Abstract
Reducing the incidence of both the degree of assistance required at calving, as well as the extent of perinatal mortality (PM) has both economic and societal benefits. The existence of heritable genetic variability in both traits signifies the presence of underlying genomic variability. The objective of the present study was to locate regions of the genome, and by extension putative genes and mutations, that are likely to be underpinning the genetic variability in direct calving difficulty (DCD), maternal calving difficulty (MCD), and PM. Imputed whole-genome single-nucleotide polymorphism (SNP) data on up to 8,304 Angus (AA), 17,175 Charolais (CH), 16,794 Limousin (LM), and 18,474 Holstein-Friesian (HF) sires representing 5,866,712 calving events from descendants were used. Several putative quantitative trait loci (QTL) regions associated with calving performance both within and across dairy and beef breeds were identified, although the majority were both breed- and trait-specific. QTL surrounding and encompassing the myostatin (MSTN) gene were associated (P < 5 × 10−8) with DCD and PM in both the CH and LM populations. The well-known Q204X mutation was the fifth strongest association with DCD in the CH population and accounted for 5.09% of the genetic variance in DCD. In contrast, none of the 259 segregating variants in MSTN were associated (P > × 10−6) with DCD in the LM population but a genomic region 617 kb downstream of MSTN was associated (P < 5 × 10−8). The genetic architecture for DCD differed in the HF population relative to the CH and LM, where two QTL encompassing ZNF613 on Bos taurus autosome (BTA)18 and PLAG1 on BTA14 were identified in the former. Pleiotropic SNP associated with all three calving performance traits were also identified in the three beef breeds; 5 SNP were pleiotropic in AA, 116 in LM, and 882 in CH but no SNP was associated with more than one trait within the HF population. The majority of these pleiotropic SNP were on BTA2 surrounding MSTN and were associated with both DCD and PM. Multiple previously reported, but also novel QTL, associated with calving performance were detected in this large study. These also included QTL regions harboring SNP with the same direction of allele substitution effect for both DCD and MCD thus contributing to a more effective simultaneous selection for both traits.
Keywords: calving difficulty, genome-wide association, mortality, pleiotropy, sequence, single-nucleotide polymorphism
Introduction
The association between the extent of assistance required at calving and subsequent performance has been extensively documented in both dairy (Dematawewa and Berger, 1997; Berry et al., 2007; Eaglen et al., 2011) and beef (Drennan and Berry, 2006) cattle; similarly, the association between perinatal mortality (PM) and subsequent performance has been reported in several cattle populations (Berry et al., 2007). Calving dystocia has been associated with reduced subsequent reproductive performance (Dematawewa and Berger, 1997; Berry et al., 2007; Eaglen et al., 2011), reduced milk production (Dematawewa and Berger, 1997; Berry et al., 2007; Eaglen et al., 2011), greater body condition score loss postcalving (Berry et al., 2007), as well as compromised health (Berry et al., 2007) in dairy cattle; the association in dairy cattle between dystocia and both calf performance and survival has also been documented (Peeler et al., 1994; Meyer et al., 2001; Berry et al., 2007). While a reduction in performance obviously affects farm profit, the periparturient period is also known to be a very stressful period in the lifetime of, not only the mature female, but also the progeny (Eaglen et al., 2011). Hence, reducing the extent of assistance required during the birthing process, as well as reducing PM rates, should not only improve herd profit but also the well-being status of the herd. Therefore, all potentially useful strategies to ameliorate these repercussions should be thoroughly investigated. Breeding program, with or without the exploitation of genomic information, as an approach to reduce the reliance on calving assistance or reducing the risk of PM is one such strategy.
The extent of genetic variability underlying phenotypic differences in calving dystocia has been well-documented in both dairy (Eaglen and Bijma, 2009) and beef cattle (Mujibi and Crews, 2009; Crowley et al., 2011); similarly, genetic variation in PM in cattle is also known to exist (Crowley et al., 2011; Ring et al., 2018). While periparturient management strategies are known to influence the risk of calving difficulty, direct heritability estimates for calving dystocia in cattle have been documented to range from 0.08 to 0.24 (Eaglen and Bijma, 2009; Mujibi and Crews, 2009; Crowley et al., 2011) with reported maternal heritability estimates ranging from 0.02 to 0.10 (Eaglen and Bijma, 2009; Mujibi and Crews, 2009; Crowley et al., 2011). Direct and maternal heritability estimates for PM in cattle have been reported to range from 0.01 to 0.03 (Crowley et al., 2011; Ring et al., 2018). Hence, being able to explain the additive genetic variation in both calving difficulty (i.e., direct and maternal) and PM through genomic markers, or the causal mutations themselves, could not only aid breeding schemes attempting to reduce the incidence of both, but such information could also facilitate optimized mating programs as a tool to minimize the likelihood of requiring assistance at calving and/or of PM.
The objective of the present study was to use imputed whole-genome sequence (WGS) data from a large population of cattle in an attempt to locate regions of the bovine genome thought to harbor genes and polymorphisms contributing to the genetic variability in both direct (DCD) and maternal calving difficulty (MCD) as well as PM. Of particular interest in the present study was the exploitation of data from multiple breeds in an attempt to quantify if the associated regions persisted across breeds, but also if commonalities existed between the detected genomic regions for the three traits investigated thus aiding in the resolving of known genetic antagonisms (Crowley et al., 2011).
Materials and Methods
The data used in the present study originated from a preexisting database managed by the Irish Cattle Breeding Federation (ICBF). Therefore, it was not necessary to obtain animal care and use committee approval in advance of conducting this study.
Phenotypes
In Ireland, calving difficulty is subjectively scored by producers on a linear scale of 1 to 4 at the time of birth, where 1 = no calving assistance; 2 = slight assistance (assistance by one person, without needing to use a calf puller); 3 = considerable assistance (assistance by one person using a calf puller or more than one person); 4 = veterinary assistance (including caesarean). Perinatal mortality is also recorded as a binary variable by producers, and indicates whether the calf died within a 24-h period after birth.
Estimated breeding values (EBVs) for DCD and MCD, as well as direct PM with their respective reliability estimates were obtained from the ICBF database from the December 2018 national genetic evaluation. The DCD and MCD breeding value estimates were generated in a direct-maternal multibreed genetic evaluation, whereas PM estimates were generated in a univariate multibreed model (i.e., no maternal genetic component); the heritability of each trait in the respective models was 9% for DCD, 2% for MCD, and 2% for PM. Of the animals with generated EBVs, only purebred (i.e., ≥87.5% of a single breed) genotyped sires of any of the four numerically largest breeds were retained for analysis; Angus (AA), Charolais (CH), Holstein-Friesian (HF), and Limousin (LM). The effective record contribution (ERC) of each sire, taking into consideration the genotyped animals, was estimated using the Harris and Johnston (1998) method; only animals with an ERC ≥ 1 were retained for use in the present study. Deregression of the EBVs was completed using the secant method with a full animal model pedigree file. The number of animals per trait after edits is detailed in Table 1, as well as the median ERC.
Table 1.
The number of animals included per trait and breed
| Trait | AA | CH | HF | LM | All |
|---|---|---|---|---|---|
| Direct calving difficulty | |||||
| Number of animals | 8,304 | 17,175 | 18,191 | 16,794 | 60,464 |
| Median ERC | 7.31 | 13.38 | 2.23 | 13.57 | 8.40 |
| Number of SNP after edits | 14,778,952 | 16,091,418 | 14,331.494 | 16,144,687 | 16,713,783 |
| Maternal calving difficulty | |||||
| Number of animals | 7,013 | 11,792 | 16,265 | 12,683 | 47,753 |
| Median ERC | 2.79 | 3.49 | 3.79 | 4.12 | 3.56 |
| Number of SNP after edits | 14,799,631 | 16,131,655 | 14,380,942 | 16,1814,561 | 16,498,270 |
| Direct mortality | |||||
| Number of animals | 8,104 | 16,446 | 18,474 | 15,873 | 58,897 |
| Median ERC | 6.76 | 10.87 | 3.35 | 11.32 | 7.48 |
| Number of SNP after edits | 14,791,231 | 16,088,788 | 14,348,548 | 16,148,558 | 16,487,386. |
Genotype data
Genotypes of 60,464 sires with calving EBVs were imputed to WGS as part of a larger data set of 638,662 genotyped animals from multiple breeds as detailed previously by Purfield et al. (2019a). All animals included in the present study were genotyped on a variety of genotype panels including the Illumina High Density (HD; n = 3,042; 777,962 SNP), Illumina Bovine SNP50 (n = 1,094; 54,001 SNP), the custom Irish Dairy and Beef (IDB) V1 (n = 14,001; 16,622 SNP), the IDBV2 (n = 26,284; 16,223 SNP), or the IDBV3 (n = 16,043; 52,445 SNP) genotype panels. All 638,662 genotyped animals had a call rate ≥ 90% and only autosomal SNP, SNP with a known chromosome and position on UMD 3.1, and only SNP with a call rate ≥ 90% were retained within each panel.
Prior to imputation to WGS, all genotyped animals of the larger 638,662 genotyped data set were first imputed to HD using a two-step approach in FImpute (Sargolzaei et al., 2014); this involved imputing all IDB-genotyped animals to the Bovine SNP50 density and subsequently imputing all resulting genotypes, including the Bovine SNP50 genotypes, to HD using a multibreed reference population of 5,504 HD-genotyped animals. Imputation of all 638,662 HD-imputed animals to WGS was then undertaken using a reference population of 2,333 Bos taurus animals of multiple breeds from Run6.0 of the 1000 Bull Genomes Project. Imputation of the HD genotypes to WGS was achieved by firstly phasing all 638,662 HD-imputed animals using Eagle (Loh et al., 2016; version 2.3.2) and subsequently imputing all animals to WGS using minimac3 (Das et al., 2016).
Regions with possible poor WGS imputation accuracy (n = 687,137 SNP) were identified using a data set of 147,309 verified parent–progeny relationships from the 638,662 genotyped data set as previously described by Purfield et al. (2019a) and subsequently removed from the data set. In addition, all SNP with a minor allele frequency (MAF) < 0.005 across all animals were removed for the multibreed analysis and all SNP with a MAF < 0.005 within each breed were removed for the respective within-breed analysis. The number of SNP that remained for analysis within each breed is detailed in Table 1.
Genome-wide association analyses
Whole-genome association analyses were performed within each breed separately, as well as in a data set of all breeds combined, using an animal linear mixed model in Wombat (Meyer and Tier, 2012). The VanRaden Method I (2008) was used to create a genomic relationship matrix based on just the imputed autosomal SNP from the edited HD panel (n = 642,153 SNP). All imputed sequence SNP, scored as 0, 1, or 2, were included individually as a fixed effect covariate in the model 1 at a time. Breed was also included as a fixed effect for the multibreed analyses. Each dependent variable was also weighted using the approach outlined by Garrick et al. (2009);
where wi is the weighting factor of the ith deregressed EBV, h2 is the heritability estimate for the trait in question, is the reliability of the ith deregressed EBV, and c is the proportion of genetic variance not accounted by the SNP and set at 0.9 for analyses thus allowing each SNP to attribute up to 10% of the genetic variance. Test statistics for all SNP were obtained and converted into their corresponding P-values. The genomic inflation factor was estimated within each breed and ranged from 0.99 in the AA population to 1.08 in the LM population, whereas the across-breed inflation factor ranged from 0.98 for MCD to 1.07 for DCD. A genome-wide SNP significance threshold of P ≤ 5 × 10−8 and a suggestive threshold of P ≤ 1 × 10−6 were applied to each analysis (Pe’er et al., 2008). The proportion of the genetic variance attributable to individual SNP was calculated as 2pqa2/σ 2, where p was the major allele frequency, q was the MAF, a was the estimated allele substitution effect, and σ 2 was the genetic variance for the respective trait.
Defining quantitative trait loci
The definition of a quantitative trait loci (QTL) was undertaken as outlined in detail by Purfield et al. (2019a) and McGovern et al. (2019). All SNP surpassing the suggestive significance threshold (P ≤ 1 × 10−6) were used for defining QTL regions associated with each trait separately. To estimate the QTL start and end positions, all SNP within a 5-Mb window and in strong linkage disequilibrium (LD) (r2 of ≥ 0.7) with each suggestively associated SNP were considered to be part of the QTL. Overlapping QTL were merged together and considered as the same QTL. To limit the number of false-positive QTL, a minimum of two suggestively associated SNP had to be present in the QTL region. Genes within and overlapping each QTL were identified using NCBI map viewer (http://www.ncbi.nlm.nih.gov/mapview) and Ensembl (http://ensemble.org) based on the bovine UMD 3.1 build. Candidate genes were chosen from the QTL region based on previous literature and their biological function. If no gene resided in the QTL region, genes within 250 kb of the start and end position of the QTL were considered as putative candidate genes. Previously reported cattle QTL were obtained from the animal QTLdb (http://www.animalgenome.org/cgi-bin/QTLdb/index). In addition, to identify QTL present in more than one breed, each chromosome was split into 500-kb windows and each window that contained a SNP with a P ≤ 1 × 10−4 present in two or more breeds was considered a putative across-breed QTL. A similar approach was used to detect QTL common to all three calving traits. This threshold was previously applied by Tenghe et al. (2016) when detecting across-trait QTL thus enabling the identification of putative across-breed genomic regions with less stringency.
Results
Genotypes from more than 40 million sequence variants were imputed for the 60,464 sires from the four cattle breeds which represented a total of 5,866,712 calving events from their descendants used in the estimation of their EBVs.
Direct calving difficulty
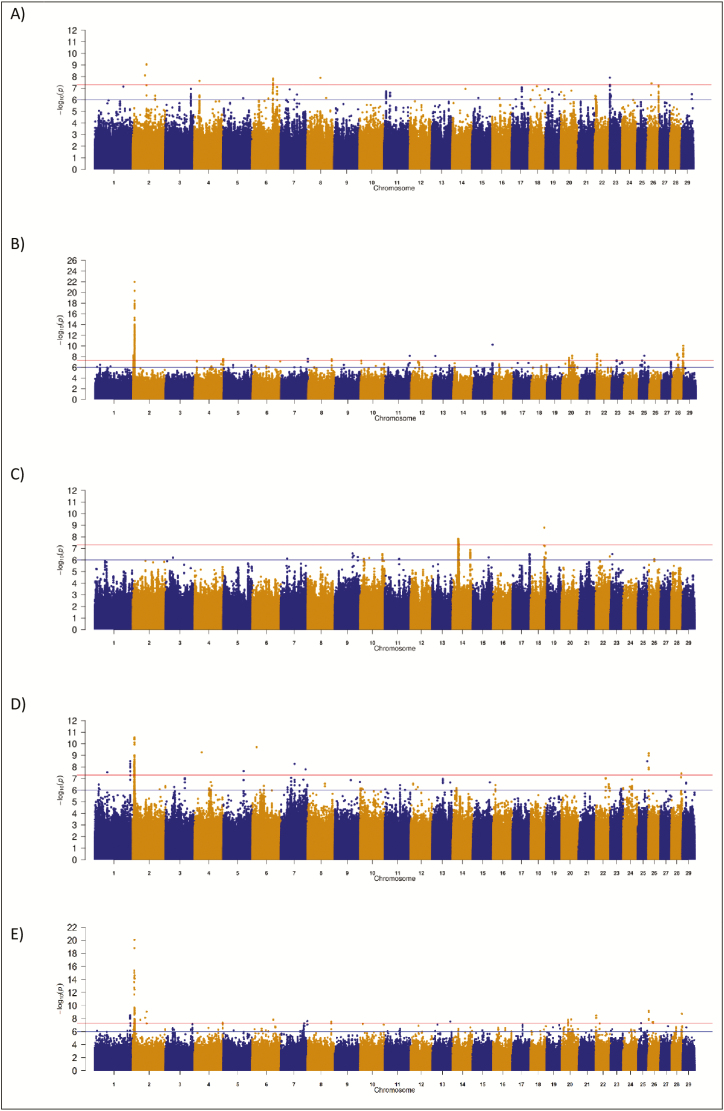
As more genome-wide significant SNP were associated with DCD than both MCD and PM, only QTL regions containing genome-wide significant (P < 5 × 10−8) SNP are listed in Table 2 for DCD. The number of significant QTL regions associated with DCD differed per breed ranging from 2 QTL regions in the HF and LM populations to 17 in the CH population (Table 2). The strongest association with DCD resided on Bos taurus autosome (BTA)2 in the CH population where a QTL region encompassing myostatin (MSTN) contained 146 significantly associated (P < 5 × 10−8) SNP (Figure 1B). The fifth strongest association within this QTL on BTA2 in the CH population was the well-known Q204X stop-gain mutation within MSTN which had a P-value of 1.42 × 10−14 and explained 5.09% of the genetic variance in DCD. In total, 44 MSTN-annotated SNP within this QTL spanning from 58.72 to 67.21 Mb were significantly associated (P < 5 × 10−8) with DCD in the CH population. Overall, there were 26 CH animals within the edited data set that were homozygous for the T allele at the Q204X mutation and these had greater (P < 0.001) calving difficulty (mean DCD EBV 0.482, SD 0.461) than the 15,452 noncarriers of the Q204X mutation (mean DCD EBV of 0.182, SD 0.214) (Supplementary Figure 1). Similar to CH, the strongest association within the LM population was also on BTA2, but it was 617 kb downstream of the MSTN. Indeed none of the 259 SNP within MSTN segregating in the LM were associated (P > 1 × 10−6) with DCD in the LM population; the A allele of the F94L (rs11065568) mutation in MSTN (P = 1.54 × 10−4) had a positive allele substitution effect on DCD.
Table 2.
QTL associated with direct calving difficulty1
| Breed | BTA | Start | End | No. SNP | Strongest SNP | Strongest SNP position | + Allele | + Allele freq | P-value | SNP effect MCD | SNP effect PM | No. genes | Candidate gene(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | 2 | 58317099 | 58398267 | 4 | rs799051476a | 58388382 | G | 0.012 | 8.76 × 10−10 | ++ | ++ | 0 | N/A |
| 4 | 21958676 | 22090437 | 38 | rs449164295b | 22090437 | T | 0.029 | 2.36 × 10−8 | −− | ++ | 1 | ETV1 | |
| 6 | 88842546 | 88935317 | 9 | rs800690355a | 88935317 | A | 0.007 | 1.52 × 10−8 | ++ | ++ | 0 | GC | |
| CH | 2 | 1850243 | 1938014 | 44 | rs110820498a | 1925667 | T | 0.090 | 5.74 × 10−9 | − | ++ | 1 | PLEKHB2 |
| 2 | 3201163 | 3241531 | 14 | rs384824032a | 3241507 | T | 0.240 | 6.09 × 10−9 | − | ++ | 0 | N/A | |
| 2 | 3887357 | 3902265 | 20 | rs210421829a | 3899839 | A | 0.457 | 5.53 × 10−9 | − | ++ | 0 | N/A | |
| 2 | 5905672 | 6720629 | 192 | rs482419628a | 6630781 | G | 0.054 | 1.09 × 10−22 | −− | ++ | 9 | MSTN | |
| 2 | 6727404 | 7212998 | 260 | rs799943285b | 6808074 | G | 0.058 | 4.95 × 10−21 | −− | ++ | 1 | SLC40A1 † | |
| 2 | 7334284 | 7678545 | 6 | rs798702162a | 7609482 | G | 0.051 | 8.61 × 10−13 | −− | ++ | 1 | COL3A1 | |
| 4 | 118379848 | 118960619 | 12 | rs377892980d | 118932719 | C | 0.990 | 2.80 × 10−8 | − | ++ | 4 | RNF32 † , LMBR1, NOM1 | |
| 7 | 111633395 | 111639100 | 2 | rs383826293a | 111633395 | T | 0.008 | 2.49 × 10−8 | − | ++ | 0 | N/A | |
| 8 | 99124662 | 99444787 | 4 | .a | 99124662 | C | 0.988 | 3.04 × 10−8 | ++ | + | 2 | KLF4 | |
| 15 | 80425653 | 80425659 | 2 | rs42805962d | 80425653 | C | 0.008 | 5.81 × 10−11 | −− | −− | 1 | ENSBTAG00000035988 † | |
| 20 | 27639892 | 28292172 | 14 | .a | 27888972 | G | 0.011 | 1.74 × 10−8 | − | ++ | 1 | ISL1 | |
| 20 | 40735668 | 41092503 | 7 | rs717218519b | 41009804 | G | 0.013 | 6.01 × 10−9 | ++ | ++ | 1 | NPR3 † | |
| 22 | 988356 | 1889045 | 11 | .a | 1889045 | A | 0.007 | 3.55 × 10−9 | NA | − | 8 | SLC4A7 | |
| 23 | 21839465 | 21839742 | 4 | rs460572227b | 21839717 | T | 0.007 | 4.87 × 10−8 | − | ++ | 1 | ENSBTAG00000046237 † | |
| 25 | 22436613 | 22497098 | 2 | rs385805439c | 22458425 | T | 0.005 | 6.56 × 10−9 | NA | + | 1 | RBBP6 † | |
| 28 | 20486363 | 20541765 | 14 | rs208597252a | 20506931 | C | 0.012 | 3.23 × 10−9 | −− | −− | 0 | N/A | |
| 28 | 24368554 | 24389785 | 3 | rs210629062a | 24368554 | C | 0.035 | 3.61 × 10−8 | ++ | + | 0 | N/A | |
| 28 | 45109768 | 45388748 | 24 | rs476163771a | 45359784 | A | 0.986 | 8.80 × 10−11 | ++ | −− | 0 | TMEM72 | |
| HF | 14 | 24847507 | 27124281 | 262 | rs134489103d | 24953585 | T | 0.968 | 1.51 × 10−8 | + | − | 17 | PLAG1 |
| 18 | 58141989 | 58407174 | 8 | rs381577268d | 58141989 | T | 0.025 | 1.63 × 10−9 | + | − | 4 | ZNF613 † | |
| LM | 1 | 147046621 | 147071689 | 5 | .b | 147046683 | G | 0.009 | 3.21 × 10−9 | − | + | 1 | SLC19A1 † |
| 2 | 5751482 | 5769372 | 52 | rs211366913d | 5751482 | G | 0.048 | 6.19 × 10−9 | − | ++ | 1 | MFSD6 † | |
| 2 | 6692259 | 6837692 | 94 | rs137213740a | 6696029 | T | 0.909 | 7.69 × 10−11 | −− | ++ | 1 | SLC40A1 | |
| 5 | 85007482 | 85162856 | 3 | rs111563671c | 85162856 | T | 0.972 | 2.29 × 10−8 | + | + | 1 | KRAS † | |
| 7 | 57666736 | 58298162 | 4 | rs133717945a | 57666736 | A | 0.067 | 5.43 × 10−9 | + | + | 1 | ENSBTAG00000019739 | |
| 26 | 2392863 | 2461384 | 5 | rs111672216a | 2461384 | T | 0.007 | 6.67 × 10−10 | ++ | − | 0 | N/A | |
| 28 | 40647337 | 42372750 | 2 | rs378942567a | 42372750 | G | 0.023 | 3.57 × 10−8 | − | ++ | 16 | PPYR1 |
1No. SNP is the number of SNP within the QTL with a P-value ≤ 5 × 10−8. P-value is the P-value of strongest SNP association. + Allele is the allele that had a positive SNP effect on direct calving difficulty. SNP effect MCD is the QTL-lead SNP’s direction effect for maternal calving difficulty; + represents SNP effects in the same direction, ++ represents the same direction and large SNP effect size for MCD, i.e., a SNP effect size in the top 20%, − represents opposing SNP effects and −− represents opposing SNP effect directions that have a large SNP effect size. N/A (Non-applicable) was if the SNP wasn’t present in that dataset due to allele frequency. SNP effect PM similar to MCD but represents the SNP effects for PM. No. of genes is the number of genes in the QTL. If no gene was present in the QTL the nearest functional candidate gene within 250 kb was chosen. aintergenic, bintronic, cupstream gene variant, ddownstream gene variant, †gene in which the most significant SNP within the QTL was identified in.
Figure 1.
Manhattan plots for direct calving difficulty in (A) Angus, (B) Charolais, (C) Holstein Friesian, (D) Limousin, and (E) across all breeds. The red line indicates the genome-wide significance threshold of P ≤ 5 × 10−8 and the blue is the suggestive significance threshold of P ≤ 1 × 10−6.
The strongest association in the AA population for DCD was on BTA6 in the QTL spanning from 88.67 to 88.94 Mb which contained the GC gene which encodes for the vitamin D-binding protein; no SNP within GC was significantly or suggestively associated with DCD with the strongest association (rs211646036) having a P-value of 5.73 × 10−4. The strongest association within the HF population was rs381577268 (P-value = 1.63 × 10−9), a downstream variant of zinc finger protein 613 (ZNF613) located in a QTL positioned between 58.14 and 58.41 Mb on BTA18; the SNP accounted for 1.32% of the genetic variance in DCD (Table 2). Within the edited data set, 19 animals were homozygous for the T allele of the rs381577268 SNP and these animals had greater (P < 0.001) EBVs (i.e., worse) for DCD (mean EBV 0.118, SD 0.148) than both heterozygous (n = 865; mean EBV 0.002, SD 0.102) and homozygous CC sires (n = 17,307; mean EBV −0.056, SD 0.069) (Supplementary Figure 2). The allele frequency of the T allele was, nonetheless, low within the HF population with a frequency of 0.025. Similar low frequencies were also observed in the beef populations where the T allele had a frequency of 0.007 and 0.005 in the AA and CH populations, respectively, and below the MAF threshold for inclusion in the LM population (MAF = 0.002). Despite rs381577268 segregating within each of the beef populations investigated, neither it, nor any other variant within ZNF613, was associated with DCD in these breeds. However, several SNP from 57.88 to 57.98 Mb in the CH population and from 58.01 to 58.23 Mb in the LM population did have P-values ranging from 5.64 × 10−5 to 4.81 × 10−3 for DCD.
Another plausible QTL was also identified in the HF population on BTA14 in the region of 24.48 to 28.12 Mb encompassing the pleomorphic adenoma gene 1 (PLAG1) gene. In total, 58 SNP within this QTL were significantly associated with DCD and the most significant SNP, rs134489103, was only 53 kb upstream of PLAG1; 21 PLAG1-annotated SNP were segregating in the HF population and their P-values ranged from 8.81 × 10−7 to 9.01 × 10−6. This QTL on BTA14 is most likely a dairy-specific QTL as none of the 835 (CH) to 1219 (AA) segregating variants within or 250 kb up/downstream of PLAG1 were associated with DCD in the beef populations analyzed (P > 0.023).
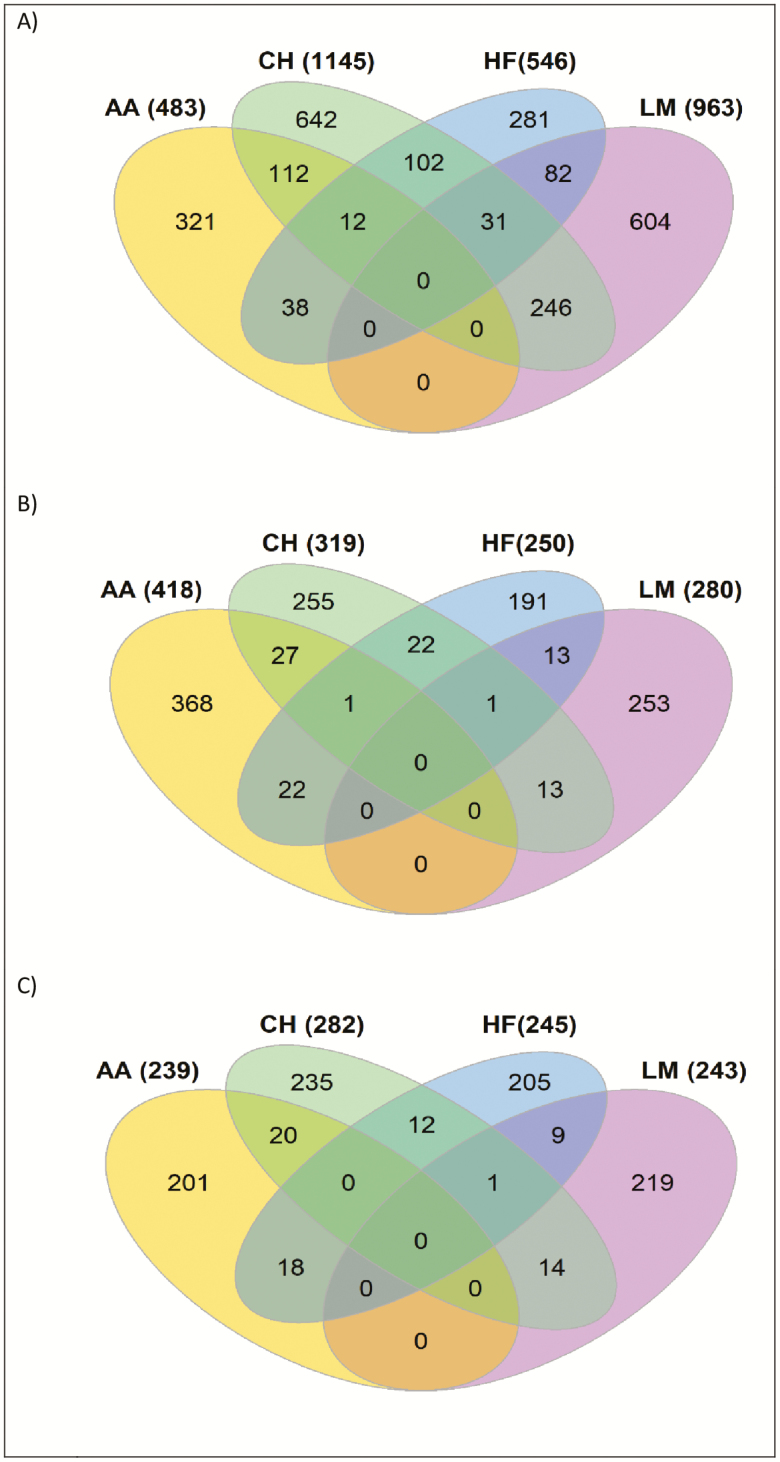
The across-breed analyses involving all 60,464 sires with breed fitted as a fixed effect for DCD identified 115 significant and 289 suggestively associated SNP. Twenty-seven QTL across 18 BTAs where identified, of which the strongest QTL association was located on BTA2 encompassing MSTN (Supplementary Table 1). Multiple novel QTL associations were identified, but the allele frequency of the lead variant within these QTL tended to be quite low; of the 27 QTL identified, the lead variant within 14 of the QTL had a MAF < 0.01. As genomic regions rather than individual SNP may be influencing DCD across all breeds, overlapping 500-kb windows that contained at least one SNP with a P-value < 1 × 10−4 within each breed were identified (Figure 2). The majority of the 500-kb windows harboring a SNP with a P-value < 1 × 10−4 were unique to a single breed and no window was significant (P < 1 × 10−4) in all four breeds for DCD. Despite this, 31 windows (out of a total of 2,150 unique windows across all breeds) were shared between the CH, LM, and HF populations; these were spread across multiple autosomes (BTA1, 3, 4, 5, 7, 9, 10, 15, 18, 23, 24, and 28) and include plausible candidate genes such as INSIG1 on BTA4, NUDT12 on BTA7, GMDS on BTA23, and GRID which encodes GluD1 on BTA28. In addition, a further 12 genomic windows across 11 autosomes were shared between the AA, LM, and HF populations and included the candidate genes zinc finger E-box-binding homeobox 2 (ZEB2) on BTA2, PDCD5 on BTA18, and ARHAP5 and AKAP6 on BTA21. Although a large proportion of windows were shared between the AA and CH populations, and between the CH and LM populations, no windows were shared among all three beef breeds.
Figure 2.
The number of overlapping 500-kb windows that contained a SNP with a P-value < 10−4 for (A) direct calving difficulty, (B) maternal calving difficulty, and (C) PM. Yellow represents Angus (AA), green is Charolais (CH), blue is Holstein-Friesian (HF), and pink is Limousin (LM).
Maternal calving difficulty
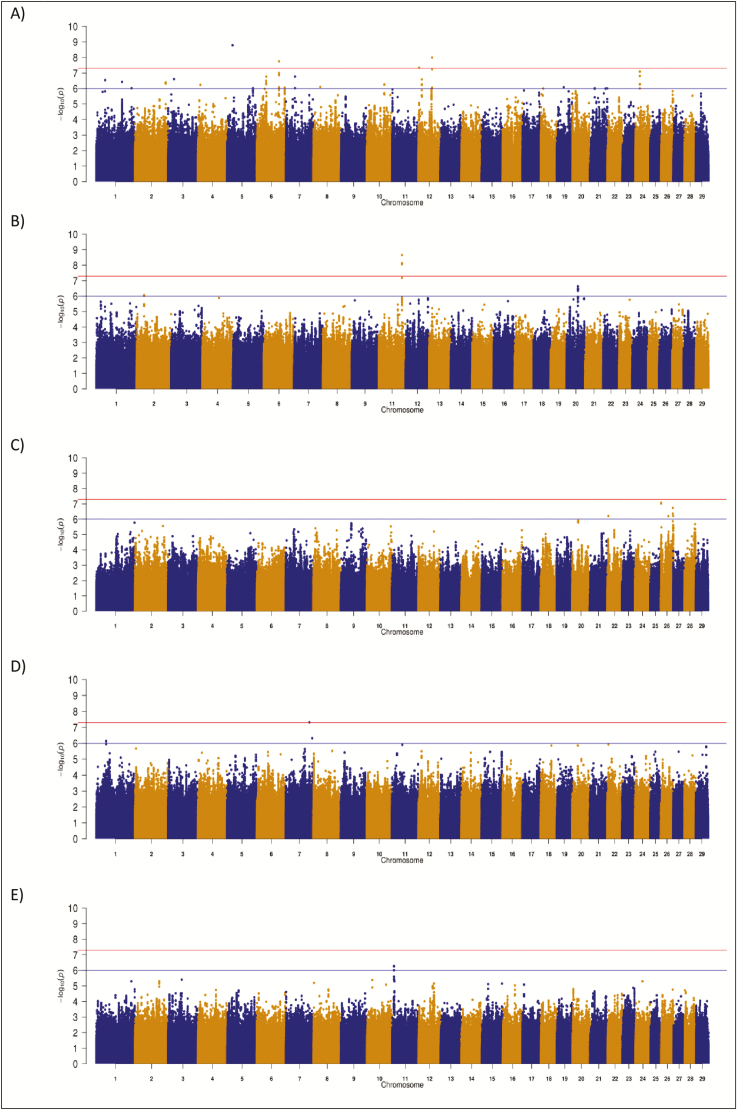
Genome-wide significant associations with MCD were detected in all three beef breeds but not in the HF population. The AA population had the greatest number of QTL associated with MCD; in total, 12 QTL regions contained SNP that were suggestively (P < 1 × 10−6) associated with MCD (Table 3). The strongest association within the AA population was an intergenic variant, rs798343691, on BTA5 (P = 1.66 × 10−9) (Figure 3), although no other SNP in LD (r2 ≥ 0.7) with this SNP in the AA was suggestively associated with MCD and, as such, it was not considered a putative QTL region. The QTL region containing the strongest association within the AA population was on BTA12 spanning from 56.67 to 56.68 Mb where one SNP was significantly associated with MCD and four others were suggestively associated with MCD; no putative candidate gene within 250 kb upstream or downstream was identified. Several intronic variants within the candidate gene prostate androgen-regulated mucin-like protein 1 (PARM1) were suggestively associated with MCD in the QTL from 91.60 to 91.63 Mb on BTA6 but, of the two segregating missense variants within PARM1 (rs111027720 and rs207780467) captured within the data set, neither were significant (P-value ≥ 0.1). In addition, a QTL region surrounding the well-known stature-associated genes, non-SMC condensin I complex subunit G (NCAPG) and ligand-dependent nuclear receptor corepressor-like (LCORL), was also associated with MCD in AA, although none of the 241 and 553 segregating SNP actually within each of the genes, respectively, were suggestively associated with MCD. The strongest association with MCD in the CH population was on BTA11, where 11 variants within a QTL from 92.40 to 92.47 Mb were significantly associated with MCD. No candidate genes were identified within the QTL boundaries although both GGTA1 and DAB2IP were located within 250 kb of the QTL. The most significant variant within this QTL (rs380146645) in the CH population explained 3.76% of the genetic variance in MCD but the favorable C allele associated with an improvement in MCD was near-fixation (allele frequency = 0.994). In total, three QTL were identified to be suggestively associated with MCD in the HF population, all of which were located on BTA26; only one QTL on BTA1 was identified in the LM population (Table 3). Plausible candidate genes identified in these QTL include GLRX3 and DPYSL4 in the HF population and EPHA6 in the LM population.
Table 3.
QTL associated with maternal calving difficulty1
| Breed | BTA | Start | End | No. SNP | Strongest SNP | Strongest SNP position | + Allele | + Allele freq | P-value | SNP effect DCD | SNP effect PM | No. genes | Candidate gene(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | 1 | 36828793 | 36860576 | 2 | a. | 36828793 | C | 0.008 | 2.89 × 10−7 | −− | −− | 1 | EPHA3 |
| 1 | 144496949 | 144546610 | 2 | rs479310525b | 144545050 | T | 0.029 | 9.61 × 10−7 | −− | − | 1 | PDE9A † | |
| 2 | 126264896 | 126316073 | 6 | rs482380239b | 126278844 | C | 0.989 | 4.09 × 10−7 | + | − | 1 | FGR † | |
| 4 | 10350276 | 10372066 | 3 | rs523829414c | 10369692 | A | 0.005 | 5.84 × 10−7 | + | − | 1 | HEPACAM2 † | |
| 5 | 105753443 | 105755633 | 2 | rs209426521a | 105753443 | T | 0.015 | 9.60 × 10−7 | + | + | 0 | N/A | |
| 6 | 38518710 | 39614235 | 4 | rs111701266a | 39097535 | G | 0.007 | 1.71 × 10−7 | −− | + | 7 | NCAPG,LCORL | |
| 6 | 91602424 | 91626735 | 7 | rs210873983b | 91602424 | A | 0.011 | 1.76 × 10−8 | ++ | ++ | 1 | PARM1 † | |
| 7 | 37616120 | 37616155 | 2 | rs378671192a | 37616155 | A | 0.006 | 1.71 × 10−7 | ++ | −− | 0 | N/A | |
| 10 | 72286947 | 72287008 | 3 | rs381793861a | 72286947 | T | 0.009 | 5.59 × 10−7 | − | ++ | 0 | N/A | |
| 12 | 14636353 | 14960925 | 3 | rs800390196a | 14750727 | A | 0.014 | 2.63 × 10−7 | + | + | 2 | TSC22D1 | |
| 12 | 56675034 | 56682826 | 5 | rs209495875a | 56676660 | A | 0.044 | 1.04 × 10−8 | + | − | 0 | N/A | |
| 24 | 19405971 | 19427409 | 5 | rs109333678a | 19405971 | G | 0.995 | 8.17 × 10−8 | ++ | −− | 0 | N/A | |
| CH | 2 | 29919882 | 29920135 | 3 | rs380146645a | 29919882 | G | 0.017 | 8.67 × 10−7 | −− | − | 0 | N/A |
| 11 | 92402757 | 92405438 | 12 | rs445896050a | 92402757 | T | 0.006 | 2.26 × 10−9 | −− | + | 0 | N/A | |
| 20 | 44514471 | 44520630 | 6 | rs379731054a | 44518024 | A | 0.014 | 2.35 × 10−7 | −− | −− | 0 | N/A | |
| HF | 26 | 1759408 | 2501334 | 3 | rs133176675a | 1765944 | A | 0.924 | 8.48 × 10−8 | ++ | −− | 0 | N/A |
| 26 | 49916091 | 49918252 | 2 | rs110755029a | 49918252 | C | 0.788 | 1.83 × 10−7 | ++ | + | 0 | GLRX3 | |
| 26 | 51542180 | 51546414 | 3 | rs209816010b | 51542180 | T | 0.892 | 6.16 × 10−7 | ++ | + | 1 | STK32C † | |
| LM | 1 | 40539352 | 40546791 | 2 | rs380343675a | 40539352 | C | 0.032 | 7.11 × 10−7 | −− | + | 0 | EPHA6 |
1No. SNP is the number of SNP within the QTL with a P-value ≤ 1 × 10−6. P-value is the P-value of strongest SNP association. + Allele is the allele that had a positive SNP effect on maternal calving difficulty. SNP effect DCD is the QTL-lead SNP’s direction effect for direct calving difficulty; + represents SNP effects in the same direction, ++ represents the same direction and large SNP effect size for DCD, i.e., a SNP effect size in the top 20%, − represents opposing SNP effects and −− represents opposing SNP effect directions that have a large SNP effect size. N/A (Non-applicable) was if the SNP wasn’t present in that dataset due to allele frequency. SNP effect PM similar to DCD but represents the SNP effects for PM. No. of genes is the number of genes in the QTL. If no gene was present in the QTL the nearest functional candidate gene within 250 kb was chosen. aintergenic, bintronic, cdownstream gene variant, †gene in which the most significant SNP within the QTL was identified in.
Figure 3.
Manhattan plots for maternal calving difficulty in (A) Angus, (B) Charolais, (C) Holstein Friesian, (D) Limousin, and (E) across all breeds. The red line indicates the genome-wide significance threshold of P ≤ 5 × 10−8 and the blue is the suggestive significance threshold of P ≤ 1 × 10−6.
Only one QTL region associated with MCD was identified in the across-breed analysis of all 47,753 sires; this QTL which was not previously identified in any of the within-breed analyses contained two suggestively associated intronic SNP which were located in solute carrier family 9 member A2 (SLC9A2) on BTA11 (Supplementary Table 1). No genomic window was associated with MCD across all breeds in the window-based analyses. Two separate genomic windows on BTA7 were, however, associated with MCD in the AA, CH, and HF populations and in the CH, HF, and LM populations, respectively; candidate genes within these windows include ENSBTAG00000003086 in the AA, CH, and HF populations and CANX, CBY3, HNRNPH1, and ADAMTS2 in the CH, HF, and LM populations.
Direct PM
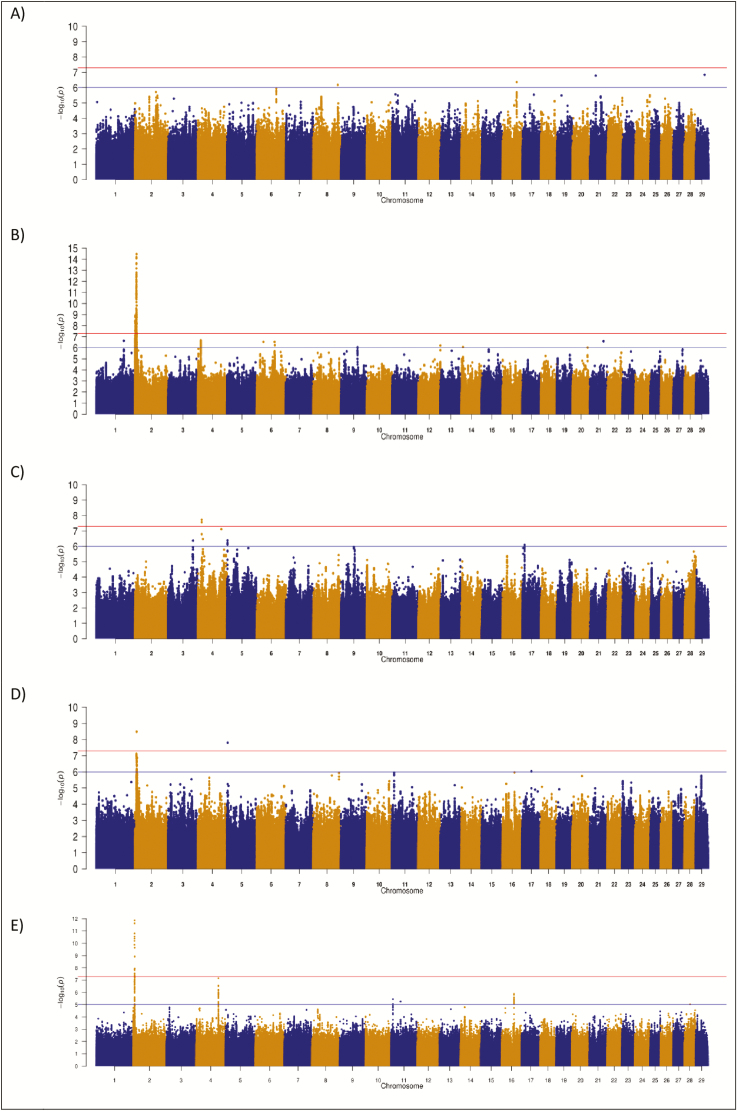
Several genomic regions were significantly associated with PM in the CH, HF, and LM populations; no region was significant in the AA (Figure 4). The strongest association for PM was on BTA2 in the CH population, where a QTL spanning from 5.38 to 6.94 Mb encompassing MSTN contained 335 significantly associated SNP; the Q204X stop-gain mutation was the 26th strongest association with a P-value of 3.54 × 10−12. The frequency of the Q204X T allele was 0.051. Homozygous carriers of the Q204X T allele (n = 22) had a worse (P < 0.0001) EBV for mortality (mean EBV 0.006, SD 0.015) than homozygous (n = 14,844) noncarriers (mean EBV −0.0002, SD 0.013) (Supplementary Figure 3). Aside from MSTN, a putative QTL was also identified on BTA4 in the CH population, where 60 SNP within the plausible candidate gene pyruvate dehydrogenase kinase 4 (PDK4) were suggestively associated with PM (Table 4). In total, three missense variants within PDK4 were captured within the 1000 Bull data set, but only one (rs109626492) had a MAF > 0.005 in the CH population and this SNP was not associated with PM (P = 0.46); of the remaining two missense variants, rs515875763 was fixed for the nondeleterious allele and rs522912661, which had a MAF of 3.04 × 10−5, was included in a post hoc analysis but was not associated with PM (P = 0.014). The strongest QTL association with PM in the HF population was also on BTA4, albeit 4 Mb further downstream from the end of the identified CH QTL; the nearest gene to this HF QTL was RPA3 located 398 kb upstream and closer to the CH QTL.
Figure 4.
Manhattan plots for direct mortality in (A) Angus, (B) Charolais, (C) Holstein Friesian, (D) Limousin, and (E) across all breeds. The red line indicates the genome-wide significance threshold of P ≤ 5 × 10−8 and the blue is the suggestive significance threshold of P ≤ 1 × 10−6.
Table 4.
QTL associated with direct PM1
| Breed | BTA | Start | End | No. SNP | Strongest SNP | Strongest SNP position | + Allele | + Allele freq | P-value | SNP effect DCD | SNP effect MCD | No. genes | Candidate gene(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | 8 | 100644490 | 100645055 | 2 | rs797916567b | 100644490 | A | 0.016 | 6.69 × 10−7 | ++ | − | 1 | PTPN3 † |
| 16 | 58808269 | 58818335 | 2 | rs134283619b | 58814914 | A | 0.019 | 4.38 × 10−7 | + | −− | 1 | RFWD2 † | |
| CH | 2 | 301926 | 957129 | 18 | rs43598129a | 900492 | G | 0.050 | 1.42 × 10−7 | ++ | − | 3 | HERC2, NIPA1,OCA2 |
| 2 | 1007617 | 1226503 | 5 | rs458371570a | 1110887 | A | 0.039 | 3.79 × 10−9 | ++ | − | 2 | CYFIP1, TUBGCP5 | |
| 2 | 1296023 | 3143209 | 125 | rs110820498a | 1925667 | T | 0.090 | 1.31 × 10−9 | ++ | − | 4 | N/A | |
| 2 | 3278825 | 3626278 | 28 | rs461349284a | 3626278 | A | 0.051 | 4.89 × 10−8 | ++ | − | 0 | N/A | |
| 2 | 4580833 | 4602350 | 7 | rs383237760a | 4602350 | A | 0.774 | 4.46 × 10−7 | ++ | + | 0 | N/A | |
| 2 | 5305742 | 6938016 | 654 | rs799943285a | 6808074 | G | 0.056 | 6.56 × 10−15 | ++ | −− | 17 | MSTN | |
| 2 | 7334284 | 7609482 | 5 | rs798834436a | 7488750 | T | 0.052 | 1.54 × 10−7 | ++ | −− | 1 | COL3A1 | |
| 2 | 7773097 | 7776214 | 3 | rs210662731a | 7773097 | T | 0.369 | 3.39 × 10−7 | ++ | − | 0 | GULP1 | |
| 4 | 12723521 | 12787299 | 139 | rs381171531a | 12733539 | C | 0.040 | 2.09 × 10−7 | − | − | 2 | PDK4 | |
| 6 | 73569965 | 73700948 | 2 | rs43473352c | 73636267 | G | 0.935 | 2.93 × 10−7 | + | + | 2 | HOPX | |
| 9 | 68152800 | 68267416 | 8 | rs379327303a | 68152800 | A | 0.988 | 8.85 × 10−7 | −− | + | 1 | LAMA2 | |
| 14 | 8591593 | 8591601 | 2 | rs526183014 | 8591601 | A | 0.552 | 8.44 × 10−7 | + | + | 0 | ZFAT | |
| HF | 3 | 101909079 | 102183746 | 11 | rs378988599b | 102122935 | T | 0.818 | 4.30 × 10−7 | + | − | 3 | C1orf228,TMEM53,RNF220 † |
| 4 | 16001571 | 16013815 | 4 | rs209481751a | 16013815 | A | 0.539 | 1.87 × 10−8 | + | + | 0 | N/A | |
| 5 | 1492436 | 1503881 | 4 | rs438879818b | 1503881 | T | 0.086 | 4.11 × 10−7 | − | + | 1 | TBC1D15 † | |
| 17 | 8589800 | 8600386 | 2 | rs382728302a | 8600386 | A | 0.040 | 8.11 × 10−7 | + | − | 0 | N/A | |
| LM | 2 | 6481284 | 6751879 | 47 | rs110953984a | 6655139 | A | 0.031 | 7.76 × 10−8 | ++ | −− | 6 | MSTN |
| 2 | 7503983 | 7562999 | 21 | rs210765106a | 7505936 | A | 0.103 | 1.19 × 10−7 | ++ | −− | 0 | N/A | |
| 2 | 7625971 | 7819581 | 11 | rs378228653b | 7787549 | T | 0.931 | 2.40 × 10−7 | ++ | −− | 2 | GULP1 † | |
| 2 | 8147499 | 8314779 | 14 | rs800119023a | 8268605 | A | 0.010 | 3.63 × 10−7 | ++ | −− | 0 | N/A | |
| 2 | 9497001 | 9502488 | 4 | rs382982814a | 9497001 | T | 0.057 | 1.38 × 10−7 | ++ | −− | 2 | ZSWIM2, | |
| 5 | 2155306 | 2167134 | 2 | rs378053936b | 2167134 | C | 0.118 | 1.56 × 10−8 | + | − | 2 | TRHDE † |
1No. SNP is the number of SNP within the QTL with a P-value ≤ 1 × 10−6. P-value is the P-value of strongest SNP association. + Allele is the allele that had a positive SNP effect on maternal calving difficulty. SNP effect DCD is the QTL-lead SNP’s direction effect for direct calving difficulty; + represents SNP effects in the same direction, ++ represents the same direction and large SNP effect size for DCD, i.e., a SNP effect size in the top 20%, − represents opposing SNP effects and −− represents opposing SNP effect directions that have a large SNP effect size. N/A (Non-applicable) was if the SNP wasn’t present in that dataset due to allele frequency. SNP effect MCD is similar to DCD but represents the SNP effects for maternal calving difficulty. No. of genes is the number of genes in the QTL. If no gene was present in the QTL the nearest functional candidate gene within 250 kb was chosen. aintergenic, bintronic, cdownstream gene variant, †gene in which the most significant SNP within the QTL was identified in.
In total, four QTL regions were associated with PM across all breeds (Supplementary Table 1); three were located on BTA2 from 5.97 to 7.61 Mb, of which the QTL from 5.97 to 6.63 Mb encompassed MSTN was the strongest association with PM. The across-breed analyses identified a novel QTL on BTA4, downstream of the gene ZNF800 which had not been previously identified in any of the within-breed analyses. Similar to DCD and MCD, no genomic region associated with PM was common to all four breeds and only one 500-kb window was associated with PM in three breeds; this region was located on BTA4 within the CH, HF, and LM populations with PDK4 being the plausible candidate gene. The minimal overlap identified across breeds suggests that a greater proportion of SNP with a P-value < 10−4 are breed-specific associations in comparison to those associated with either DCD or MCD.
Direct PM adjusted for calving difficulty
Upon inclusion of the EBV for DCD as a covariate in the statistical model for the association analysis relating PM to each SNP in the CH population, the QTL on BTA2 encompassing MSTN remained associated with PM although it reduced in significance (Supplementary Table 2). In total, five QTL were identified in the CH population using this approach, of which three were previously identified in the PM analysis suggesting that these QTL contribute still to the incidence of PM, irrespective of a difficult calving. Of most interest, however, was the novel QTL on BTA15 from 29.26 to 29.88 Mb which contained four suggestively associated SNP with PM adjusted for DCD with sodium voltage-gated channel beta subunit 4 (SCN4B) being the likely putative candidate gene; the minor allele (G) of the strongest SNP in this QTL was associated with a reduction in PM and had a moderate allele frequency of 0.153. No missense variant within SCN4B was segregating within the CH population in the current data set although several intronic variants within SCN4B did have P-values ranging from 5.66 × 10−6 to 9.75 × 10−5.
Pleiotropic associations
The number of SNP associated (i.e., P-value < 10−4) with more than one calving trait differed by breed; no SNP was associated with more than one trait in the HF population, 5 SNP were pleiotropic in the AA population, 116 were pleiotropic in the LM population, and 882 SNP were pleiotropic in the CH population (Supplementary Figure 4). Of the 882 SNP identified in the CH population that were associated with both DCD and PM, all but one were located on BTA2 residing between 1.22 and 8.07 Mb and all had the same SNP effect direction for both CD and PM. This included the Q204X mutation where the minor T allele was associated with an increase (i.e., worsening) in both DCD and PM but was not associated (P = 0.11) with MCD. The remaining SNP identified in the CH population was rs384777136, an intergenic variant on BTA10 whose minor allele (MAF = 0.012) increased both DCD (P = 2.35 × 10−5) and PM (P = 4.87 × 10−5). Similar to the CH population, the overlap between DCD and PM in the LM population was primarily on BTA2 spanning from 5.94 to 9.49 Mb with 111 SNP in this region having the same effect direction for both traits and a P-value < 10−4. Only two SNP, both located on BTA2, were pleiotropic with a P-value < 10−4 across all the three calving traits in the LM population; the minor alleles at both SNP were associated with a deterioration in DCD and PM but an improvement in MCD. The minor allele at SNP rs799053464 on BTA5 which had a P-value < 10−4 for both DCD and MCD in the AA population also differed in SNP effect direction for both traits. Overlap between DCD and PM was also detected in the AA population on BTA8 in the 13.45 to 13.47 Mb region where four intergenic SNP had P-values ranging from 6.31 × 10−5 to 9.94 × 10−5 for DCD and from 2.78 × 10−5 to 4.21 × 10−5 for PM; the minor allele for all four SNP, which had a mean allele frequency of 0.015, was associated with a decrease (i.e., better) in EBV for all three calving traits.
More overlapping genomic regions among all three calving traits were detected when 500-kb windows rather than individual SNP were considered to identify pleiotropic genomic regions. Genomic regions that contained a SNP with a P-value < 10−4 associated with all calving performance traits existed in each breed, although the degree of overlap differed by breed (Figure 5); overlap ranged from one 500-kb window in the HF to seven regions in each of the AA and CH populations. The seven genomic windows in the AA population were located across five chromosomes (BTA6, 7, 13, 22, and 24) with multiple underlying plausible candidate genes including HPGDS (BTA6), RBMS3 (BTA22), NCPC, and MYO5B (BTA24). The MCD-associated SNP in each window was in antagonistic pleiotrophy with either DCD or PM in six of the seven windows. Five of the seven windows identified in the CH population to be associated with all three calving traits were located in the first 4.5 Mb of BTA2, whereas the remaining two windows were located on BTA12 and BTA22 encompassing GPC6 and LZTFL1, respectively. The only window identified in the HF to be associated with all three calving traits was located on BTA7 where proline rich 16 (PRR16) was a plausible candidate gene; each SNP with a P-value < 10−4 within the window for each of the calving traits had the same SNP effect direction for all three traits.
Figure 5.
The number of 500-kb windows containing an associated SNP with a P-value < 10−4 that overlapped across traits within each breed. Each window had to contain a SNP with an unadjusted P-value < 10−4. (A) Angus, (B) Charolais, (C) Holstein-Friesian, and (D) Limousin. Pink represents direct calving difficulty (DCD), green represents maternal calving difficulty (MCD), and blue represents PM.
A greater percentage of window overlap was evident between DCD and PM rather than with MCD; for example, in the CH, 46.74% of the PM-associated windows were also associated with DCD while only 10.37% were associated with MCD. Despite detecting window overlap across all traits, the majority of genomic windows were trait-specific. As the majority of SNP were trait-specific, the allele substitution effects with a P-value < 10−4 for an individual calving trait were investigated across each of the traits (Supplementary Figure 5). A greater percentage of SNP with a P-value < 10−4 for DCD had opposing SNP substitution effects with MCD than those with PM. For example, only 31.04% of the SNP with a P-value < 10−4 for DCD in the CH population had allele substitution effects which were opposite in sign for PM, whereas 59.88% of SNP had opposing allele substitution effects for MCD.
Discussion
This association study relating the bovine genome to calving performance, which is one of the largest studies to date, identified multiple underlying QTL both within and across dairy and beef breeds; the majority of the associations, however, appear to be both breed- and trait-specific. The existence of breed-specific SNP effects and trait-specific SNP effects for calving performance traits has been previously documented (Purfield et al., 2015; Tiezzi et al., 2018); thus, highlighting the complexity of the genomic features influencing the expression of these traits. The high proportion of trait-specific QTL identified in the present study was unexpected given the existence of a moderate negative genetic correlation between the direct and maternal components of calving difficulty (Crowley et al., 2011); this correlation manifests itself as a small easy calving female calf, once mature, having a greater likelihood of experiencing a difficult calving due to her reduced size and pelvic opening dimensions (Philipsson, 1979). Therefore, genomic regions common to both DCD and MCD were expected. Understanding the genomic architecture influencing the direct and maternal components of calving difficulty separately and the known genetic antagonism (Crowley et al., 2011) may aid in effective simultaneous selection for both traits as the genetic linkage across traits could be potentially resolved during meiosis once the underlying causal mutations are known. However, additional analyses with the new bovine reference genome ARS-UCD 1.2 and the ever-increasing quantity of gene expression analyses would be required to functionally characterize any putative candidate gene identified in the present study.
Antagonistic pleiotropy
A limited number of pleiotropic SNP were identified in the beef populations, although they were primarily located on BTA2 either within or near to the MSTN gene; these SNP were, however, only associated with DCD and PM and not with MCD. Heavier cattle birth weight has been previously reported for double-muscle carriers which ultimately lead to an increase in the incidence of calving difficulty and PM within South Devon cattle (Wiener et al., 2002).
In contrast to previous calving association studies (Thomasen et al., 2008; Sahana et al., 2011), no pleiotropic SNP was identified in the HF population despite the known strong genetic correlation between DCD and PM (Steinbock et al., 2003) and the moderate negative correlation between DCD and MCD (Eaglen and Bijma, 2009). The inability to identify pleiotropic calving SNP is most likely a reflection of the polygenic architecture in the HF, which is also reflected in the low heritability of MCD and PM in HF populations (Eaglen and Bijma, 2009) in comparison to beef populations (Mujibi and Crews, 2009). In addition, difficult calving bulls are simply not tolerated within the dairy industry due to the adverse effects of a difficulty calving on both milk performance and subsequent fertility (Dematawewa and Berger, 1997; Berry et al., 2007; Eaglen et al., 2011). Therefore, only a limited number of SNP of large effect was associated with calving performance in the HF population in comparison to the beef populations in the present study. The smaller SNP effect size and the limited role of major genes in calving performance in the HF population may be why a greater percentage of SNP had the same SNP effect direction for DCD and MCD than the beef populations; in total 83.01% of the 7,533 SNP identified in the HF population (Supplementary Figure 4) had the same SNP effect direction for both DCD and MCD, whereas in comparison 37.97% and 31.72% of SNP associated with DCD in the CH and LM SNP shared effect directions with MCD.
As the characteristics of a difficult calving are associated with an increased risk of PM (Mee et al., 2008) a greater proportion of SNP sharing the same SNP effect direction is likely and this was corroborated in the beef populations used in the present study. The use of 500-kb windows identified additional putative pleiotropic genomic regions associated with calving performance within each of the breeds but the antagonistic pleiotropy that existed between DCD and MCD remained in the beef populations. Although antagonistic pleiotropy was detected between DCD and MCD, QTL associated with DCD where the lead SNP had a small opposing SNP effect on MCD compared to that for DCD were identified (Table 1) and vice versa (Table 2). This suggests that genetic gain for improved calving performance could be achieved by identifying trait-specific QTL of large effect that have a minimal effect on their correlated trait.
The role of BTA18 in calving difficulty
Chromosome 18 has long been associated with calving difficulty in multiple Holstein populations (Cole et al., 2009, 2011; Sahana et al., 2011; Höglund et al., 2012; Purfield et al., 2014, 2015) suggesting that a true QTL association impacting calving ability exists on this autosome. Indeed the same genomic region has also been associated with multiple fertility-related traits and body conformation traits (Abo-Ismail et al., 2017; Muller et al., 2017; Doyle et al., 2020a). Despite several studies proposing the same genomic region on BTA18, consistency is lacking on the suggested underlying candidate gene. Cole et al. (2009) was the first to propose that the pleiotropic effects of this QTL may be due to the QTL involvement in embryo development and prolonged gestation thus increasing calving difficulty, and subsequently impacting fertility performance. This theory was further substantiated by recent research from Fang et al. (2019) who proposed ZNF613 as a plausible candidate gene within this region that was likely causing a lengthening of gestation within a large U.S. Holstein population of 27,214 Holstein bulls. Fang et al. (2019) demonstrated strong evidence that it was the methylation pattern of the second intron of ZNF613 that was lengthening gestation and found this pattern to be also associated with sire calving ease, body depth, and conception rate in Holsteins. The downstream variant of ZNF613, rs381577268, which was the strongest association with DCD in our HF population, was also the strongest association for gestation length in a HF population overlapping in animals used in the present study (Purfield et al., 2019b). Animals homozygous for the T allele at this position in the present study had a 4.7-d longer gestation length and, on average, a worse (P < 0.001) EBVs for DCD and PM (P < 0.05) than homozygous noncarriers (Purfield et al., 2019b). Despite the strong evidence corroborating ZNF613 as the plausible candidate gene, limited genetic gain in calving difficulty is likely to be achieved through its targeted selection in a breeding program as the frequency of the favorable (i.e., C) allele was near-fixation in both Irish (0.98) and U.S. Holstein populations (0.93). This QTL on BTA18 is also likely to have a limited role in improving across-breed genomic evaluations, as several studies involving other dairy breeds such as Jersey (Tiezzi et al., 2018), Fleckvieh (Pausch et al., 2011), and Brown Swiss (Frischknecht et al., 2017; Tiezzi et al., 2018) failed to associate this QTL with calving performance. In addition, no SNP within ZNF613 was suggestively associated with DCD in the any of the beef populations investigated in the present study. However, this may be due to a difference in the linkage phase between the imputed alleles and causal mutations in each of the breeds, as the marginal significance of SNP (P > 1 × 10−6) just upstream of this QTL in the CH and LM populations does suggest that a QTL extending from the 57.9 to 58.2 Mb on BTA18 influences DCD in multiple breeds, albeit to a different extent.
Additionally known calving performance QTL
The role of MSTN in calving performance has long since been established (Casas et al., 1999; Bellinge et al., 2005). In the present study, QTL regions containing MSTN were associated with both DCD and PM in the CH population, whereas in the LM population, the QTL associated with these traits were located just downstream of MSTN. The well-known stop-gain Q204X mutation within the CH population contributed to 5.09% of the genetic variance in DCD. Interestingly the F94L mutation in MSTN, which is known to result in a moderate muscle hypertrophy phenotype in the LM population, was not associated with any calving performance trait in the present study. This is not unexpected as previous studies have failed to detect a difference in birth weight in LM between F94L carriers vs. noncarriers (Esmailizadeh et al., 2008). This is also further substantiated by the lack of an association of the F94L mutation with DCD within the CH population; in total 282 homozygous F94L carriers existed within the CH population and the P-value for F94L was 0.189. The nonsignificant association of F94L with calving performance suggests that the targeted selection of this mutation for increased carcass performance (Esmailizadeh et al., 2008) within both the CH and LM populations can be achieved without the adverse effects of greater dystocia; the same, however, is not true for Q204X.
The known strong genetic correlation between calving difficulty and both birth weight and body size (Johanson and Berger, 2003) implies that genomic regions and candidate genes previously associated with cattle height should possibly be in common with those associated with calving performance in the present study. The PLAG1 gene, which has been previously associated with height in both humans (Gudbjartsson et al., 2008) and cattle (Pryce et al., 2011; Bouwman et al., 2018), as well as live weight in cattle (Littlejohn et al., 2012; Nishimura et al., 2012), was indeed associated with DCD in the HF population used in the present study. Although PLAG1 has been associated with calving performance in multiple breeds, such as Fleckvieh (Pausch et al., 2011) as well as U.S. Gelbvieh and Simmental (Saatchi et al., 2014), this QTL region on BTA14 was only associated with DCD in the HF population in the present study despite the segregation of variants within 250 kb up/downstream of PLAG1 in each of the beef populations; this included the seven PLAG1 variants identified by Karim et al. (2011) which had MAFs ranging from 0.014 to 0.024 in the AA population, from 0.011 to 0.013 in the CH population, and from 0.032 to 0.035 in the LM population. Interestingly, a QTL on BTA6, which contained the stature-related genes, NCAPG and LCORL, was also associated with MCD but only within the AA population. This region has been previously associated with calving performance in several populations (Olsen et al., 2010; Bongiorni et al., 2012; Saatchi et al., 2014) and was the strongest QTL association for height of withers within an Irish AA population (Doyle et al., 2020b). The minor alleles of the four associated SNP (P < 1 × 10−6) identified within this QTL (Table 3) were below the MAF threshold for inclusion in the present study in the remaining breeds suggesting this QTL is of greater importance in the AA population which is characterized as being phenotypically smaller than the CH, HF, and LM populations (Bouwman et al., 2018).
Novel associations
Although several major well-known genes attributed to calving performance were confirmed in the present study, several novel candidate genes are also proposed. Interestingly, PDK4 which was suggestively associated with PM in the CH population, and identified as a putative across-breed association in the window analyses in the CH, HF, and LM populations, has been documented to play a role in the glucose homeostasis during the embryo development in both pigs (Lan et al., 2009) and cattle (Lonergan et al., 2016). A 16-bp deletion within PDK4 has also been attributed to dilated cardiomyopathy within the Doberman Pinscher canine breed, which ultimately results in their sudden death as their hearts are unable to generate sufficient energy for long-term healthy function (Bolfer et al., 2016). The allele frequency of the favorable allele of the strongest SNP in this QTL (rs381171531), which was associated with a reduction in PM, was highly prevalent (0.959) in the CH population; nonetheless, some genetic gain could still be made through its targeted selection while having limited impact on both DCD and MCD.
The strongest association in the AA population for DCD encompassed the GC gene which encodes the vitamin D-binding protein. Although the calf itself cannot generate vitamin D during gestation, it is suggested that this QTL is impacting DCD by affecting the calf’s birth weight. Studies in humans have associated several GC variants with a lighter birth weight in infants due to a reduction in circulating vitamin D levels (Chun et al., 2017). Interestingly, this region has been previously associated with gestation length in Holstein-Friesians (Maltecca et al., 2009) and GC was identified as a putative candidate gene for suckling traits in beef cattle (Michenet et al., 2016). The association of variants downstream of the GC gene suggests that it is the regulatory expression of GC that is influencing DCD rather than a disruptive mutation. Another putative candidate gene identified in the AA population was PARM1 which contained several intronic variants that were suggestively associated with MCD. PARM1 has been previously shown to play a role in the development of the blastocyst during early embryonic development (Zolini et al., 2019) but it is unclear what role it could have on MCD. The across-breed analyses of 47,753 sires also identified another putative candidate gene associated with MCD on BTA11; SLC9A2 has been shown to be differentially regulated over the course of gestation in humans and may play a role in intrauterine growth restriction (Johansson et al., 2002). The candidate gene, ZEB2 which was identified as a candidate gene for DCD in the AA, LM, and HF populations from the window analyses has been previously associated with birth weight in Brahman cattle (Martinez et al., 2017) and its deletion has also been documented to cause polled and multisystemic syndrome in CH cattle (Capitan et al., 2012).
Novel candidate genes associated with all three calving traits included PRR16 which was identified in the HF population. Largen, the product of PRR16, has been shown to be essential in maintaining cell size homeostasis with overexpression of this gene in transgenic mice resulted in embryonic death (Yamamoto et al., 2014). PRR16 is highly expressed in both human placental endothelial cells and also in the bovine endometrium. Although it has been shown to regulate cell size, PRR16’s role in calving performance is unclear but may affect performance through the regulation of mitochondrial functions and mRNA translation.
Across-breed associations
Despite the majority of detected associations being trait-specific, common genomic regions that were associated with calving performance across the different breeds were identified and could help in possibly facilitating across-breed genomic predictions. The greatest percentage of overlap was for DCD between the CH and LM populations with 311 SNP surrounding MSTN having the same sign of allele substitution effect for DCD. Saatchi et al. (2014) previously reported that major genes such as MSTN were pleiotropic for DCD in multiple cattle breeds and that these QTL also explain the majority of genetic variance in birth, weanling, yearling, and carcass weight. Aside from well-known major genes, multiple novel genes such as ZEB2 (DCD), SLC9A2 (MCD), and PDK4 (PM) were also identified, but the proportion of genetic variance explained by these candidates was minimal thus reaffirming that the majority of variance in calving performance is attributed to the additive (and possibly multiplicative) effect of many polymorphisms of small effect. Therefore, to accommodate breed-specific major genes of large effect, it is suggested that genomic prediction algorithms should be potentially partitioned into a within-breed component and an across-breed component for calving performance.
Mortality adjusted for calving difficulty
The association between dystocia and PM in cattle is well-known (Mee et al., 2008). Little, however, is known about the prevalence of PM in unassisted calving events. There are many possible causes for such mortality events including congenetial defects, placental dysfunction (Berglund et al., 2003), infections (Givens and Marley, 2008), precalving nutrition (Mee et al., 2013), or possibly management issues such as age at first calving (Mee et al., 2008). Indeed several lethal recessive mutations and haplotypes have been identified in cattle in recent years (VanRaden et al., 2011; Fritz et al., 2013; Daetwyler et al., 2014; Adams et al., 2016). Three well-known attributers to calf mortality include complex vertebral malformation, bovine leukocyte adhesion deficiency, and brachyspina, and although the causative mutation within each gene is known, none of these mutations were included in the present data set and no segregating variant within their causative genes had a P-value < 10−4 for PM. Interestingly, the causative mutations identified in the HF lethal haplotypes HH1 (rs448942533) and HH3 (rs456206907) were segregating within our HF population but were not significant for PM with P-values of 0.83 and 0.54, respectively.
Several QTL regions in the present study were associated with mortality adjusted for genetic merit for DCD in the CH population but only two QTL had not been previously identified in the PM analysis. The continued association of QTL surrounding MSTN and PDK4 suggests that these candidate genes remain associated with PM even when calving difficulty is accounted for. SCN4B, which was identified as a novel putative candidate gene for PM not associated with dystocia, has been previously associated with atrial fibrillation in humans (Li et al., 2013). Although further investigation into the putative role of SCN4B in PM within the CH population is needed, the moderate allele frequency of the G allele (MAF = 0.153) for the strongest SNP association within this QTL suggests genetic improvement could be made if selected upon.
Designer matings using genomic information
Prediction of genetic merit for calving performance traits using genomic selection methodology, like most traits, is generally undertaken at a macro level by summing the genomic feature effects across the entire genome (Hayes et al., 2009). No distinction is generally made between the biological basis as to why a particular parent may generate calves with a greater than average risk of either calving difficulty or mortality, and how that biology complements the genomic features of its mate. Feto-pelvic disproportion is one of the most common risk factors associated with dystocia in cattle (Mee et al., 2008). Hence, mating of individuals compatible on feto-pelvic dimensions based on specific genomic predictions could help advert the risk of calving dystocia and, by association (Berry et al., 2007), calf mortality. One clear example of the potential of exploiting underlying genomic information is that related to mutations within the MSTN gene and risk of dystocia. MSTN, which is also known as growth differentiation factor 8 (GDF-8), is expressed in developing and mature skeletal muscle (McPherron and Lee, 1997); MSTN activity represses skeletal growth. Therefore, animals, especially with the mutant nt821 variant, exhibit muscular hypertrophy (Grobet et al., 1997). Wiener et al. (2002) reported a greater calving difficulty in cattle carrying the nt821 (they called this del11) mutation. While the nt821 mutation in the MSTN gene has a clear major gene effect, one of the motivations in the present study was to identify other potential variants associated with animal conformation which, when summed together, could provide a more in-depth assessment of the likely contributing factors to eventual dystocia. Upperman et al. (2019) proposed a strategy for recommending matings among beef animals carrying lethal mutations; a similar strategy could be deployed for calving performance but instead of the characteristic of interest being a lethal mutation, it could be genetic predisposition to dystocia or PM. Mates both with a high genetic risk, and underpinned by the same biological rational, could then be avoided. Other than MSTN, several other genes such as PLAG1, GC, and NCAPG/LCORL were associated with body size and such knowledge could help assess the likelihood of dystocia in a future mating given the genotypes of both candidate mates.
Being able to disentangle to genetic architecture of calving performance traits, as well as being useful in mating programs, can also be useful in breeding schemes. Many traits in breeding goals where selection in the same direction is favored can be antagonistic. The objective of an efficient breeding program is to identify animals that excel in both traits, despite the antagonistic correlation. Genetic correlations are due to either pleiotropy or linkage. Falconer and Mackay (1996) suggested that simultaneous selection for two traits in the same direction will, over time, cause an antagonistic genetic correlation to develop and this will be due to pleiotropy. While sometimes disputed (Robinson, 1996), most studies in cattle (Crowley et al., 2011) have reported an antagonistic genetic correlation between DCD and MCD. The favored direction of selection for both traits is the same. Hence, of interest is the ability to identify genomic regions with allele substitution effects for both traits in the same direction, or with an estimated effect for one of the traits but not for the other. The strongest SNP within several QTL in the present study (Tables 1–3) had the same SNP effect direction across all calving traits and of those QTL that had opposing effects, the SNP effect was small for the other calving traits.
Conclusions
Several genomic regions associated with calving performance in dairy and beef cattle were identified in the present study although the majority of these were unique to a given breed or trait. Major genes previously documented to be associated with calf body size such as MSTN, PLAG1, and NCAPG/LCORL were associated with calving performance and should be exploited within genomic mating programs to predict the likelihood of a dystocia event, and, where necessary minimize the risk. Novel putative candidates such as PARM1, PDK4, and SLC9A2 were also identified, although the proportion of genetic variance explained by these candidates was small. This substantiates the theory that calving performance traits are complex quantitative traits under the influence of many polymorphisms each of small effect. Antagonistic pleiotropy between DCD and MCD was detected, although several QTL regions that had the same SNP effect direction across both traits were identified suggesting simultaneous genetic improvement for both traits through targeted selection is feasible once the genomic architecture is known.
Supplementary Material
Acknowledgments
Funding from the European Union’s Horizon 2020 research and innovation program – GenTORE – under grant agreement no. 727213 is greatly appreciated, as is the funding from the Science Foundation Ireland principal investigator award grant (14/IA/2576) and Research Centers (16/RC/3835; VistaMilk). We gratefully acknowledge the 1000 Bull Genomes Consortium for providing accessibility to whole-genome sequence data which was used in this study and to the Irish Cattle Breeding Federation for providing access to genotype and phenotype data.
Glossary
Abbreviations
- AA
Angus
- BTA
Bos taurus autosome
- CH
Charolais
- DCD
direct calving difficulty
- EBV
estimated breeding value
- ERC
effective record contribution
- HD
high density
- HF
Holstein-Friesian
- ICBF
Irish Cattle Breeding Federation
- IDB
Irish Dairy and Beef
- LCORL
ligand-dependent nuclear receptor corepressor-like
- LD
linkage disequilibrium
- LM
Limousin
- MAF
minor allele frequency
- MCD
maternal calving difficulty
- MSTN
myostatin
- NCAPG
non-SMC condensin I complex subunit G
- PARM1
prostate androgen-regulated mucin-like protein 1
- PDK4
pyruvate dehydrogenase kinase 4
- PLAG1
pleomorphic adenoma gene 1
- PM
perinatal mortality
- PRR16
proline rich 16
- QTL
quantitative trait loci
- SCN4B
sodium voltage-gated channel beta subunit 4
- SLC9A2
solute carrier family 9 member A2
- SNP
single-nucleotide polymorphism
- WGS
whole-genome sequence
- ZEB2
zinc finger E-box-binding homeobox 2
- ZNF613
zinc finger protein 613
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Abo-Ismail M. K., Brito L. F., Miller S. P., Sargolzaei M., Grossi D. A., Moore S. S., Plastow G., Stothard P., Nayeri S., and Schenkel F. S.. . 2017. Genome-wide association studies and genomic prediction of breeding values for calving performance and body conformation traits in Holstein cattle. Genet. Sel. Evol. 49:82. doi: 10.1186/s12711-017-0356-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams H. A., Sonstegard T. S., VanRaden P. M., Null D. J., Van Tassell C. P., Larkin D. M., and Lewin H. A.. . 2016. Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J. Dairy Sci. 99:6693–6701. doi: 10.3168/jds.2015-10517 [DOI] [PubMed] [Google Scholar]
- Bellinge R. H., Liberles D. A., Iaschi S. P., O’Brien P. A., and Tay G. K.. . 2005. Myostatin and its implications on animal breeding: a review. Anim. Genet. 36:1–6. doi: 10.1111/j.1365-2052.2004.01229.x [DOI] [PubMed] [Google Scholar]
- Berglund B., Steinbock L., and Elvander M.. . 2003. Causes of stillbirth and time of death in Swedish Holstein calves examined post mortem. Acta Vet. Scand. 44:111–120. doi: 10.1186/1751-0147-44-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. P., Lee J. M., Macdonald K. A., and Roche J. R.. . 2007. Body condition score and body weight effects on dystocia and stillbirths and consequent effects on postcalving performance. J. Dairy Sci. 90:4201–4211. doi: 10.3168/jds.2007-0023 [DOI] [PubMed] [Google Scholar]
- Bolfer L., Estrada A. H., Winter B., Taggart K., Lourenco F., Terada N., Byrne B. J., Conlon T. J., and Pacak C. A.. . 2016. The role of the pyruvate dehydrogenase kinase 4 (PDK4) gene mutation in the development of dilated cardiomyopathy in dobermanpinschers and potential application of cardiac gene therapy. Mol. Ther. 24:S70–S71. doi: 10.1016/S1525-0016(16)32989-6 [DOI] [Google Scholar]
- Bongiorni S., Mancini G., Chillemi G., Pariset L., and Valentini A.. . 2012. Identification of a short region on chromosome 6 affecting direct calving ease in Piedmontese cattle breed. PLoS One 7:e50137. doi: 10.1371/journal.pone.0050137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman A. C., Daetwyler H. D., Chamberlain A. J., Ponce C. H., Sargolzaei M., Schenkel F. S., Sahana G., Govignon-Gion A., Boitard S., Dolezal M., . et al. 2018. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 50:362–367. doi: 10.1038/s41588-018-0056-5 [DOI] [PubMed] [Google Scholar]
- Capitan A., Allais-Bonnet A., Pinton A., Marquant-Le Guienne B., Le Bourhis D., Grohs C., Bouet S., Clément L., Salas-Cortes L., Venot E., . et al. 2012. A 3.7 Mb deletion encompassing ZEB2 causes a novel polled and multisystemic syndrome in the progeny of a somatic mosaic bull. PLoS One 7:e49084. doi: 10.1371/journal.pone.0049084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas E., Keele J. W., Fahrenkrug S. C., Smith T. P., Cundiff L. V., and Stone R. T.. . 1999. Quantitative analysis of birth, weaning, and yearling weights and calving difficulty in Piedmontese crossbreds segregating an inactive myostatin allele. J. Anim. Sci. 77:1686–1692. doi: 10.2527/1999.7771686x [DOI] [PubMed] [Google Scholar]
- Chun S. K., Shin S., Kim M. Y., Joung H., and Chung J.. . 2017. Effects of maternal genetic polymorphisms in vitamin D-binding protein and serum 25-hydroxyvitamin D concentration on infant birth weight. Nutrition 35:36–42. doi: 10.1016/j.nut.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Cole J. B., VanRaden P. M., O’Connell J. R., Van Tassell C. P., Sonstegard T. S., Schnabel R. D., Taylor J. F., and Wiggans G. R.. . 2009. Distribution and location of genetic effects for dairy traits. J. Dairy Sci. 92:2931–2946. doi: 10.3168/jds.2008-1762 [DOI] [PubMed] [Google Scholar]
- Cole J. B., Wiggans G. R., Ma L., Sonstegard T. S., Lawlor T. J. Jr, Crooker B. A., Van Tassell C. P., Yang J., Wang S., Matukumalli L. K., . et al. 2011. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics 12:408. doi: 10.1186/1471-2164-12-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J. J., Evans R. D., Mc Hugh N., Kenny D. A., McGee M., Crews D. H. Jr, and Berry D. P.. . 2011. Genetic relationships between feed efficiency in growing males and beef cow performance. J. Anim. Sci. 89:3372–3381. doi: 10.2527/jas.2011-3835 [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Capitan A., Pausch H., Stothard P., van Binsbergen R., Brøndum R. F., Liao X., Djari A., Rodriguez S. C., Grohs C., . et al. 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46:858–865. doi: 10.1038/ng.3034 [DOI] [PubMed] [Google Scholar]
- Das S., Forer L., Schönherr S., Sidore C., Locke A. E., Kwong A., Vrieze S. I., Chew E. Y., Levy S., McGue M., . et al. 2016. Next-generation genotype imputation service and methods. Nat. Genet. 48:1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dematawewa C. M., and Berger P. J.. . 1997. Effect of dystocia on yield, fertility, and cow losses and an economic evaluation of dystocia scores for Holsteins. J. Dairy Sci. 80:754–761. doi: 10.3168/jds.s0022-0302(97)75995-2 [DOI] [PubMed] [Google Scholar]
- Doyle J. L., Berry D. P., Veerkamp R. F., Carthy T. R., Evans R. D., Walsh S. W., and Purfield D. C.. . 2020a. Genomic regions associated with muscularity in beef cattle differ in five contrasting cattle breeds. Genet. Sel. Evol. 52:2. doi: 10.1186/s12711-020-0523-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. L., Berry D. P., Veerkamp R. F., Carthy T. R., Walsh S. W., Evans R. D., and Purfield D. C.. . 2020b. Genomic regions associated with skeletal type traits in beef and dairy cattle are common to regions associated with carcass traits, feed intake and calving difficulty. Front. Genet. 11:20. doi: 10.3389/fgene.2020.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan M. J., and Berry D. P.. . 2006. Factors affecting body condition score, live weight and reproductive performance in spring-calving suckler cows. Irish J. Agr. Food Res. 45:25–38. [Google Scholar]
- Eaglen S. A., and Bijma P.. . 2009. Genetic parameters of direct and maternal effects for calving ease in Dutch Holstein-Friesian cattle. J. Dairy Sci. 92:2229–2237. doi: 10.3168/jds.2008-1654 [DOI] [PubMed] [Google Scholar]
- Eaglen S. A., Coffey M. P., Woolliams J. A., Mrode R., and Wall E.. . 2011. Phenotypic effects of calving ease on the subsequent fertility and milk production of dam and calf in UK Holstein-Friesian heifers. J. Dairy Sci. 94:5413–5423. doi: 10.3168/jds.2010-4040 [DOI] [PubMed] [Google Scholar]
- Esmailizadeh A. K., Bottema C. D., Sellick G. S., Verbyla A. P., Morris C. A., Cullen N. G., and Pitchford W. S.. . 2008. Effects of the myostatin F94L substitution on beef traits. J. Anim. Sci. 86:1038–1046. doi: 10.2527/jas.2007-0589 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., and Mackay T. F. C.. . 1996. Introduction to quantitative genetics. 4th ed. Longman, Harlow, United Kingdom. [Google Scholar]
- Fang L., Jiang J., Li B., Zhou Y., Freebern E., Vanraden P. M., Cole J. B., Liu G. E., and Ma L.. . 2019. Genetic and epigenetic architecture of paternal origin contribute to gestation length in cattle. Commun. Biol. 2:100. doi: 10.1038/s42003-019-0341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht M., B. Bapst, F. R. Seefried, H. Signer-Hasler, D. Garrick, C. Stricker, R. Fries, I. Russ, J. Solkner, A. Bieber,et al. 2017. Genome-wide association studies of fertility and calving traits in Brown Swiss cattle using imputed whole-genome sequences. BMC Genomics. 18:910. doi: 10.1186/s12864-017-4308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S., A. Capitan, A. Djari, S. C. Rodriguez, A. Barbat, A. Baur, C. Grohs, B. Weiss, M. Boussaha, D. Esquerré,et al. 2013. Detection of haplotypes associated with prenatal death in dairy cattle and identification of deleterious mutations in GART, SHBG and SLC37A2. PLoS One 8:e65550. doi: 10.1371/journal.pone.0065550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D. J., Taylor J. F., and Fernando R. L.. . 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41:55. doi: 10.1186/1297-9686-41-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens D. M., and Marley M. S. D.. . 2008. Infectious causes of embryonic and fetal mortality. Theriogenology 70:270–285. doi: 10.1016/j.theriogenology.2008.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobet L., Martin L. J., Poncelet D., Pirottin D., Brouwers B., Riquet J., Schoeberlein A., Dunner S., Ménissier F., Massabanda J., . et al. 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17:71–74. doi: 10.1038/ng0997-71 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D. F., Walters G. B., Thorleifsson G., Stefansson H., Halldorsson B. V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., . et al. 2008. Many sequence variants affecting diversity of adult human height. Nat. Genet. 40:609–615. doi: 10.1038/ng.122 [DOI] [PubMed] [Google Scholar]
- Harris B., and Johnson D.. . 1998. Approximate reliability of genetic evaluations under an animal model. J. Dairy Sci. 81:2723–2728. doi: 10.3168/jds.S0022-0302(98)75829-1 [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. J., and Goddard M. E.. . 2009. Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92:433–443. doi: 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- Höglund J. K., Guldbrandtsen B., Lund M. S., and Sahana G.. . 2012. Analyzes of genome-wide association follow-up study for calving traits in dairy cattle. BMC Genet. 13:71. doi: 10.1186/1471-2156-13-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson J. M., and Berger P. J.. . 2003. Birth weight as a predictor of calving ease and perinatal mortality in Holstein cattle. J. Dairy Sci. 86:3745–3755. doi: 10.3168/jds.S0022-0302(03)73981-2 [DOI] [PubMed] [Google Scholar]
- Johansson M., Glazier J. D., Sibley C. P., Jansson T., and Powell T. L.. . 2002. Activity and protein expression of the Na+/H+ exchanger is reduced in syncytiotrophoblast microvillous plasma membranes isolated from preterm intrauterine growth restriction pregnancies. J. Clin. Endocrinol. Metab. 87:5686–5694. doi: 10.1210/jc.2002-020214 [DOI] [PubMed] [Google Scholar]
- Karim L., Takeda H., Lin L., Druet T., Arias J. A., Baurain D., Cambisano N., Davis S. R., Farnir F., Grisart B., . et al. 2011. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43:405–413. doi: 10.1038/ng.814 [DOI] [PubMed] [Google Scholar]
- Lan J., Lei M. G., Zhang Y. B., Wang J. H., Feng X. T., Xu D. Q., Gui J. F., and Xiong Y. Z.. . 2009. Characterization of the porcine differentially expressed PDK4 gene and association with meat quality. Mol. Biol. Rep. 36:2003–2010. doi: 10.1007/s11033-008-9411-4 [DOI] [PubMed] [Google Scholar]
- Li R. G., Wang Q., Xu Y. J., Zhang M., Qu X. K., Liu X., Fang W. Y., and Yang Y. Q.. . 2013. Mutations of the SCN4B-encoded sodium channel β4 subunit in familial atrial fibrillation. Int. J. Mol. Med. 32:144–150. doi: 10.3892/ijmm.2013.1355 [DOI] [PubMed] [Google Scholar]
- Littlejohn M., Grala T., Sanders K., Walker C., Waghorn G., Macdonald K., Coppieters W., Georges M., Spelman R., Hillerton E., . et al. 2012. Genetic variation in PLAG1 associates with early life body weight and peripubertal weight and growth in Bos taurus. Anim. Genet. 43:591–594. doi: 10.1111/j.1365-2052.2011.02293.x [DOI] [PubMed] [Google Scholar]
- Loh P. R., Palamara P. F., and Price A. L.. . 2016. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 48:811–816. doi: 10.1038/ng.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan P., Fair T., Forde N., and Rizos D.. . 2016. Embryo development in dairy cattle. Theriogenology 86:270–277. doi: 10.1016/j.theriogenology.2016.04.040 [DOI] [PubMed] [Google Scholar]
- Maltecca C., Weigel K. A., Khatib H., Cowan M., and Bagnato A.. . 2009. Whole-genome scan for quantitative trait loci associated with birth weight, gestation length and passive immune transfer in a Holstein x Jersey crossbred population. Anim. Genet. 40:27–34. doi: 10.1111/j.1365-2052.2008.01793.x [DOI] [PubMed] [Google Scholar]
- Martínez R., Bejarano D., Gómez Y., Dasoneville R., Jiménez A., Even G., Sölkner J., and Mészáros G.. . 2017. Genome-wide association study for birth, weaning and yearling weight in Colombian Brahman cattle. Genet. Mol. Biol. 40:453–459. doi: 10.1590/1678-4685-GMB-2016-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern S. P., Purfield D. C., Ring S. C., Carthy T. R., Graham D. A., and Berry D. P.. . 2019. Candidate genes associated with the heritable humoral response to Mycobacterium avium ssp. paratuberculosis in dairy cows have factors in common with gastrointestinal diseases in humans. J. Dairy Sci. 102:4249–4263. doi: 10.3168/jds.2018-15906 [DOI] [PubMed] [Google Scholar]
- McPherron A. C., and Lee S. J.. . 1997. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 94:12457–12461. doi: 10.1073/pnas.94.23.12457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee J. F., Berry D. P., and Cromie A. R.. . 2008. Prevalence of, and risk factors associated with, perinatal calf mortality in pasture-based Holstein-Friesian cows. Animal 2:613–620. doi: 10.1017/S1751731108001699 [DOI] [PubMed] [Google Scholar]
- Mee J. F., Grant J., Sánchez-Miguel C., and Doherty M.. . 2013. Pre-calving and calving management practices in dairy herds with a history of high or low bovine perinatal mortality. Animals (Basel) 3:866–881. doi: 10.3390/ani3030866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C. L., Berger P. J., Koehler K. J., Thompson J. R., and Sattler C. G.. . 2001. Phenotypic trends in incidence of stillbirth for Holsteins in the United States. J. Dairy Sci. 84:515–523. doi: 10.3168/jds.S0022-0302(01)74502-X [DOI] [PubMed] [Google Scholar]
- Meyer K., and Tier B.. . 2012. “SNP Snappy”: a strategy for fast genome-wide association studies fitting a full mixed model. Genetics 190:275–277. doi: 10.1534/genetics.111.134841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michenet A., Barbat M., Saintilan R., Venot E., and Phocas F.. . 2016. Detection of quantitative trait loci for maternal traits using high-density genotypes of Blonde d’Aquitaine beef cattle. BMC Genet. 17:88. doi: 10.1186/s12863-016-0397-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujibi F. D., and Crews D. H. Jr. 2009. Genetic parameters for calving ease, gestation length, and birth weight in Charolais cattle. J. Anim. Sci. 87:2759–2766. doi: 10.2527/jas.2008-1141 [DOI] [PubMed] [Google Scholar]
- Müller M. P., Rothammer S., Seichter D., Russ I., Hinrichs D., Tetens J., Thaller G., and Medugorac I.. . 2017. Genome-wide mapping of 10 calving and fertility traits in Holstein dairy cattle with special regard to chromosome 18. J. Dairy Sci. 100:1987–2006. doi: 10.3168/jds.2016-11506 [DOI] [PubMed] [Google Scholar]
- Nishimura S., Watanabe T., Mizoshita K., Tatsuda K., Fujita T., Watanabe N., Sugimoto Y., and Takasuga A.. . 2012. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in Japanese Black cattle. BMC Genet. 13:40. doi: 10.1186/1471-2156-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen H. G., Hayes B. J., Kent M. P., Nome T., Svendsen M., and Lien S.. . 2010. A genome wide association study for QTL affecting direct and maternal effects of stillbirth and dystocia in cattle. Anim. Genet. 41:273–280. doi: 10.1111/j.1365-2052.2009.01998.x [DOI] [PubMed] [Google Scholar]
- Pausch H., Flisikowski K., Jung S., Emmerling R., Edel C., Götz K. U., and Fries R.. . 2011. Genome-wide association study identifies two major loci affecting calving ease and growth-related traits in cattle. Genetics 187:289–297. doi: 10.1534/genetics.110.124057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeler E. J., Otte M. J., and Esslemont R. J.. . 1994. Interrelationships of periparturient diseases in dairy-cows. Vet. Rec. 134:129–132. doi: 10.1136/vr.134.6.129 [DOI] [PubMed] [Google Scholar]
- Pe’er I., Yelensky R., Altshuler D., and Daly M. J.. . 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32:381–385. doi: 10.1002/gepi.20303 [DOI] [PubMed] [Google Scholar]
- Philipsson J. 1979. Breeding for calving performance. Martinus Nijhoff, The Hague, The Netherlands. [Google Scholar]
- Pryce J. E., Hayes B. J., Bolormaa S., and Goddard M. E.. . 2011. Polymorphic regions affecting human height also control stature in cattle. Genetics 187:981–984. doi: 10.1534/genetics.110.123943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D. C., Bradley D. G., Evans R. D., Kearney F. J., and Berry D. P.. . 2015. Genome-wide association study for calving performance using high-density genotypes in dairy and beef cattle. Genet. Sel. Evol. 47:47. doi: 10.1186/s12711-015-0126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D. C., Bradley D. G., Kearney J. F., and Berry D. P.. . 2014. Genome-wide association study for calving traits in Holstein-Friesian dairy cattle. Animal 8:224–235. doi: 10.1017/S175173111300195X [DOI] [PubMed] [Google Scholar]
- Purfield D. C., Evans R. D., and Berry D. P.. . 2019a. Reaffirmation of known major genes and the identification of novel candidate genes associated with carcass-related metrics based on whole genome sequence within a large multi-breed cattle population. BMC Genomics 20:720. doi: 10.1186/s12864-019-6071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield D. C., Evans R. D., Carthy T. R., and Berry D. P.. . 2019b. Genomic regions associated with gestation length detected using whole-genome sequence data differ between dairy and beef cattle. Front. Genet. 10:1068. doi: 10.3389/fgene.2019.01068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring S. C., McCarthy J., Kelleher M. M., Doherty M. L., and Berry D. P.. . 2018. Risk factors associated with animal mortality in pasture-based, seasonal-calving dairy and beef herds. J. Anim. Sci. 96:35–55. doi: 10.1093/jas/skx072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. L. 1996. Models which might explain negative correlations between direct and maternal genetic effects. Livest. Prod. Sci. 45:111–122. doi: 10.1016/0301-6226(96)00002-4 [DOI] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., and Garrick D. J.. . 2014. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics 15:442. doi: 10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahana G., Guldbrandtsen B., and Lund M. S.. . 2011. Genome-wide association study for calving traits in Danish and Swedish Holstein cattle. J. Dairy Sci. 94:479–486. doi: 10.3168/jds.2010-3381 [DOI] [PubMed] [Google Scholar]
- Sargolzaei M., Chesnais J. P., and Schenkel F. S.. . 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 15:478. doi: 10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbock L., Näsholm A., Berglund B., Johansson K., and Philipsson J.. . 2003. Genetic effects on stillbirth and calving difficulty in Swedish Holsteins at first and second calving. J. Dairy Sci. 86:2228–2235. doi: 10.3168/jds.S0022-0302(03)73813-2 [DOI] [PubMed] [Google Scholar]
- Tenghe A. M. M., Bouwman A. C., Berglund B., Strandberg E., de Koning D. J., and Veerkamp R. F.. . 2016. Genome-wide association study for endocrine fertility traits using single nucleotide polymorphism arrays and sequence variants in dairy cattle. J. Dairy Sci. 99:5470–5485. doi: 10.3168/jds.2015-10533 [DOI] [PubMed] [Google Scholar]
- Thomasen J. R., Guldbrandtsen B., Sørensen P., Thomsen B., and Lund M. S.. . 2008. Quantitative trait loci affecting calving traits in Danish Holstein cattle. J. Dairy Sci. 91:2098–2105. doi: 10.3168/jds.2007-0602 [DOI] [PubMed] [Google Scholar]
- Tiezzi F., Arceo M. E., Cole J. B., and Maltecca C.. . 2018. Including gene networks to predict calving difficulty in Holstein, Brown Swiss and Jersey cattle. BMC Genet. 19:20. doi: 10.1186/s12863-018-0606-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upperman L. R., Kinghorn B. P., MacNeil M. D., and Van Eenennaam A. L.. . 2019. Management of lethal recessive alleles in beef cattle through the use of mate selection software. Genet. Sel. Evol. 51:36. doi: 10.1186/s12711-019-0477-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- VanRaden P. M., Olson K. M., Null D. J., and Hutchison J. L.. . 2011. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. J. Dairy Sci. 94:6153–6161. doi: 10.3168/jds.2011-4624 [DOI] [PubMed] [Google Scholar]
- Wiener P., Smith J. A., Lewis A. M., Woolliams J. A., and Williams J. L.. . 2002. Muscle-related traits in cattle: the role of the myostatin gene in the South Devon breed. Genet. Sel. Evol. 34:221–232. doi: 10.1186/1297-9686-34-2-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Gandin V., Sasaki M., McCracken S., Li W., Silvester J. L., Elia A. J., Wang F., Wakutani Y., Alexandrova R., . et al. 2014. Largen: a molecular regulator of mammalian cell size control. Mol. Cell 53:904–915. doi: 10.1016/j.molcel.2014.02.028 [DOI] [PubMed] [Google Scholar]
- Zolini A. M., Negrón-Pérez V. M., and Hansen P. J.. . 2019. Importance of prostate androgen-regulated mucin-like protein 1 in development of the bovine blastocyst. BMC Dev. Biol. 19:15. doi: 10.1186/s12861-019-0195-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.