356–366. doi: 10.1016/j.chom.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Guo Y., Pan Y. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 2020;2:1–6. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.F.W., Kok K.H., Zhu Z. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N., Shang J., Jiang S. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front Microbiol. 2020;11:1–19. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y.-R., Cao Q.-D., Hong Z.-S. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y., Du J., Su H. Identification of diverse bat alphacoronaviruses and betacoronaviruses in china provides new insights into the evolution and origin of coronavirus-related diseases. Front Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Z., Xu Y., Bao L. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:1–28. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu A., Peng Y., Huang B. Commentary Genome Composition and Divergence of the Novel Coronavirus ( 2019-nCoV ) Originating in China. Cell Host Microbe. 2020:1–4. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C., Liu Z., Chen Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angeletti S., Benvenuto D., Bianchi M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020:1–5. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X., Wu C., Li X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;213:54–63. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liangsheng Z., Jian-Rong Yang Z.Z., Lin Z. Genomic variations of SARS-CoV-2 suggest multiple outbreak sources of transmission. medRxiv Prepr. 2020;1–9 [Google Scholar]
- 21.Forster P., Forster L., Renfrew C. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J.-M., Jan S.S., Wei X. Evidence of the Recombinant Origin and Ongoing Mutations in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) bioRxiv Prepr. 2020:1–17. [Google Scholar]
- 23.Siddell S.G. The Coronaviridae. 1995. Coronavirus Gene Expression; p. 38. [Google Scholar]
- 24.Coutard B., Valle C., de Lamballerie X. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:1–8. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S. Drug and vaccine design against Novel Coronavirus (2019-nCoV) spike protein through Computational approach. Preprints. 2020:1–16. [Google Scholar]
- 27.Neuman B.W., Adair B.D., Yoshioka C. Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. J Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia S., Zhu Y., Liu M. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoeman D., Fielding B.C. Coronavirus envelope protein: Current knowledge. Virol J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glowacka I., Bertram S., Muller M.A. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the perfusion conformation. Science. 2020;(80):1–9. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan Y., Shang J., Graham R. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol. 2020:1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pradhan P., Pandey A.K., Mishra A. Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag. bioRxiv Prepr. 2020:1–14. [Google Scholar]
- 34.Huang Q., Yu L., Petros A.M. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 35.V’kovski P., Gerber M., Kelly J. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife. 2019;8:1–30. doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan X., Hao Q., Mu Y. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int J Biochem Cell Biol. 2006;38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Zeng Y., Ye L., Zhu S. The nucleocapsid protein of SARS-associated coronavirus inhibits B23 phosphorylation. Biochem Biophys Res Commun. 2008;369:287–291. doi: 10.1016/j.bbrc.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q., Li C., Zhang Q. Interactions of SARS Coronavirus Nucleocapsid Protein with the host cell proteasome subunit p42. Virol J. 2010;7:1–8. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X., Pan J., Tao J. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surjit M., Liu B., Chow V.T.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou B., Liu J., Wang Q. The Nucleocapsid Protein of Severe Acute Respiratory Syndrome Coronavirus Inhibits Cell Cytokinesis and Proliferation by Interacting with Translation Elongation Factor 1. J Virol. 2008;82:6962–6971. doi: 10.1128/JVI.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo H., Chen Q., Chen J. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–2628. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S., Kang S., Yang M. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. bioRxiv Prepr. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C.K., Sue S.C., Yu T.H. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C.-K., Hsu Y.-L., Chang Y.-H. Multiple Nucleic Acid Binding Sites and Intrinsic Disorder of Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein: Implications for Ribonucleocapsid Protein Packaging. J Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C.K., Hou M.H., Chang C.F. The SARS coronavirus nucleocapsid protein - Forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo H., Ye F., Chen K. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 50.Chen C.Y., Chang C.K., Chang Y.W. Structure of the SARS Coronavirus Nucleocapsid Protein RNA-binding Dimerization Domain Suggests a Mechanism for Helical Packaging of Viral RNA. J Mol Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruch T.R., Machamer C.E. The Hydrophobic Domain of Infectious Bronchitis Virus E Protein Alters the Host Secretory Pathway and Is Important for Release of Infectious Virus. J Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: A small membrane protein with multiple functions. Cell Mol Life Sci. 2007;64:2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: Roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta - Biomembr. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen X., Xue J.H., Yu C.Y. Small envelope protein E of SARS: Cloning, expression, purification, CD determination, and bioinformatics analysis. Acta Pharmacol Sin. 2003;24:505–511. [PubMed] [Google Scholar]
- 56.Gupta M.K., Vemula S., Donde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2020;0:1–17. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta-Biomembr. 2018;1860:1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bianchi M., Benvenuto D., Giovanetti M. Sars-CoV-2 Envelope and Membrane proteins: differences from closely related proteins linked to cross-species transmission? Preprints. 2020:1–14. doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tseng Y.T., Wang S.M., Huang K.J. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J Biomed Sci. 2014;21:1–11. doi: 10.1186/1423-0127-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Y., Hogue B.G. Role of the Coronavirus E Viroporin Protein Transmembrane Domain in Virus Assembly. J Virol. 2007;81:3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi Complex-Targeting Signal in the Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. J Virol. 2011;85:5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teoh K.-T., Siu Y.-L., Chan W.-L. The SARS Coronavirus E Protein Interacts with PALS1 and Alters Tight Junction Formation and Epithelial Morphogenesis. Mol Biol Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arndt A.L., Larson B.J., Hogue B.G. A Conserved Domain in the Coronavirus Membrane Protein Tail Is Important for Virus Assembly. J Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neuman B.W., Kiss G., Kunding A.H. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tseng Y.T., Wang S.M., Huang K.J. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J Biol Chem. 2010;285:12862–12872. doi: 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang X., Gao J., Zheng H. The Membrane Protein of SARS-CoV Suppresses NF-kB Activation. J Med Virol. 2007;79:1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsoi H., Li L., Chen Z.S. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1PKB/Akt signalling. Biochem J. 2014;464:439–447. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y., Liu L. The Membrane Protein of Severe Acute Respiratory Syndrome Coronavirus Functions as a Novel Cytosolic Pathogen-Associated Molecular Pattern To Promote Beta Interferon Induction via a Toll. MBio. 2016;7 doi: 10.1128/mBio.01872-15. e01872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.