Abstract
Serial crystallography (SX) provides an opportunity to observe the molecular dynamics of macromolecular structures at room temperature via pump-probe studies. The delivery of crystals embedded in a viscous medium via an injector or syringe is widely performed in synchrotrons or X-ray free-electron laser facilities with low repetition rates. Various viscous media have been developed; however, there are cases in which the delivery material undesirably interacts chemically or biologically with specific protein samples, or changes the stability of the injection stream, depending on the crystallization solution. Therefore, continued discovery and characterization of new delivery media is necessary for expanding future SX applications. Here, the preparation and characterization of new polysaccharide (wheat starch (WS) and alginate)-based sample delivery media are introduced for SX. Crystals embedded in a WS or alginate injection medium showed a stable injection stream at a flow rate of < 200 nL/min and low-level X-ray background scattering similar to other hydrogels. Using these media, serial millisecond crystallography (SMX) was performed, and the room temperature crystal structures of glucose isomerase and lysozyme were determined at 1.9–2.0 Å resolutions. WS and alginate will allow an expanded application of sample delivery media in SX experiments.
Keywords: serial crystallography, sample delivery, viscous medium, wheat starch, alginate, SMX, SFX, background scattering
1. Introduction
Traditional X-ray crystallography plays an important role in structural biology, as it allows detailed understanding of chemical and biological functions and, ultimately, the mechanisms underlying them [1,2]. It also provides insight into drug design or rational engineering of industrially relevant enzymes [3,4]. However, it has experimental limitations, including radiation damage, cryogenic temperatures, and static structures [5,6], which can be addressed by the recently emerging serial crystallography (SX) technique [7]. X-ray free electron lasers (XFELs) with ultrafast pulse-duration and synchrotron X-rays reduce radiation damage during data collection [8,9,10,11]. Among them, the SX experiment at synchrotron enables low-dose data collection, since the crystal samples’ X-ray exposure time is shorter than in the conventional crystallographic approach [8]. These techniques can be beneficially applied to observe radiation-sensitive metalloproteins or structural flexibility at room temperature, and time-resolved studies [7,8,12,13,14]. In addition, the intense X-ray facilitates the determination of the crystal structure from crystals of a few microns or smaller. Thus, SX has scientific advantages over traditional X-ray crystallography; however, it requires a large number of crystal samples and a sample delivery method that continuously delivers crystals to the X-ray interaction point [15]. Various sample delivery systems, such as injectors [16,17,18], syringes [19,20,21], fixed target scans [22,23,24,25,26], microfluidic devices [27,28], capillaries [29,30], sample extractors [31], and polyimide tube-based containers (quasi 1D fixed target) [32], have been applied to serial femtosecond crystallography (SFX) or serial millisecond crystallography (SMX) experiments. Among them, the delivery of crystals embedded in a viscous medium produced stable injection streams, even at very low flow rates, and was widely applicable in SX in synchrotrons or XFEL facilities with low repetition rates [33].
Sample delivery media can be classified into either amphiphilic, hydrophobic, or hydrophilic materials [33]. Amphiphilic sample delivery medium, such as lipidic cubic phases (LCPs) [17], and hydrophobic sample delivery media, such as greases [19,34,35,36] and shortening [37], produce a stable injection stream with high viscosity while exhibiting certain background scattering rings from the delivery material. Hydrophilic delivery materials can be further classified into sugar-based media, such as agarose [38], hyaluronic acid (HA) [34], hydroxyethyl cellulose (HEC) [35], and sodium carboxymethyl cellulose (NaCMC) [39], and polymer-based media, such as Pluronic F-127 [39], poly(ethylene oxide) [14], and polyacrylamide (PAM) [40]. Although typical sugar-based delivery media reveal very low levels of X-ray background scattering [34,35,38,39] and have the advantage of improving the signal-to-noise ratio (SNR) when compared with other sample delivery media [34,35,38,39], they can be recognized as substrates by certain protein crystal samples, resulting in biological and chemical reactions that can dissolve or damage crystal samples [33]. For example, agarose or cellulose-derived delivery materials can react with agarase or cellulase crystal samples. Therefore, continuous development of delivery media is necessary for extensive applications in future SX studies.
Starch is the main source of energy for humans and is stored as a carbohydrate reserve in plants [41]. It is almost entirely composed of linear amylose and its branched counterpart, amylopectin [42]. These polysaccharides consist of α-(1,4)-linked D-glucose residue chains interconnected via α-(1,6)-glucosidic linkages [41]. Alginates are abundant in nature and primarily found in marine brown algae or capsular polysaccharides in soil bacteria [43]. They belong to a family of unbranched binary copolymers and consist of (1,4)-linked β-d-mannuronic acid and α-l-guluronic acid chains in widely varying compositions and sequences [43]. Alginates have an almost temperature-independent sol/gel transition in the presence of multivalent cations (e.g., Ca2+), which is applicable in the immobilization of living cells [43]. Since these materials have oligosaccharides with different chemical structures than those previously reported, they can provide an expanded opportunity for use as a delivery medium for sugar-binding protein crystals in SX experiments.
Here, the preparation and characterization of wheat starch (WS) and alginate as sample delivery media are introduced for SX experiment. Both delivery media produced stable injection streams at flow rates <200 nL/min and generated very low-level X-ray background scattering. SMX experiments using WS and alginate as delivery media were performed, and the room temperature structures of glucose isomerase and lysozyme were determined at 1.9–2.0 Å resolutions.
2. Results
2.1. Characterization of the Starch Injection Matrix
Typically, polysaccharides can become gelatinized when the temperature is lowered after swelling at high temperature [44]. Additionally, they can remain viscous via hydrogen bonding and van der Waals interactions between chains [45]. In previous reports, the sugar-based delivery medium, agarose, was used in gel form at lower temperatures following melting at high temperatures [38], whereas highly viscous HA, HEC, and NaCMC were prepared by increasing their concentrations, then used as delivery media [34,35,39]. Based on this background, it was expected that WS and alginate could be used as delivery materials in gel or high-viscosity states.
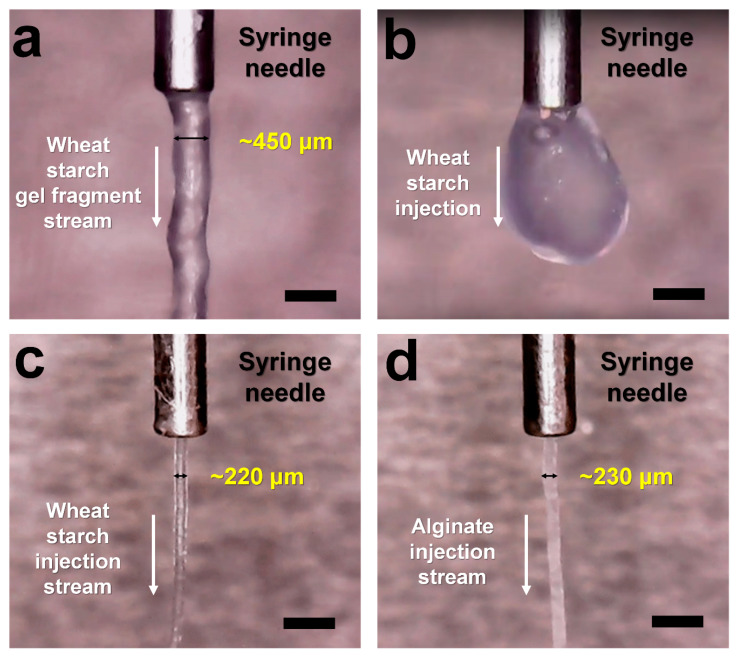
WS and alginate gels were first prepared by raising and lowering the temperature, then used as delivery materials. WS and alginate (5% w/v) were melted above 100 °C and left at room temperature to form gels. However, as temperature was lowered, water from the gel evaporated and adhered to the upper tube cap, indicating that the gel had a concentration higher than 5%. Meanwhile, WS and alginate gel structures are difficult to break; hence, mixing them directly to embed the crystal sample into the gel may cause physical damage to the sample, thereby increasing the mosaicity of the crystal and reducing the diffraction intensity. In order to solve this problem, as in the preparation of the PAM injection matrix [40], WS and alginate gels were fragmented, and the crystal samples were embedded onto the gel fragment. After melting the WS and alginate at >100 °C, the WS and alginate solutions were transferred into 100 μL syringes using a pipette and left at room temperature for 10 min. After gelatinization, the WS and alginate gels were mixed and fragmented by pushing the plungers back and forth in a dual syringe setup. Next, these gel fragments were gently mixed with lysozyme crystal samples using the same setup. Crystal samples embedded in WS or alginate gel fragments were able to produce a continuous injection stream through the syringe needle at a flow rate of 200 nL/min (Figure 1a). The behavior of this injection stream is expected to be used to deliver crystal samples; however, as mentioned above, the evaporation of water from the gel during the process of raising and lowering the temperature can change the final gel concentration. Thus, this approach was not considered suitable in terms of reproducibility of the delivery medium preparation. Meanwhile, the previous study used low-melting agarose [38], which may have little effect on the evaporation of water. As a result, the melting temperature of sugar is considered to be important when gelling sugar and using it as a delivery medium.
Figure 1.
Snapshot of the injection stream of (a) 5% (w/v) WS gel fragment, (b) 5% (w/v) WS, (c) 14% (w/v) WS, and (d) 14% (w/v) alginate. WS and alginate delivery media were extruded from a syringe needle with an inner diameter of 168 μm at room temperature. The scale bar indicates 718 μm.
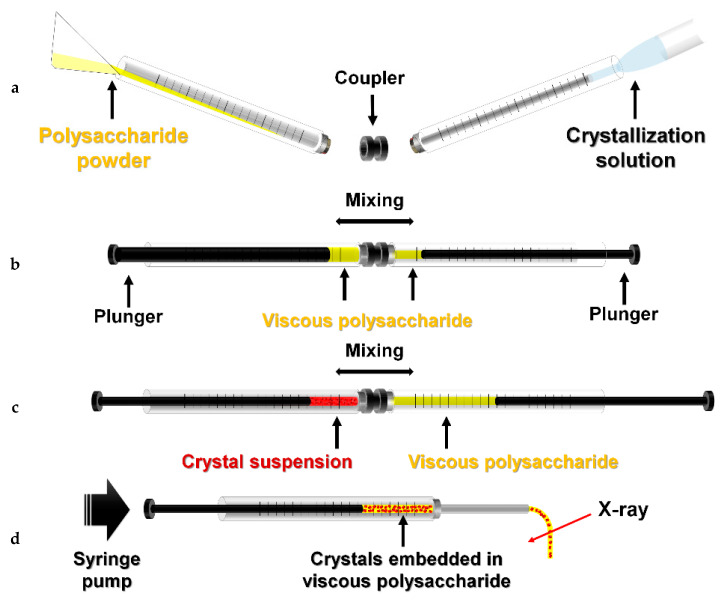
Next, highly viscous WS and alginate were prepared by increasing the concentrations, then used as delivery media. During sample preparation, it was difficult to transfer highly viscous WS and alginate prepared in microcentrifuge tubes into syringes using pipettes, and the use of spatulas would lead to large sample losses. Therefore, as previously reported for the preparation of the poly(ethylene oxide) (PEO) delivery medium [14], WS or alginate powder was added directly into a 250 μL syringe with an internal diameter of 2.3 mm, and the crystallization solution to dissolve the powder was placed in a 100 μL syringe (Figure 2a). Both syringes were connected to the coupler then mixed back and forth > 50 times using a plunger (Figure 2b). The viscous delivery medium was transferred to the 100 μL syringe, and the 250 μL syringe was removed. The 100 μL syringe containing the crystal suspension was connected to the syringe containing the delivery medium and gently mixed back and forth 30 times using the plunger (Figure 2c). The crystal samples embedded in the delivery medium were transferred to one syringe, and the partner syringe and coupler were removed. After connecting the needle to the syringe containing the crystals, the delivery medium containing crystal samples was delivered to an X-ray interaction point using a syringe pump (Figure 2d).
Figure 2.
Preparation of WS and alginate delivery media for SX. (a) Wheat starch (WS) or alginate powder was added to a 250 μL syringe and crystallization solution to a 100 μL syringe. (b) WS or alginate was mixed with crystallization solution in a dual syringe setup. (c) Viscous WS or alginate was mixed with the crystal suspension. (d) After connecting the syringe needle, crystals embedded in the viscous medium were delivered to an X-ray interaction point using a syringe pump.
WS or alginate (5–20% w/v) was prepared, and the injection stability was initially screened using a syringe with a 168 μm inner diameter (ID) needle at a flow rate of 200 nL/min. At WS and alginate concentrations less than 8%, the injection stream was unstable or often presented drop-like formations at the tip of the needle (Figure 2b). WS and alginate showed stable injection streams at concentrations above 12%. In this SMX experiment, a 20% (w/v) WS or alginate delivery medium was mixed with the crystal suspension in a 3.5:1.5 ratio in a dual syringe setup, and the final 14% (w/v) WS or alginate delivery medium was used in the SMX experiment. The diameters of the injection stream flows of WS and alginate were ~220 and ~230 μm, respectively, despite passing through a needle with a 168 μm inner diameter (Figure 2c,d).
2.2. Measurement of Background Scattering
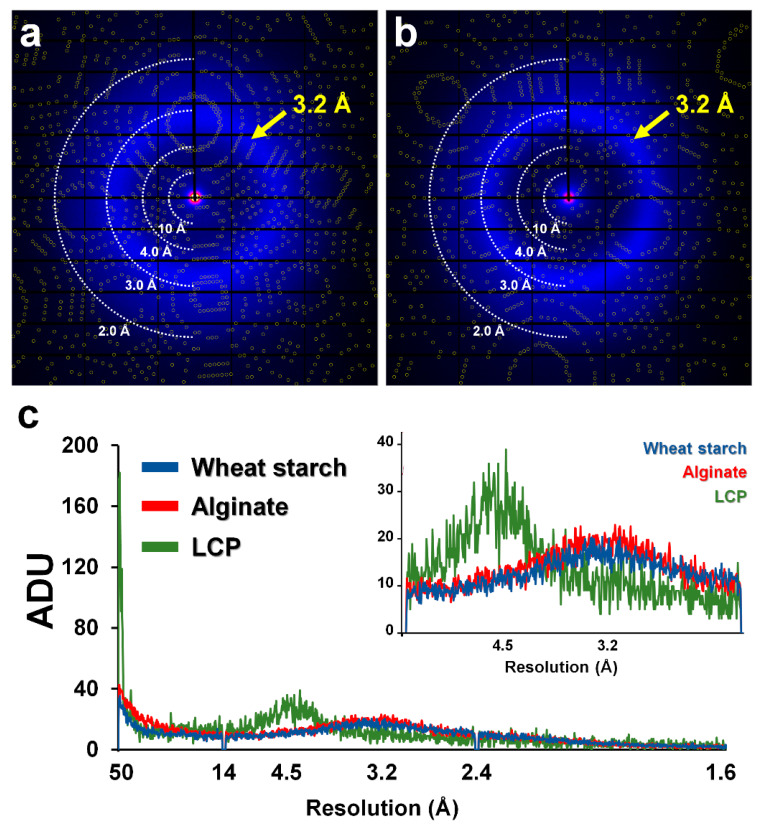
X-ray exposed crystal embedded sample delivery could produce a diffraction pattern of the crystal sample, as well as background scattering of the delivery material [33]. X-ray background scattering of the delivery medium could affect the SNR and diffraction data quality; hence, it is an important criterion during the selection of delivery media for SX experiments [33,38]. Since the viscous state of WS and alginate maintains unregulated hydrogen bonds between polysaccharides, it is expected to show very weak or no scattering that does not produce significant scattering at certain angles. To prove this, the X-ray background scattering of the WS and alginate delivery media was measured. The 14% (w/v) WS and alginate injection streams were delivered to X-rays via a syringe needle with a 168 μm ID. The diameters of WS and alginate injection streams were approximately 220 μm and 230 μm, respectively. Average intensities were analyzed from the beam stopper to 1.6 Å resolution. The highest intensity, approximately 40 ADU (analog-to-digital unit), was observed at 100 Å around the beam stopper for both the WS and (Figure 3a) and alginate (Figure 3b) delivery media. Moreover, both media showed diffused scattering of < 20 ADU in the 3.2–3.4 Å region, which is considered as solvent scattering from the crystals and the delivery media. Next, the background scattering of WS and alginate was compared with LCP delivery medium, which is widely used in SFX research. The diameter of LCP used for background scattering measurements was approximately 190 μm, which was delivered from a syringe with a 168 μm ID. LCP showed high background scattering of > 170 ADU at 100 Å around the beam stopper and also produced >30 ADU levels of background scattering at 4–4.5 Å, which is considered as scattering from lipid packing (Figure 3c). LCP showed higher intensity for background scattering around 4.5 Å resolution than WS or alginate, while it was lower than WS or alginate around 3.2 Å resolution. Not surprisingly, the LCP used in the experiment was composed of 60% (w/v) monoolein, whereas WS and alginate had a concentration of 14% (w/v). In other words, the proportion of solvent in the LCP injection stream was 40%, whereas the proportion of solvent in the WS and alginate injection streams was 86%; thus, the higher water scattering intensity at 3.2 Å for WS and alginate compared to LCP was expected. Nevertheless, overall background scattering was lower for the WS and alginate injection streams than for LCP.
Figure 3.
Measurement of X-ray background scattering of WS and alginate. Diffraction pattern of glucose isomerase embedded in 10% (w/v) (a) WS and (b) alginate delivery media. The circles show predicted positions of Bragg peaks by CrystFEL. (c) 2D profile of the average scattering of WS (blue), alginate (red), and LCP (green). (Inset) Magnified view of background scattering in the range 14–2.4 Å. Yellow arrows indicate 3.2 Å resolution area.
2.3. Application of WS and Alginate Injection Media for SMX
To demonstrate that WS and alginate can be applied to SX experiments as sample delivery media, SMX experiments were performed at a synchrotron (Table 1). Glucose isomerase (GI) and lysozyme crystals were embedded in the WS or alginate, and they were extruded from syringe needle with a 168 μm ID to X-ray beam path at a flow rate of 200 nL/min at room temperature.
Table 1.
X-ray data collection and refinement statistics.
| Data Collection | Glucose Isomerase Embedded in WS | Lysozyme Embedded in WS | Glucose Isomerase Embedded in Alginate | Lysozyme Embedded in Alginate |
|---|---|---|---|---|
| Energy (eV) | 12,659 | |||
| Expose time | 100 ms | |||
| Space group | I222 | P43212 | I222 | P43212 |
| Cell dimension (Å) | ||||
| a | 94.49 | 79.36 | 94.09 | 79.45 |
| b | 100.19 | 79.36 | 99.88 | 79.45 |
| c | 103.40 | 38.63 | 103.00 | 38.47 |
| Collected images | 81,798 | 34,484 | 25,000 | 4563 |
| Hits images | 8361 | 4231 | 10,268 | 3968 |
| Indexed images | 7732 | 3789 | 9946 | 1506 |
| Indexed pattern | 8281 | 4383 | 11,289 | 2066 |
| Resolution (Å) | 72.5-2.00 (2.07-2.00) |
80.0-2.00 (2.07-2.00) |
72.5-2.00 (2.07-2.00) |
80.0-1.90 (1.97-1.90) |
| Unique reflections | 33,248 (3247) | 8808 (844) | 32,970 (3233) | 10,209 (989) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 148.5 (101.5) | 126.6 (88.4) | 241.9 (167.8) | 158.1 (112.7) |
| SNR | 4.01 (1.59) | 4.04 (1.59) | 4.19 (1.28) | 3.91 (1.31) |
| CC | 0.9755 (0.5966) | 0.9637 (0.6351) | 0.9817 (0.4492) | 0.9672 (0.4418) |
| CC* | 0.9937 (0.8644) | 0.9907 (0.8814) | 0.9953 (0.7873) | 0.9916 (0.7828) |
| Rpslit (%) a | 17.22 (65.64) | 17.06 (66.17) | 18.99 (87.37) | 17.70 (85.37) |
| Wilson B factor (Å2) | 28.23 | 32.20 | 22.48 | 27.89 |
| Refinement | ||||
| Resolution (Å) | 70.29-2.00 | 56.13-2.00 | 71.70-2.00 | 56.18-1.90 |
| Rwork/Rworkb | 17.05/21.30 | 18.41/23.33 | 19.23/22.58 | 18.96/23.82 |
| R.m.s. deviations | ||||
| Bond length (Å) | 0.007 | 0.006 | 0.007 | 0.008 |
| Bond angle (°) | 0.860 | 0.838 | 0.836 | 0.970 |
| B factors (Å) | ||||
| Protein | 29.59 | 38.61 | 26.29 | 33.55 |
| Ligands | 38.88 | 47.50 | 40.40 | 41.15 |
| Ramachandran (%) | ||||
| Preferred | 95.72 | 95.80 | 96.22 | 95.97 |
| Allowed | 3.74 | 4.20 | 3.24 | 4.03 |
| Outliers | 0.53 | 0 | 0.54 | 0 |
Values for the outer shell are given in parentheses. a Rsplit = . b Rwork is defined as Σ||Fobs|-|Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively; and Rfree represents the R factor for the test set (10% of the data). CC refers to Pearson’s correlation coefficient, and CC* = .
For GI embedded in WS, a total of 81,798 images were collected, and hit images, including diffraction patterns, totaled 8361 images with a hit rate of 10.22%. The data processing resulted in a GI diffraction pattern from a total of 7732 images with an indexing rate of 92.47% (Figure 3A). In the indexed image, a total of 8281 diffraction patterns, including multi-crystal hit patterns, were obtained. Data were processed up to 2.0 Å, with completeness, SNR, Rsplit, and CC* of 100, 4.01, 17.22, and 0.9937, respectively. Rwork and Rfree of the final model structure were 17.05% and 21.30%, respectively. For lysozyme embedded in WS, a total of 34,484 images were collected, and hit images, including diffraction patterns, totaled 4231 images at a hit rate of 12.27%. The data processing resulted in a lysozyme diffraction pattern from a total of 3789 images with an indexing rate of 89.55%. In the indexed image, a total of 4383 diffraction patterns, including multi-crystal hit patterns, were obtained. Indexed diffraction data were processed up to 2.0 Å, with completeness, SNR, Rsplit, and CC* of 100, 4.04, 17.06, and 0.9907, respectively. Rwork and Rfree of the final model structure were 18.41% and 23.33%, respectively.
For GI embedded in alginate, a total of 25,000 images were collected, and hit images, including diffraction patterns, totaled 10,268 images with a hit rate of 41.07%. The data processing resulted in a GI diffraction pattern from a total of 9946 images with an indexing rate of 96.86% (Figure 3B). In the indexed image, a total of 11,289 diffraction patterns, including multi-crystal hit patterns, were obtained. Data were processed up to 2.0 Å, with completeness, SNR, Rsplit, and CC* of 100, 4.19, 18.99, and 0.9953, respectively. Rwork and Rfree of the final model structure were 19.23% and 22.58%, respectively. For lysozyme embedded in alginate, a total of 4563 images were collected, and hit images, including diffraction patterns, totaled 3968 images at a hit rate of 86.96%. The data processing resulted in a lysozyme diffraction pattern from a total of 1506 images with an indexing rate of 37.95%. In the indexed image, a total of 2066 diffraction patterns, including multi-crystal hit patterns, were obtained. Indexed diffraction data were processed up to 1.9 Å, with completeness, SNR, Rsplit, and CC* of 100, 3.91, 17.70, and 0.9916, respectively. Rwork and Rfree of the final model structure were 18.96% and 23.82%, respectively.
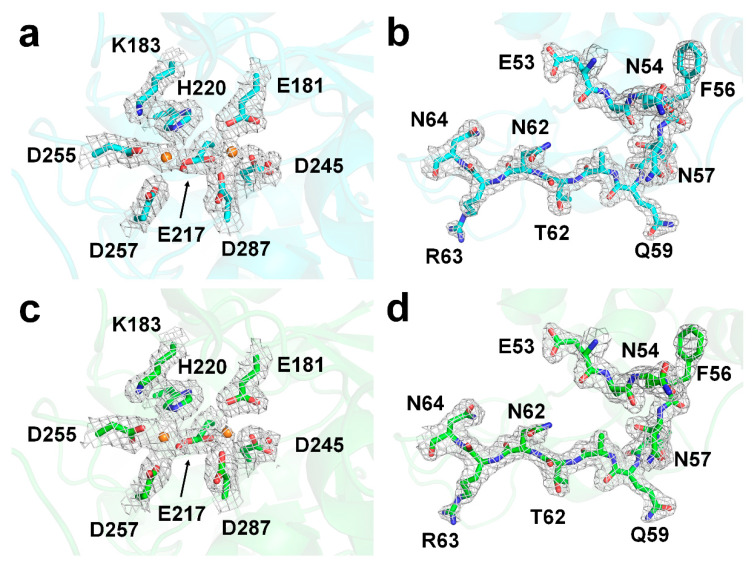
The electron density maps for the entire protein structure were clearly visible (Figure 4). In lysozyme delivered in WS and alginate, no negative fo-fc electron density maps were assumed to have significant radiation damage in four disulfide bonds (C24-C145, C48-C133, C82-C98, and C94-C112). The active site of GI contains two metal-binding sites, which are important for substrate recognition and enzyme activity [46,47]. For GI delivered in WS and alginate, there was no significant negative fo-fc electron density map, indicating no significant radiation damage.
Figure 4.
2Fo-Fc electron density map (grey mesh, 1.2 σ) of the active sites of (a) glucose isomerase and (b) lysozyme delivered in WS (cyan), and (c) glucose isomerase and (d) lysozyme delivered in alginate (green).
3. Discussion
Here, the preparation and characterization of WS and alginate delivery media were reported for SX experiments. Although alginate performed well as a delivery medium in this experiment, it could form a sol/gel and may be unsuitable as a delivery material when Ca2+ is present in the crystallization solution. The chemical structures of sugar-based delivery media reported to date are shown in Table 2. When selecting a sugar-based delivery material, it may be possible to first consider whether chemical interactions between the target protein and the selected delivery material exist based on the chemical structure of the delivery material. To confirm crystal damage caused by the interaction between selected polysaccharide and the crystal sample, a mixture of the crystal sample and the delivery medium was incubated at room temperature for several hours, followed by a crystal morphology evaluation under a microscope or by crystal diffraction quality evaluation by exposing it to X-rays. If the reported polysaccharide-based delivery medium is unsuitable due to specific or nonspecific interaction with the target protein, it is possible to prepare a new viscous delivery material using a polysaccharide substance composed of non-interacting sugars. Although the degree of viscosity varies depending on individual polysaccharide properties, general linear polysaccharides, such as WS, alginate, HA, HEC, and NaCMC, form viscous states by increasing the hydrogen bonds between sugar chains with increasing concentration [45]. These new viscous sugars also present very low levels of background scattering without scattering at certain angles because they exist as random hydrogen bonds between polysaccharide chains. Meanwhile, the crystallization solution of the crystal samples in the practical experiment will be diverse. In actual experiments, the interaction between a specific composition of the crystallization solution and the selected polysaccharide-based delivery material can produce an unstable injection stream. In this case, other polysaccharide-based delivery materials, which do not react with a crystal sample and are not affected by the crystallization solution’s viscosity, may be alternatively used. Therefore, this study extends the pool of viscous crystal delivery media useful in SX experiments.
Table 2.
Chemical structure of the polysaccharide-based viscous delivery media.
| Delivery Medium | Chemical Structure | Component | Reference |
|---|---|---|---|
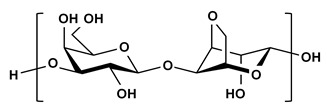
| Agarose |
|
β-(1-4)-(3,6)-anhydro-L-galactose and α-(1-3)-D-galactose | [34] |
| Hyaluronic acid (HA) |
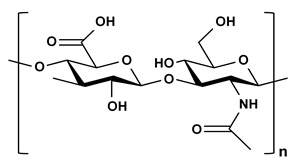
|
β-d-(1-3) glucuronic acid and β-d-(1-4)-N-acetylglucosamine | [32] |
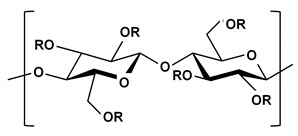
| Hydroxyethyl cellulose (HEC) |
|
R is H or hydroxyl ethyl group (–CH2CH2–OH) | [31] |
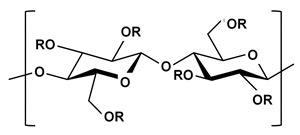
| Sodium carboxymethyl cellulose (NaCMC) |
|
R is H or carboxymethyl groups (–CH2–COOH) | [35] |
| Wheat starch (WS) |
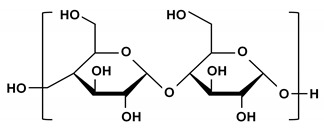
|
Polymer of glucose | This study |
| Alginate |
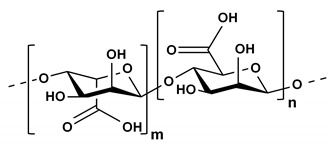
|
(1,4)-linked β-d-mannuronic acid and α-l-guluronic acid | This study |
4. Materials and Methods
4.1. Protein Crystal Preparation
GI from Streptomyces rubiginosus was purchased from Hampton Research (HR7-098, Aliso Viejo, CA, USA), and was supplied in crystalline form and directly used for the SX experiment without post-crystallization, as previously reported [24]. The size and density of GI were < 60 × 60 × 40 μm3 and 3 × 107 crystals/mL, respectively. Lysozyme from chicken egg white was purchased from Sigma-Aldrich (L6876, St. Louis, MO, USA) and crystallized as previously reported [40]. The size and density of lysozyme were < 60 × 60 × 40 μm3 and 3 × 107 crystals/mL, respectively.
4.2. Preparation of Starch and Aginate Injection Matrix
Unmodified wheat starch (WS, S5127) and alginate (A2033) were purchased from Sigma-Aldrich (St. Louis, MO, USA). WS (14 mg) or alginate powder (14 mg) was added to a 250 μL syringe (Hamilton, 81065, Reno, NV, USA) and connected via coupler with another 100 μL syringe (Hamilton, 81065) containing crystallization solution (70 μL). The syringe plungers were moved gently back and forth more than 50 times. After removing the 250 μL syringe, the 100 μL syringe containing the protein crystal suspension (30 μL) was connected via a coupler to a 100 μL syringe containing the delivery medium. The syringe plungers were moved gently back and forth more than 20 times, and the crystals were embedded in delivery medium. After moving the mixture to one syringe, the coupler and empty syringe were removed, and the syringe needle was connected. The syringe containing the crystals embedded in WS or alginate was installed into a Fusion Touch 100 syringe pump (CHEMYX, Stafford, TX, USA), and the plunger was pushed through the motor drive to deliver the sample [21].
4.3. Data Collection
SMX experiments using a WS injection matrix were performed at the 11C beamline, PLS-II, Pohang, Korea [48]. The X-ray beam size at the sample position was approximately 4 (vertical) × 8 (horizontal) μm2 (FWHM). The photon flux was 1.3 × 1012 photons/s, and the wavelength was 0.9795. Crystals embedded in WS or alginate were delivered via a syringe pump in horizontal direction to avoid beamline interference [21]. The inner diameter of the syringe needle was 168 μm, and the sample was delivered at a flow rate of 200–300 nL/min. Crystals were X-ray exposed for 100 ms. Diffraction images were corrected on a Pilatus 6M with 10 Hz readout at room temperature.
4.4. Data Processing and Structure Determination
Images containing the diffraction pattern were filtered using the Cheetah program [49]. The diffraction pattern of the hit image was processed using CrystFEL [50]. The phases of GI and lysozyme were obtained by molecular replacement using phase-MR in Phenix [51], using the crystal structures of GI (PDB code 5ZYD) [47] and lysozyme (PDB code 6IG6) [40] as the search models. The model was built using COOT [52], individual proteins were refined using Phenix.refinement in PHENIX [53], geometry was validated using MolProbity [54], and all structural figures were produced with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA). The structure factor and coordinate files have been deposited in the Protein Data Bank (Available online: www.rcsb.org/) under the PDB code 7BVL (glucose isomerase delivered in WS), 7BVM (lysozyme delivered in WS), 7BVN (glucose isomerase delivered in alginate), and 7BVO (lysozyme delivered in alginate). Diffraction images have been deposited to CXIDB under ID 124 (glucose isomerase delivered in WS), 125 (lysozyme delivered in WS), 126 (glucose isomerase delivered in alginate), and 127 (lysozyme delivered in alginate).
4.5. Background Scattering Analysis
WS, PAM, and LCP (60% (w/v) monoolein) media were delivered to the X-ray interaction point through a syringe needle with a 168 μm ID. The photon flux of X-rays was 1.3 × 1012 photons/s, and exposure was for 100 ms. Background scattering of the delivery media was analyzed using ADXV (Available online: https://www.scripps.edu/tainer/arvai/adxv.html).
Acknowledgments
I would like to thank the beamline staff at 11C beamline at Pohang Accelerator Laboratory for their assistance with data collection; and Global Science experimental Data hub Center (GSDC) at the Korea Institute of Science and Technology Information (KISTI) for computational support.
Abbreviations
| SX | serial crystallography |
| SMX | serial millisecond crystallography |
| SFX | serial femtosecond crystallography |
| WS | wheat starch |
| ADU | analog-to-digital unit |
| GI | glucose isomerase |
Funding
This work was supported by a Korea University grant and funded by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MOE) (NRF-2017R1D1A1B03033087 and NRF-2017M3A9F6029736).
Conflicts of Interest
The author declares no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Egli M. Diffraction techniques in structural biology. Curr. Protoc. Nucleic. Acid. Chem. 2016;65:7.13.1–7.13.41. doi: 10.1002/cpnc.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su X.D., Zhang H., Terwilliger T.C., Liljas A., Xiao J., Dong Y. Protein Crystallography from the Perspective of Technology Developments. Cryst. Rev. 2015;21:122–153. doi: 10.1080/0889311X.2014.973868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomes A., Chruszcz M., Gustchina A., Minor W., Mueller G.A., Pedersen L.C., Wlodawer A., Chapman M.D. 100 Years later: Celebrating the contributions of x-ray crystallography to allergy and clinical immunology. J. Allergy Clin. Immun. 2015;136:29–37. doi: 10.1016/j.jaci.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blundell T.L. Protein crystallography and drug discovery: Recollections of knowledge exchange between academia and industry. IUCrJ. 2017;4:308–321. doi: 10.1107/S2052252517009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holton J.M., Frankel K.A. The minimum crystal size needed for a complete diffraction data set. Acta. Cryst. D. 2010;66:393–408. doi: 10.1107/S0907444910007262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taberman H. Radiation Damage in Macromolecular Crystallography—An Experimentalist’s View. Crystals. 2018;8:157. doi: 10.3390/cryst8040157. [DOI] [Google Scholar]
- 7.Standfuss J., Spence J. Serial crystallography at synchrotrons and X-ray lasers. IUCrJ. 2017;4:100–101. doi: 10.1107/S2052252517001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinert T., Olieric N., Cheng R., Brunle S., James D., Ozerov D., Gashi D., Vera L., Marsh M., Jaeger K., et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017;8:542. doi: 10.1038/s41467-017-00630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlichting I. Serial femtosecond crystallography: The first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J., Kim H.Y., Park J., Kim S., Kim S., Rah S., Lim J., Nam K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick-Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018;25:289–292. doi: 10.1107/S1600577517016186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogly P., James D., Wang D., White T.A., Zatsepin N., Shilova A., Nelson G., Liu H., Johansson L., Heymann M., et al. Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ. 2015;2:168–176. doi: 10.1107/S2052252514026487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebrahim A., Moreno-Chicano T., Appleby M.V., Chaplin A.K., Beale J.H., Sherrell D.A., Duyvesteyn H.M.E., Owada S., Tono K., Sugimoto H., et al. Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins. IUCrJ. 2019;6:543–551. doi: 10.1107/S2052252519003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spence J.C.H. Approaches to time-resolved diffraction using an XFEL. Faraday Discuss. 2014;171:429–438. doi: 10.1039/C4FD00025K. [DOI] [PubMed] [Google Scholar]
- 14.Martin-Garcia J.M., Conrad C.E., Nelson G., Stander N., Zatsepin N.A., Zook J., Zhu L., Geiger J., Chun E., Kissick D., et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ. 2017;4:439–454. doi: 10.1107/S205225251700570X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martiel I., Muller-Werkmeister H.M., Cohen A.E. Strategies for sample delivery for femtosecond crystallography. Acta. Cryst. D Struct. Biol. 2019;75:160–177. doi: 10.1107/S2059798318017953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePonte D.P., Weierstall U., Schmidt K., Warner J., Starodub D., Spence J.C.H., Doak R.B. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D. 2008:41. doi: 10.1088/0022-3727/41/19/195505. [DOI] [Google Scholar]
- 17.Weierstall U., James D., Wang C., White T.A., Wang D.J., Liu W., Spence J.C.H., Doak R.B., Nelson G., Fromme P., et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierra R.G., Gati C., Laksmono H., Dao E.H., Gul S., Fuller F., Kern J., Chatterjee R., Ibrahim M., Brewster A.S., et al. Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II. Nat Methods. 2016;13:59–62. doi: 10.1038/nmeth.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugahara M., Mizohata E., Nango E., Suzuki M., Tanaka T., Masudala T., Tanaka R., Shimamura T., Tanaka Y., Suno C., et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat Methods. 2015;12:61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 20.Berntsen P., Hadian Jazi M., Kusel M., Martin A.V., Ericsson T., Call M.J., Trenker R., Roque F.G., Darmanin C., Abbey B. The serial millisecond crystallography instrument at the Australian Synchrotron incorporating the “Lipidico” injector. Rev. Sci. Instrum. 2019;90:085110. doi: 10.1063/1.5104298. [DOI] [PubMed] [Google Scholar]
- 21.Park S.Y., Nam K.H. Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography. J. Synchrotron Radiat. 2019;26:1815–1819. doi: 10.1107/S160057751900897X. [DOI] [PubMed] [Google Scholar]
- 22.Hunter M.S., Segelke B., Messerschmidt M., Williams G.J., Zatsepin N.A., Barty A., Benner W.H., Carlson D.B., Coleman M., Graf A., et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2014;4:6026. doi: 10.1038/srep06026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roedig P., Ginn H.M., Pakendorf T., Sutton G., Harlos K., Walter T.S., Meyer J., Fischer P., Duman R., Vartiainen I., et al. High-speed fixed-target serial virus crystallography. Nat Methods. 2017;14:805–810. doi: 10.1038/nmeth.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D., Baek S., Park J., Lee K., Kim J., Lee S.J., Chung W.K., Lee J.L., Cho Y., Nam K.H. Nylon mesh-based sample holder for fixed-target serial femtosecond crystallography. Sci. Rep. 2019;9:6971. doi: 10.1038/s41598-019-43485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen A.E., Soltis S.M., Gonzalez A., Aguila L., Alonso-Mori R., Barnes C.O., Baxter E.L., Brehmer W., Brewster A.S., Brunger A.T., et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc. Natl. Acad. Sci. USA. 2014;111:17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roessler C.G., Kuczewski A., Stearns R., Ellson R., Olechno J., Orville A.M., Allaire M., Soares A.S., Héroux A. Acoustic methods for high-throughput protein crystal mounting at next-generation macromolecular crystallographic beamlines. J. Synchrotron Radiat. 2013;20:805–808. doi: 10.1107/S0909049513020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteiro D.C.F., Vakili M., Harich J., Sztucki M., Meier S.M., Horrell S., Josts I., Trebbin M. A microfluidic flow-focusing device for low sample consumption serial synchrotron crystallography experiments in liquid flow. J. Synchrotron Radiat. 2019;26:406–412. doi: 10.1107/S1600577519000304. [DOI] [PubMed] [Google Scholar]
- 28.Calvey G.D., Katz A.M., Pollack L. Microfluidic Mixing Injector Holder Enables Routine Structural Enzymology Measurements with Mix-and-Inject Serial Crystallography Using X-ray Free Electron Lasers. Anal. Chem. 2019;91:7139–7144. doi: 10.1021/acs.analchem.9b00311. [DOI] [PubMed] [Google Scholar]
- 29.Stellato F., Oberthur D., Liang M., Bean R., Gati C., Yefanov O., Barty A., Burkhardt A., Fischer P., Galli L., et al. Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ. 2014;1:204–212. doi: 10.1107/S2052252514010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam K.H. Stable sample delivery in viscous media via a capillary for serial crystallography. J. Appl. Cryst. 2020;53:45–50. doi: 10.1107/S1600576719014985. [DOI] [Google Scholar]
- 31.Mathews I.I., Allison K., Robbins T., Lyubimov A.Y., Uervirojnangkoorn M., Brunger A.T., Khosla C., DeMirci H., McPhillips S.E., Hollenbeck M., et al. The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography. Biochemistry. 2017;56:4751–4756. doi: 10.1021/acs.biochem.7b00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee D., Park S., Lee K., Kim J., Park G., Nam K.H., Baek S., Chung W.K., Lee J.-L., Cho Y., et al. Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography. J. Appl. Cryst. 2020;53:477–485. doi: 10.1107/S1600576720002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam K.H. Sample Delivery Media for Serial Crystallography. Int. J. Mol. Sci. 2019;20:1094. doi: 10.3390/ijms20051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugahara M., Song C.Y., Suzuki M., Masuda T., Inoue S., Nakane T., Yumoto F., Nango E., Tanaka R., Tono K., et al. Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Sci. Rep. 2016;6:24484. doi: 10.1038/srep24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugahara M., Nakane T., Masuda T., Suzuki M., Inoue S., Song C.Y., Tanaka R., Nakatsu T., Mizohata E., Yumoto F., et al. Hydroxyethyl cellulose matrix applied to serial crystallography. Sci. Rep. 2017;7:703. doi: 10.1038/s41598-017-00761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugahara M., Motomura K., Suzuki M., Masuda T., Joti Y., Numata K., Tono K., Yabashi M., Ishikawa T. Viscosity-adjustable grease matrices for serial nanocrystallography. Sci. Rep. 2020;10:1371. doi: 10.1038/s41598-020-57675-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam K.H. Shortening injection matrix for serial crystallography. Sci. Rep. 2020;10:107. doi: 10.1038/s41598-019-56135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conrad C.E., Basu S., James D., Wang D., Schaffer A., Roy-Chowdhury S., Zatsepin N.A., Aquila A., Coe J., Gati C., et al. A novel inert crystal delivery medium for serial femtosecond crystallography. Iucrj. 2015;2:421–430. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacsova G., Grunbein M.L., Kloos M., Barends T.R.M., Schlesinger R., Heberle J., Kabsch W., Shoeman R.L., Doak R.B., Schlichting I. Viscous hydrophilic injection matrices for serial crystallography. IUCrJ. 2017;4:400–410. doi: 10.1107/S2052252517005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J., Park S., Kim J., Park G., Cho Y., Nam K.H. Polyacrylamide injection matrix for serial femtosecond crystallography. Sci. Rep. 2019;9:2525. doi: 10.1038/s41598-019-39020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertoft E. Understanding Starch Structure: Recent Progress. Agronomy. 2017;7:56. doi: 10.3390/agronomy7030056. [DOI] [Google Scholar]
- 42.Zobel H.F. Molecules to Granules: A Comprehensive Starch Review. Starch - Stärke. 1988;40:44–50. doi: 10.1002/star.19880400203. [DOI] [Google Scholar]
- 43.Draget K.I., Smidsrød O., Skjåk-Bræk G., Vandamme E.J., De Baets S., Steinbüchel A. Biopolymers Online. Wiley-VCH; Weinheim, Germany: 2005. Alginates from Algae. [Google Scholar]
- 44.Tako M. The Principle of Polysaccharide Gels. Adv. Biosci. Biotechnol. 2015;06:22–36. doi: 10.4236/abb.2015.61004. [DOI] [Google Scholar]
- 45.Tester R.F., Karkalas J. Encyclopedia of Food Sciences and Nutrition. Academic Press; Cambridge, UK: 2003. CARBOHYDRATES | Interactions with Other Food Components; pp. 875–881. [Google Scholar]
- 46.Bae J.E., Hwang K.Y., Nam K.H. Structural analysis of substrate recognition by glucose isomerase in Mn2+ binding mode at M2 site in S. rubiginosus. Biochem. Biophys. Res. Commun. 2018;503:770–775. doi: 10.1016/j.bbrc.2018.06.074. [DOI] [PubMed] [Google Scholar]
- 47.Bae J.E., Kim I.J., Nam K.H. Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode. Biochem. Biophys. Res. Commun. 2017;493:666–670. doi: 10.1016/j.bbrc.2017.08.134. [DOI] [PubMed] [Google Scholar]
- 48.Park S.Y., Ha S.C., Kim Y.G. The Protein Crystallography Beamlines at the Pohang Light Source II. Biodesign. 2017;5:30–34. [Google Scholar]
- 49.Barty A., Kirian R.A., Maia F.R., Hantke M., Yoon C.H., White T.A., Chapman H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Cryst. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White T.A., Mariani V., Brehm W., Yefanov O., Barty A., Beyerlein K.R., Chervinskii F., Galli L., Gati C., Nakane T., et al. Recent developments in CrystFEL. J. Appl. Cryst. 2016;49:680–689. doi: 10.1107/S1600576716004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta. Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta. Cryst. D Biol. Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta. Cryst. D Biol. Cryst. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]