Abstract
Objective
miR-381 is implicated in the occurrence and development of various cancers, yet its role in head and neck squamous cell carcinoma (HNSCC) remains largely unknown. This study sought to research the direct target of miR-381 in HNSCC and investigate their roles in cancer progression.
Methods
miRNA and mRNA expression files of HNSCC were accessed from TCGA database and then processed for differential analysis. Bioinformatics databases were employed to predict the target mRNAs of the potential miRNA. qRT-PCR was conducted to determine the expression levels of the target miRNA and mRNA. Then, a series of in vitro experiments like CCK-8, colony formation assay, wound healing assay and transwell assay were performed to detect cell proliferation, migration and invasion. Dual-luciferase reporter gene assay was carried out for the further validation of the targeted relationship between the miRNA and mRNA.
Results
miR-381 was observed to be greatly down-regulated in HNSCC cells, and its overexpression could inhibit cell proliferation, migration and invasion. Besides, dual-luciferase reporter gene assay confirmed that STC2 was a direct target of miR-381, and their expression levels were reversely correlated. Moreover, rescue experiments demonstrated that overexpressing STC2 could rescue the inhibitory effect of miR-381 overexpression on cell proliferation, migration and invasion. Also, we verified that miR-381/STC2 exerted its function on HNSCC proliferation by mediating the FAK/PI3K/Akt/mTOR signaling pathway.
Conclusion
miR-381 suppresses cell proliferation, migration and invasion in HNSCC through targeting STC2, and participates in HNSCC development probably via the FAK/PI3K/Akt/mTOR signaling pathway.
Keywords: miR-381, STC2, FAK/PI3K/Akt/mTOR, HNSCC
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer globally which occurs in oral, oropharynx, larynx or hypopharynx.1 Characterized by a high rate of incidence and mortality, HNSCC accounts for nearly 3% of the total malignancies, with a poor outcome in locally advanced stages and a 5-year overall survival rate approximately as 50%.2–4 Although great effort has been made to exploit the underlying molecular mechanisms in HNSCC occurrence and development, most studies focus their attention on protein-coding genes, and the dysregulation of miRNAs as well as their role in HNSCC remains elusive.5
MicroRNAs (miRNAs) are small non-coding RNAs that mediate gene expression at a post-transcriptional level.6 A recent study has suggested that miRNAs are key players involved in tumor occurrence, and clarifying the expression pattern of miRNAs as well as investigating their roles in cancer might be crucial for early diagnosis improvement, prognosis prediction and specific therapeutic methods mining.7 miR-381 is located on chromosome 14q32.31,8 and has been reported to be altered in various cancers. For example, miR-381 is found to be down-regulated in gastric cancer,9 lung cancer,10 cervical cancer,11 epithelial ovarian cancer12 and colorectal cancer.13 While in glioma and pituitary adenoma,14 miR-381 is up-regulated and implicated in cell proliferation and invasion. All these reports suggest the important role of miR-381 in cancer occurrence and development. However, the role of miR-381 in HNSCC has not been studied.
In the present study, we described the expression pattern of miR-381 in HNSCC, and clarified its function on cell biological behaviors as well as the underlying mechanism. We found that miR-381 was remarkably decreased in HNSCC, and was capable of inhibiting cell proliferation, migration and invasion via targeting STC2. Our study makes it possible that miR-381 can be used as a potential therapeutic target in HNSCC treatment.
Materials and Methods
Bioinformatics Analysis
Data of miRNA-seq (cancer: n=518, adjacent normal: n=44) and mRNA-seq (cancer: n=495, adjacent normal: n=44) in HNSCC were accessed from the TCGA database (https://portal.gdc.cancer.gov/). Differential analysis was carried out using the “edgeR” package, with the threshold set as |logFC|>2 and padj<0.05. miRDB (http://mirdb.org/miRDB/index.html) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) two databases were applied for target prediction for the differentially expressed miRNA (DEmiRNA) we interested in, and the potential mRNA was screened by applying a Venn diagram. Survival analysis was performed on the miRNA in the TCGA-HNSCC dataset combined with the corresponding clinical information.
Cell Culture
Human normal oral epithelial cell line HOEC (BNCC340217), HNSCC cell lines SAS (ATCC64403), FaDu (ATCCTCP-1012), TCA8113 (BNCC100733), SCC-4 (BNCC340434) and embryonic kidney cell line HEK-293T (ATCCCRL-1573) were all purchased from BeNa Culture Collection (BNCC; Beijing, China). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), and maintained under a humidified environment in 5% CO2 at 37 °C.
Vector Construction and Cell Transfection
miR-381 mimic, oe-STC2 and their negative controls (NC-mimic and oe-NC) were all obtained from GenePharma (Shanghai, China). Pre-miR-381 sequence was amplified with the forward primer designed as 5ꞌ-CGTGAATGATAGTGAGGAAC-3ꞌ and the reverse primer as 5ꞌ-GTGAACGATTTGCCACACACA-3ꞌ, and then inserted into the PLKO.3G vector. STC2-flag vector was constructed using the pCMV-3tag-3a plasmid for access of STC2 overexpression. Then, pCMV-STC2 (oe-STC2) and pCMV-3tag-3a (oe-NC) were transfected into cells using the Lipofectamine® 2000 reagent (Invitrogen, Carlsbad, USA), respectively. The RNA concentration was set as 50 nM, and the total RNA and proteins were extracted after 48 h of transfection.
qRT-PCR
Total RNA of FaDu cells was isolated using the GenElute mRNA Miniprep kit (Sigma-Aldrich, St Louis, MO). Then the RNA samples were reversely transcribed into miR-381 cDNA by applying a One Step miRNA cDNA Synthesis Kit (HaiGene, Harbin, China) and into STC2 cDNA through a DNA Reverse Transcription Kit (Applied Biosystems). Thereafter, real-time quantitative PCR (qRT-PCR) was conducted using the miSYBR-Green PCR Kit (TransGen Biotech, China), following the manufacturer’s instructions. U6 and GAPDH were taken as the endogenous miRNA and mRNA regulators. Primer sequences were listed in Table 1, and 2−ΔΔCt was employed for the normalization of miR-381 and STC2 expression levels.
Table 1.
Primer Sequences for qRT-PCR
| Gene | Forward | Reverse |
|---|---|---|
| miR-381 | 5ꞌ-TACTTAAAGCGAGGTTGCCCTT −3ꞌ | 5ꞌ-GGCAAGCTCTCTGTGAGTA-3ꞌ |
| STC2 | 5ꞌ-TGAAATGTAAGGCCCACGCT-3ꞌ | 5ꞌ-CGAGGTGCAGAAGCTCAAGA-3ꞌ |
| U6 | 5ꞌ-CTCGCTTCGGCAGCACA-3ꞌ | 5ꞌ-AACGCTTCACGAATTTGCGT-3ꞌ |
| GAPDH | 5ꞌ-ACAACTTTGGTATCGTGGAAGG-3ꞌ | 5ꞌ-GCCATCACGCCACAGTTTC-3ꞌ |
Western Blot
Transfected cells were washed with cold phosphate-buffered saline (PBS) for preparation. RIPA lysis buffer (Thermo Fisher Scientific, MA, USA) was applied for extraction of total proteins, and bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA) was used for protein quantification. Subsequently, the proteins were treated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for separation, and then transferred onto the nitrocellulose membranes (ZY-160FP; Zeye Bio Co., Ltd., Shanghai, China). After being blocked in 5% BSA/TBST for 2 h, the membranes were incubated with primary rabbit polyclonal antibodies overnight at 4 °C, followed by horseradish peroxidase (HRP) -labeled secondary antibody (ab205718; 1:1000) for hybridization at room temperature for 2 h. The primary antibodies comprised STC2 (ab255610; 1:1000), p (phosphorylated)-PI3K (ab182651; 1:1000), PI3K (ab32089; 1:1000), p-AKT (ab38449; 1:500), AKT (ab8805; 1:500), p-mTOR (ab109268; 1:2000) and mTOR (ab2732; 1:2000). Electrochemiluminescence (ECL; ECL808-25, Biomiga Inc, San Diego, USA) was applied for protein bands visualization, and the band density was then measured using the Image J software (ver.1.44; http://rsbweb.nih.gov/ij/index.html).
CCK-8
96-well plates were used for cell seeding at a density of 6×103 cells/well. After transfection for 1, 2 and 3 days, 10 μL of reagent supplied by the cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added per well for 2 h of incubation at 37 °C, respectively. The absorbance value at 450 nm was read by an enzyme-labeled instrument (Multiskan MK3; Thermo Fisher Scientific, MA, USA) for the determination of cell viability.
Colony Formation Assay
Transfected cells were cultured in 6-well plates (2×102 cells/well) and the mediums were replaced every 3 days. Colonies visible were firstly washed with PBS, then processed for 10 min of fixation in methanol and for staining by 0.1% crystal violet (Sigma-Aldrich). Thereafter, the colonies were observed under a microscope and calculated.
Wound Healing Assay
When cells confluency reached around 80%, the tip of a 200 ul pipette was used to make a scratch across the monolayer of the well center. Then the cells were washed with PBS three times, and continuously cultured for 24 h with fresh mediums. Images were captured under an inverted microscope and the wound closure rate was calculated.
Transwell Assay
Cells were initially digested, centrifuged and resuspended in serum-free mediums for starvation treatment (105 cells/mL). Approximately 2×104 cells were seeded into the Matrigel matrix-coated (Corning, Corning, NY) upper chambers, and the DMEM containing 10% FBS was added into the lower chambers. After 24 h of incubation at 37 °C, non-invaded cells were wiped off by applying a cotton swab, whereas cells invaded into the lower chambers were stained using 0.1% crystal violet. Five fields were chosen at random for statistical analysis.
Dual-Luciferase Reporter Gene Assay
Wild-type STC2 (STC2-wt) and mutant STC2 (STC2-mt) constructs were generated through cloning the STC2 3ꞌ-UTR-wt and STC2 3ꞌ-UTR-mt into the pGL3 luciferase vector, and then co-transfected with miR-381 mimic or NC mimic into HEK-293T cells. Dual-Glo Firefly Luciferase Assay Kit (Promega) was employed to treat cells and Lumino Skan Ascentluminometer (Thermo Fisher Scientific, Waltham, MA, USA) was applied to determine the luciferase activity. The luciferase activity was presented as a ratio of Firefly luciferase intensity to Renilla luciferase intensity.
Statistical Analysis
SPSS 22.0 software (IBM Corp. Armonk, NY, USA) was used for data processing, and the GraphPad Prism 6 (La Jolla, CA) was applied for graphic demonstration. Measurement data were expressed as mean ± standard deviation (SD). Student’s t-test and one-way ANOVA were conducted for comparative analysis between two groups and among multiple groups, respectively. All results were representative of at least three independent experiments. P<0.05 was considered statistically significant.
Results
miR-381 Is Down-Regulated in HNSCC Tissues and Cells
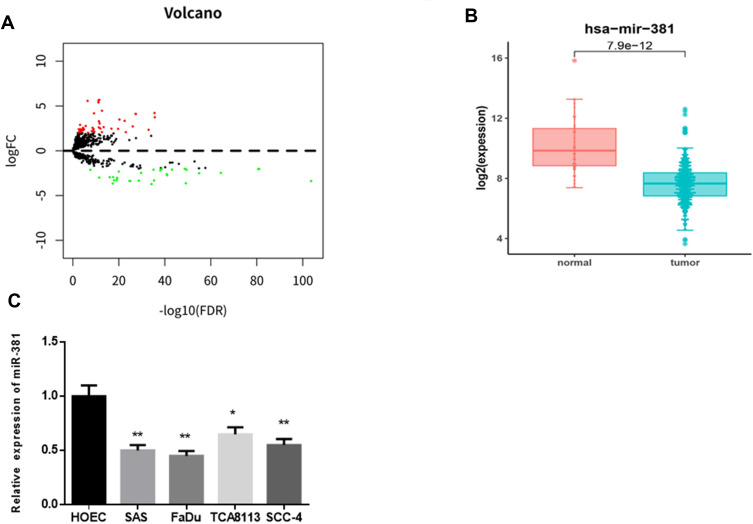
In all, 79 DEmiRNAs were identified via differential analysis using the “edgeR” package, consisting of 47 up-regulated miRNAs and 32 down-regulated miRNAs (Figure 1A). As indicated by bioinformatics analysis shown in Figure 1B, miR-381 was significantly down-regulated in HNSCC tissues. It has been reported that miR-381 participates in the malignant progression of various cancers, such as breast cancer and ovarian cancer.11,15 Thus, miR-381 was used as the target miRNA for follow-up analysis. Thereafter, miR-381 was detected at a cellular level by qRT-PCR, finding that miR-381 was greatly decreased in HNSCC cell lines SAS, FaDu, TCA8113 and SCC-4 relative to that in the human normal oral epithelial cell line HOEC (Figure 1C).
Figure 1.
miR-381 is down-regulated in HNSCC tissues and cells. (A) Differential analysis was performed on the miRNAs as shown in a volcano plot. Red refers to the up-regulated miRNAs and green refers to the down-regulated miRNAs. (B) The expression level of miR-381 in the TCGA-HNSCC dataset was analyzed and shown in a boxplot. Then (C) miR-381 was detected in human normal oral epithelial cell HOEC and HNSCC cell lines (*p<0.05; **p<0.01).
miR-381 Overexpression Inhibits Cell Proliferation, Migration and Invasion in HNSCC
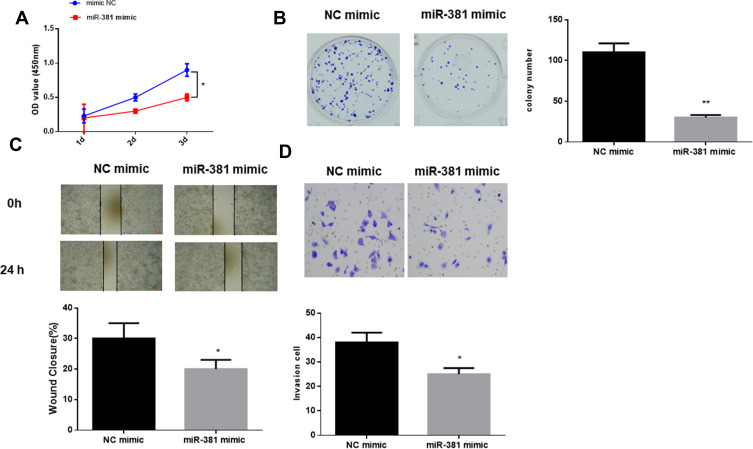
To clarify the role of miR-381 in HNSCC cell biological behaviors, miR-381 mimic and NC mimic were transfected into FaDu cells, respectively. CCK-8 and colony formation assay suggested that cells transfected with miR-381 mimic had significantly reduced viability and colony number relative to the cells with NC mimic (Figure 2A and B). Besides, wound healing and transwell assays were performed and revealed the relative poorer migration and invasion abilities in the cells with miR-381 mimic by comparison with the negative control (Figure 2C and D). Taken together, miR-381 overexpression was responsible for the inhibition of cell proliferation, migration and invasion in HNSCC.
Figure 2.
miR-381 overexpression inhibits cell proliferation, migration and invasion in HNSCC. miR-381 mimic and NC mimic were used to transfect FaDu cells. After transfection, the cells were harvested for the determination of (A) cell viability, (B) colony forming ability, (C) cell migration and (D) invasion via a series of in vitro experiments (*p<0.05; **p<0.01).
STC2 Is a Direct Target of miR-381
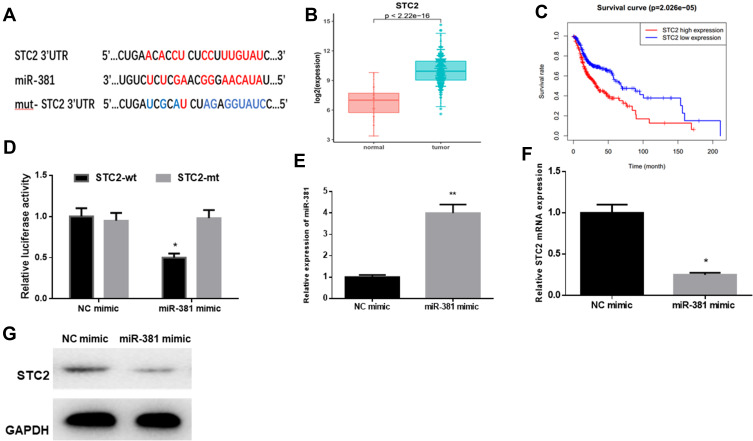
starBase database was employed to predict the target genes of miR-381, discovering that miR-381 potentially bound with the 3ꞌ-UTR within STC2 (Figure 3A). In the meantime, STC2 was observed to be highly-expressed in HNSCC tissues via bioinformatics analysis, and negatively associated with miR-381 expression (Figure 3B). For survival analysis, patients were divided into high expression and low expression groups based on the median level of STC2, and it was found that patients with high STC2 had a poorer prognosis relative to patients with low expression (Figure 3C). To further verify the targeted relationship between miR-381 and STC2, dual-luciferase reporter gene assay was conducted and indicated that the luciferase activity in the cells transfected with STC2-wt+miR-381 mimic was significantly decreased relative to the cells with STC2-wt+NC mimic, while there was no remarkable difference in the cells with STC2-mt (Figure 3D). Furthermore, miR-381 mimic was transiently transfected into FaDu cells for miR-381 overexpression construction. As indicated by qRT-PCR and Western blot, STC2 was greatly reduced in both mRNA and protein levels in cells with miR-381 overexpression relative to that in cells with the negative control (Figure 3E–G). Collectively, STC2 was a direct target of miR-381 in HNSCC.
Figure 3.
STC2 is a direct target of miR-381 in HNSCC. (A) miR-381 was predicted to bind with the 3ꞌ-UTR within STC2 using the starBase database. (B) Bioinformatics analysis showed that STC2 was highly expressed in HNSCC tissues, and (C) Kaplan–Meier survival analysis suggested that high STC2 expression was closely correlated with poor prognosis. STC2-wt and STC2-mt were co-transfected with miR-381 mimic or NC mimic into FaDu cells, respectively, and (D) dual-luciferase reporter gene assay was conducted to further verify the targeted relationship between miR-381 and STC2. Thereafter, FaDu cells were transfected with miR-381 mimic or NC mimic alone, then collected for detection of (E) miR-381 and (F, G) STC2 by qRT-PCR and Western blot (*p<0.05; **p<0.01).
miR-381 Suppresses Cell Proliferation, Migration and Invasion in HNSCC via Targeting STC2
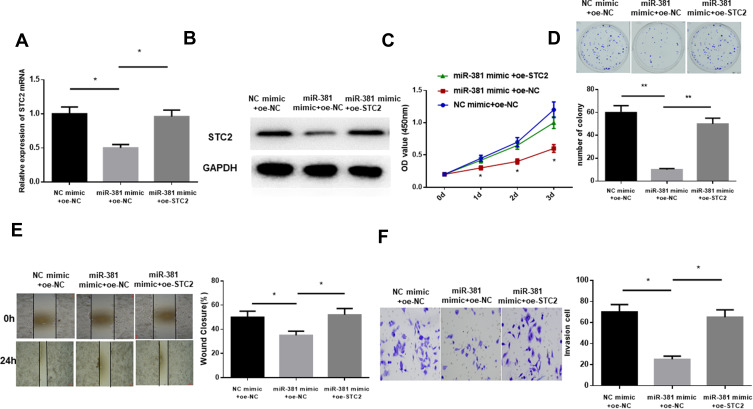
As abovementioned, STC2 was a target of miR-381. To explore the underlying mechanism of miR-381/STC2 in HNSCC, miR-381 mimic+oe-NC, miR-381 mimic+oe-STC2 and NC mimic+oe-NC were transfected into FaDu cells. qRT-PCR and Western blot were performed to assay the transfection efficiency, showing that STC2 was significantly decreased after miR-381 was overexpressed, but gradually increased back when it was concurrently overexpressed (Figure 4A and B). Moreover, CCK-8 and colony formation assay revealed that miR-381 overexpression could play an inhibitory role in cell proliferation and colony forming ability through down-regulating STC2, as indicated by the higher OD value and more colonies in the miR-381 mimic+oe-STC2 group relative to the miR-381 mimic+oe-NC group (Figure 4C and D). Similarly, cell migration and invasion could also be suppressed by miR-381 overexpression, but such inhibitory effect could be attenuated while STC2 was simultaneously overexpressed, as detected by wound healing and transwell assays (Figure 4E and F). Overall, the above findings elucidated that miR-381 targeted reduced STC2, thereby inhibiting cell proliferation, migration and invasion in HNSCC.
Figure 4.
miR-381 inhibits cell proliferation, migration and invasion in HNSCC via targeting STC2. miR-381 mimic+oe-NC, miR-381 mimic+oe-STC2 and NC mimic+oe-NC were transfected into FaDu cells, and (A) qRT-PCR and (B) Western blot were carried out to test STC2 in mRNA and protein levels. Then the cells were collected for cell (C) viability and (D) colony forming ability determination via CCK-8 and colony formation assay, as well as for (E) cell migration and (F) invasion examination by wound healing and transwell assays (*p<0.05; **p<0.01).
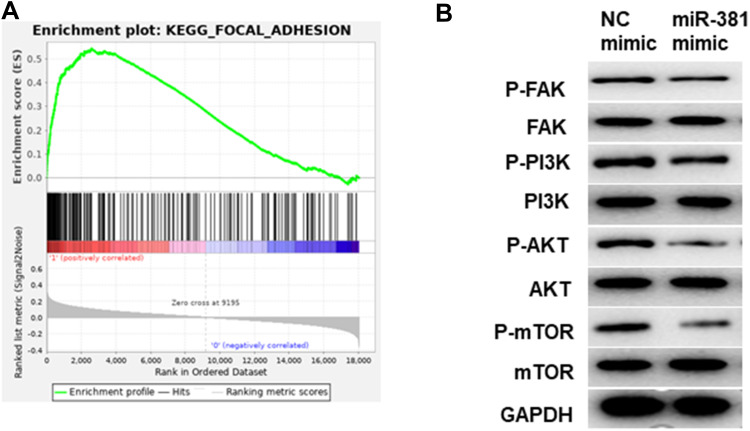
miR-381/STC2 Mediates HNSCC Cell Proliferation via the FAK/PI3K/Akt/mTOR Signaling Pathway
GSEA enrichment analysis was performed to further illuminate the regulatory role of miR381/STC2 in HNSCC cell proliferation, showing that STC2-mediated genes were mainly activated in the focal adhesion kinase (FAK) signaling pathway (Figure 5A). PI3K (phosphatidylinositol 3-kinase)/Akt (protein kinase B)/mTOR (mammalian target of rapamycin) is the most common downstream pathway of FAK. Thus, protein levels of FAK, PI3K, Akt, mTOR and their phosphorylated forms (p-) were detected using Western blot. As shown in Figure 5B, p-FAK, p-PI3K, p-Akt and p-mTOR were all reduced in the cells transfected with miR-381 mimic relative to the negative control. This result demonstrated that miR-381/STC2 might function on HNSCC cell proliferation via the FAK/PI3K/Akt/mTOR signaling pathway.
Figure 5.
miR-381/STC2 mediates HNSCC cell proliferation via the FAK/PI3K/Akt/mTOR signaling pathway. (A) GSEA enrichment analysis was conducted and found that STC2-mediated genes were mainly enriched in FAK signaling pathway. (B) Western blot was performed to detect FAK/PI3K/Akt/mTOR signaling pathway-related proteins.
Discussion
Despite the great progress in cancer treatment, the overall survival of HNSCC remains very poor, which urges us to explore novel therapeutic methods for efficacy improvement.16 Increasing evidence has suggested that miRNAs are vital regulators involved in tumorigenesis and development17,18 and they can be used as biomarkers for cancer diagnosis and prognosis.19 At present, several miRNAs that are associated with HNSCC diagnosis and prognosis have been identified, such as miR-21,20 miR-451,21 miR-37522 and miR-93.23 However, these miRNAs have not been widely recognized.
In the present study, miR-381 was identified and found to be significantly down-regulated in HNSCC cells relative to the normal oral epithelial cells, which is in agreement with the reports on breast cancer,24 cervical cancer,11 colorectal cancer13 and other cancers. This revealed that miR-381 might have the potential serving as a tumor suppressor in HNSCC. Xia et al12 discovered that miR-381 plays an inhibitory role in cell proliferation, migration and invasion in epithelial ovarian cancer through inhibiting YY1. While in oral squamous cell carcinoma, miR-381-3p can result in the inhibition of cell proliferation, cell cycle arrest and the promotion of cell apoptosis via directly targeting FGFR2.25 All these findings elucidate that miR-381 targets diverse genes in different cancer types.
This study found that miR-381 exerted its inhibitory role cell proliferation, colony forming, migration and invasion in HNSCC by acting as a tumor suppressor. Besides, miR-381 was investigated to target the 3ꞌ-UTR within STC2 and exhibit a reverse correlation with STC2 expression. Prior studies have revealed that STC2 is able to potentiate cell proliferation and metastasis in colorectal cancer26 and hepatocellular carcinoma.27 Besides, it has been identified as a target of miR-184 in glioblastoma multiforme, and miR-184 can inhibit tumor cell proliferation, migration and invasion via suppressing STC2 expression. Hence, we hypothesized that miR-381 might modulate STC2 expression to realize its regulatory role in HNSCC. Meanwhile, we conducted rescue experiments and found that STC2 overexpression could attenuate the miR-381-induced inhibition of cell proliferation, migration and invasion, suggesting that STC2 was a positive regulator in HNSCC, which was consistent with the previous study.28
FAK is implicated in the fundamental processes of normal and cancer cells via its kinase activity and scaffolding function.29 Activated FAK can lead to the phosphorylation of downstream PI3K, thereby activating Akt.30 Akt, a downstream effector of PI3K, can participate in various basic cellular responses and has been proved to play a pivotal role in cancer occurrence.31 mTOR is the downstream target of Akt and is capable of mediating diverse cell functions by means of integrating multiple pathways-derived signals.32 A previous study believes that FAK/PI3K/Akt/mTOR signaling pathway is crucial in cell proliferation, differentiation, survival and tumor cell metastasis.33 In our study, we assayed the levels of FAK/PI3K/Akt/mTOR signaling pathway-associated proteins, and discovered that miR-381/STC2 exerted its negative role in HNSCC cell proliferation via the FAK/PI3K/Akt/mTOR signaling pathway.
In conclusion, our study explored the expression of miR-381 in HNSCC cells, and confirmed that miR-381 had the ability to suppress cell proliferation, migration and invasion in HNSCC via targeting STC2. These results shed light on the involvement of miR-381 in HNSCC development, which provides a novel candidate target in the future HNSCC treatment.
Highlights
miR-381 is down-regulated in HNSCC;
miR-381 inhibits cell proliferation, migration and invasion in HNSCC;
STC2 is a direct target of miR-381 in HNSCC;
miR-381/STC2 participates in HNSCC development via the FAK/PI3K/Akt/mTOR axis.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. doi: 10.1038/nrc2982 [DOI] [PubMed] [Google Scholar]
- 2.Peitzsch C, Nathansen J, Schniewind SI, Schwarz F, Dubrovska A. Cancer stem cells in head and neck squamous cell carcinoma: identification, characterization and clinical implications. Cancers (Basel). 2019;11(5):616. doi: 10.3390/cancers11050616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moy JD, Moskovitz JM, Ferris RL. Biological mechanisms of immune escape and implications for immunotherapy in head and neck squamous cell carcinoma. Eur J Cancer. 2017;76:152–166. doi: 10.1016/j.ejca.2016.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee JC, Khuri FR, Shin DM. Advances in chemoprevention of head and neck cancer. Oncologist. 2004;9:302–311. doi: 10.1634/theoncologist.9-3-302 [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Chen D, Cabay RJ, Wang A, Crowe DL, Zhou X. Role of microRNA-138 as a potential tumor suppressor in head and neck squamous cell carcinoma. Int Rev Cell Mol Biol. 2013;303:357–385. doi: 10.1016/B978-0-12-407697-6.00009-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koshizuka K, Hanazawa T, Arai T, et al. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev. 2017;36:525–545. doi: 10.1007/s10555-017-9692-y [DOI] [PubMed] [Google Scholar]
- 7.Saad MA, Kuo SZ, Rahimy E, et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol Cancer. 2015;14(1):181. doi: 10.1186/s12943-015-0452-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formosa A, Markert EK, Lena AM, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33(44):5173–5182. doi: 10.1038/onc.2013.451 [DOI] [PubMed] [Google Scholar]
- 9.Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29. doi: 10.1186/s13046-017-0499-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang R-S, Zheng Y-L, Zhao J, Chun X. microRNA-381 suppresses the growth and increases cisplatin sensitivity in non-small cell lung cancer cells through inhibition of nuclear factor-κB signaling. Biomed Pharmacother. 2018;98:538–544. doi: 10.1016/j.biopha.2017.12.092 [DOI] [PubMed] [Google Scholar]
- 11.Shang A, Zhou C, Bian G, et al. miR-381-3p restrains cervical cancer progression by downregulating FGF7. J Cell Biochem. 2019;120:778–789. doi: 10.1002/jcb.27438 [DOI] [PubMed] [Google Scholar]
- 12.Xia B, Li H, Yang S, Liu T, Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. 2016;37:9157–9167. doi: 10.1007/s13277-016-4805-8 [DOI] [PubMed] [Google Scholar]
- 13.He X, Wei Y, Wang Y, et al. MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther. 2016;9:1231–1239. doi: 10.2147/OTT.S99228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, Liu X, Wang Z, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32. doi: 10.1016/j.brainres.2011.03.034 [DOI] [PubMed] [Google Scholar]
- 15.Wu M, Fan B, Guo Q, et al. Knockdown of SETDB1 inhibits breast cancer progression by miR-381-3p-related regulation. Biol Res. 2018;51(1):39. doi: 10.1186/s40659-018-0189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nohata N, Hanazawa T, Kinoshita T, et al. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer. 2013;108(8):1648–1658. doi: 10.1038/bjc.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Hu W, Li G, et al. Inhibition of miR-9-5p suppresses prostate cancer progress by targeting StarD13. Cell Mol Biol Lett. 2019;24(1):20. doi: 10.1186/s11658-019-0145-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pishkari S, Paryan M, Hashemi M, Baldini E, Mohammadi-Yeganeh S. The role of microRNAs in different types of thyroid carcinoma: a comprehensive analysis to find new miRNA supplementary therapies. J Endocrinol Invest. 2018;41:269–283. doi: 10.1007/s40618-017-0735-6 [DOI] [PubMed] [Google Scholar]
- 19.Zhai H, Ju J. Implications of microRNAs in colorectal cancer development, diagnosis, prognosis, and therapeutics. Front Genet. 2011;2. doi: 10.3389/fgene.2011.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arantes LMRB, Laus AC, Melendez ME, et al. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;8(6):9911–9921. doi: 10.18632/oncotarget.14253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Zhang G, Wu Z, et al. MicoRNA-451 is a novel tumor suppressor via targeting c-myc in head and neck squamous cell carcinomas. J Cancer Res Ther. 2015;11(Suppl 6):C216–C221. doi: 10.4103/0973-1482.168189 [DOI] [PubMed] [Google Scholar]
- 22.Wang P, Xu L, Li L, et al. The microRNA-375 as a potentially promising biomarker to predict the prognosis of patients with head and neck or esophageal squamous cell carcinoma: a meta-analysis. Eur Arch Otorhinolaryngol. 2019;276(4):957–968. doi: 10.1007/s00405-019-05325-8 [DOI] [PubMed] [Google Scholar]
- 23.Li G, Ren S, Su Z, et al. Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumour Biol. 2015;36(5):3949–3956. doi: 10.1007/s13277-015-3038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Y, Xu W, Zhao W, et al. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–433. doi: 10.1016/j.biopha.2016.12.051 [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Ruan H, Hu X, Cao A, Song L. miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res. 2017;7:913–922. [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Zhou X, Fang Z, Pan Z. Effect of STC2 gene silencing on colorectal cancer cells. Mol Med Rep. 2019;20:977–984. doi: 10.3892/mmr.2019.10332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Wu K, Sun Y, et al. STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration in vitro. BMB Rep. 2012;45:629–634. doi: 10.5483/bmbrep.2012.45.11.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S, Ji Q, Chang B, et al. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8(4):5976–5991. doi: 10.18632/oncotarget.13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132–149. doi: 10.1016/j.pharmthera.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 30.Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 31.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Y, Dodhia S, Su GH. Dysregulations in the PI3K pathway and targeted therapies for head and neck squamous cell carcinoma. Oncotarget. 2017;8(13):22203–22217. doi: 10.18632/oncotarget.14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neoh C-A, Wu W-T, Dai G-F, et al. Flaccidoxide-13-acetate extracted from the soft coral cladiella kashmani reduces human bladder cancer cell migration and invasion through reducing activation of the FAK/PI3K/AKT/mTOR signaling pathway. Molecules. 2017;23(1):58. doi: 10.3390/molecules23010058 [DOI] [PMC free article] [PubMed] [Google Scholar]