Abstract
Objective
This study aimed to investigate the effect of Wnt/β-catenin signal pathway mediated by miR-342-5p targeting CBX2 gene on the proliferation, metastasis, invasion and apoptosis of ovarian cancer cells, and to explore its related regulatory mechanism.
Methods
Human normal ovarian epithelial cell line IOSE80, human ovarian cancer cell line SKOV3 and OVCAR3 were the subjects. Software were used to predict the binding site of miR-342-5p targeting CBX2 gene. The proliferation rate of ovarian cancer cells was detected by MTT method; the cell viability of each group was observed by colony formation test; the apoptosis of cells in each group was detected by flow cytometry; the invasive ability of cells was determined by transwell test, and the migration ability of cells was detected by scratch test. The mRNA expression levels of miR-342-5p, CBX2, Wnt1, β-catenin, C-myc and Cyclin D1 were measured by qRT-PCR. Also, Western blot was used to determine the protein expression levels of CBX2, Wnt1, β-catenin, C-myc and Cyclin D1.
Results
CBX2 was identified as the target gene of miR-342-5p. MTT test results showed that miR-342-5p could significantly inhibit the proliferation of SKOV3 and OVCAR3 cells, colony formation assay results indicated that the viability of SKOV3 and OVCAR3 cells transfected with miR-342-5p decreased significantly, and flow cytometry results suggested that miR-342-5p could promote the apoptosis of SKOV3 and OVCAR3 cells. Also, the results of transwell showed that miR-342-5p could significantly inhibit the invasive ability of SKOV3 and OVCAR3 cells, and the results of scratch assay suggested that miR-342-5p could significantly inhibit the migration of SKOV3 and OVCAR3 cells. Moreover, qRT-PCR and Western blot results indicated that the mRNA and protein expression levels of CBX2, Wnt1, β-catenin, C-myc and Cyclin D1 decreased in SKOV3 and OVCAR3 cells transfected with miR-342-5p, while the mRNA expression levels of miR-342-5p increased significantly (P<0.05).
Conclusion
MiR-342-5p targeted gene is CBX2, which can significantly reduce the proliferation, invasion, migration and viability of ovarian cancer cell lines SKOV3 and OVCAR3, and promote their apoptosis. The mechanism may be related to the mediation of Wnt/β-catenin signal pathway and down-regulation of the related genes expression.
Keywords: miR-342-5p, CBX2, ovarian cancer, Wnt/β-catenin signal pathway
Introduction
Ovarian cancer, the world’s deadliest gynecological malignancy, accounts for 5% of cancer deaths in women. In 2018, 22,240 new cases of ovarian cancer were diagnosed in the United States.1,2 Also, the worldwide incidence of ovarian cancer remains very high, and the 5-year survival rate is still less than 30%, despite the rapid development of treatments, including chemotherapy and surgery, over the past few decades. Therefore, more attention has been paid to the molecular biological mechanism of the occurrence and development of ovarian cancer.3 An important malignant marker of human cancer is the maintenance of proliferative signals and the activation of invasion and metastasis.4 Inhibiting the endless proliferation and activation of invasion and metastasis in cancer cells is the basic method to solve ovarian cancer, so exploring the molecular mechanism of malignant growth and metastasis may provide new treatment strategies for ovarian cancer.5
miRNA, a cellular regulatory factor, participates in many cellular regulatory processes and is closely related to many processes of cancer cells, such as cancer cell cycle, apoptosis, autophagy and oxidative stress. MiR-342 gene is located in the third intron region of Evl (Ena/VASP-like) gene, and two miRNAs are produced during biosynthesis, namely miR-342-3p and miR-342-5p. Previous studies have demonstrated that miR-342-3p plays a role of tumor suppressor gene in cervical cancer by targeting FOXM1 to down-regulate.6 Bitaraf et al pointed out that miR-342-5p is significantly down-regulated in breast cancer tissues and can be used as a potential biomarker.7 The experiments of Liu et al have indicated that miR-342-5p inhibits the growth, migration and invasion of osteosarcoma cells by targeting Wnt7b.8 Some studies have also suggested that miR-342-5p has predicted binding sites in the 3ʹUTR of the three genes (TCF7, MSI1 and PAX5) involved in Wnt signal transduction. MiR-342-5p inhibits the expression of luciferase gene constructors of these genes 3ʹUTR and down-regulates the protein expression of TCF7 transcription factors, which can mediate the classical Wnt pathway.9 These studies predict that miR-342-5p influences the proliferation, metastasis and invasion of ovarian cancer cells, but its specific regulatory mechanism remains unclear.
PcG protein complex, an important epigenetic regulatory factor, has attracted more and more attention because of its important role in stem cell differentiation, cell development, senescence, tumor and many other biological processes. CBX family protein is an important component of PcG protein complex. CBX2 gene is an important transcriptional regulatory factor.10 According to gene search in NCBI (https://www.ncbi.nlm.nih.gov/), CBX2 gene is located on chromosome 17 (17q25.3) in human (Homo sapiens) with the full length of 11397 bp consisting of 7 exons and encoding 1211 amino acids. Also, in normal female tissues, the expression level of CBX2 in ovary is the highest. Previous studies have shown that CBX2 inhibits some female signal pathways by regulating Foxl2 and Wnt4 signal pathways.11 Zheng et al12 found that overexpression of CBX2 in breast cancer promotes tumor progression through the PI3K/AKT signaling pathway. Mao et al13 found that CBX2 may be a target for the treatment of hepatocellular carcinoma. Clermont et al14 found that CBX2 can be used as a drug target for prostate cancer. The occurrence and development of tumors are regulated by abnormally activated cellular signaling pathways, in which Wnt/β-catenin signaling pathway is involved in a variety of human tumorigenesis. There are four main pathways of Wnt signal pathway: Wnt/β-catenin pathway, Wnt/Ca2+ pathway, Wnt cell plane polarization pathway and intracellular pathway that regulates spindle direction and asymmetric cell division. Currently, the most studied pathway is Wnt/β-catenin pathway (classical Wnt signal pathway).15 Wnt/β-catenin signal pathway plays a key role in the regulation of cell pluripotency and malignant transformation, and is closely related to tumor proliferation, metastasis and other biological functions.16 The abnormal activation of Wnt/β-catenin signal pathway leads to the abnormal expression of β-catenin, which gradually increases in normal ovary, benign ovarian tumor, borderline ovarian tumor and ovarian cancer tissues. The positive expression of β-catenin is correlated with the depth of tumor infiltration and lymph node metastasis. Some scholars have also found that Wnt signal pathway is activated in ovarian cancer stem cells, which promotes the entry of β-catenin into the cell nucleus and acts on TCF and LEF transcription factors to induce the growth, differentiation and metastasis of ovarian cancer stem cells.17 Previous studies have shown that CBX2 can stabilize the testicular pathway by inhibiting Wnt signal.18 All in all, whether there is a correlation between miR-342-5p and CBX2, and whether CBX2 gene can mediate Wnt signal pathway to affect the proliferation, metastasis and invasion of ovarian cancer cells need to be further studied.
Materials and Methods
Clinical Tissue Samples
Ovarian cancer tissues and its adjacent tissues with normal histology were collected from patients with ovarian cancer who underwent resection in our hospital from 2018 to 2020. All the collected tissues were preserved at −80°C. Also, this study was approved by the Medical ethics committee of Henan Provincial People’s Hospital and obtained the written informed consent of each patient before the operation.
Cell Culture
Human normal ovarian epithelial cell line (IOSE80) and human ovarian cancer cell line (SKOV3 and OVCAR3) were purchased from Shanghai Cell Bank of Chinese Academy of Medical Sciences. All cells were cultured in DMEM medium (Hyclone; Logan, UT) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin, and the medium was placed in a moist incubator containing 5% CO2 at 37°C.
Grouping and Modeling
Tissue Samples
The ovarian cancer tissues were tumor group, while the non-tumor adjacent tissues were normal control group.
Cell Samples
Human normal ovarian epithelial cells were IOSE80 group, while human ovarian cancer cells were divided into SKOV3 group and OVCAR3 group.
Transfection of Human Ovarian Cancer Cells
SKOV3 and OVCAR3 cells grown logarithmically were inoculated into two six-well plates with 1.5 × 105 cells per well. According to the LipofectamineTM2000 (Invitrogen, USA) instructions, the Mimic miR-342-5p (Shanghai Jemma Company, China) and the Mimic NC (Shanghai Jemma Company, China) were mixed with lipofectamine2000 in OPTI-MEM, respectively, to obtain the Mimic and Mimic NC transfection mixture and let them stand for 20 min, then they were added to the cells. After transfection at 37°C with 5% CO2 for 6 h, the transfection medium was replaced with medium containing serum for further culture. According to the experimental design, SKOV3 cell lines were divided into SKOV3-blank group, SKOV3-NC group and SKOV3 group, and OVCAR3 cell lines were divided into OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group.
Database Retrieval of miR-342-5p Target Gene
The biomedical databases such as miRDB, TargetScan and TarBase were searched to screen 22 predictive target genes of miR-342-5p, among which CBX2 was selected as the experimental object of this study, and its binding sites were predicted.
qRT-PCR to Detect the mRNA Expression of miR-342-5p and CBX2
The mRNA expression levels of miR-342-5p and CBX2 in ovarian cancer tissues, para-cancerous tissues, ISOE80 group, SKOV3 group and OVCAR3 group were detected by qRT-PCR. While the mRNA expression levels of miR-342-5p, CBX2, Wnt1, β-catenin, C-myc and Cyclin D1 in SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group were detected. The total RNA was extracted from the samples with Trizol reagent (Invitrogen, Carlsbad, Calif., USA), which was then reverse transcribed to complementary DNA (cDNA), followed by SYBR Green Real-Time PCR (Takara, Dalian, China). The thermal cycles were as follows: 95°C for 30 s, 95°C for 5 s lasting 40 cycles, and 60°C for 35 s. Finally, Ct method was used for relative quantitative analysis. The primer sequences used are shown in Table 1.
Table 1.
qRT-PCR Primer Sequences
| mRNA Name | Forward Primer | Reverse Primer |
|---|---|---|
| miR-342-5p | 5ʹ-TCTCACACAGAAATCGCACCCGT-3ʹ | 5ʹ-CGGAGGGGTGCTATCTGTGATTGAG-3ʹ |
| U6 | 5ʹ-ACGAATTTGCGTGTCATCC-3ʹ | 5ʹ-CAGCACATATACTAAAATTGGAAGG-3ʹ |
| CBX2 | 5ʹ-GCTCCCAGCTGTTATGTTTGG-3ʹ | 5ʹ-GCTCCCAGCTGTTATGTTTGG-3ʹ |
| Wnt1 | 5ʹ-GGCCGAAGTCAATGTTGTCG-3ʹ | 5ʹ-CAACCGAGGCTGTCGAGAAA-3ʹ |
| β-catenin | 5ʹ-TGTTCTTGAGTGAAGGACTGA-3ʹ | 5ʹ-ATTTGATGGAGTTGGACATGG-3ʹ |
| C-myc | 5ʹ-GGGAGGCTGCTGGTTTTCC-3ʹ | 5ʹ-TACAACACCCGAGCAAGGAC-3ʹ |
| Cyclin D1 | 5ʹ-CTCCTCGCACTTCTGTTCCTC-3ʹ | 5ʹ-GATGCCAACCTCCTCAACGAC-3ʹ |
Dual-Luciferase Reporter Assay
SKOV3 and OVCAR3 cells were inoculated in 96-well plates with 1.5 × 104 cells per well and 100 μL per well. The mimic, target gene 3ʹUTR double reporter gene vector and LipofectamineTM2000 reagent were diluted with OPTI-MEM medium, respectively. After 5 min, the three were mixed and let stand for 20 min. The mixture of mimic and wild-type vector, mimic and mutant vector, mimic NC and wild-type vector, mimic NC and mutant vector were respectively prepared in this system. Before adding 50 μL of mixture, 50 μL of medium was sucked out from each well. After 8 h, each well was replaced with a fresh medium of 100 μL. When the reporter gene cell lysis buffer was prepared, 100 μL was added to each well to fully lyse the cells. After centrifugation, the supernatant was taken to detect the activity of luciferase in each group of cells.
MMT Method to Detect Cell Proliferation Rate
The cell proliferation rate was detected by MMT method in SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group. The cells of each group were inoculated with 1 × 104 cells per well on a 96-well plate, and 3 repeated wells were set up in each group. After 1, 2 and 3 days of culture, 2 μL MTT reagent (5 mg/mL; Sigma, St. Louis, USA) was added to each well, and then incubated for 4 h. 150mL DMSO was added after sucking out the original medium. Finally, the absorbance of the sample was determined at 492nm by microplate spectrophotometer (Thermo, Spectronic, Madison, WI, USA).
Cell Colony Formation Experiment
The cell viability of SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group was observed by cell colony formation assay. Individual cells of each group were obtained with 0.25% trypsin and stored in RPMI-1640 medium containing 10% FBS. The culture medium was diluted to make 10mL medium containing 100 cells, and petri dishes were gently rotated to evenly distribute the cells. After that, the petri dishes were cultured in a humid cell incubator at 37°C and 5% CO2 for 1–2 weeks, and then the supernatant was removed. Furthermore, the petri dishes were rinsed with PBS twice and 4% paraformaldehyde was added to fix the cells. After 15 min, paraformaldehyde was removed, and an appropriate amount of GIMSA staining agent was added. Waiting for 10–30 min, the dye was rinsed off with running water. The petri dishes were inverted to take photos, when they became dry.
Flow Cytometry to Detect Apoptosis
SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group were digested with trypsin and washed with PBS twice, then the cells of each group were collected. After that, Annexin V apoptosis detection reagent FITC (E. BioSoCion, USA) with Propidium Iodide was used to stain the cells at room temperature. At the end, apoptosis was analyzed by flow cytometry (BDBiosciences, Franklin Lakes, NJ).
Transwell Assay to Detect the Invasive Ability of Cells
Transwell assay was performed on SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group to detect the cell invasion ability. The cells of each group (5 × 104) were placed in the upper chamber of the transwell chambers (Corning Incorporated, USA), and RPMI-1640 and FBS were added to the lower chamber. After 48 h, the cells on the surface of the chamber membrane were removed with a cotton swab, while the invading cells under the chamber membrane were fixed and stained with crystal violet. After drying, the analysis was carried out under the optical microscope.
Scratch Test to Test the Ability of Migration
The logarithmic growth phase cells of each group were diluted with DMEM containing 10% FBS and inoculated in a 6-well plate. When the cell fusion reached 80%, the bottom of the wells was scratched with the tip of a 20 μL sterile pipette. After rinsing with PBS twice, the FBS-free medium containing 20 μg/mL mitomycin C was added to weaken the interference of cell proliferation. The healed wound was photographed at 0 and 24 h after the scratch. The scratch area was measured by inverted microscope. S0 was the scratch area at 0 h and S1 was the scratch area at 24 h. Mobility = (S1-S0)/S0 × 100%.
Western Blot to Detect the Expression of Protein in Cells
RIPA lysis buffer was added to the cells of SKOV3-blank group, SKOV3-NC group, SKOV3 group, OVCAR3-blank group, OVCAR3-NC group and OVCAR3 group and centrifuged. The supernatant was collected as the total protein of each group, and the protein concentration was determined by BCA method. After SDS-PAGE electrophoresis, the separated protein samples were transferred to PVDF membrane, sealed with 5% skimmed milk powder for 1.5 h, and rinsed with PBST for 3 times. It was then incubated with primary antibody at 4°C overnight. After being washed by PBST for 3 times, it was sealed with the secondary antibody at room temperature for 1.5 h. The membrane was washed with PBST for 5 times, and ECL was used for color development. The protein expression levels of CBX2 (ab80044, abcam), Wnt1 (ab15251, abcam), β-catenin (ab16051, abcam), C-myc (ab32072, abcam) and Cyclin D1 (ab16663, abcam) in each group were detected.
Statistical Analysis
The experimental results were expressed as mean ±SD, and the statistical analysis was conducted by SPSS 21.0 software. T-test of two independent samples was used for the comparison of quantitative data between two groups, the quantitative data of three or more groups were compared by one-way ANOVA, and the pairwise comparison were conducted by q-SNK method to determine the statistical difference of pairwise comparison between multiple groups. P<0.05 means that the difference was statistically significant.
Results
Predictive Results of miR-342-5p Target Gene in Ovarian Cancer
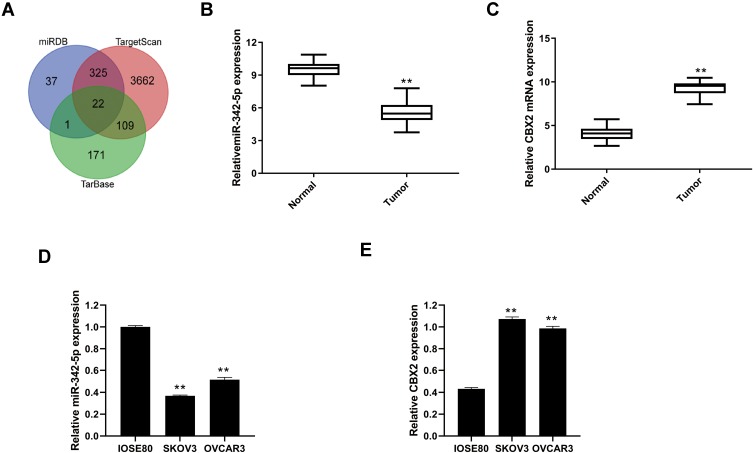
Through the database search of miRDB, TargetScan and TarBase, 22 predicted target genes were obtained from the intersection of the Wayne diagram (Figure 1A).
Figure 1.
Prediction of miR-342-5p target gene by searching on miRDB, TargetScan and TarBase. (A) Wayne diagram of miR-342-5p target gene predicted by database search of miRDB, TargetScan and TarBase. (B) The mRNA expression levels of miR-342-5p in ovarian cancer tissues and paracancerous tissues. (C) The mRNA expression levels of CBX2 in ovarian cancer tissues and paracancerous tissues. (D) The mRNA expression levels of miR-342-5p in IOSE80, SKOV3 and OVCAR3 cells. (E) The mRNA expression levels of CBX2 in IOSE80, SKOV3 and OVCAR3 cells. **P < 0.01 vs Normal group or IOSE80 group.
The mRNA expression levels of miR-342-5p and CBX2 in ovarian cancer tissues and non-tumor adjacent tissues were detected by qRT-PCR. The results indicated that compared with normal group, the mRNA expression level of miR-342-5p was significantly lower and CBX2 was significantly higher in tumor group (Figure 1B and C). qRT-PCR was also used to detect the mRNA expression levels of miR-342-5p and CBX2 in IOSE80 group, SKOV3 group and OVCAR3 group. The results (Figure 1D and E) showed that compared with IOSE80 group, the mRNA expression level of CBX2 in SKOV3 group and OVCAR3 group was significantly increased, while the mRNA expression level of miR-342-5p was significantly decreased, with statistically significant differences (P<0.05).
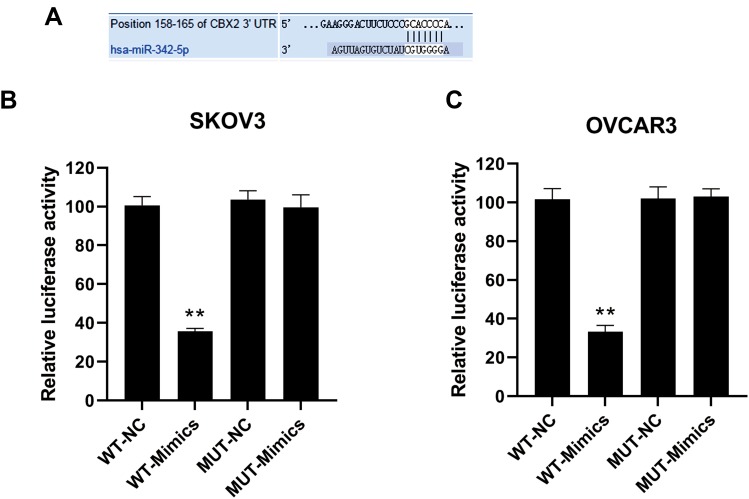
Identification of miR-342-5p Target Gene in Ovarian Cancer
The binding sites of miR-342-5p to its target gene CBX2 were predicted by software shown in Figure 2A. The results of dual-luciferase reporter assay (Figure 2B and C) suggested that in SKOV3-wild type and OVCAR3-wild type groups, the luciferase activity of miR-342-5p agonist group was significantly lower than that of the control group, while in the mutant groups, there was no significant change in the luciferase activity between the miR-342-5p agonist group and the control group. This indicated that the co-transfection of miR-342-5p mimics significantly inhibited the luciferase activity of CBX2-WT vector, but did not inhibit the luciferase activity of CBX2-MUT vector, suggesting that CBX2 was the target gene of miR-342-5p.
Figure 2.
The binding site of miR-342-5p to its target gene CBX2 predicted by software and the results of dual-luciferase reporter assay. (A) The predicted binding sites of miR-342-5p target gene CBX2. (B) Dual-luciferase reporter assay was used to verify the targeting relationship between miR-342-5p and CBX in transfected SKOV3 cells. (C) The targeting relationship between miR-342-5p and CBX was verified by dual-luciferase reporter assay in transfected OVCAR3 cells. **P < 0.01 vs NC group.
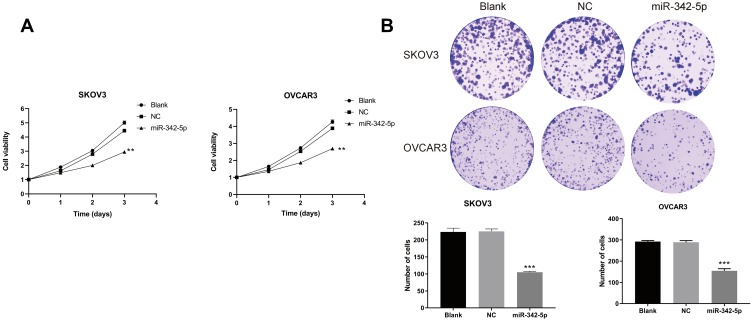
Cell Activity Detection
The results of MTT assay (Figure 3A) indicated that in the two ovarian cancer cell lines SKOV3 and OVCAR3, there was no significant difference in cell viability between the Blank group and the NC group at 24, 48 and 72 h. While the cell viability of SKOV3 group and OVCAR3 group transfected with miR-342-5p was significantly lower than that of NC group (P<0.05). The results of cell colony formation assay (Figure 3B) suggested that there was no significant difference in the number of cells between Blank group and NC group in the two cell lines. Compared with NC group, the number of cells in SKOV3 group and OVCAR3 group decreased significantly (P<0.05).
Figure 3.
MTT method to determine cell viability and cell colony formation test to detect cell cloning ability. (A) The viability of SKOV3 and OVCAR3 cells detected by MTT. (B) The cloning ability of SKOV3 and OVCAR3 cells detected by cell colony formation test. **P < 0.01 vs NC group, ***P < 0.001 vs NC group.
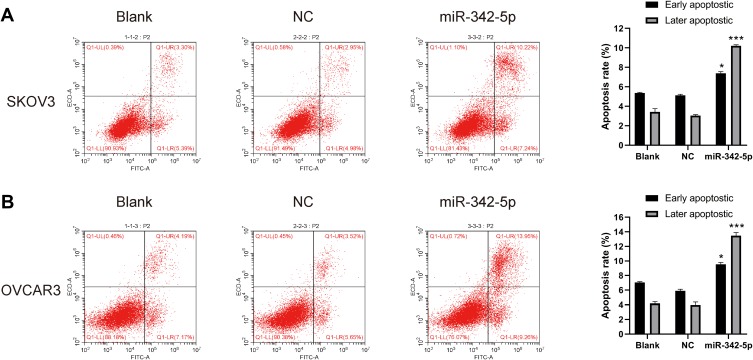
Detection of Apoptosis by Flow Cytometry
The results of flow cytometry indicated that in SKOV3 and OVCAR3 cell lines, there was no significant difference in apoptosis rate between Blank group and NC group. While the apoptosis rate in SKOV3 and OVCAR3 group was significantly higher than that in NC group (P<0.05) (Figure 4A and B).
Figure 4.
Cell apoptosis detected by flow cytometry. (A) The apoptosis rate of SKOV3 cells and (B) the apoptosis rate of OVCAR3 cells. *P < 0.05 vs NC group, ***P < 0.001 vs NC group.
The Results of Cell Invasion and Migration
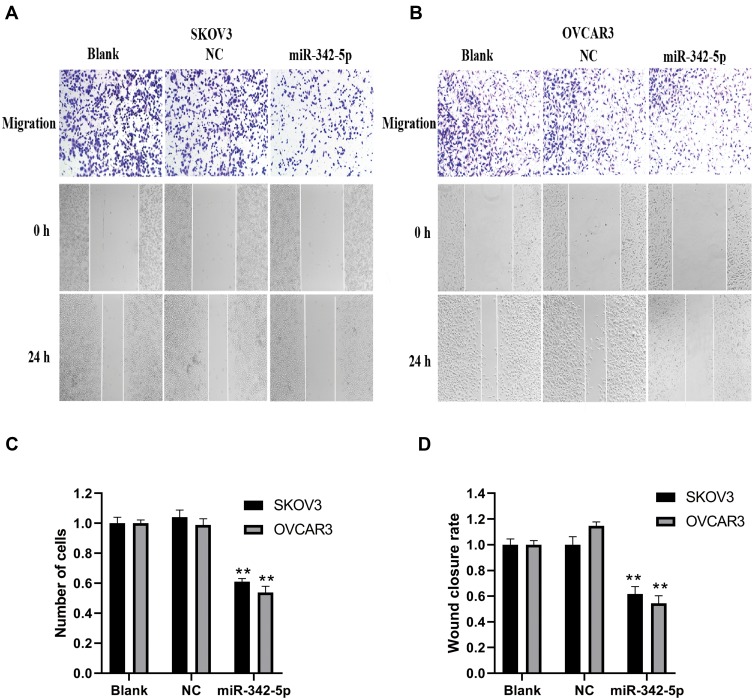
The results of transwell assay and scratch test are explained as follows (Figure 5A and B). The results of transwell assay (Figure 5C) found that there was no significant difference in invasion ability between Blank group and NC group, while the invasion ability of SKOV3 and OVCAR3 groups was significantly lower than that of NC group (P<0.05). The results of scratch test (Figure 5D) showed that there was no significant difference in migration and cell number between Blank group and NC group in SKOV3 and OVCAR3 cell lines after 24 h of scratch. Compared with NC group, the migration ability, cell number and fusion degree of SKOV3 group and OVCAR3 group transfected with miR-342-5p were significantly decreased (P<0.01).
Figure 5.
Detection of cell invasion by transwell assay and cell migration by scratch test. (A) Transwell assay was used to detect the invasive ability of SKOV3 and OVCAR3 cells. (B) Scratch test was applied to detect the migration ability of SKOV3 and OVCAR3 cells. (C) The statistical chart shows the number of cell invasion of SKOV3 and OVCAR3. (D) The statistical chart shows the cell mobility of SKOV3 and OVCAR3. **P < 0.01 vs NC group.
Detection of mRNA and Protein Expression Levels
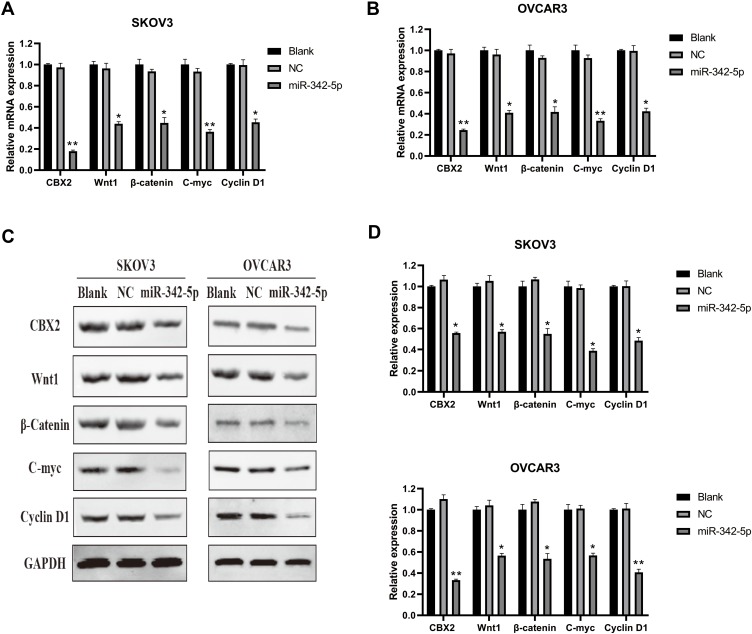
The results of qRT-PCR detection (Figure 6A and B) indicated that there was no significant difference in CBX2, Wnt1, β-Catenin, C-myc and Cyclin D1 mRNA between Blank group and NC group in SKOV3 and OVCAR3 cell lines, while the mRNA expression levels of CBX2, Wnt1, β-catenin, C-myc and Cyclin D1 in SKOV3 and OVCAR3 cells transfected with miR-342-5p were significantly lower than those in NC group (P<0.05).
Figure 6.
The results of relative expression of mRNA and proteins detected by qRT-PCR and WB. (A, B) The mRNA expressions of CBX2, Wnt1, β-Catenin, C-myc and Cyclin D1 in SKOV3 and OVCAR3 cells were detected by qRT-PCR. (C, D) The protein expressions of CBX2, Wnt1, β-Catenin, C-myc and Cyclin D1 in SKOV3 and OVCAR3 cells were detected by Western Blot. *P < 0.05 vs NC group, **P < 0.01 vs NC group.
Western Blot results (Figure 6C and D) suggested that there were no significant differences in the protein expression of CBX2, Wnt1, β-Catenin, C-myc and Cyclin D1 between Blank group and NC group in SKOV3 and OVCAR3 cell lines. Compared with the NC group, the protein expression levels of CBX2, Wnt1, β-Catenin, C-myc and Cyclin D1 in SKOV3 and OVCAR3 cells transfected with miR-342-5p were decreased, with statistically significant difference (P < 0.05).
Discussion
Ovarian cancer is the deadliest gynecological malignant tumor in the world. Inhibiting the unlimited proliferation, invasion and metastasis of ovarian cancer cells is the basic method to treat ovarian cancer. Therefore, exploring the molecular mechanism of malignant growth and metastasis to provide new treatment strategies for ovarian cancer has become one of the research hotspots. miRNA is a class of endogenous non-coding RNAs with a length of about 20 to 24 nucleotides. It could directly control the expression of individual proteins and entire proteomes related to important biological processes through affecting regulatory factors of pre-mRNA, and it plays an important role in cell growth, differentiation, programmed cell death and the regulation of lipid metabolism pathways.19–21 Moreover, many miRNAs are related to the occurrence, development and spread of cancer.22,23 miRNA, as a key regulator, is involved in almost all cellular processes in mammals and other multicellular organisms, regardless of its carcinogenic or anticancer roles.20 Current studies24 have shown that various types of malignant tumors are associated with miRNA disorders, and about 50% of the known human miRNA are susceptible to cancer. Therefore, increasing studies focus on the mechanism of these miRNAs’ role in cancer, and their target genes and signal pathways, so as to provide a research basis for diagnosis and treatment of cancer. Previous studies have shown that miR-342-5p can be significantly down-regulated in breast cancer,6 and also inhibit the growth, migration and invasion of osteosarcoma cells.8 Then, whether the miRNA is differentially expressed in ovarian malignant tumors and whether the proliferation, invasion and metastasis of cancer cells are caused by the change of the miRNA content are needed to be identified. Also, if the miRNA is artificially added, will it affect ovarian cancer cells; and what targeting gene and signaling pathways does the miRNA use to regulate ovarian cancer cells? In this study, through a series of experiments such as qRT-PCR, flow cytometry and Western Blot, we found that miR-342-5p can significantly reduce the proliferation, invasion, migration and viability of ovarian cancer cell lines SKOV3 and OVCAR3 by targeting gene CBX2, and promote their apoptosis. The mechanism may be related to mediating Wnt signal pathway and down-regulating the expression of related genes.
In general, miRNA is thought to bind to downstream target genes and inhibit gene transcription or induce gene degradation.25 In this study, CBX2 was confirmed to be the target gene of miR-342-5p by dual-luciferase reporter gene detection. The results of qRT-PCR detection in ovarian cancer tissues and non-tumor adjacent tissues showed that the mRNA expression level of miR-342-5p was significantly lower, while the mRNA expression level of CBX2 was significantly higher in tumor group. Also, in the qRT-PCR detection results of IOSE80 group, SKOV3 group and OVCAR3 group, the mRNA expression level of miR-342-5p in SKOV3 group and OVCAR3 group was significantly lower than that in IOSE80 group, and the mRNA expression level of CBX2 was significantly higher than that in IOSE80 group, which indicates that the down-regulation of miR-342-5p may lead to the increase of mRNA level of CBX2, suggesting that miR-342-5p may suppress ovarian cancer cells by binding to the downstream target gene CBX2 and inhibiting gene transcription or inducing gene degradation.
Furthermore, the results of cell colony formation assay and transwell assay suggested that the cell viability and invasive ability of SKOV3 and OVCAR3 groups transfected with miR-342-5p were significantly decreased. The apoptosis rate detected by flow cytometry was significantly increased in SKOV3 and OVCAR3 groups. The results of scratch test showed that the migration ability of cell, the number of cells and the degree of fusion decreased significantly in SKOV3 and OVCAR3 groups. These results suggested that miR-342-5p can inhibit the growth and metastasis of ovarian cancer cells in vitro.
To explore the potential mechanism of miR-342-5p in regulating the growth and metastasis of ovarian cancer cells, this study was focused on the expression of Wnt signal pathway. The expression of a series of target genes, including cell proliferation regulatory genes, developmental control genes and tumorigenesis-related genes, such as C-myc, CyclinD1, MMPs and VEGF, was conducted by classical Wnt signal transduction pathway, which could lead to abnormal cell proliferation and even cell carcinogenesis.26–29 It has been found that in liver cancer and gastric cancer, survivin and β-catenin may affect tumor transformation, promote cell proliferation and inhibit apoptosis through the cyclin family and Wnt signal pathway.30 C-myc is a proto-oncogene, which is not only an ectopic gene but also an adjustable gene. It can stimulate cell proliferation and induce apoptosis. C-myc is the first gene identified as the target of the Wnt signal pathway. After the action of β-catenin/Tef-4 complex, it is activated and leads to tumorigenesis.31 In this study, the mRNA and protein expression of C-myc were decreased in SKOV3 and OVCAR3 cells transfected with miR-342-5p, indicating that miR-342-5p may down-regulate the expression of C-myc gene to have anti-cancer effect. CyclinD1, a type of Cyclin gene, plays an important role in the progression of G1-S in the cell cycle. Inhibiting CyclinD1 expression can block the cells from G1 phase to S phase, while overexpressing CyclinD1 can shorten G1 phase and accelerate cell proliferation. CyclinD1 is the downstream regulatory target gene of Wnt/β-catenin. The increased expression of β-catenin in the cytoplasm activates CyclinD1 to promote cell division and growth, resulting in uncontrolled cell proliferation and tumorigenesis. The results of Western Blot detection indicated that the mRNA and protein expression levels of β-catenin and Cyclin D1 in miR-342-5p-transfected SKOV3 and OVCAR3 cells decreased, indicating that miR-342-5p may down-regulate the expression of Cyclin D1 by regulating β-catenin to inhibit tumorigenesis. Also, some experimental results32 also suggest that down-regulation of Wnt1 gene by siRNA interference technology can arrest the cell cycle in G2/M phase, thereby reducing the proliferation ability of cancer cells and promoting the apoptosis of tumor stem cells. The results of flow cytometry found that Wnt1 promotes the occurrence and development of ovarian cancer by disrupting the cell cycle. Also, the mRNA and protein expression of Wnt1 decreased in SKOV3 and OVCAR3 cells transfected with miR-342-5p, confirming that miR-342-5p may also decrease the proliferation ability of cancer cells by down-regulating the expression of Wnt1, thereby having the anti-cancer effect.
The Wnt pathway is a complex network, and the occurrence and metastasis of tumor are related to the structural and functional changes of its signal elements at all levels. In this study, the expression of miR-342-5p in ovarian cancer SKOV3 and OVCAR3 cell lines and its effect on the expression of other genes were detected, which demonstrated that miR-342-5p could down-regulate the expression of a series of target genes in classical Wnt signal pathway, such as C-myc, CyclinD1, so as to affect the proliferation, migration and apoptosis of cancer cells. This study also confirmed that the highest expression of CBX2 in ovarian content was correlated with miR-342-5p, and there was differential expression between ovarian tumor tissue and normal tissue. However, whether CBX2 is involved in the regulation of the Wnt signal pathway and thus has an impact on the proliferation, metastasis and invasion of ovarian cancer cells requires further experimental design to verify the expression of relevant target genes.
In conclusion, through database search and in vitro determination, the expression level of miR-342-5p decreases during the occurrence of ovarian tumors, which reduces the inhibition of the transcription of its target gene CBX2, resulting in an increase in the expression level of CBX2. Transfection of miR-342-5p into SKOV3 and OVCAR3 cancer cells can inhibit their growth, proliferation and metastasis in vitro and the mechanism is related to the effect on Wnt signal pathway and down-regulation of C-myc, CyclinD1 and other genes expression, which could help elucidate the molecular regulatory mechanism of miR-342-5p in ovarian tumor tissues and explore new therapeutic strategies for ovarian cancer. However, this study only discovered a small part of the regulatory mechanism of malignant behavior of ovarian cancer, and the relationship between the miR-342-5p target gene CBX2 and malignant growth and metastasis of ovarian cancer needs to be further studied.
Acknowledgements
Thank you for the technical support of Guangzhou Yujia Biotechnology Co., Ltd.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou MF, Ling J, Wu QY, Zhang CX. Long non-coding RNA PVT1 functions as an oncogene in ovarian cancer via upregulating SOX2. Eur Rev Med Pharmacol Sci. 2018;22(21):7183–7188. doi: 10.26355/eurrev_201811_16251 [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 5.Zhang R, Shi H, Ren F, et al. MicroRNA-338-3p suppresses ovarian cancer cells growth and metastasis: implication of Wnt/catenin beta and MEK/ERK signaling pathways. J Exp Clin Cancer Res. 2019;38(1):494. doi: 10.1186/s13046-019-1494-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588(17):3298–3307. doi: 10.1016/j.febslet.2014.07.020 [DOI] [PubMed] [Google Scholar]
- 7.Bitaraf A, Babashah S, Garshasbi M. Aberrant expression of a five-microRNA signature in breast carcinoma as a promising biomarker for diagnosis. J Clin Lab Anal. 2020;34(2):e23063. doi: 10.1002/jcla.23063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Wang Z, Zhou X, et al. miR-342-5p inhibits osteosarcoma cell growth, migration, invasion, and sensitivity to Doxorubicin through targeting Wnt7b. Cell Cycle. 2019;18(23):3325–3336. doi: 10.1080/15384101.2019.1676087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrdlickova R, Nehyba J, Bargmann W, Bose HJ. Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One. 2014;9(2):e86990. doi: 10.1371/journal.pone.0086990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jangal M, Lebeau B, Witcher M. Beyond EZH2: is the polycomb protein CBX2 an emerging target for anti-cancer therapy? Expert Opin Ther Targets. 2019;23(7):565–578. doi: 10.1080/14728222.2019.1627329 [DOI] [PubMed] [Google Scholar]
- 11.Eid W, Opitz L, Biason-Lauber A. Genome-wide identification of CBX2 targets: insights in the human sex development network. Mol Endocrinol. 2015;29(2):247–257. doi: 10.1210/me.2014-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng S, Lv P, Su J, Miao K, Xu H, Li M. Overexpression of CBX2 in breast cancer promotes tumor progression through the PI3K/AKT signaling pathway. Am J Transl Res. 2019;11(3):1668–1682. [PMC free article] [PubMed] [Google Scholar]
- 13.Mao J, Tian Y, Wang C, et al. CBX2 regulates proliferation and apoptosis via the phosphorylation of YAP in hepatocellular carcinoma. J Cancer. 2019;10(12):2706–2719. doi: 10.7150/jca.31845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clermont PL, Crea F, Chiang YT, et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2019;29(1):44–65. doi: 10.1016/j.tcb.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijayakumar S, Liu G, Rus IA, et al. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/beta-catenin target gene, CDC25A. Cancer Cell. 2011;19(5):601–612. doi: 10.1016/j.ccr.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfankuchen DB, Baltes F, Batool T, Li JP, Schlesinger M, Bendas G. Heparin antagonizes cisplatin resistance of A2780 ovarian cancer cells by affecting the Wnt signaling pathway. Oncotarget. 2017;8(40):67553–67566. doi: 10.18632/oncotarget.18738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Moreno SA, Lin YT, Futtner CR, Salamone IM, Capel B, Maatouk DM. CBX2 is required to stabilize the testis pathway by repressing Wnt signaling. PLoS Genet. 2019;15(5):e1007895. doi: 10.1371/journal.pgen.1007895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: a new player enters the game. Oncotarget. 2015;6(7):4562–4568. doi: 10.18632/oncotarget.2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slotwinski R, Lech G, Slotwinska SM. MicroRNAs in pancreatic cancer diagnosis and therapy. Cent Eur J Immunol. 2018;43(3):314–324. doi: 10.5114/ceji.2018.80051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Yu X, Shen J, Wu WK, Chan MT. MicroRNA expression and its clinical implications in Ewing’s sarcoma. Cell Prolif. 2015;48(1):1–6. doi: 10.1111/cpr.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos J, Gil DCR, Medeiros R. Dysregulation of cellular microRNAs by human oncogenic viruses - implications for tumorigenesis. Biochim Biophys Acta Gene Regul Mech. 2018;1861(2):95–105. doi: 10.1016/j.bbagrm.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 23.Santos J, Peixoto DSS, Costa NR, Gil DCR, Medeiros R. The role of microRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers (Basel). 2018;10(12):493. doi: 10.3390/cancers10120493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braicu OL, Budisan L, Buiga R, et al. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225–4238. doi: 10.2147/OTT.S137107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. doi: 10.1126/science.1111444 [DOI] [PubMed] [Google Scholar]
- 26.Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3(12):1546–1556. doi: 10.18632/oncotarget.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dilek FH, Topak N, Tokyol C, Akbulut G, Dilek ON. Beta-Catenin and its relation to VEGF and cyclin D1 expression in pT3 rectosigmoid cancers. Turk J Gastroenterol. 2010;21(4):365–371. doi: 10.4318/tjg.2010.0122 [DOI] [PubMed] [Google Scholar]
- 28.Yoshioka S, King ML, Ran S, et al. WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/beta-catenin pathway. Mol Cancer Res. 2012;10(3):469–482. doi: 10.1158/1541-7786.MCR-11-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai Y, Wu R, Schwartz DR, et al. Role of beta-catenin/T-cell factor-regulated genes in ovarian endometrioid adenocarcinomas. Am J Pathol. 2002;160(4):1229–1238. doi: 10.1016/S0002-9440(10)62550-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gougelet A, Colnot S. A complex interplay between Wnt/beta-catenin signalling and the cell cycle in the adult liver. Int J Hepatol. 2012;2012:816125. doi: 10.1155/2012/816125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jingushi K, Nakamura T, Takahashi-Yanaga F, et al. Differentiation-inducing factor-1 suppresses the expression of c-Myc in the human cancer cell lines. J Pharmacol Sci. 2013;121(2):103–109. doi: 10.1254/jphs.12204FP [DOI] [PubMed] [Google Scholar]
- 32.Liao YJ, Yin XL, Deng Y, Peng XW. PRC1 gene silencing inhibits proliferation, invasion, and angiogenesis of retinoblastoma cells through the inhibition of the Wnt/beta-catenin signaling pathway. J Cell Biochem. 2019;120(10):16840–16852. doi: 10.1002/jcb.28942 [DOI] [PubMed] [Google Scholar]